Abstract
Interest in lipolysis and the metabolism of triglyceride-rich lipoproteins was recently reignited by the discovery of severe hypertriglyceridemia (chylomicronemia) in glycosylphosphatidylinositol-anchored high density lipoprotein–binding protein 1 (GPIHBP1)-deficient mice. GPIHBP1 is expressed exclusively in capillary endothelial cells and binds lipoprotein lipase (LPL) avidly. These findings prompted speculation that GPIHBP1 serves as a binding site for LPL in the capillary lumen, creating “a platform for lipolysis.” More recent studies have identified a second and more intriguing role for GPIHBP1—picking up LPL in the subendothelial spaces and transporting it across endothelial cells to the capillary lumen. Here, we review the studies that revealed that GPIHBP1 is the LPL transporter and discuss which amino acid sequences are required for GPIHBP1–LPL interactions. We also discuss the human genetics of LPL transport, focusing on cases of chylomicronemia caused by GPIHBP1 mutations that abolish GPIHBP1’s ability to bind LPL, and LPL mutations that prevent LPL binding to GPIHBP1.
Keywords: chylomicrons, endothelial cells, lymphocyte antigen 6 proteins, hypertriglyceridemia, chylomicronemia
The outline for the lipolytic processing of lipoproteins has been established for decades (1, 2). Triglyceride-rich lipoproteins are secreted by intestinal enterocytes and hepatocytes and enter the systemic circulation. The triglycerides are then hydrolyzed by lipoprotein lipase (LPL) along the luminal surface of capillaries, mainly within heart, adipose tissue, and skeletal muscle, releasing fatty acids for use by parenchymal cells (1). When LPL activity is absent, either as a consequence of defects in LPL or its cofactor apolipoprotein (apo-) CII, the lipolysis of triglyceride-rich lipoproteins is blocked, interfering with the delivery of nutrients to parenchymal cells and leading to markedly elevated triglyceride levels in the plasma (chylomicronemia) (3–5). In recent years, this schema has been refined with the identification of proteins that regulate the efficiency of lipolysis (e.g., apo-CIII, apo-AV, ANGPTL3, ANGPTL4, LMF1) (6–12).
But despite significant progress, key elements in LPL-mediated lipoprotein processing remained stubborn mysteries. One was how LPL is attached to the luminal face of capillaries. For more than 50 years, it has been recognized that LPL can be released from its intravascular binding sites with heparin (13, 14), a sulfated glycosaminoglycan. That finding suggested that LPL might be attached to heparan sulfate proteoglycans (HSPGs) that coat the surface of capillaries. Supporting this notion was the fact that LPL contains several positively charged heparin-binding domains (15–17), the fact that LPL binds to HSPGs (18–20), and the fact that LPL binding to endothelial cells can be reduced by treating the cells with enzymes that cleave HSPGs (21). Despite these observations and a fairly strong consensus that HSPGs were likely required for LPL binding, this notion was never particularly satisfying, largely because HSPGs are expressed in many cell types, including the parenchymal cells from which LPL is secreted, and it was never clear why LPL would end up binding preferentially to the HSPGs in the capillary lumen.
Another mystery was the mechanism by which LPL is transported to the capillary lumen. LPL is synthesized by adipocytes and myocytes and secreted into the surrounding interstitial spaces, but LPL's ability to process lipoprotein triglycerides depends on its ability to reach the capillary lumen. Obunike et al. (22) investigated LPL transport across endothelial cells grown on transwell filters and concluded that LPL was carried across cells by the VLDL receptor and HSPGs. Against the idea that the VLDL receptor is important, however, was the fact that VLDL receptor deficiency in mice does not lead to significant hypertriglyceridemia (23).
During the past few years, there has been progress in solving both mysteries—the binding site for LPL within capillaries and the mechanism by which LPL is transported to the capillary lumen. We now know that glycosylphosphatidylinositol-anchored high density lipoprotein–binding protein 1, (GPIHBP1), a GPI-anchored protein of capillary endothelial cells, binds LPL in the subendothelial spaces and transports it to the capillary lumen (24). The binding of LPL to GPIHBP1 is highly specific; even single amino acid substitutions in GPIHBP1 or LPL can abolish their interaction, leading to impaired lipolysis and hypertriglyceridemia (25–29). We discuss each of these topics in this review.
GPIHBP1, a member of the Ly6 family of proteins
GPIHBP1 was initially identified by Ioka et al. (30) using expression cloning. They generated a cDNA library from the livers of low density lipoprotein receptor–deficient mice and then screened for cDNA clones that conferred upon Chinese hamster ovary (CHO) cells the ability to bind high-density lipoproteins (HDL) that had been labeled with a fluorescent dye (1,1-dioctadecyl-3,3,3,3-tetramethylindocarbocyanine perchlorate, or DiI). They identified two cDNA clones, one encoding scavenger receptor class B, type 1 (SR-BI), and the other encoding GPIHBP1. Identifying SR-B1 in this screen was not a surprise, given SR-B1’s well-established HDL-binding properties (31), but GPIHBP1 was a novel molecule.
Mouse GPIHBP1 contains 228 amino acids specifying an N-terminal signal peptide, an acidic domain (where 17 of 25 consecutive residues are glutamate or aspartate) (32, 33), a cysteine-rich lymphocyte antigen 6 (Ly6) motif, and a carboxyl-terminal hydrophobic sequence that triggers the addition of a GPI anchor (30, 32, 33). The signal peptide is cleaved within the endoplasmic reticulum, and the carboxyl-terminal hydrophobic sequence is removed during the addition of a GPI anchor; thus, mature GPIHBP1 has two noteworthy protein domains—the acidic domain (amino acids 24–48 in mouse GPIHBP1) and the cysteine-rich Ly6 domain (from Cys-63 to Cys-135 in mouse GPIHBP1). The Ly6 domain in mouse GPIHBP1 contains a single N-linked glycosylation site (Asn-76), while human GPIHBP1 has two (Asn-78 and Asn-82) (34). Glycosylation of these sites is important for efficient trafficking of GPIHBP1 to the cell surface (34).
In their initial paper, Ioka et al. (30) pointed out that GPIHBP1 is a member of the Ly6 family of proteins. In humans and mice, there are ∼25–30 Ly6 proteins (35, 36). Several have been studied extensively, for example UPAR (the urokinase-type plasminogen activator receptor) (37), CD59 (an inhibitor of the complement cascade) (38), and SLURP1 (a keratinocyte protein that is mutated in Mal de Meleda, a recessive palmoplantar keratoderma) (39), but many Ly6 family members have no known function (40). The defining feature of this family of proteins is an ∼80–amino acid “Ly6 domain” containing 10 cysteines—all arranged in a characteristic spacing pattern. Each of the cysteines is disulfide-linked (41), with the first cysteine forming a disulfide bond with the fifth cysteine, the second with the third, the fourth with the sixth, the seventh with the eighth, and the ninth with the tenth (41–43). The disulfide binding creates a three-fingered domain, with each finger consisting mainly of β-pleated sheets (35, 44–46). Most Ly6 family members contain a single Ly6 motif, but several, for example, C4.4A and Heldisin, have two (47), and UPAR has three (37). Most Ly6 proteins are GPI-anchored, but SLURP1 and SLURP2 (48) have no GPI anchor and are secreted from cells. Apart from the conserved three-fingered structural motif, most Ly6 proteins are quite distinct in their amino acid sequences. GPIHBP1’s Ly6 domain has almost no amino acid sequence homology with other Ly6 family members, aside from the 10 conserved cysteines.
Ioka et al. (30) found that GPIHBP1 expression in CHO cells led to uptake of DiI-labeled HDL, although the amount of uptake was significantly less than in SR-B1–transfected cells. Treating GPIHBP1-transfected cells with phosphatidylinositol-specific phospholipase C (PIPLC, an enzyme that cleaves the GPI anchor and releases GPI-anchored proteins from the cell surface) abolished the ability of the cells to bind HDL (30). GPIHBP1 appeared to mediate selective uptake of lipids by cells (i.e., lipid components of HDL were taken up by cells, but degradation of 125I-labeled HDL proteins was negligible). Unlike SR-B1, GPIHBP1 did not increase cholesterol efflux to HDL in the medium (30).
Northern blots by Ioka et al. (30) showed that GPIHBP1 was expressed highly in the heart (30). They also performed in situ hybridization studies and concluded that GPIHBP1 was expressed in cardiac myocytes, the sinusoidal endothelium and Kupffer cells in the liver, alveolar macrophages of the lung, and the bronchial epithelium. Based on the expression of GPIHBP1 in scavenger cells (e.g., Kupffer cells, alveolar macrophages), along with GPIHBP1’s ability to promote the uptake of HDL lipids, they concluded that GPIHBP1 likely plays a role in “the initial entry of HDL cholesterol into scavenger cells for further transportation of cholesterol” (30). They added that knockout mice would be important for defining protein function in vivo.
The paper by Ioka et al. (30) was important for uncovering GPIHBP1, but it contained several findings that subsequently could not be confirmed. Ironically, one was that GPIHBP1-transfected cells actually bind HDL. Using several independent assay systems, Gin et al. (49) found no evidence that GPIHBP1 binds HDL. What explains this discrepancy is not entirely clear, but it is noteworthy that the experiments by Ioka et al. (30) were performed with CHO cells, and those cells produce LPL (50), a protein that binds avidly to GPIHBP1 (33). Also, Beigneux et al. (33) demonstrated that GPIHBP1 binds apo-AV; thus, it is possible that the HDL binding observed by Ioka et al. (30) was due to apo-AV in their HDL preparations. Another questionable result in the initial manuscript by Ioka et al. (30) was the apparent synthesis of GPIHBP1 by macrophages and cardiomyocytes (as judged by in situ hybridization) (30). Antibodies against GPIHBP1 were not available when Ioka et al. (30) identified GPIHBP1; hence, they were unable to confirm their findings with immunohistochemistry. Later, Beigneux et al. (33) and Davies et al. (24) showed that GPIHBP1 is expressed only in capillary endothelial cells, as judged by immunohistochemical studies with GPIHBP1-specific monoclonal and polyclonal antibodies. Also, Gpihbp1 transcripts are absent in mouse peritoneal macrophages, as judged by quantitative RT-PCR (L. G. Fong, S. G. Young, unpublished observations).
Identification of a role for GPIHBP1 in lipolysis
The function of GPIHBP1 in lipid metabolism began to unfold in 2007 with the analysis of Gpihbp1 knockout mice (33). On a chow diet, Gpihbp1−/− mice have milky plasma with plasma triglyceride levels of 2000–5000 mg/dl and cholesterol levels of 300–900 mg/dl (33). The creation of GPIHBP1 knockout mice was not specifically inspired by Ioka et al. (30) but instead occurred in a screen, initiated by Genentech scientists, of knockout mice for 472 genes encoding secreted and membrane proteins (51). The presence of hypertriglyceridemia in Gpihbp1 knockout mice prompted efforts to evaluate the function of GPIHBP1 and its role in triglyceride metabolism (33). Not surprisingly, nearly all of the triglycerides and cholesterol in the plasma of Gpihbp1−/− mice was contained in d < 1.006 g/ml lipoproteins, and the diameter of the triglyceride-rich lipoproteins in Gpihbp1−/− mice was far greater than in wild-type mice. The plasma apo-B48 levels in Gpihbp1−/− mice were also elevated and the clearance of retinyl palmitate was markedly delayed (33), implying that the lipolytic processing of triglyceride-rich lipoproteins was impaired.
Two other observations by Beigneux et al. (33) were consistent with a role for GPIHBP1 in lipolysis. The first was that GPIHBP1-transfected cells bind LPL avidly (Kd = 3.6 × 10−8 M) (33). The second was that GPIHBP1 is located exclusively in capillary endothelial cells (33), and importantly, is expressed along the capillary lumen (24) (Fig. 1). High levels of GPIHBP1 expression were observed in heart and brown adipose tissue, tissues known to be crucial for the processing of triglyceride-rich lipoproteins (24, 33). These findings led Beigneux et al. (33) to suggest that GPIHBP1 serves as a binding site for LPL within capillaries—a “platform for lipolysis.” Beigneux et al. (33) also speculated that GPIHBP1 might play a particularly important role in the metabolism of apo-B48–containing lipoproteins, but follow-up studies by Weinstein et al. (52) showed that this was not the case. Weinstein et al. (52) bred Gpihbp1−/− mice that synthesize exclusively apo-B48 (Gpihbp1Apob−/−48/48) as well as Gpihbp1−/− mice that synthesize exclusively apo-B100 (Gpihbp1Apob−/−100/100) and showed that the hyperlipidemia was equally severe in both groups of mice.
Fig. 1.
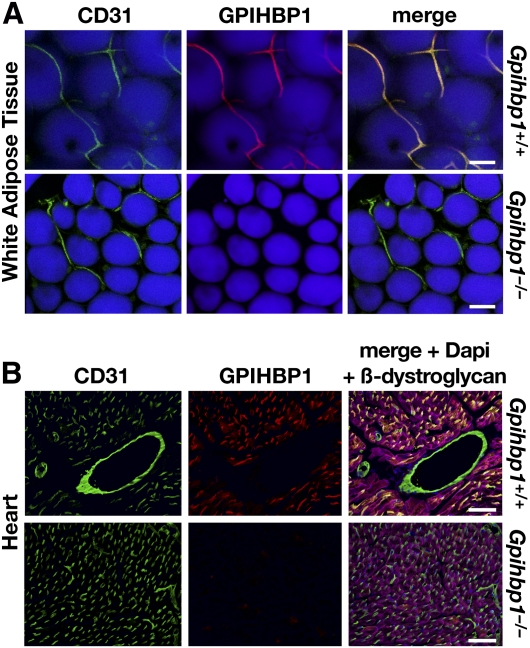
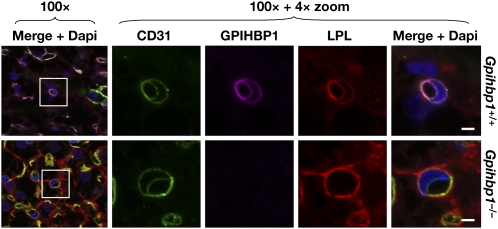
GPIHBP1 is located in capillary endothelial cells. A: Confocal immunofluorescence microscopy images of white adipose tissue showing that GPIHBP1 is located in capillaries of wild-type but not Gpihbp1−/− mice. Mice were injected intravenously with Alexa555-labeled anti-GPIHBP1 (red) and hamster anti-CD31 antibodies and then perfused extensively with PBS. Fixed adipose explants were stained with FITC-labeled anti-hamster antibodies (green) and BODIPY (boron-dipyrromethane) to label adipocytes (blue). Scale bar, 100 µm. B: Confocal images of heart from the same wild-type and Gpihbp1−/− mice, showing GPIHBP1 staining in the capillaries of wild-type but not Gpihbp1−/− mice. Tissues were stained with an antibody against β-dystroglycan (magenta), a plasma membrane protein of myocytes. Scale bar, 100 µm. Reproduced, with permission, from Davies et al. (24).
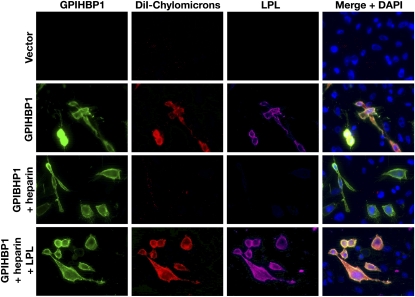
Beigneux et al. (33) also found that GPIHBP1-expressing CHO cells bind both apo-AV–phospholipid disks and chylomicrons (d < 1.006 lipoproteins from Gpihbp1−/− mice). The ability of apo-AV to bind to GPIHBP1 was not particularly surprising, given that apo-AV contains a strong heparin-binding domain (53). The binding of chylomicrons to GPIHBP1-expressing CHO cells was intriguing and raised the possibility that other heparin-binding proteins (e.g., apo-E, apo-B) might be capable of binding to GPIHBP1, but this does not appear to be the case (49). Instead, chylomicron binding to GPIHBP1-expressing CHO cells appears to be mediated by the hamster LPL produced by those cells. Several observations support this conclusion. First, chylomicrons no longer bind to GPIHBP1-transfected CHO cells after the cells are treated with heparin, but binding could be restored by incubating cells with human LPL (Fig. 2). Second, chylomicrons do not bind to GPIHBP1-transfected Chinese hamster lung (CHL) fibroblasts (which express only trace amounts of LPL) (49), but these cells acquire the ability to bind chylomicrons after they are incubated with LPL. Third, chylomicrons do not bind to CHO cells that express mutant GPIHBP1 proteins with Ly6 domain missense mutations that abolish LPL binding. Interestingly, those mutations, which leave the acidic domain intact, have no effect on apo-AV binding (49).
Fig. 2.
Immunofluorescence microscopy studies showing that chylomicron binding to GPIHBP1-transfected CHO cells requires LPL. CHO pgsA-745 cells were transfected with empty vector or S-protein–tagged wild-type GPIHBP1. After washing with PBS, the cells were incubated with DiI-labeled chylomicrons (red). Hamster LPL secreted by CHO pgsA-745 cells or exogenously added human LPL was detected on the surface of GPIHBP1-transfected CHO cells with monoclonal antibody 5D2 (87, 88) (purple), and GPIHBP1 was detected with an antibody against the S-protein tag (green). The binding of DiI-labeled chylomicrons to GPIHBP1-transfected cells was also assessed in cells that had been incubated with heparin and in heparin-treated cells that had been washed and then incubated with human LPL. Reproduced, with permission, from Gin et al. (49).
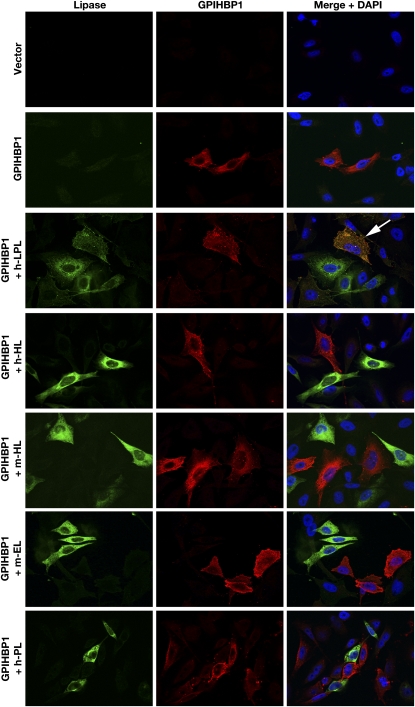
The fact that GPIHBP1 binds LPL raised the question of whether GPIHBP1 might also bind other members of the lipase family [hepatic lipase (HL), endothelial lipase (EL), pancreatic lipase (PL)]. Recent studies by Gin et al. (49) showed that this is not the case. Using a cell-based immunofluorescence microscopy assay, they showed that GPIHBP1 binds LPL avidly but does not bind HL, EL, or PL (Fig. 3). Similar findings were obtained with cell-based Western blot assays (49). Whether the other lipases have dedicated binding partners on the cell surface is not known.
Fig. 3.
Immunofluorescence microscopy experiments showing that LPL, but not other lipase family members, binds to GPIHBP1-transfected CHO cells. CHO-K1 cells were transiently transfected with an S-protein–tagged GPIHBP1 or V5-tagged versions of different lipases [human LPL (h-LPL), human hepatic lipase (h-HL), mouse hepatic lipase (m-HL), mouse endothelial lipase (m-EL), human pancreatic lipase (h-PL)]. The cells that had been transiently transfected with GPIHBP1 were mixed with cells that had been transiently transfected with one of the four different lipases. The next day, the cells were permeabilized and stained for GPIHBP1 (red) and for lipases (green). Only LPL bound to GPIHBP1-expressing cells (arrow points to a “yellow” GPIHBP1-expressing cell with LPL on its surface). Reproduced, with permission, from Gin et al. (49).
The plasma triglyceride levels in adult Gpihbp1−/− mice on a chow diet, 2000–5000 mg/dl, are similar to those in adult mice with LPL deficiency (Lpl−/−) (54, 55), suggesting that the level of LPL-mediated lipoprotein processing in Gpihbp1−/− mice is negligible. But the phenotypes of Gpihbp1−/− and Lpl−/− mice differ significantly during the suckling phase. The plasma triglyceride levels in suckling Gpihbp1−/− mice are relatively low, ∼120 mg/dl (33). In contrast, newborn Lpl−/− mice have plasma triglyceride levels of ∼20,000 mg/dl and die within 24 h (56). Why are suckling Gpihbp1−/− and Lpl−/− mice so different? The answer likely rests with the production of LPL in the liver of suckling mice (33). Beigneux et al. (33) pointed out that the emergence of severe hypertriglyceridemia in Gpihbp1−/− mice is temporally related to waning LPL expression in the liver (33). Further supporting this idea is the observation that interventions that increase hepatic LPL expression in adult Gpihbp1−/− mice lower their plasma triglyceride levels, while interventions that lower hepatic LPL expression increase plasma triglyceride levels (57). But why would hepatic LPL expression lower triglyceride levels in the absence of GPIHBP1? The answer, we suspect, relates to the fact that the capillaries of the liver are fenestrated, making it possible for hepatic LPL to interact with plasma lipoproteins in the absence of GPIHBP1-mediated LPL transport.
Two recent studies have begun to address whether GPIHBP1 affects LPL catalytic activity. A study by Sonnenburg et al. (58) reported that a soluble version of GPIHBP1 (one lacking the GPI anchor) is effective in binding LPL but that it did not activate LPL catalytic activity. Interestingly, however, they found that GPIHBP1-bound LPL was not susceptible to inhibition by ANGPTL3 and ANGPTL4. In a subsequent study, Liu et al. (59) reported that ANGPTL3 inactivates LPL by increasing its susceptibility to endoproteolytic cleavage by proprotein convertases (59). In their hands, soluble GPIHBP1 did not protect LPL from this cleavage event.
GPIHBP1rsquos role in transporting LPL to the capillary lumen
After discovering that Gpihbp1−/− mice have chylomicronemia, the idea that GPIHBP1 might be crucial for the proper localization of LPL within capillaries gradually picked up steam. Weinstein et al. (60) found that Gpihbp1−/− mice, despite manifesting severe chylomicronemia, had entirely normal amounts of LPL in tissue extracts. In the same study, they examined the kinetics of LPL entry into the plasma after an intravenous injection of heparin. In wild-type mice, plasma LPL levels peaked within 1–3 min after a heparin injection, but in Gpihbp1−/− mice, LPL entered the plasma more slowly and the levels peaked 15 min after the heparin injection. Also, an intravenous injection of Intralipid released LPL into the plasma in wild-type mice, but not in Gpihbp1−/− mice (60). Together, these observations suggested that the LPL in Gpihbp1−/− mice might be mislocalized, perhaps in an extravascular compartment where the enzyme would be irrelevant to the processing of triglyceride-rich lipoproteins in the plasma.
To test the notion that LPL was mislocalized in Gpihbp1−/− mice, Davies et al. (24) used LPL-specific antibodies and confocal immunofluorescence microscopy to study LPL localization in mouse tissues. In wild-type mice, most of the LPL in tissues was in capillaries, colocalizing with GPIHBP1 and CD31 (an endothelial cell marker) (Fig. 4). In Gpihbp1−/− mice, LPL localization was markedly different: almost none of the LPL was inside capillaries and instead was distributed around parenchymal cells (in close proximity to collagen IV on the outside of adipocytes in brown adipose tissue and β-dystroglycan on the surface of myocytes in skeletal muscle and heart) (Fig. 5). Of note, when GPIHBP1 is absent, the LPL in the interstitium is immobilized. The immobilization of interstitial LPL can be inferred from the immunohistochemical findings (Fig. 5) and by the fact that tissue stores of LPL are quite normal in Gpihbp1−/− mice while plasma LPL levels are very low (60). Precisely which molecules in the interstitium bind LPL in Gpihbp1−/− mice is unknown, but heparan sulfate proteoglycans would be a good possibility, particularly since the LPL in Gpihbp1−/− mice can be released into the plasma with heparin (60). We suspect that “post-secretion” binding of LPL in the interstitium is important in normal physiology—to prevent newly secreted LPL from disappearing into the lymph and possibly to serve as an LPL reservoir for GPIHBP1-mediated transport. Presumably, GPIHBP1 binds LPL with much greater affinity than other molecules in the subendothelial spaces, resulting in net movement of LPL to endothelial cells.
Fig. 4.
GPIHBP1 and LPL are found in capillaries. A, B: Confocal images showing the binding of GPIHBP1-, LPL-, and CD31-specific antibodies to brown adipose tissue (BAT) from a wild-type mouse. Images were taken with a 63× objective; (A) close-up (scale bar, 50 µm) and (B) wide-field (scale bar, 100 µm). C: Confocal images showing the binding of GPIHBP1-, LPL-, and β-dystroglycan-specific antibodies to heart tissue from a wild-type mouse (scale bar, 25 µm). Reproduced, with permission, from Davies et al. (24).
Fig. 5.
5. Mislocalization of LPL in tissues of Gpihbp1−/− mice. A: Confocal microscopy showing the binding of GPIHBP1-, LPL-, and collagen IV–specific antibodies to brown adipose tissue (BAT) from wild-type and Gpihbp1−/− mice. Scale bars, 100 µm. B, C: Confocal microscopy showing the binding of CD31-, LPL-, and β-dystroglycan-specific antibodies to heart (B) and skeletal muscle (C) of wild-type and Gpihbp1−/− mice. The scale bars show a distance of 100 µm (skeletal muscle) or 50 µm (heart). Reproduced, with permission, from Davies et al. (24).
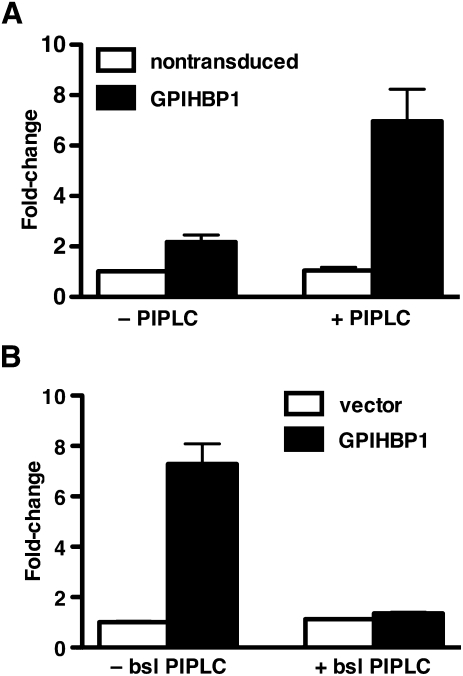
The fact that LPL was mislocalized in Gpihbp1−/− mice raised the possibility that GPIHBP1 might be involved in LPL transport to the capillary lumen. But if GPIHBP1 were involved, it would need to be located at both the basolateral and apical surfaces of endothelial cells. We were initially skeptical that this would be the case, given that GPI-anchored proteins are generally targeted to the apical face of polarized cells (61). However, several lines of evidence showed that GPIHBP1 is indeed located on both the basolateral and apical surfaces of endothelial cells (24). First, when endothelial cells are cultured on transwell filters, GPIHBP1 can be released from both the basolateral and apical plasma membrane with PIPLC (24). Second, confocal microscopy revealed that GPIHBP1 is located on both the basolateral and apical surfaces of cultured endothelial cells. Third, and most importantly, when capillaries in brown adipose tissue were examined by confocal microscopy, GPIHBP1 was present in similar amounts on the basolateral and luminal face of the cells (on both sides of endothelial cell nuclei) (Fig. 6) (24).
Fig. 6.
Confocal immunofluorescence microscopy of brown adipose tissue from a wild-type mouse, showing that GPIHBP1 and CD31 are located at both the basolateral and the apical surfaces of capillary endothelial cells. To visualize both the apical and basolateral surfaces of capillaries, cross-sections of capillaries containing an endothelial cell nucleus (blue) were identified (boxed areas) and viewed at high magnification. Both GPIHBP1 (red) and CD31 (green) were expressed at the apical (arrowheads) and basolateral (arrows) surfaces of endothelial cells. Scale bar, 2.5 μm. Reproduced, with permission, from Davies et al. (24).
Having acquired evidence that GPIHBP1 is located on both sides of endothelial cells, Davies et al. (24) investigated whether GPIHBP1 moves across cells. They first asked if GPIHBP1-expressing endothelial cells grown on transwell filters are capable of transporting a GPIHBP1-specific monoclonal antibody from the basolateral medium to the apical face of the cells. Indeed, this was the case; a GPIHBP1-specific monoclonal antibody was transported across GPIHBP1-expressing endothelial cells while an irrelevant antibody was not. The movement of the GPIHBP1-specific antibody across cells was abolished when GPIHBP1 at the basolateral surface of the cell was clipped off with PIPLC (Fig. 7).
Fig. 7.
GPIHBP1-transduced endothelial cells, but not nontransduced endothelial cells, are capable of transporting a GPIHBP1-specific monoclonal antibody from the basolateral medium to the apical surface of endothelial cell monolayers. A: Bar graph quantifying the amount of GPIHBP1-specific 11A12 antibody found in the apical medium of endothelial cell monolayers. In these studies, an IRDye800-labeled 11A12 antibody and an IRDye680-labeled anti-goat IgG control antibody were added to the basolateral chamber and incubated for 3 h at 37°C. The apical surface was treated with either PIPLC (1 U/ml; 30 min) or PBS, and both apical and basolateral media were dot blotted, scanned, and quantified with an Odyssey scanner. The bar graph shows the fold change (mean ± SEM) compared with untreated nontransduced cells. B: Assessing the ability of PIPLC in the basolateral medium to block the transport of antibody 11A12 from the basolateral medium to the apical surface of endothelial cell monolayers. IRDye800-labeled antibody 11A12 and IRDye680-labeled anti-goat IgG antibody were added to the basolateral chamber in the presence or absence of PIPLC (1 U/ml) and incubated for 1.5 h at 37°C. The apical surface was treated with PIPLC (1 U/ml; 30 min) and the apical medium was dot blotted, scanned, and quantified with the Odyssey scanner. The bar graph shows the fold change (mean ± SEM) compared with untreated vector-transduced cells. Reproduced, with permission, from Davies et al. (24).
Using similar experimental approaches, Davies et al. (24) showed that GPIHBP1 also transports LPL (both a V5-tagged human LPL as well as native bovine LPL) from the basolateral medium to the apical surface of endothelial cells (where it could be released into the apical medium with heparin). The transport of LPL to the apical face of cells was abolished by adding PIPLC or heparin to the basolateral medium. Also, no LPL transport was observed in endothelial cells that expressed a mutant form of GPIHBP1 that lacked the capacity to bind LPL.
Davies et al. (24) also showed that GPIHBP1 functions as a transporter in vivo. They injected a rat monoclonal antibody against GPIHBP1, along with an irrelevant rabbit antibody, into the quadriceps of wild-type and Gpihbp1−/− mice. Immunohistochemistry studies revealed that the injected antibodies quickly distributed throughout the extravascular spaces around myocytes. In as little as 20 min, however, the GPIHBP1-specific rat monoclonal antibody (but not the irrelevant rabbit antibody) associated with endothelial cells in wild-type mice and could be detected in the capillary lumen after an intravenous injection of an Alexa 568–labeled anti–rat IgG. No transport of the GPIHBP1-specific antibody to the capillary lumen was observed in Gpihbp1−/− mice.
The mislocalization of LPL in Gpihbp1−/− mice, along with experiments showing that GPIHBP1 moves LPL across endothelial cells, strongly suggested that GPIHBP1 is essential for delivering LPL to the capillary lumen. Confocal immunofluorescence microscopy studies have provided strong support for this idea (24). When cross-sections of capillaries from wild-type mice were examined, LPL was present along the luminal surface of capillaries, colocalizing with GPIHBP1 and CD31. When capillaries from Gpihbp1−/− mice were imaged, no LPL could be found in the capillary lumen (Fig. 8).
Fig. 8.
Confocal microscopy showing the binding of CD31-, GPIHBP1- and LPL-specific antibodies to brown adipose tissue of a wild-type mouse and a Gpihbp1−/− mouse. Images were taken with a 100× objective without optical zoom or with 4× optical zoom. To visualize both the apical and basolateral surfaces of capillaries, cross-sections of capillaries containing an endothelial cell nucleus (blue) were identified (boxed areas) and viewed at high magnification. GPIHBP1 (purple), LPL (red), and CD31 (green) were detected at the apical and basolateral surfaces of endothelial cells in wild-type mice, but no LPL was present on the apical (luminal) surface of capillaries in Gpihbp1−/− mice. Scale bar, 2.5 µm. Reproduced, with permission, from Davies et al. (24).
Structural features of GPIHBP1 and LPL required for their interaction
Gin et al. (62) showed that GPIHBP1’s N-terminal acidic domain is important for binding LPL. A rabbit antiserum against the acidic domain blocked LPL binding. Also, a mutant GPIHBP1 in which the aspartates and glutamates in the second half of the acidic domain (residues 38–48) were changed to alanine lacked the ability to bind LPL. These findings suggested that electrostatic interactions are crucial for GPIHBP1–LPL interactions. Consistent with this notion, the binding of LPL to GPIHBP1 could be blocked by polyaspartate, polyglutamate, or heparin (62). Also, LPL binding to GPIHBP1 was eliminated when positively charged residues in LPL's carboxyl-terminal heparin-binding domain (K403, R405, K407, K413, K414) were changed to alanine (62).
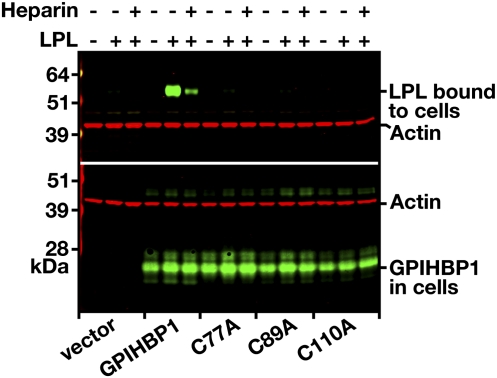
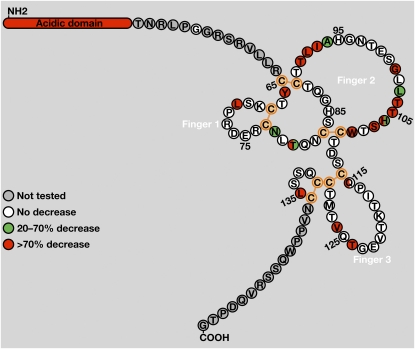
Multiple lines of evidence indicate that GPIHBP1’s Ly6 domain also plays a role in LPL binding. For example, Gin et al. (62) showed that a chimeric protein containing GPIHBP1’s acidic domain and the Ly6 domain of another Ly6 family member (CD59) was incapable of binding LPL. Later, Beigneux et al. (42) showed that changing any of the 10 cysteines in human GPIHBP1’s Ly6 domain to alanine abolished the ability of GPIHBP1 to bind LPL (Fig. 9). Similar findings were obtained with a cell-free assay. Thus, intact disulfide bonds—and the maintenance of the three-fingered structure of GPIHBP1’s Ly6 domain—is critical for LPL binding. In a more recent study, Beigneux et al. (26) changed each amino acid in human GPIHBP1’s Ly6 domain to alanine and then assessed the ability of the mutant proteins to bind LPL. Aside from the conserved cysteines, they identified 12 residues that are crucial for LPL binding, nine of which were clustered in finger 2 of GPIHBP1’s three-fingered Ly6 domain (Fig. 10) (26). These GPIHBP1 mutants also lacked the capacity to transport LPL from the basolateral to the apical surface of cultured endothelial cells (26). The fact that the Ly6 domain proved to be important for LPL binding was not entirely unexpected, given that the Ly6 domains of other Ly6 proteins, for example, those of UPAR and CD59, are crucial for ligand binding (37, 63). In the case of UPAR, many of the residues involved in ligand binding are located within finger 2 of the Ly6 domain (37, 47, 64–66), as was the case for GPIHBP1 (26).
Fig. 9.
Reduced binding of human LPL to cells expressing mutant versions of GPIHBP1 with cysteine-to-alanine mutations in the Ly6 domain. CHO pgsA-745 cells were electroporated with an S-protein–tagged wild-type GPIHBP1 construct or mutant constructs with cysteine-to-alanine mutations. Twenty-four hours after the electroporation, cells were incubated with V5-tagged human LPL in the presence or absence of heparin (500 U/ml). After washing the cells, cell extracts were prepared and the level of LPL bound to the cells was assessed by western blotting with a V5-tag–specific antibody. Simultaneously, the level of GPIHBP1 in cell extracts was assessed by Western blotting with an antibody against the S-protein tag. Actin was used as a loading control. Reproduced, with permission, from Beigneux et al. (42).
Fig. 10.
Amino acid residues in GPIHBP1’s Ly6 domain required for LPL binding. A: The amino acid sequence of human GPIHBP1 from immediately after the acidic domain (Thr-51) to the GPI-anchoring site (Gly-151). This schematic illustrates GPIHBP1’s five disulfide bonds (orange) and the three fingers of the Ly6 domain [based on the crystal structures of CD59 (89) and UPAR (37)]. In human GPIHBP1, the predicted disulfide bonds are C65–C89, C68–C77, C83–C110, C114–C130, C131–C136. In mouse GPIHBP1, the corresponding disulfide bonds are C63–C88, C66–C75, C81–C109, C113–C129, C130–C135. Replacement of the amino acids highlighted in green with alanine led to a moderate reduction in LPL binding (20–70%), while replacement of residues highlighted in red resulted in a marked reduction in LPL binding (>70%). For a subset of residues, a second amino acid substitution (aside from alanine) was assessed. Residues highlighted with two colors indicate cases where the effect of the alanine substitution (on the right) differed from the effect of the other amino acid substitution (on the left). Reproduced, with permission, from Beigneux et al. (26).
The discovery that two distinct domains within GPIHBP1 are important for LPL binding prompted Beigneux and coworkers to propose that there might be two corresponding domains within LPL required for binding to GPIHBP1 (26, 42), one interacting with GPIHBP1’s acidic domain and a second interacting with the Ly6 domain. It seems likely that LPL's positively charged heparin-binding domains would interact with GPIHBP1’s acidic domain. The LPL residues that would interact with GPIHBP1’s Ly6 domain have not been identified with certainty, but a stretch of residues located downstream from the carboxyl-terminal heparin-binding domain might be involved. Recent studies by Voss et al. (28) showed that mutations of Cys-418 and Glu-421 abolish human LPL binding to GPIHBP1 without significantly affecting LPL binding to heparin. Furthermore, a monoclonal antibody against chicken LPL (cLPL) with an epitope between amino acids 416 and 435 blocked binding of cLPL to GPIHBP1. Additional mutagenesis studies revealed that changing cLPL residues 421–425, 426–430, or 431–435 to alanine virtually abolishes cLPL binding to GPIHBP1 without significant effects on cLPL binding to heparin. Of note, Cys-418 and Cys-438 are linked by a disulfide bond (67), and earlier studies suggested that the intervening loop might be important for dimerization of LPL. The studies by Voss et al. (28) suggest that the same loop may be important for LPL's ability to bind GPIHBP1.
GIHBP1 expression patterns
GPIHBP1 is expressed exclusively in capillary endothelial cells, while LPL is expressed in parenchymal cells. In general, however, the tissue pattern of GPIHBP1 expression is similar to that of LPL. For example, Gpihbp1 transcripts are found at high levels in heart and brown adipose tissue and at moderate levels in skeletal muscle, mirroring the pattern of Lpl expression in tissues (33).
Gpihbp1 expression in the mouse is regulated by feeding state. In their initial study, Beigneux et al. (33) found that Gpihbp1 expression increases in skeletal muscle after a fast and returns to fed levels 6 h after refeeding. Later, Davies et al. (68) reported that fasting mice have higher levels of Gpihbp1 expression in heart, brown adipose tissue, and white adipose tissue. The fasted mice had higher levels of Lpl expression in heart and brown adipose tissue, but lower Lpl expression in white adipose tissue. The changes in Lpl expression with fasting make sense, as they seemingly promote fuel delivery to vital organs (and away from storage) during periods of energy deprivation. The “rationale” for increased Gpihbp1 expression in white adipose tissue during fasting is not clear. One possibility is that the increase in Gpihbp1 expression in adipose tissue of fasted mice serves an as-yet-unidentified function unrelated to lipolysis. Another is that increased Gpihbp1 expression in the white adipose tissue of fasted mice somehow facilitates the turnover of LPL.
Whether the “fasting/refeeding regulation” of GPIHBP1 is physiologically important is uncertain. The changes in expression with fasting and refeeding were two-fold or less, and studies of humans suggest that half-normal amounts of GPIHBP1 have no significant impact on triglyceride levels (25, 27). Also, at this point, we caution that all data on GPIHBP1 regulation is derived from the mouse; whether GPIHBP1 expression in humans is regulated in a similar fashion is unknown.
While the tissue pattern of Gpihbp1 expression is fairly similar to that of Lpl, there are several tissues where the expression pattern is discordant. First, the capillaries of the brain—an organ that uses glucose for fuel—express almost no Gpihbp1, while certain regions of the brain, notably the hippocampus, express large amounts of Lpl (69–73). The explanation for this discrepancy is unknown, but one potential explanation is that LPL in the brain is not transported into capillaries and functions in the extravascular compartment. A second tissue where levels of Gpihbp1 and LPL transcripts are discrepant is the lung. Gpihbp1 is expressed at high levels in the lung, while Lpl transcript levels are very low. Interestingly, however, the lung contains a significant amount of LPL protein. We suspect that the LPL found in the lung originates in other tissues and then is “scavenged” from the plasma by GPIHBP1 inside lung capillaries. In support of this idea, the levels of LPL in lung extracts are lower in Gpihbp1−/− mice than in wild-type mice (74). Also, Lpl−/− mice harboring a muscle-specific human LPL transgene have no human LPL transcripts in the lung but have significant amounts of human LPL protein in the lung (74). Moreover, when a human LPL adenovirus was injected into wild-type mice, significant amounts of human LPL could be detected in the lung. In these experiments, the vast majority of the LPL adenovirus was targeted to the liver, but low amounts of human LPL transcripts could be found in other tissues. The level of human LPL transcripts in the lung were lower than in skeletal muscle, but the LPL protein levels in the lung were high (∼15% of those in liver) while LPL protein levels in muscle were low (<1% of those in liver) (M. Weinstein, L. G. Fong, S. G. Young, unpublished observations).
Because GPIHBP1 is expressed in capillary endothelial cells, Olafsen, Fong, and coworkers (74) reasoned that it might be possible to gain insights into the relative amounts of GPIHBP1 in different tissues by injecting radiolabeled GPIHBP1-specific antibodies intravenously and then assessing antibody distribution by positron emission tomography scanning. Indeed, when an 124I-labeled GPIHBP1-specific monoclonal antibody was injected into wild-type mice, it was taken up by GPIHBP1-expressing endothelial cells in mouse tissues and quickly disappeared from the plasma. Interestingly, the liver removed most of the antibody, followed by the lung, but significant amounts were taken up by brown adipose tissue and heart. A sizeable amount of the GPIHBP1-specific monoclonal antibody persisted in tissues for >72 h, suggesting that the GPIHBP1 in capillary endothelial cells could have a long half-life.
Finding high levels of GPIHBP1 expression in capillary endothelial cells in mouse tissues led us to suspect that we might find high levels of GPIHBP1 expression in endothelial cell lines, but this was not the case. Davies et al. (68) were unable to detect Gpihbp1 expression (as judged by qRT-PCR) in rat heart microvascular endothelial cells (68), human umbilical vein endothelial cells (68), or bovine aortic endothelial cells (unpublished observations). They also isolated primary mouse endothelial cells from white adipose tissue and found that Gpihbp1 transcripts disappear after a single passage. Why Gpihbp1 expression is silenced in cultured endothelial cells is unknown. Gpihbp1 is not expressed in undifferentiated mouse embryonic stem cells, but expression is activated when the embryonic stem cells are differentiated into embryoid bodies. In embryoid bodies, GPIHBP1 is found in CD31-positive endothelial cells surrounding beating cardiomyocytes (68).
Relatively little is known about the transcription factors that regulate Gpihbp1 expression, but it seems likely that PPARγ plays a role. Administering a PPARγ agonist to mice markedly upregulates Gpihbp1 expression in mouse tissues, while PPARα and PPARδ agonists have no effect (68). Gpihbp1 expression in embryoid bodies is also upregulated when a PPARγ agonist is added to the medium. Of note, a PPARγ binding site is located immediately upstream from exon 1 of Gpihbp1, and that site exhibits activity in a luciferase reporter assay. Also, Gpihbp1 transcript levels in brown and white adipose tissue are higher in wild-type mice than in endothelial cell–specific PPARγ knockout mice, suggesting that PPARγ regulates Gpihbp1 expression levels in vivo (68).
Hypertriglyceridemia due to defective GPIHBP1ndashLPL interactions
The discovery of chylomicronemia in Gpihbp1−/− mice (33) prompted efforts to uncover GPIHBP1 mutations in humans with hypertriglyceridemia. Wang and Hegele (75) screened 160 human subjects with chylomicronemia and identified two siblings who were homozygous for a G56R substitution in GPIHBP1. This mutation was not found in normolipidemic controls. Several family members who were heterozygous for the G56R mutation had milder forms of hypertriglyceridemia. Because the substitution of an arginine for a glycine is not a conservative change and because Gly-56 is conserved in many mammalian species, Wang and Hegele (75) suggested that the G56R substitution was responsible for the chylomicronemia. Subsequent studies by Gin et al. (76) and Franssen, Young, and coworkers (25) examined the functional relevance of the G56R mutation. They expressed GPIHBP1 harboring the G56R mutation in CHO cells and found that it reached the cell surface efficiently and bound LPL as well as wild-type GPIHBP1. Finding that the GPIHBP1-G56R bound LDL normally raised doubts about its relevance to hypertriglyceridemia, although one could conceivably argue that the development of a knock-in mouse model (or the analysis of additional human families) would be necessary to assess the significance of the mutation in a definitive fashion. In any case, the report by Wang and Hegele (75) showed that GPIHBP1 mutations are rare, even in selected subjects with severe and unexplained hypertriglyceridemia.
Over the past few years, seven GPIHBP1 missense mutations that eliminate LPL binding have been identified in patients with chylomicronemia (Q115P, C65Y, C65S, C68G, C68Y, C89F, and G175R) (25, 27, 43, 77). All affected patients had two mutant alleles; heterozygotes were invariably normolipidemic. Thus, GPIHBP1 deficiency in humans is a recessive syndrome [as it is in mice (33)].
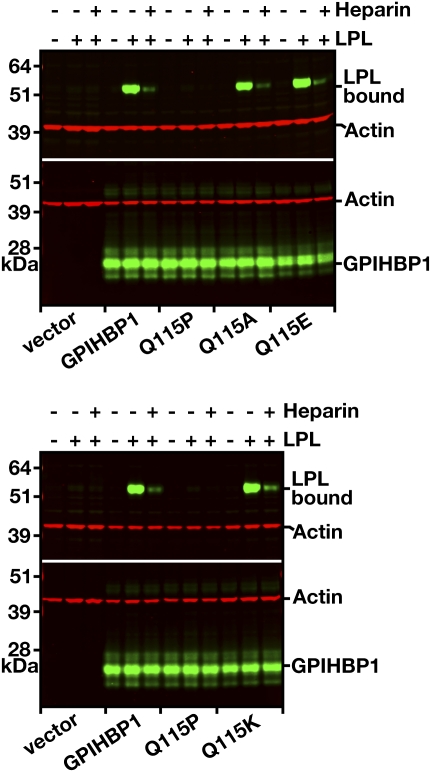
The first missense mutation with unequivocal functional consequences was a Q115P substitution (43), located in finger 3 of GPIHBP1’s three-fingered Ly6 motif. This mutation was identified by screening 60 patients with severe hypertriglyceridemia in whom coding-sequence mutations in LPL, APOC2, or APOA5 had been excluded (43). A young man with lifelong chylomicronemia and a history of hepatosplenomegaly and lipemia retinalis was homozygous for a Q115P substitution in GPIHBP1. On a low-fat diet, the plasma triglyceride level was 744 mg/dl. When GPIHBP1-Q115P was expressed in CHO cells, it reached the cell surface but could not bind LPL (Fig. 11), providing a likely mechanism for the patient's hyperlipidemia. Interestingly, introducing a lysine or a glutamate into residue 115 did not disrupt LPL binding (lysine is found in the dog GPIHBP1 sequence, while glutamate is found in the platypus sequence) (Fig. 11) (26).
Fig. 11.
Assessing the effects of mutations in Gln-115 of GPIHBP1 on GPIHBP1’s ability to bind LPL. CHO-K1 cells were electroporated with wild-type GPIHBP1 or mutant GPIHBP1 proteins containing mutations of Gln-115 (Q115A, Q115E, Q115K, Q115P). All constructs contained an S-protein tag. On the morning after the electroporation, the cells were incubated with V5-tagged human LPL (± heparin, 500 U/ml) for 2 h at 4°C. After washing the cells, Western blots of cell lysates were performed with a V5-specific antibody (to assess binding of LPL to cells) and an antibody against the S-protein tag (to assess GPIHBP1 expression levels). Actin was used as a loading control. Reproduced, with permission, from Beigneux et al. (26).
A homozygous C65Y mutation in GPIHBP1 was identified in a 3-year-old boy with chylomicronemia (25). The proband had no mutations in LPL, LMF1, APOC2, or APOA5 (25). The patient had plasma triglyceride levels of 1,575 mg/dl and a history of lipemia retinalis and pancreatitis. The proband's three siblings and his parents (who were first cousins) were heterozygous for the C65Y mutation and were normolipemic. When GPIHBP1-C65Y was expressed in CHO cells, it reached the cell surface but lacked the ability to bind LPL (25).
The C65S and C68G mutations were identified in a Swedish family in which three of four siblings had chylomicronemia (33). All three carried both a C65S allele and a C68G allele. The plasma triglycerides in the three compound heterozygotes were markedly elevated (798–2570 mg/dl), and several had experienced bouts of pancreatitis. The father carried the C65S mutation and the mother the C68G mutation; both were normolipidemic. Studies with transfected cells revealed that GPIHBP1-C65S and GPIHBP1-C68G reached the cell surface, but neither were capable of binding LPL (27). LPL mass and activity levels were normal in the adipose tissue of affected subjects—a finding that is consistent with the normal stores of LPL in tissues of Gpihbp1−/− mice (60). Interestingly, the authors examined the breast milk of one of the C65S/C68G compound heterozygotes. The LPL mass and activity levels in the milk of the compound heterozygote were as high or higher than in the milk from unaffected individuals, even though the lipid content of the milk was low and there was a shift toward medium-chain and saturated fatty acids (suggesting that the milk lipids were produced from de novo lipogenesis) (27). Similar abnormalities in the lipid content of milk have been observed in the setting of LPL deficiency (78, 79). Observing normal or elevated LPL levels in the milk was an intriguing finding and showed that GPIHBP1 defects do not impair LPL secretion by the mammary epithelium.
Coca-Prieto et al. (29) analyzed five patients with childhood-onset chylomicronemia. Four were homozygous for a common mutation in the catalytic domain of LPL (G188E), but one had a C68Y mutation in GPIHBP1. The effect of the C68Y mutation on LPL binding was not reported, but given the results of other studies (25, 27, 42), it is overwhelmingly likely that the C68Y mutation abolishes LPL binding.
Most recently, Charrière et al. (77) identified GPIHBP1 defects in two subjects among 376 unrelated patients with chylomicronemia (plasma triglycerides >1300 mg/dl) and no significant mutations in LPL, APOC2, or APOA5. The first patient, a young child with triglyceride levels >1700 mg/dl and a history of pancreatitis, had a de novo C89F mutation in one GPIHBP1 allele and a second mutant allele with a deletion of the entire GPIHBP1 gene (77). The second patient, a 35-year-old man with pancreatitis and severe chylomicronemia (plasma triglycerides, 2,280 mg/dl), was homozygous for a G175R mutation in GPIHBP1. The G175R mutation was located in the carboxyl-terminal hydrophobic domain that is normally replaced by the GPI anchor; presumably, that mutation interferes with efficient replacement of the hydrophobic domain by the GPI anchor. Heterozygotes in both pedigrees were normolipidemic. In cell transfection experiments, they found reduced amounts of the C89F and G175R at the cell surface and reduced amounts of LPL binding. In the case of the G175R mutation, reduced LPL binding appeared to be fully explained by reduced amounts of the protein at the cell surface. A very intriguing finding in the report by Charrière et al. (77) was that a commonly occurring C14F polymorphism in the signal peptide of GPIHBP1 was more frequent in the patients with chylomicronemia than in normolipidemic controls (P < 0.001) (77).
Gpihbp1−/− mice have low levels of LPL in preheparin plasma samples (60), and the same appears to be the case in humans with GPIHBP1 defects. The preheparin LPL mass levels in the three C65S/C68G compound heterozygotes were quite low, only 5–15% of those in normolipemic controls (27).
When Gpihbp1−/− mice were given intravenous heparin (50 U, ∼1450 U/kg), LPL mass and activity levels in the plasma at the 15-min time point were similar to those in wild-type mice (60). In contrast, LPL levels in the postheparin plasma are low in humans with GPIHBP1 defects. After an intravenous injection of heparin (50 U/kg), plasma LPL mass levels in the Q115P homozygote were ∼10% of those in controls (33). Similarly, the levels of LPL mass in the postheparin plasma of the three C65S/C68G compound heterozygotes were low, only ∼5% of those in the normolipidemic controls (27). Postheparin LPL activity levels were also extremely low or undetectable in the C65Y, C68Y, and G175R homozygotes and in the C89F patient (25, 29, 77). Of note, postheparin LPL levels were normal in the siblings with the G56R mutation (75), a mutation that had no effect on LPL binding.
It is not clear why the postheparin plasma LPL levels in patients with GPIHBP1 defects are so low, while they were high in Gpihbp1−/− mice, but one factor might be the dose of heparin used in humans, which was far lower than that used in the mouse experiments (60). Another potential explanation would be that mouse LPL is more readily released from its binding sites, compared with human LPL.
In the Gpihbp1−/− mice, the postheparin LPL levels reached levels similar to those in wild-type mice (60), but the kinetics of LPL entry into the plasma were delayed. Limited evidence suggests that this also occurs in humans with GPIHBP1 defects. In the C65S/C68G compound heterozygotes (27), LPL mass increased slowly for 20 to 60 min after heparin (25, 27). When the Q115P homozygote was infused with heparin for 6 h, LPL also entered the plasma slowly (25). The entry of LPL into the plasma of Gpihbp1−/− mice was accompanied by reduced plasma triglyceride levels (60). There is some evidence that this also occurs in humans. After administering heparin to the Q115P homozygote, active LPL entered the plasma and the triglyceride levels fell from 1,780 to 534 mg/dl (25). The entry of hepatic lipase into the plasma after heparin appears to be normal in both GPIHBP1-deficient humans (25, 27) and Gpihbp1−/− mice (33).
The discovery of point mutations in GPIHBP1 that abolish its ability to bind LPL suggested that there might be mirror image mutations in LPL that would abolish LPL's ability to bind to GPIHBP1. To address this possibility, Voss et al. (28) took a second look at LPL mutations that had been previously identified in patients with chylomicronemia. Two mutations, C418Y and E421K, were of particular interest because they were distant from LPL's N-terminal catalytic domain and because they were reported to have little or no effect on LPL's catalytic activity (80, 81).
The C418Y mutation was initially identified in a 30-year-old male with severe hypertriglyceridemia (plasma triglycerides, 2,368 mg/dl) and a history of pancreatitis (81). DNA sequencing revealed that the patient carried both an I194T mutation, which is known to abolish LPL catalytic activity (82), and a C418Y mutation. The patient's postheparin plasma contained a significant amount of LPL mass, but no LPL activity. COS cell transfection studies showed that the C418Y mutation had little effect on LPL catalytic activity (81).
The E421K mutation was found in a 24-year-old woman who succumbed to hypertriglyceridemia-related pancreatitis during pregnancy (80). The plasma triglyceride level was 7,632 mg/dl. The patient carried a single mutant LPL allele with an E421K mutation. Transfection experiments revealed that this mutation had no significant effects on enzymatic activity. Family members who carried the E421K mutation had mild hypertriglyceridemia and mildly reduced levels of LPL activity in their postheparin plasma (80).
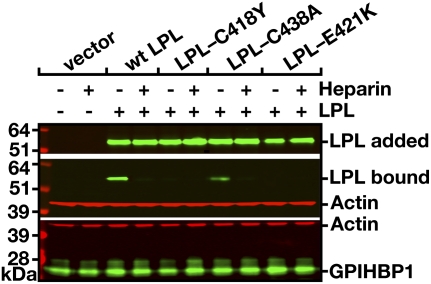
Voss et al. (28) hypothesized that the C418Y and E421K mutations (80, 81) might cause hypertriglyceridemia by abolishing LPL's ability to bind to GPIHBP1. They began by confirming that the enzymatic specific activities of the mutant LPLs were normal, and they further showed that the mutations had little effect on LPL binding to heparin. They then showed, with both a cell-based western blot binding assay (Fig. 12) and an immunofluorescence microscopy assay (Fig. 13), that both the C418Y and E421K mutations abolished LPL's ability to bind to GPIHBP1 (28). These mutations also abolished binding to GPIHBP1 when they were introduced into mouse or chicken LPL. Also, neither C418Y-LPL nor E421K-LPL was capable of binding to soluble GPIHBP1 in a cell-free assay. Other mutations known to abolish LPL catalytic activity, for example, I194T and S132G (82, 83), had no effect on LPL's ability to bind to GPIHBP1 (28).
Fig. 12.
Western blot assessing binding of wild-type human LPL, LPL-C418Y, LPL-C438A, and LPL-E421K to GPIHBP1-transfected CHO-K1 cells in the presence or absence of heparin (500 U/ml). After a 2-h incubation, cells were washed extensively, and cell extracts were prepared for Western blotting with GPIHBP1- and LPL-specific antibodies. β-actin levels were measured as a loading control. The top panel shows the amount of LPL present in the conditioned medium added to the GPIHBP1-transfected CHO-K1 cells. Reproduced, with permission, from Voss et al. (28).
Fig. 13.
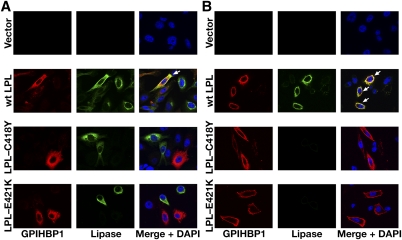
13. Immunofluorescence microscopy assay of LPL binding to GPIHBP1. CHO-K1 cells that had been transfected with S-protein–tagged human GPIHBP1 were mixed with CHO-K1 cells that had been transfected with wild-type human LPL, LPL-C418Y, or LPL-E421K and then plated on a coverslip. After a 2-h incubation at 37°C, permeabilized (A) and nonpermeabilized (B) cells were stained for GPIHBP1 with an antibody against the S-protein tag (red) and LPL with antibody 5D2 (green). Cell nuclei were stained with DAPI (blue). Wild-type LPL secreted by LPL-transfected cells was captured by neighboring GPIHBP1-expressing cells; thus, LPL and GPIHBP1 colocalize in the merged image (arrows). LPL-C418Y and LPL-E421K did not bind to GPIHBP1; hence, no colocalization was observed. Reproduced, with permission, from Voss et al. (28).
Voss et al. (28) predicted that C418Y-LPL and E421K-LPL would lack the ability to be transported across endothelial cells. Indeed, when C418Y-LPL and E421K-LPL were added to the basolateral medium of confluent GPIHBP1-expressing endothelial cells, they were not transported to the apical face of the cells. Their inability to be transported across endothelial cells likely explains the low postheparin LPL levels in carriers of the E421K mutation (80) and the absence of catalytically active LPL in the postheparin plasma of the C418Y/I194T compound heterozygote (81).
The C418Y mutation was intriguing because Cys-418 is involved in a disulfide bond with Cys-438 (67). Although this disulfide linkage is not important for LPL's catalytic activity (84), the inability of LPL-C418Y to bind to GPIHBP1 raised the possibility that the C418–C438 disulfide bond might be absolutely required for GPIHBP1 binding. This is apparently not the case, however, because LPL-C438A and LPL-C438Y manifested some, albeit reduced, ability to bind to GPIHBP1 (28).
The properties of the C418Y and E421K mutants implied that sequences in the carboxyl terminus of LPL are important for GPIHBP1 binding. Supporting this notion are the observations that a monoclonal antibody with an epitope in this region blocks LPL binding to GPIHBP1, and the fact that other mutations in this region abolish LPL's ability to bind to GPIHBP1 (28). It is tempting to speculate that this carboxyl-terminal domain (residues ∼418–435), which is located downstream from LPL's principal heparin-binding domain, interacts with GPIHBP1’s Ly6 domain.
We speculate that “two-domain interactions” between GPIHBP1 and LPL are physiologically important. We imagine that GPIHBP1’s acidic domain functions as a lasso, binding LPL's heparin-binding domains and pulling LPL away from the HSPGs in the interstitium. Once GPIHBP1’s acidic domain has dislodged the LPL, its Ly6 domain would engage LPL's carboxyl terminus, leading to higher affinity and perhaps nearly irreversible interactions. In this fashion, GPIHBP1 on endothelial cells could serve to sweep up interstitial LPL and transport it to the capillary lumen. We consider this two-domain model to be attractive, but fully evaluating domain–domain interactions will likely require cocrystallization of LPL and GPIHBP1.
Perspectives
Recent studies on GPIHBP1 solved a longstanding riddle in plasma triglyceride metabolism—how LPL that is secreted by parenchymal cells reaches the capillary lumen (Fig. 14) (24, 33). And follow-up investigations (25, 27, 28, 43) yielded fresh insights into human hyperlipidemia, revealing that some patients develop chylomicronemia as a result of mutations that interfere with GPIHBP1–LPL interactions (Fig. 14). But despite the progress, many topics remain to be investigated. For example, while GPIHBP1 is clearly crucial for transporting LPL across endothelial cells, mechanisms of lipolysis at the cell surface still need to be defined. Are GPIHBP1 and LPL located largely or exclusively in caveolae? And if so, is the clustering of LPL on the cell surface required for efficient lipolysis? Based on the known catalytic activity of LPL and the short half-life of chylomicrons in the plasma, Olivecrona et al. (85) calculated that a single LPL molecule would be utterly incapable of cleaving all of the triglycerides in a single chylomicron particle. They concluded that many molecules of LPL must act simultaneously on each particle. In the future, it will be important to determine if GPIHBP1 somehow facilitates LPL clustering on the cell surface—so that lipolysis can proceed rapidly.
Fig. 14.
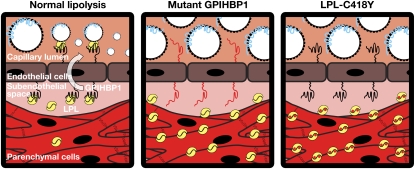
Schematic depicting the localization of LPL in the setting of normal lipolysis, in the setting of a GPIHBP1 mutant that cannot bind LPL (e.g., GPIHBP1-C65Y) (25), or in the setting of a LPL mutant (e.g., LPL-C418Y) that lacks the ability to bind GPIHBP1 (28). When LPL cannot bind to GPIHBP1, LPL does not reach the capillary lumen, resulting in defective lipolysis and chylomicronemia.
Also, mechanisms of GPIHBP1 and LPL movement across cells still need to be defined. Do GPIHBP1 and LPL move across endothelial cells in vesicles derived from caveolae? Is the movement of LPL and GPIHBP1 unidirectional (basolateral to apical) or bidirectional? Are other cellular proteins, aside from GPIHBP1, crucial for moving GPIHBP1 and LPL across cells? Might some cases of hypertriglyceridemia in humans be caused by defects in the machinery for moving vesicles across cells? Could some cases of acquired hypertriglyceridemia be caused by endothelial cell dysfunction and sluggish GPIHBP1/LPL transport? Are there polymorphisms in LPL that increase binding to GPIHBP1 and augment LPL transport to the capillary lumen? These questions need to be answered. In addition, more work is needed to determine if GPIHBP1 modulates the catalytic activity of one or both of LPL's partner monomers, or if GPIHBP1 is relevant to the inhibition of LPL activity by ANGPTL3 and ANGPTL4.
Also needed is a better understanding of LPL sequences that interact with GPIHBP1. Amino acids 416–435 appear to be important (28), but no one knows if other sequences play a role. Also, the fact that LPL is secreted as a homodimer needs to be considered. Do both partner monomers bind to GPIHBP1 simultaneously? Or is it possible that LPL behaves as a functional heterodimer (86) and that only one of the two monomers binds to GPIHBP1? We know that denatured LPL cannot bind to GPIHBP1 (62), but it will be important to determine if a properly folded carboxyl-terminal fragment of LPL retains the capacity to bind to GPIHBP1. If so, would it be possible to purify such a fragment and define its structure? Ultimately, it would be highly desirable to crystallize a GPIHBP1–LPL complex.
The discovery of GPIHBP1 has cast a spotlight on other lingering mysteries in lipolysis. Immunocytochemistry studies have shown that GPIHBP1 is located only in the smallest capillaries of adipose tissue and striated muscle, and that it is absent from larger blood vessels (Figs. 1, 4) (24). What factors regulate this expression pattern? Are paracrine signals from parenchymal cells involved? Or is the distinctive expression pattern of GPIHBP1 “hardwired” by specific DNA enhancer elements? Also, little information exists on the molecular interactions that lead chylomicrons to marginate within capillaries—so that lipolysis can proceed. Does chylomicron margination depend on interactions between apolipoproteins and HSPGs on the luminal surface of capillaries, or does it depend solely on interactions of chylomicron lipids with GPIHBP1-bound LPL? If LPL is the key factor in chylomicron margination within capillaries, would chylomicrons simply “flow on by” when GPIHBP1 is absent? In any case, it will be important to define which molecular interactions—and which protein sequences—are involved in chylomicron margination within capillaries.
The discovery of GPIHBP1 also raises questions about LPL expression in the brain. LPL is expressed highly in certain regions of the brain, as judged by in situ hybridization (69–73), but there is little GPIHBP1 in the brain. What underlies this discrepancy? Is there another LPL transporter in the brain? Or could LPL have an extravascular role in the brain?
GPIHBP1 is easily identified in genomic databases of all mammals, including the egg-laying platypus (32), but no one has yet identified GPIHBP1 in other vertebrates (e.g., fish, amphibians, birds). The fact that GPIHBP1 is apparently absent in other vertebrates is surprising because those species also produce triglyceride-rich lipoproteins and express a highly conserved LPL protein. What could explain the apparent absence of GPIHBP1? No one knows, but it is possible to imagine several possibilities. Perhaps endothelial cells of other vertebrates synthesize some LPL, rendering an LPL transporter less important. Or perhaps another LPL transporter, with little or no homology to GPIHBP1, exists in other vertebrates. Still another possibility is that the LPL in other vertebrates is not transported at all and remains trapped in the extravascular spaces. Rather than moving LPL to the intravascular compartment where lipoproteins are located (the “mammalian strategy”), perhaps other vertebrates move lipoproteins to the extravascular compartment where LPL resides. Regardless of which explanation applies, we suggest that GPIHBP1 may be a defining characteristic of mammals. And we would further suggest that GPIHBP1’s appearance in mammals could be related to another defining feature of mammals—nursing the young. Nursing requires the secretion of prodigious amounts of triglycerides into the milk, and that in turn depends on extraction of triglycerides from triglyceride-rich lipoproteins. Perhaps GPIHBP1 expression—and moving LPL to the capillary lumen—improves the efficiency of triglyceride metabolism and simultaneously facilitates the production of milk fat by the mammary gland.
Footnotes
Abbreviations:
- ANGPTL
- angiopoietin-like protein
- BODIPY
- boron-dipyrromethane
- CHO
- Chinese hamster ovary
- cLPL
- chicken lipoprotein lipase
- DiI
- 1,1-dioctadecyl-3,3,3,3-tetramethylindocarbocyanine perchlorate
- EL
- endothelial lipase
- GPI
- glycosylphosphatidylinositol
- GPIHBP1
- glycosylphosphatidylinositol-anchored high density lipoprotein–binding protein 1
- HSPG
- heparan sulfate proteoglycan
- Ly6
- Lymphocyte Antigen 6
- PIPLC
- phosphatidylinositol-specific phospholipase C
- SR-BI
- scavenger receptor class B, type 1
This work was supported by a Scientist Development Award from the American Heart Association, National Office (to A.P.B.), R01 HL094732 (to A.P.B.), P01 HL090553 (to S.G.Y.), and R01 HL087228 (to S.G.Y.). The authors have declared that no conflict of interest exists.
References
- 1.Havel R. J., Kane J. P. 2001. Introduction: Structure and metabolism of plasma lipoproteins. The Metabolic and Molecular Bases of Inherited Disease. Scriver C. R., Beaudet A. L., Sly W. S., Valle D., Childs B., Kinzler K. W., Vogelstein B., McGraw-Hill, New York: 2705–2716 [Google Scholar]
- 2.Havel R. J. 2010. Triglyceride-rich lipoproteins and plasma lipid transport. Arterioscler. Thromb. Vasc. Biol. 30: 9–19 [DOI] [PubMed] [Google Scholar]
- 3.Brunzell J. D., Deeb S. S. 2001. Familial lipoprotein lipase deficiency, apo C–II deficiency, and hepatic lipase deficiency. The Metabolic and Molecular Bases of Inherited Disease. Scriver C. R., Beaudet A. L., Sly W. S., Valle D., Childs B., Kinzler K. W., Vogelstein B., McGraw-Hill, New York: 2789–2816 [Google Scholar]
- 4.Havel R. J., Gordon R. S., Jr 1960. Idiopathic hyperlipemia: metabolic studies in an affected family. J. Clin. Invest. 39: 1777–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Connelly P. W., Maguire G. F., Little J. A. 1987. Apolipoprotein CIISt. Michael. Familial apolipoprotein CII deficiency associated with premature vascular disease. J. Clin. Invest. 80: 1597–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pennacchio L. A., Olivier M., Hubacek J. A., Krauss R. M., Rubin E. M., Cohen J. C. 2002. Two independent apolipoprotein A5 haplotypes influence human plasma triglyceride levels. Hum. Mol. Genet. 11: 3031–3038 [DOI] [PubMed] [Google Scholar]
- 7.Ito Y., Azrolan N., O'Connell A., Walsh A., Breslow J. L. 1990. Hypertriglyceridemia as a result of human apo CIII gene expression in transgenic mice. Science. 249: 790–793 [DOI] [PubMed] [Google Scholar]
- 8.Ginsberg H. N., Le N-A., Goldberg I. J., Gibson J. C., Rubinstein A., Wang-Iverson P., Norum R., Brown W. V. 1986. Apolipoprotein B metabolism in subjects with deficiency of apolipoproteins CIII and AI. Evidence that apolipoprotein CIII inhibits catabolism of triglyceride-rich lipoproteins by lipoprotein lipase in vivo. J. Clin. Invest. 78: 1287–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshida K., Shimizugawa T., Ono M., Furukawa H. 2002. Angiopoietin-like protein 4 is a potent hyperlipidemia-inducing factor in mice and inhibitor of lipoprotein lipase. J. Lipid Res. 43: 1770–1772 [DOI] [PubMed] [Google Scholar]
- 10.Köster A., Chao Y. B., Mosior M., Ford A., Gonzalez-DeWhitt P. A., Hale J. E., Li D., Qiu Y., Fraser C. C., Yang D. D., et al. 2005. Transgenic angiopoietin-like (angptl)4 overexpression and targeted disruption of angptl4 and angptl3: regulation of triglyceride metabolism. Endocrinology. 146: 4943–4950 [DOI] [PubMed] [Google Scholar]
- 11.Koishi R., Ando Y., Ono M., Shimamura M., Yasumo H., Fujiwara T., Horikoshi H., Furukawa H. 2002. Angptl3 regulates lipid metabolism in mice. Nat. Genet. 30: 151–157 [DOI] [PubMed] [Google Scholar]
- 12.Péterfy M., Ben-Zeev O., Mao H. Z., Weissglas-Volkov D., Aouizerat B. E., Pullinger C. R., Frost P. H., Kane J. P., Malloy M. J., Reue K., et al. 2007. Mutations in LMF1 cause combined lipase deficiency and severe hypertriglyceridemia. Nat. Genet. 39: 1483–1487 [DOI] [PubMed] [Google Scholar]
- 13.Korn E. D. 1955. Clearing factor, a heparin-activated lipoprotein lipase. II. Substrate specificity and activation of coconut oil. J. Biol. Chem. 215: 15–26 [PubMed] [Google Scholar]
- 14.Robinson D. S., French J. E. 1960. Heparin, the clearing factor lipase, and fat transport. Pharmacol. Rev. 12: 241–263 [PubMed] [Google Scholar]
- 15.Davis R. C., Wong H., Nikazy J., Wang K., Han Q., Schotz M. C. 1992. Chimeras of hepatic lipase and lipoprotein lipase. Domain localization of enzyme-specific properties. J. Biol. Chem. 267: 21499–21504 [PubMed] [Google Scholar]
- 16.Hata A., Ridinger D. N., Sutherland S., Emi M., Shuhua Z., Myers R. L., Ren K., Cheng T., Inoue I., Wilson D. E., et al. 1993. Binding of lipoprotein lipase to heparin. Identification of five critical residues in two distinct segments of the amino-terminal domain. J. Biol. Chem. 268: 8447–8457 [PubMed] [Google Scholar]
- 17.Sendak R. A., Melford K., Kao A., Bensadoun A. 1998. Identification of the epitope of a monoclonal antibody that inhibits heparin binding of lipoprotein lipase: new evidence for a carboxyl-terminal heparin-binding domain. J. Lipid Res. 39: 633–646 [PubMed] [Google Scholar]
- 18.Cheng C. F., Oosta G. M., Bensadoun A., Rosenberg R. D. 1981. Binding of lipoprotein lipase to endothelial cells in culture. J. Biol. Chem. 256: 12893–12898 [PubMed] [Google Scholar]
- 19.Lookene A., Chevreuil O., Ostergaard P., Olivecrona G. 1996. Interaction of lipoprotein lipase with heparin fragments and with heparan sulfate: stoichiometry, stabilization, and kinetics. Biochemistry. 35: 12155–12163 [DOI] [PubMed] [Google Scholar]
- 20.Lookene A., Savonen R., Olivecrona G. 1997. Interaction of lipoproteins with heparan sulfate proteoglycans and with lipoprotein lipase. Studies by surface plasmon resonance technique. Biochemistry. 36: 5267–5275 [DOI] [PubMed] [Google Scholar]
- 21.Shimada K., Gill P. J., Silbert J. E., Douglas W. H., Fanburg B. L. 1981. Involvement of cell surface heparin sulfate in the binding of lipoprotein lipase to cultured bovine endothelial cells. J. Clin. Invest. 68: 995–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Obunike J. C., Lutz E. P., Li Z., Paka L., Katopodis T., Strickland D. K., Kozarsky K. F., Pillarisetti S., Goldberg I. J. 2001. Transcytosis of lipoprotein lipase across cultured endothelial cells requires both heparan sulfate proteoglycans and the very low density lipoprotein receptor. J. Biol. Chem. 276: 8934–8941 [DOI] [PubMed] [Google Scholar]
- 23.Frykman P. K., Brown M. S., Yamamoto T., Goldstein J. L., Herz J. 1995. Normal plasma lipoproteins and fertility in gene-targeted mice homozygous for a disruption in the gene encoding very low density lipoprotein receptor. Proc. Natl. Acad. Sci. USA. 92: 8453–8457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davies B. S. J., Beigneux A. P., Barnes R. H., II, Tu Y., Gin P., Weinstein M. M., Nobumori C., Nyrén R., Goldberg I. J., Olivecrona G., et al. 2010. GPIHBP1 is responsible for the entry of lipoprotein lipase into capillaries. Cell Metab. 12: 42–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franssen R., Young S. G., Peelman F., Hertecant J., Sierts J. A., Schimmel A. W. M., Bensadoun A., Kastelein J. J. P., Fong L. G., Dallinga-Thie G. M., et al. 2010. Chylomicronemia with low postheparin lipoprotein lipase levels in the setting of GPIHBP1 defects. Circ. Cardiovasc. Genet. 3: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beigneux A. P., Davies B. S., Tat S., Chen J., Gin P., Voss C. V., Weinstein M. M., Bensadoun A., Pullinger C. R., Fong L. G., et al. 2011. Assessing the role of the glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 (GPIHBP1) three-finger domain in binding lipoprotein lipase. J. Biol. Chem. 286: 19735–19743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olivecrona G., Ehrenborg E., Semb H., Makoveichuk E., Lindberg A., Hayden M. R., Gin P., Davies B. S. J., Weinstein M. M., Fong L. G., et al. 2010. Mutation of conserved cysteines in the Ly6 domain of GPIHBP1 in familial chylomicronemia. J. Lipid Res. 51: 1535–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voss C. V., Davies B. S., Tat S., Gin P., Fong L. G., Pelletier C., Mottler C. D., Bensadoun A., Beigneux A. P., Young S. G. 2011. Mutations in lipoprotein lipase that block binding to the endothelial cell transporter GPIHBP1. Proc. Natl. Acad. Sci. USA. 108: 7980–7984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coca-Prieto I., Kroupa O., Gonzalez-Santos P., Magne J., Olivecrona G., Ehrenborg E., Valdivielso P. 2011. Childhood-onset chylomicronaemia with reduced plasma lipoprotein lipase activity and mass: identification of a novel GPIHBP1 mutation. J Intern. Med. 270: 224–228 [DOI] [PubMed] [Google Scholar]
- 30.Ioka R. X., Kang M-J., Kamiyama S., Kim D-H., Magoori K., Kamataki A., Ito Y., Takei Y. A., Sasaki M., Suzuki T., et al. 2003. Expression cloning and characterization of a novel glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein, GPI-HBP1. J. Biol. Chem. 278: 7344–7349 [DOI] [PubMed] [Google Scholar]
- 31.Acton S., Rigotti A., Landschulz K. T., Xu S., Hobbs H. H., Krieger M. 1996. Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science. 271: 518–520 [DOI] [PubMed] [Google Scholar]
- 32.Young S. G., Davies B. S., Fong L. G., Gin P., Weinstein M. M., Bensadoun A., Beigneux A. P. 2007. GPIHBP1: an endothelial cell molecule important for the lipolytic processing of chylomicrons. Curr. Opin. Lipidol. 18: 389–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beigneux A. P., Davies B., Gin P., Weinstein M. M., Farber E., Qiao X., Peale P., Bunting S., Walzem R. L., Wong J. S., et al. 2007. Glycosylphosphatidylinositol-anchored high density lipoprotein–binding protein 1 plays a critical role in the lipolytic processing of chylomicrons. Cell Metab. 5: 279–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beigneux A. P., Gin P., Davies B. S., Weinstein M. M., Bensadoun A., Ryan R. O., Fong L. G., Young S. G. 2008. Glycosylation of Asn-76 in mouse GPIHBP1 is critical for its appearance on the cell surface and the binding of chylomicrons and lipoprotein lipase. J. Lipid Res. 49: 1312–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levitin F., Weiss M., Hahn Y., Stern O., Papke R. L., Matusik R., Nandana S. R., Ziv R., Pichinuk E., Salame S., et al. 2008. PATE gene clusters code for multiple, secreted TFP/Ly-6/uPAR proteins that are expressed in reproductive and neuron-rich tissues and possess neuromodulatory activity. J. Biol. Chem. 283: 16928–16939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beigneux A. P., Davies B. S., Bensadoun A., Fong L. G., Young S. G. 2009. GPIHBP1, a GPI-anchored protein required for the lipolytic processing of triglyceride-rich lipoproteins. J. Lipid Res. 50(Suppl): S57–S62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Llinas P., Le Du M. H., Gardsvoll H., Dano K., Ploug M., Gilquin B., Stura E. A., Menez A. 2005. Crystal structure of the human urokinase plasminogen activator receptor bound to an antagonist peptide. EMBO J. 24: 1655–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang Y., Fedarovich A., Tomlinson S., Davies C. 2007. Crystal structure of CD59: implications for molecular recognition of the complement proteins C8 and C9 in the membrane-attack complex. Acta Crystallogr. D Biol. Crystallogr. 63: 714–721 [DOI] [PubMed] [Google Scholar]
- 39.Favre B., Plantard L., Aeschbach L., Brakch N., Christen-Zaech S., de Viragh P. A., Sergeant A., Huber M., Hohl D. 2007. SLURP1 is a late marker of epidermal differentiation and is absent in Mal de Meleda. J. Invest. Dermatol. 127: 301–308 [DOI] [PubMed] [Google Scholar]
- 40.Mallya M., Campbell R. D., Aguado B. 2006. Characterization of the five novel Ly-6 superfamily members encoded in the MHC, and detection of cells expressing their potential ligands. Protein Sci. 15: 2244–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kieffer B., Driscoll P. C., Campbell I. D., Willis A. C., van der Merwe P. A., Davis S. J. 1994. Three-dimensional solution structure of the extracellular region of the complement regulatory protein CD59, a new cell-surface protein domain related to snake venom neurotoxins. Biochemistry. 33: 4471–4482 [PubMed] [Google Scholar]
- 42.Beigneux A. P., Gin P., Davies B. S. J., Weinstein M. M., Bensadoun A., Fong L. G., Young S. G. 2009. Highly conserved cysteines within the Ly6 domain of GPIHBP1 are crucial for the binding of lipoprotein lipase. J. Biol. Chem. 284: 30240–30247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beigneux A. P., Franssen R., Bensadoun A., Gin P., Melford K., Peter J., Walzem R. L., Weinstein M. M., Davies B. S., Kuivenhoven J. A., et al. 2009. Chylomicronemia with a mutant GPIHBP1 (Q115P) that cannot bind lipoprotein lipase. Arterioscler. Thromb. Vasc. Biol. 29: 956–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kjaergaard M., Hansen L. V., Jacobsen B., Gardsvoll H., Ploug M. 2008. Structure and ligand interactions of the urokinase receptor (uPAR). Front. Biosci. 13: 5441–5461 [DOI] [PubMed] [Google Scholar]
- 45.Galat A., Gross G., Drevet P., Sato A., Menez A. 2008. Conserved structural determinants in three-fingered protein domains. FEBS J. 275: 3207–3225 [DOI] [PubMed] [Google Scholar]
- 46.Fry B. G., Wuster W., Kini R. M., Brusic V., Khan A., Venkataraman D., Rooney A. P. 2003. Molecular evolution and phylogeny of elapid snake venom three-finger toxins. J. Mol. Evol. 57: 110–129 [DOI] [PubMed] [Google Scholar]
- 47.Kriegbaum M., Persson M., Haldager L., Alpizar-Alpizar W., Jacobsen B., Gårdsvoll H., Kjær A., Ploug M. 2011. Rational targeting of the urokinase receptor (uPAR): development of antagonists and non-invasive imaging probes. Curr. Drug Targets. Epub ahead of print. June 27, 2011 [DOI] [PubMed] [Google Scholar]
- 48.Adermann K., Wattler F., Wattler S., Heine G., Meyer M., Forssmann W. G., Nehls M. 1999. Structural and phylogenetic characterization of human SLURP-1, the first secreted mammalian member of the Ly-6/uPAR protein superfamily. Protein Sci. 8: 810–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gin P., Beigneux A. P., Voss C., Davies B. S., Beckstead J. A., Ryan R. O., Bensadoun A., Fong L. G., Young S. G. 2011. Binding preferences for GPIHBP1, a glycosylphosphatidylinositol-anchored protein of capillary endothelial cells. Arterioscler. Thromb. Vasc. Biol. 31: 176–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Olivecrona G., Vilaro S., Esko J. D., Olivecrona T. 1996. Synthesis and secretion of lipoprotein lipase in heparan sulfate-deficient Chinese hamster ovary cells. Isr. J. Med. Sci. 32: 430–444 [PubMed] [Google Scholar]
- 51.Tang T., Li L., Tang J., Li Y., Lin W. Y., Martin F., Grant D., Solloway M., Parker L., Ye W., et al. 2010. A mouse knockout library for secreted and transmembrane proteins. Nat. Biotechnol. 28: 749–755 [DOI] [PubMed] [Google Scholar]
- 52.Weinstein M. M., Yin L., Tu Y., Wang X., Wu X., Castellani L. W., Walzem R. L., Lusis A. J., Fong L. G., Beigneux A. P., et al. 2010. Chylomicronemia elicits atherosclerosis in mice. Arterioscler. Thromb. Vasc. Biol. 30: 20–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lookene A., Beckstead J. A., Nilsson S., Olivecrona G., Ryan R. O. 2005. Apolipoprotein A-V-heparin interactions: implications for plasma lipoprotein metabolism. J. Biol. Chem. 280: 25383–25387 [DOI] [PubMed] [Google Scholar]
- 54.Strauss J. G., Frank S., Kratky D., Hammerle G., Hrzenjak A., Knipping G., von Eckardstein A., Kostner G. M., Zechner R. 2001. Adenovirus-mediated rescue of lipoprotein lipase-deficient mice. Lipolysis of triglyceride-rich lipoproteins is essential for high density lipoprotein maturation in mice. J. Biol. Chem. 276: 36083–36090 [DOI] [PubMed] [Google Scholar]
- 55.Zhang X., Qi R., Xian X., Yang F., Blackstein M., Deng X., Fan J., Ross C., Karasinska J., Hayden M. R., et al. 2008. Spontaneous atherosclerosis in aged lipoprotein lipase deficient mice with severe hypertriglyceridemia on a normal chow diet. Circ. Res. 102: 250–256 [DOI] [PubMed] [Google Scholar]
- 56.Weinstock P. H., Bisgaier C. L., Aalto-Setälä K., Radner H., Ramakrishnan R., Levak-Frank S., Essenburg A. D., Zechner R., Breslow J. L. 1995. Severe hypertriglyceridemia, reduced high density lipoprotein, and neonatal death in lipoprotein lipase knockout mice. Mild hypertriglyceridemia with impaired low density lipoprotein clearance in heterozygotes. J. Clin. Invest. 96: 2555–2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weinstein M. M., Tu Y., Beigneux A. P., Davies B. S., Gin P., Voss C., Walzem R. L., Reue K., Tontonoz P., Bensadoun A., et al. 2010. Cholesterol intake modulates plasma triglyceride levels in glycosylphosphatidylinositol HDL-binding protein 1-deficient mice. Arterioscler. Thromb. Vasc. Biol. 30: 2106–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sonnenburg W. K., Yu D., Lee E. C., Xiong W., Gololobov G., Key B., Gay J., Wilganowski N., Hu Y., Zhao S., et al. 2009. GPIHBP1 stabilizes lipoprotein lipase and prevents its inhibition by angiopoietin-like 3 and angiopoietin-like 4. J. Lipid Res. 50: 2421–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu J., Afroza H., Rader D. J., Jin W. 2010. Angiopoietin-like protein 3 inhibits lipoprotein lipase activity through enhancing its cleavage by proprotein convertases. J. Biol. Chem. 285: 27561–27570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weinstein M. M., Beigneux A. P., Davies B. S., Gin P., Yin L., Estrada K., Melford K., Bishop J. R., Esko J. D., Fong L. G., et al. 2008. Abnormal patterns of lipoprotein lipase release into the plasma in GPIHBP1-deficient mice. J. Biol. Chem. 283: 34511–34518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sharom F. J., Lehto M. T. 2002. Glycosylphosphatidylinositol-anchored proteins: structure, function, and cleavage by phosphatidylinositol-specific phospholipase C. Biochem. Cell Biol. 80: 535–549 [DOI] [PubMed] [Google Scholar]
- 62.Gin P., Yin L., Davies B. S., Weinstein M. M., Ryan R. O., Bensadoun A., Fong L. G., Young S. G., Beigneux A. P. 2008. The acidic domain of GPIHBP1 is important for the binding of lipoprotein lipase and chylomicrons. J. Biol. Chem. 283: 29554–29562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bodian D. L., Davis S. J., Morgan B. P., Rushmere N. K. 1997. Mutational analysis of the active site and antibody epitopes of the complement-inhibitory glycoprotein, CD59. J. Exp. Med. 185: 507–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gårdsvoll H., Dano K., Ploug M. 1999. Mapping part of the functional epitope for ligand binding on the receptor for urokinase-type plasminogen activator by site-directed mutagenesis. J. Biol. Chem. 274: 37995–38003 [DOI] [PubMed] [Google Scholar]
- 65.Gårdsvoll H., Gilquin B., Le Du M. H., Ménèz A., Jørgensen T. J. D., Ploug M. 2006. Characterization of the functional epitope on the urokinase receptor. J. Biol. Chem. 281: 19260–19272 [DOI] [PubMed] [Google Scholar]
- 66.Lin L., Gardsvoll H., Huai Q., Huang M., Ploug M. 2010. Structure-based engineering of species selectivity in the interaction between urokinase and its receptor: implication for preclinical cancer therapy. J. Biol. Chem. 285: 10982–10992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang C. Y., Gu Z. W., Yang H. X., Rohde M. F., Gotto A. M., Jr., Pownall H. J. 1989. Structure of bovine milk lipoprotein lipase. J. Biol. Chem. 264: 16822–16827 [PubMed] [Google Scholar]
- 68.Davies B. S., Waki H., Beigneux A. P., Farber E., Weinstein M. M., Wilpitz D. C., Tai L. J., Evans R. M., Fong L. G., Tontonoz P., et al. 2008. The expression of GPIHBP1, an endothelial cell binding site for lipoprotein lipase and chylomicrons, is induced by peroxisome proliferator-activated receptor-gamma. Mol. Endocrinol. 11: 2496–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ben-Zeev O., Doolittle M. H., Singh N., Chang C. H., Schotz M. C. 1990. Synthesis and regulation of lipoprotein lipase in the hippocampus. J. Lipid Res. 31: 1307–1313 [PubMed] [Google Scholar]
- 70.Bessesen D. H., Richards C. L., Etienne J., Goers J. W., Eckel R. H. 1993. Spinal cord of the rat contains more lipoprotein lipase than other brain regions. J. Lipid Res. 34: 229–238 [PubMed] [Google Scholar]
- 71.Goldberg I. J., Soprano D. R., Wyatt M. L., Vanni T. M., Kirchgessner T. G., Schotz M. C. 1989. Localization of lipoprotein lipase mRNA in selected rat tissues. J. Lipid Res. 30: 1569–1577 [PubMed] [Google Scholar]
- 72.Vilaró S., Camps L., Reina M., Perez-Clausell J., Llobera M., Olivecrona T. 1990. Localization of lipoprotein lipase to discrete areas of the guinea pig brain. Brain Res. 506: 249–253 [DOI] [PubMed] [Google Scholar]
- 73.Yacoub L. K., Vanni T. M., Goldberg I. J. 1990. Lipoprotein lipase mRNA in neonatal and adult mouse tissues: comparison of normal and combined lipase deficiency (cld) mice assessed by in situ hybridization. J. Lipid Res. 31: 1845–1852 [PubMed] [Google Scholar]
- 74.Olafsen T., Young S. G., Davies B. S., Beigneux A. P., Kenanova V. E., Voss C., Young G., Wong K. P., Barnes R. H., 2nd, Tu Y., et al. 2010. Unexpected expression pattern for glycosylphosphatidylinositol-anchored HDL-binding protein 1 (GPIHBP1) in mouse tissues revealed by positron emission tomography scanning. J. Biol. Chem. 285: 39239–39248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang J., Hegele R. A. 2007. Homozygous missense mutation (G56R) in glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 (GPI-HBP1) in two siblings with fasting chylomicronemia (MIM 144650). Lipids Health Dis. 6: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gin P., Beigneux A. P., Davies B., Young M. F., Ryan R. O., Bensadoun A., Fong L. G., Young S. G. 2007. Normal binding of lipoprotein lipase, chylomicrons, and apo-AV to GPIHBP1 containing a G56R amino acid substitution. Biochim. Biophys. Acta. 1771: 1464–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Charrière S., Peretti N., Bernard S., Charcosset M., Sassolas A., Lachaux A., Moulin P., Marçais C. 2011. GPIHBP1 C89F neomutation and hydrophobic C-terminal domain G175R mutation in two pedigrees with severe hyperchylomicronemia. J. Clin. Endocrinol. Metab. In press [DOI] [PubMed] [Google Scholar]
- 78.Berger G. M., Spark A., Baillie P. M., Huskisson J., Stockwell G., van der Merwe E. 1983. Absence of serum-stimulated lipase activity and altered lipid content in milk from a patient with type I hyperlipoproteinaemia. Pediatr. Res. 17: 835–839 [DOI] [PubMed] [Google Scholar]
- 79.Steiner G., Myher J. J., Kuksis A. 1985. Milk and plasma lipid composition in a lactating patient with type I hyperlipoproteinemia. Am. J. Clin. Nutr. 41: 121–128 [DOI] [PubMed] [Google Scholar]
- 80.Henderson H., Leisegang F., Hassan F., Hayden M., Marais D. 1998. A novel Glu421Lys substitution in the lipoprotein lipase gene in pregnancy-induced hypertriglyceridemic pancreatitis. Clin. Chim. Acta. 269: 1–12 [DOI] [PubMed] [Google Scholar]
- 81.Henderson H. E., Hassan F., Marais D., Hayden M. R. 1996. A new mutation destroying disulphide bridging in the C-terminal domain of lipoprotein lipase. Biochem. Biophys. Res. Commun. 227: 189–194 [DOI] [PubMed] [Google Scholar]
- 82.Henderson H. E., Ma Y., Hassan M. F., Monsalve M. V., Marais A. D., Winkler F., Gubernator K., Peterson J., Brunzell J. D., Hayden M. R. 1991. Amino acid substitution (Ile194—-Thr) in exon 5 of the lipoprotein lipase gene causes lipoprotein lipase deficiency in three unrelated probands. Support for a multicentric origin. J. Clin. Invest. 87: 2005–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Emmerich J., Beg O. U., Peterson J., Previato L., Brunzell J. D., Brewer H. B., Jr., Santamarina-Fojo S. 1992. Human lipoprotein lipase. Analysis of the catalytic triad by site-directed mutagenesis of Ser-132, Asp-156, and His-241. J. Biol. Chem. 267: 4161–4165 [PubMed] [Google Scholar]
- 84.Lo J. Y., Smith L. C., Chan L. 1995. Lipoprotein lipase: role of intramolecular disulfide bonds in enzyme catalysis. Biochem. Biophys. Res. Commun. 206: 266–271 [DOI] [PubMed] [Google Scholar]
- 85.Olivecrona T., Bengtsson G., Marklund S-E., Lindahl U., Höök M. 1977. Heparin–lipoprotein lipase interactions. Fed. Proc. 36: 60–65 [PubMed] [Google Scholar]
- 86.Yuan C., Sidhu R. S., Kuklev D. V., Kado Y., Wada M., Song I., Smith W. L. 2009. Cyclooxygenase allosterism, fatty acid-mediated cross-talk between monomers of cyclooxygenase homodimers. J. Biol. Chem. 284: 10046–10055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu M. S., Ma Y., Hayden M. R., Brunzell J. D. 1992. Mapping of the epitope on lipoprotein lipase recognized by a monoclonal antibody (5D2) which inhibits lipase activity. Biochim. Biophys. Acta. 1128: 113–115 [DOI] [PubMed] [Google Scholar]
- 88.Chang S-F., Reich B., Brunzell J. D., Will H. 1998. Detailed characterization of the binding site of the lipoprotein lipase-specific monoclonal antibody 5D2. J. Lipid Res. 39: 2350–2359 [PubMed] [Google Scholar]
- 89.Fletcher C. M., Harrison R. A., Lachmann P. J., Neuhaus D. 1994. Structure of a soluble, glycosylated form of the human complement regulatory protein CD59. Structure. 2: 185–199 [DOI] [PubMed] [Google Scholar]