Abstract
A-C1 protein is the product of a tumor suppressor gene negatively regulating the oncogene Ras and belongs to the HRASLS (HRAS-like suppressor) subfamily. We recently found that four members of this subfamily expressed in human tissues function as phospholipid-metabolizing enzymes. Here we examined a possible enzyme activity of A-C1. The homogenates of COS-7 cells overexpressing recombinant A-C1s from human, mouse, and rat showed a phospholipase A1/2 (PLA1/2) activity toward phosphatidylcholine (PC). This finding was confirmed with the purified A-C1. The activity was Ca2+ independent, and dithiothreitol and Nonidet P-40 were indispensable for full activity. Phosphatidylethanolamine (PE) was also a substrate and the phospholipase A1 (PLA1) activity was dominant over the PLA2 activity. Furthermore, the protein exhibited acyltransferase activities transferring an acyl group of PCs to the amino group of PEs and the hydroxyl group of lyso PCs. As for tissue distribution in human, mouse, and rat, A-C1 mRNA was abundantly expressed in testis, skeletal muscle, brain, and heart. These results demonstrate that A-C1 is a novel phospholipid-metabolizing enzyme. Moreover, the fact that all five members of the HRASLS subfamily, including A-C1, show similar catalytic properties strongly suggests that these proteins constitute a new class of enzymes showing PLA1/2 and acyltransferase activities.
Keywords: phospholipase A/acyltransferase, glycerophospholipid, lecithin retinol acyltransferase, HRAS-like suppressor
The metabolism of phospholipids is a dynamic event regulated by numerous enzymes. This includes remodeling of the acyl chains of glycerophospholipids by deacylation and reacylation (1–3). The deacylation is catalyzed by a series of phospholipase A1s (PLA1s) and PLA2s (1, 2, 4), whereas the resultant lysophospholipids are reacylated by various acyltransferases (5). Deacylation of phospholipids also leads to the generation of many classes of lipid mediators, including eicosanoids and lysophospholipids, which show their diverse cellular actions principally through G protein-coupled receptors (6, 7).
Lecithin retinol acyltransferase (LRAT) is an enzyme responsible for the metabolic cycle of retinol (8, 9). This enzyme transfers an acyl chain at the sn-1 position of phosphatidylcholine (PC) to all-trans-retinol, resulting in the formation of retinyl ester, and constitutes the LRAT family, together with various proteins such as Caenorhabditis elegans Egl-29 and several mammalian tumor suppressors (10). In human, five tumor suppressor proteins are included in this family, and they form the HRAS-like suppressor (HRASLS) subfamily or the H-rev107 subfamily (11). The members are H-rev107 (also known as HRASLS3, HRSL3, or H-REV107-1) (12, 13), TIG3 (also known as RARRES3, RIG1 or HRASLS4) (14, 15), A-C1 (also known as HRASLS) (16), HRASLS2 (11) and Ca2+-independent N-acyltransferase (iNAT, also known as HRASLS5 or HRLP5) (17) (Fig. 1). Recently we found that H-rev107 (18, 19), TIG3 (19), HRASLS2 (19) and iNAT (17, 20) have PLA1/2 and/or acyltransferase activities. By the latter activity these proteins catalyzed N-acylation of the amino group of phosphatidylethanolamine (PE) and O-acylation of lysophospholipid, using PC as an acyl donor in both of the reactions. Enzymatic N-acylation of PE results in the formation of N-acyl-PE, known as precursors of anandamide and other bioactive N-acylethanolamines (21, 22). Duncan et al. also reported PLA2 activity of H-rev107 and referred to it as adipose-specific PLA2 (23).
Fig. 1.
The deduced amino acid sequences of the members of the HRASLS subfamily. The deduced amino acid sequences of human members of the HRASLS subfamily were aligned using the program GENETYX-MAC (version 15). Closed and shaded boxes indicate identity in all five and any three or four polypeptides, respectively. The highly conserved histidine and cysteine residues and the sequence NCEHFV are indicated by asterisks and an underline, respectively.
A-C1 was originally cloned by differential display between two mouse cell lines, embryonic fibroblast C3H10T1/2 and chondrogenic ATDC5, and was noted due to its structural similarity to the tumor suppressor H-rev107 (16). The expression of A-C1 in Ras-transformed NIH3T3 cells caused an increase in the number of flat colonies and inhibition of cell growth (16). Therefore, this gene was considered to be a tumor suppressor gene. The human homolog was later cloned (24), and the methylation of the A-C1 gene in human gastric cancers was reported (25). Because all the other members of the HRASLS subfamily show phospholipid-metabolizing activities and because histidine-30 and cysteine-119 of A-C1 correspond to the catalytic dyad of LRAT (26) (Fig. 1), we hypothesized that A-C1 also functions as an enzyme involved in phospholipid metabolism. Here, we investigate the catalytic properties of the A-C1 protein and compare them with those of the other members of the HRASLS subfamily (H-rev107, TIG3, HRASLS2, and iNAT). On the basis of the obtained results, we propose to redefine this class of tumor suppressor as a novel group of phospholipid-metabolizing enzymes.
EXPERIMENTAL PROCEDURES
Materials
[1-14C]palmitic acid, 1,2-[1′-14C]dipalmitoyl-PC, 1-palmitoyl-2-[1′-14C]arachidonoyl-PE, and 1-[1′-14C]palmitoyl-lyso PC were purchased from PerkinElmer Life Science. 1-Palmitoyl-2-[1′-14C]palmitoyl-PC, horseradish peroxidase-linked anti-mouse IgG, Hybond P, and an ECL Plus kit were from GE Healthcare. 1,2-Dipalmitoyl-PC, 1,2-dioleoyl-PE, 1-palmitoyl-lyso PC, anti-FLAG monoclonal antibody M2, anti-FLAG M2 affinity gel, FLAG peptide, snake venom PLA2 and Rhizopus arrhizus lipase were from Sigma. Dulbecco's modified Eagle's medium, Lipofectamine 2000, fetal calf serum, pEF6/myc-His vector, TRIzol, and Moloney murine leukemia virus reverse transcriptase were from Invitrogen Life Technologies. 1-Palmitoyl-2-arachidonoyl-PE was from Avanti Polar Lipids (Alabaster, AL). Human Testis Marathon-Ready™ cDNA and human MTC™ Panels I and II were from Clontech. Nonidet P-40 was from Nacalai Tesque, Inc. (Kyoto, Japan). Rhizopus delemar lipase was from Seikagaku Corp. (Tokyo, Japan). Random hexamer and Ex Taq DNA polymerase were from TaKaRa Bio, Inc. (Ohtsu, Japan). KOD-Plus DNA polymerase was from TOYOBO (Osaka, Japan). Protein assay dye reagent concentrate was from Bio-Rad, and precoated Silica Gel 60 F254 aluminum sheets (20 × 20 cm, 0.2 mm thick) for TLC were from Merck (Darmstadt, Germany). N-[14C]palmitoyl-PE was prepared from [14C]palmitic acid and 1,2-dioleoyl-PE according to the method of Schmid et al. (27). 2-Palmitoyl-lyso PC was prepared from 1,2-dipalmitoyl-PC using R. delemar lipase as described previously (28). 2-[14C]palmitoyl-lyso PC was prepared from 1-palmitoyl-2-[14C]palmitoyl-PC using R. arrhizus lipase. 1-[14C]palmitoyl-2-palmitoyl-PC was prepared from 2-palmitoyl-lyso PC and [14C]palmitic acid.
Construction of expression vectors
The cDNAs encoding C-terminally FLAG-tagged A-C1s of human, mouse, and rat were amplified by PCR with Human Testis Marathon-Ready™ cDNA, mouse brain cDNA, and rat testis cDNA, respectively, as templates. The mouse and rat cDNAs were prepared from 5 μg of total RNA using Moloney murine leukemia virus reverse transcriptase and random hexamer. The primers used were the forward primers containing the SpeI site 5′-CGCACTAGTCCAAGATGGCGTTTAATGATTGCTTCAGTTTG-3′ (human A-C1), 5′-CGCACTAGTCCAAGATGGCGGTAAATGATTGCTTC-3′ (mouse A-C1), and 5′-CGCACTAGTCCAAGATGGCGGTTAACGATTGCTTCAGTC-3′ (rat A-C1), and the reverse primers containing an in-frame FLAG sequence and the NotI site 5′-CGCGCGGCCGCCTACTTATCGTCGTCATCCTTGTAATCATAGTATTTTGCTCTTTGTCCTTTTGGAAAC-3′ (human A-C1), 5′-CGCGCGGCCGCCTACTTATCGTCGTCATCCTTGTAATCATATTTCGTTCTTTGTCTTTTGGG-3′ (mouse A-C1), and 5′-CGCGCGGCCGCCTACTTATCGTCGTCATCCTTGTAATCAGATTTTGCTCCTTGTCTTTTGGGAAAC-3′ (rat A-C1). The cDNA encoding C-terminally FLAG-tagged human FAM84B was amplified by PCR with human kidney cDNA contained in human MTC™ Panel I as a template. The primers used were the forward primer 5′-CGCGGATCCGGAAAATGGGCAACCAGGTGGAGAAATTGA-3′ containing a BamHI site and the reverse primer 5′-CGCGAATTCTCACTTATCGTCGTCATCCTTGTAATCGTGTGCCACTGCCTCTCCGTCCTCC-3′ containing an in-frame FLAG sequence and an EcoRI site. PCR was carried out with KOD-Plus DNA polymerase for 30 cycles at 95°C for 20 s, 56°C for 20 s, and 72°C for 60 s in 5% (v/v) Me2SO. The obtained DNA fragments were subcloned into the corresponding restriction enzyme sites of pEF6/myc-His vector. All constructs were sequenced in both directions using an ABI 3130 Genetic Analyzer (Applied Biosystems Life Technologies; Carlsbad, CA).
Overexpression and purification of recombinant proteins
COS-7 cells were grown at 37°C to 80% confluency in 100 mm dishes containing Dulbecco's modified Eagle's medium with 10% fetal calf serum in a humidified 5% CO2 and 95% air incubator. The expression vector harboring A-C1 or FAM84B cDNA was introduced into COS-7 cells using Lipofectamine 2000 according to the manufacturer's instruction. Forty-eight hours after transfection, cells were harvested and sonicated three times each for 3 s in 20 mM Tris-HCl (pH 7.4). For the purification of recombinant FLAG-tagged human A-C1, cytosolic fractions were prepared from the cells grown in ten 100 mm dishes by centrifugation of the homogenates at 105,000 g for 55 min at 4°C and mixed with 1 ml of anti-FLAG M2 affinity gel preequilibrated with 50 mM Tris-HCl (pH 7.4) containing 150 mM NaCl and 0.05% Nonidet P-40 (buffer A). After overnight incubation at 4°C under gentle mixing, the gel was packed into a column and washed three times each with 12 ml of buffer A. The FLAG-tagged protein was eluted with buffer A containing 0.1 mg/ml of FLAG peptide and every 0.5 ml fraction was collected. The purified recombinant human iNAT was prepared as described previously (20). The protein concentration was determined by the method of Bradford with BSA as a standard (29).
Enzyme assay
For the PLA1/2 assay, the enzyme was incubated with 200 μM 1,2-[14C]dipalmitoyl-PC (45,000 cpm) in 100 μl of 50 mM Tris-HCl (pH 8), 2 mM DTT, and 0.1% Nonidet P-40 at 37°C for 30 min. For the PE N-acylation assay, the enzyme was incubated with 200 μM 1,2-[14C]dipalmitoyl-PC (45,000 cpm) and 100 μM 1,2-dioleoyl-PE in 100 μl of 50 mM Tris-HCl (pH 9), 2 mM DTT, and 0.1% Nonidet P-40 at 37°C for 30 min. For the lyso PC O-acylation assay, the enzyme was incubated with 200 μM dipalmitoyl-PC and either 100 μM 1-[14C]palmitoyl-lyso PC (20,000 cpm) or 100 μM 2-[14C]palmitoyl-lyso PC (18,000 cpm) in 100 μl of 50 mM Tris-HCl (pH 8.0), 2 mM DTT, and 0.1% Nonidet P-40 at 37°C for 30 min. The reaction was terminated by the addition of 320 μl of a mixture of chloroform-methanol (2:1; v/v) containing 5 mM 3(2)-t-butyl-4-hydroxyanisole. After centrifugation, 100 μl of the lower fraction was spotted on a silica gel thin-layer plate (10 cm height) and developed at 4°C for 25 min either in chloroform-methanol-28% ammonium hydroxide (80:20:2; v/v) for the PE N-acylation assay or in chloroform-methanol-H2O (65:25:4; v/v) for PLA1/2 and lyso PC O-acylation assays. The distribution of radioactivity on the plate was quantified using a BAS1500 bioimaging analyzer (FUJIX Ltd., Tokyo).
Western blotting
Samples (20 μg protein) were separated by SDS-PAGE on 14% gel and electrotransferred to a hydrophobic polyvinylidene difluoride membrane (Hybond P). The membrane was blocked with PBS containing 5% dried milk and 0.1% Tween 20 (buffer B) and then incubated with anti-FLAG antibody (1:2,000 dilution) in buffer B at room temperature for 1 h, followed by incubation with HRP-labeled secondary antibody (1:4,000 dilution) in buffer B at room temperature for 1 h. FLAG-tagged proteins were visualized using an ECL Plus kit and analyzed using an LAS1000plus lumino-imaging analyzer (FUJIX Ltd.).
PCR
To examine tissue distribution of human A-C1 mRNA, human MTC™ Panels I and II were used as templates for PCR amplification with Ex Taq DNA polymerase. For the analyses of rat and mouse A-C1 mRNAs, total RNAs were isolated from various organs of Wistar/ST rats and C57BL/6 mice (Japan SLC, Inc.) using TRIzol. cDNAs were prepared from 5 μg of total RNA using Moloney murine leukemia virus reverse transcriptase and random hexamer, and subjected to PCR amplification by Ex Taq DNA polymerase. The primers used for human A-C1 were the forward primer 5′-CCCAGGAATGAGAAGACACCAACAGC-3′ and the reverse primer 5′-CCCTGTGGAAGAAATCATAAAGCGGTC-3′ (nucleotides 450–474 and 553–577, respectively, in GenBank™ accession number NM_020386) and those for human GAPDH were the forward primer 5′-CGCTGAGTACGTCGTGGAGTCCACT-3′ and the reverse primer 5′-AGCAGAGGGGGCAGAGATGATGACC-3′ (nucleotides 375–399 and 456–480 in NM_002046). The primers used for rat A-C1 were the forward primer 5′-TCCGTCCTTGTTATCAGCACTGGGC-3′ and the reverse primer 5′-GTGCTGAACACAGACTTGGCACTTG-3′ (nucleotides 221–245 and 311–335 in NM_001105871) and those for rat GAPDH were the forward primer 5′-AACTCCCATTCTTCCACCTTTGATG-3′ and the reverse primer 5′-CCTGTTGCTGTAGCCATATTCATTG-3′ (nucleotides 935–959 and 1015–1039 in NM_017008). The primers used for mouse A-C1 were the forward primer 5′-TTGCTATCAGCACTGGGCACTGTAC-3′ and the reverse primer 5′-GTGCTGAACACGGACTTAGCACTTG-3′ (nucleotides 193–217 and 276–300 in NM_013751), and those for mouse GAPDH were the forward primer 5′-AACTCCCACTCTTCCACCTTCGATG-3′ and the reverse primer 5′-CCTGTTGCTGTAGCCGTATTCATTG-3′ (nucleotides 909–933 and 989–1013 in NM_008084). The PCR conditions used were as follows: denaturation at 94°C for 20 s, annealing at 60°C for 20 s, and extension at 72°C for 20 s (30 cycles for A-C1 and 25 cycles for GAPDH). Semi-quantitative real-time PCR analysis was performed with the aid of an ABI PRISM 7000 detection system (Applied Biosystems). The primers used were the same as those for conventional PCR, and the conditions were as follows: denaturation at 95°C for 6 s and annealing and extension at 62°C for 20 s (40 cycles).
RESULTS
Functional expression of A-C1 proteins
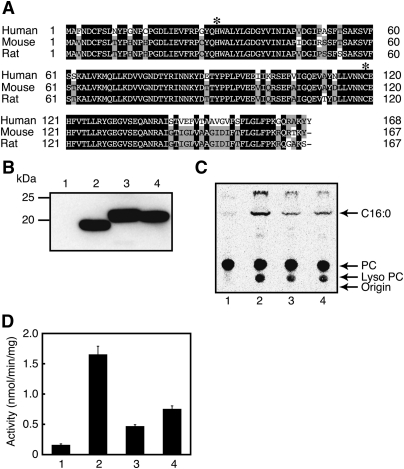
We previously cloned cDNA of A-C1 (tentatively termed RLP-2) from rat testis (GenBank™ accession number AB510983) (17). In the present study, we also cloned cDNAs of the counterparts from human testis and mouse brain (AB510981 and AB510982) based on the reported nucleotide sequences. Their sequences we determined were completely identical to those reported previously. The primary structures of A-C1 proteins were composed of 168 (human), 167 (mouse), and 167 (rat) amino acid residues, respectively (Fig. 2A). The alignment revealed their high homology to each other (85, 83, and 96% identity at amino acid level between human and mouse, between human and rat, and between mouse and rat, respectively). The putative catalytic dyad was completely conserved as histidine-30 and cysteine-119. We previously failed in the functional expression of rat A-C1 with pcDNA3.1(+) as an eukaryotic expression vector (17). We therefore constructed the pEF6/myc-His expression vector harboring the A-C1 cDNA of either human, mouse, or rat with a FLAG tag at the C terminus, and transiently expressed recombinant proteins in COS-7 cells. Based on the amino acid sequences, the molecular masses of the tagged proteins of A-C1s were calculated to be 19,745 (human), 19,778 (mouse), and 19,516 (rat) Da, respectively. When analyzed by Western blotting using anti-FLAG antibody, each cell homogenate exhibited an immunopositive band around 19–20 kDa (Fig. 2B). Although the band of human A-C1 consistently migrated a little faster than expected, the reason remained unclear.
Fig. 2.
Functional expression of human, mouse, and rat A-C1s. A: The deduced amino acid sequences of human, mouse, and rat A-C1s are shown. The sequences are aligned using the program GENETYX-MAC (version 15). Closed and shaded boxes indicate identity in all three or any two polypeptides, respectively. The highly conserved histidine and cysteine residues within the LRAT family are indicated by asterisks. B–D: COS-7 cells were transiently transfected with the insert-free vector (lane 1) or the expression vector harboring FLAG-tagged A-C1 of human (lane 2), mouse (lane 3), or rat (lane 4). The cell homogenates (20 μg protein) were analyzed by Western blotting with anti-FLAG antibody (B) or were allowed to react with 200 μM 1,2-[14C]dipalmitoyl-PC for the PLA1/2 assay, followed by separation of the products by TLC using chloroform-methanol-H2O (65:25:4; v/v) as mobile phase (C). The positions of authentic compounds on the TLC plate are indicated by arrows. C16:0, [14C]palmitic acid. The mean values ± SD of the quantified PLA1/2 activity are shown (n = 3) (D).
We next assayed the homogenates for PLA1/2 activity. When the samples were incubated with 1,2-[14C]dipalmitoyl-PC, followed by separation of the reaction products by TLC, the radioactive bands corresponding to palmitic acid and lyso PC were detected (Fig. 2C). The activities of the homogenates containing human, mouse, and rat A-C1 were 1.65, 0.47, and 0.75 nmol/min/mg of protein, respectively, whereas the endogenous activity of mock transfectant was 0.16 nmol/min/mg of protein (Fig. 2D). These results suggested that A-C1 possesses PLA1/2 activity. We examined a possible secretion of recombinant human A-C1 into the culture medium by measuring PLA1/2 activity. Consistent with the lack of the signal sequence for the secretory pathway in its primary structure, the activity was not detected in the culture medium of COS-7 cells expressing human A-C1 (data not shown). Although A-C1 was found as a tumor suppressor gene (16), its transient expression in COS-7 cells did not show an obvious effect on cell proliferation and viability (data not shown).
Characterization of the purified human A-C1
To further analyze the enzymatic properties of A-C1, we prepared cytosolic fractions from the COS-7 cell homogenate by ultracentrifugation and purified the C-terminally FLAG-tagged human A-C1 protein from the cytosol by anti-FLAG antibody-conjugated column chromatography. As analyzed by SDS-PAGE, a nearly homogenous protein band was seen around 19 kDa (Fig. 3A). The specific PLA1/2 activity of the purified protein was 182 nmol/min/mg of protein, which was 110-fold higher than that of the A-C1-expressing cell homogenate. The optimal pH was around 8 (Fig. 3B). The PLA1/2 activity increased up to 246 nmol/min/mg of protein, depending on the concentrations of the substrate PC, with an apparent Km at 80 μM (Fig. 3C). We also examined the effects of several factors on the PLA1/2 activity. The addition of 1 mM and 5 mM Ca2+ reduced the activity by 9.9% and 21.7%, respectively. On the other hand, 1 mM EDTA increased the activity by 14.3% (Fig. 3D). In the absence of the sulfhydryl reducing reagent DTT, the activity was hardly detected (Fig. 3E). In agreement with this stimulatory effect of DTT, 5 mM iodoacetate, an irreversible sulfhydryl blocker, acted as an inhibitor. The standard reaction mix also contained 0.1% Nonidet P-40 (a nonionic detergent). Removal of the detergent decreased the activity by 94.5%. These effects of Ca2+, DTT, iodoacetate, and Nonidet P-40 were similar to the catalytic properties of H-rev107, TIG3, and HRASLS2, which we reported previously (19).
Fig. 3.
Catalytic properties of the purified human A-C1. The purified protein of recombinant human A-C1 (2.0 μg) was analyzed by SDS-PAGE, followed by staining with silver nitrate (A). Marker proteins are also shown. The purified protein (0.13 μg) was assayed for the PLA1/2 activity at the indicated pH (B), with different concentrations of 1,2-[14C]dipalmitoyl-PC as a substrate (C), in the presence or absence of 1 mM and 5 mM CaCl2 and 1 mM EDTA (D), or in the presence or absence of 2 mM DTT, 0.1% Nonidet P-40, and 5 mM iodoacetate (E). The mean values ± SD are shown (n = 3). In B, the buffers used were Tris-HCl (closed circles) and glycine-NaOH (open circles) at 50 mM. In D and E, the specific enzyme activities (324 and 182 nmol/min/mg of protein) were normalized to 100%, respectively.
Because [14C]dipalmitoyl-PC that to this point we used as a substrate was radiolabeled on both sn-1 and sn-2 palmitoyl chains, we referred to the hydrolysis activity as PLA1/2 activity. To distinguish PLA1 activity from PLA2 activity, we next used [14C]PC radiolabeled only on the sn-2 palmitoyl chain (1-palmitoyl-2-[14C]palmitoyl-PC). As shown in Table 1, A-C1 showed a remarkable preference of sn-1 position over sn-2 position. A similar result was obtained using 1-[14C]palmitoyl-2-palmitoyl-PC as a substrate (data not shown). 1-Palmitoyl-2-[14C]arachidonoyl-PE was also an active substrate, and PLA1 activity was again higher than PLA2 activity (Table 1). On the other hand, lysophospholipase activities for 1-[14C]palmitoyl-lyso PC or 2-[14C]palmitoyl-lyso PC were not detected.
TABLE 1.
PLA1 and PLA2 activities of human A-C1
| PLA1 activity | PLA2 activity | PLA1/PLA2 | |
|
nmol/min/mg |
ratio | ||
| 1-Palmitoyl-2-[ C]palmitoyl-PC | 142.2 ± 5.9 | 16.0 ± 9.0 | 8.9 |
| 1-Palmitoyl-2-[ C]arachidonoyl-PE | 91.1 ± 6.3 | 13.5 ± 0.6 | 6.7 |
The purified recombinant human A-C1 (0.13 μg protein) was allowed to react with the indicated glycerophospholipids at 200 μM. Mean values ± SD are shown (n = 3).
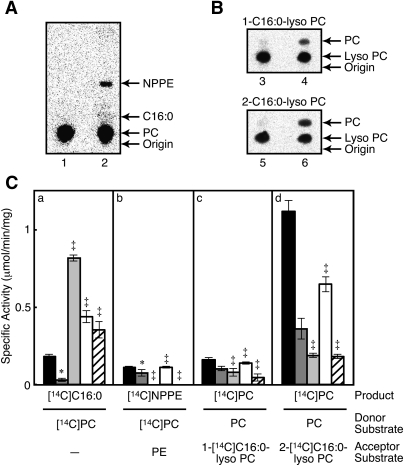
We next examined transacylation activities of the purified A-C1. When [14C]dipalmitoyl-PC and nonradioactive dioleoyl-PE were used as a donor substrate and an acceptor substrate, respectively, a radioactive band corresponding to N-palmitoyl-PE was detected on the TLC plate (Fig. 4A). In the absence of nonradioactive PE, this band was not detected (not shown). These results showed that A-C1 possesses an N-acyltransferase activity for PE. Moreover, when the protein was allowed to react with nonradioactive dipalmitoyl-PC and radioactive lyso PC (either 1-[14C]palmitoyl-lyso PC or 2-[14C]palmitoyl-lyso PC) as a donor substrate and an acceptor substrate, respectively, 14C-labeled PC was formed from both of the lyso PCs (Fig. 4B). 2-[14C]palmitoyl-lyso PC was a much more active substrate than 1-[14C]palmitoyl-lyso PC, as shown in Fig. 4C. With the aid of our previous results (19, 20), we compared catalytic properties of human A-C1, iNAT, H-rev107, HRASLS2, and TIG3, all of which belong to the HRASLS subfamily (Fig. 4C). All the purified recombinant proteins showed PLA1/2 activity, the highest being with H-rev107 and the lowest with iNAT. As for the PE N-acyltransferase activity, iNAT, HRASLS2, and A-C1 showed similar levels of activities, whereas H-rev107 and TIG3 were much less active. These five proteins also showed O-acyltransferase activities toward both 1-[14C]palmitoyl-lyso PC and 2-[14C]palmitoyl-lyso PC. The latter lyso PC was consistently a more-active acceptor substrate than the former lyso PC. In particular, A-C1 and HRASLS2 showed high O-acyltransferase activities.
Fig. 4.
PE N-acylation and lyso PC O-acylation activities of human A-C1 and comparison of catalytic activities among the HRASLS subfamily members. A, B: The purified human A-C1 (0.13 μg) was assayed for PE N-acylation activity with 200 μM 1,2-[14C]dipalmitoyl-PC and 100 μM nonradioactive PE (lane 2) or for PC O-acylation activity with 200 μM nonradioactive PC and 100 μM of either 1-[14C]palmitoyl-lyso PC (lane 4) or 2-[14C]palmitoyl-lyso PC (lane 6). The substrates were also incubated with buffer alone as controls (lanes 1, 3, and 5 for lanes 2, 4, and 6, respectively). The mobile phases used for TLC were chloroform-methanol-28% ammonium hydroxide (80:20:2; v/v) (A) and chloroform-methanol-H2O (65:25:4; v/v) (B), respectively. The positions of authentic compounds on the TLC plates are indicated by arrows. C: The purified human A-C1 (black), iNAT (dark gray), H-rev107 (light gray), HRASLS2 (white), and TIG3 (stripe) were allowed to react with 200 μM 1,2-[14C]dipalmitoyl-PC alone for PLA1/2 activity (panel a), with 200 μM 1,2-[14C]dipalmitoyl-PC and 100 μM nonradioactive PE for PE N-acylation activity (panel b), with 200 μM nonradioactive PC and 100 μM 1-[14C]palmitoyl-lyso PC for lyso PC O-acylation activity (panel c), or with 200 μM nonradioactive PC and 100 μM 2-[14C]palmitoyl-lyso PC for lyso PC O-acylation activity (panel d). Mean values ± SD are shown (n = 3). The symbols * and ‡ indicate that the data are cited from our previous articles (20) and (19), respectively. NPPE, N-palmitoyl-PE; C16:0, palmitic acid or palmitoyl.
Metabolic labeling of A-C1-expressing cells
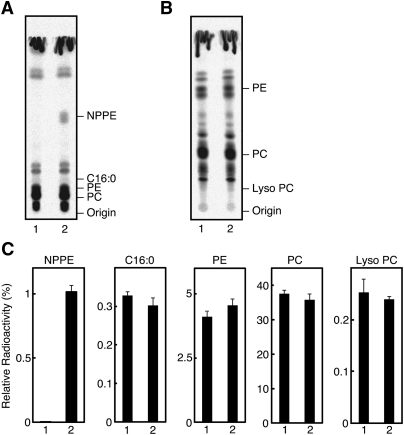
We metabolically labeled the COS-7 cells transiently expressing human A-C1 with [14C]palmitic acid. When the extracted lipids were analyzed by TLC, we detected a clear radioactive band corresponding to N-palmitoyl-PE (Fig. 5A, C). This band was not observed in the control COS-7 cells. These results suggested that A-C1 actually functions as N-acyltransferase in the living cells. On the other hand, we did not see an obvious change in the levels of radioactive bands corresponding to free palmitic acid and lyso PC (expected products of PLA1/2) (Fig. 5A–C). The levels of bands corresponding to PC and PE (potential substrates of PLA1/2) were also unaltered. These findings may be explained by a low PLA1/2 activity of A-C1 in the living cells. Another possibility is that the produced [14C]palmitic acid and [14C]lyso PC are quickly incorporated into phospholipids.
Fig. 5.
Metabolic labeling of A-C1-expressing cells with [14C]palmitic acid. The COS-7 cells transiently expressing human A-C1 (lane 2) and control COS-7 cells (lane 1) were grown to 80% confluency and were metabolically labeled with [14C]palmitic acid (1.6 μCi) for 18 h. Total lipids were then extracted and separated by TLC using chloroform-methanol-28% ammonium hydroxide (80:20:2; v/v) (A) or chloroform-methanol-H2O (65:25:4; v/v) (B) as mobile phase. The positions of authentic compounds on the TLC plate are indicated. NPPE, N-palmitoyl-PE; C16:0, palmitic acid. Relative radioactivities of the indicated compounds are shown (mean values ± SD, n = 3) (C). The radioactive band corresponding to authentic NPPE might include not only N-[14C]palmitoyl-PE but also N-acyl-O-[14C]palmitoyl-PE.
Tissue distribution of A-C1
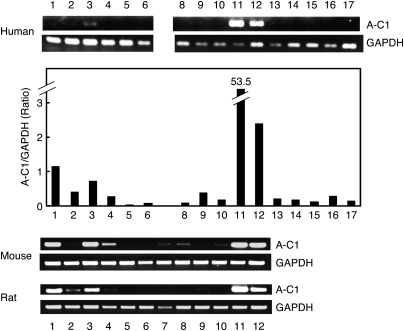
To examine tissue distribution of A-C1 in human, mouse, and rat, reverse transcriptase-PCR was employed (Fig. 6). In human, the levels of A-C1 mRNA were by far the highest in testis and skeletal muscle, followed by brain and heart. The expression levels of human A-C1 were also examined by semi-quantitative real-time PCR. The highest A-C1/GAPDH ratio was found in testis (53.5), followed by skeletal muscle (2.4), brain (1.1), and heart (0.7). Other human tissues showed lower levels. The dominant expression in these four tissues was also observed with mouse and rat. Low levels of its expression were detected in lung, stomach, kidney, and colon of mouse, and thymus, lung, and small intestine of rat. Such a relatively high expression in the limited tissues was similar to the dominant expression of iNAT in testis (17, 20, 30), but was different from ubiquitous expressions of H-rev107 and TIG3 (19).
Fig. 6.
The expression of A-C1 mRNA in human, mouse, and rat tissues. The expression of A-C1 mRNA in various human, mouse, and rat tissues was examined by reverse transcriptase-PCR. The housekeeping gene GAPDH was used as a control. The expression of A-C1 mRNA in human was also examined by semi-quantitative real-time PCR and was shown in terms of A-C1/GAPDH ratio. 1, brain; 2, thymus; 3, heart; 4, lung; 5, liver; 6, spleen; 7, stomach; 8, kidney; 9, small intestine; 10, colon; 11, testis; 12, skeletal muscle; 13, pancreas; 14, prostate; 15, ovary; 16, placenta; 17, peripheral leukocytes.
Lack of PLA1/2 activity in FAM84B
FAM84A and FAM84B were found as human genes upregulated in some tumors (31, 32). The deduced amino acid sequences of these two genes are homologous to those of the HRASLS subfamily members as shown in a phylogenetic tree (Fig. 7A) and by comparison with the sequence of human A-C1 (Fig. 7B). The two sequences exhibit 43.5% identity to each other (Fig. 7B). However, serine is substituted for the cysteine residue forming the catalytic dyad in these proteins. We cloned cDNA of FAM84B from human testis and constructed the expression vector. Although transient expression of the FLAG-tagged recombinant protein in COS-7 cells was confirmed by Western blotting using anti-FLAG antibody, the cell homogenates did not show a significant PLA1/2 activity. The same procedure was applied to cDNA cloning and expression of human FAM84A. However, we failed in its expression for unknown reasons.
Fig. 7.
The phylogenetic tree of the LRAT family members and the deduced amino acid sequences of human FAM84A and FAM84B. A: The phylogenetic tree composed of human LRAT, FAM84A, FAM84B, and the HRASLS subfamily members was constructed using the program GENETYX-MAC (version 15). B: The deduced amino acid sequences of human A-C1, FAM84A, and FAM84B were aligned using the program GENETYX-MAC (version 15). Closed and shaded boxes indicate identity in all three or any two polypeptides, respectively. The highly conserved histidine and cysteine residues and the sequence NCEHFV of A-C1 are indicated by asterisks and an underline, respectively.
DISCUSSION
A-C1 was discovered as a mouse protein that shows homology with H-rev107, a class II tumor suppressor, and that inhibits growth of Ras-transformed NIH3T3 cells (16). Its human homolog was also cloned (24). Our present studies revealed that this protein is capable of catalyzing PLA1/2-like hydrolysis, PE N-acylation, and lyso PC O-acylation. In these three reactions, PC was consistently used as an acyl donor, whereas water, the amino group of PE, and the hydroxyl group of lyso PC were used as acyl acceptors, respectively. The enzymatic properties were similar to those of other tumor suppressors belonging to the HRASLS subfamily (iNAT, H-rev107, HRASLS2, and TIG3) (19, 20). All of these proteins required DTT and Nonidet P-40 for full activities, were sensitive to the inhibition by iodoacetate, and preferred esterolysis at the sn-1 position to that at sn-2 position. As compared with the other members, A-C1 showed a relatively high lyso PC O-acyltransferase activity, and its PE N-acylation activity was as high as those of iNAT and HRASLS2. Metabolic labeling of A-C1-expressing COS-7 cells with [14C]palmitic acid revealed actual generation of N-palmitoyl-PE by A-C1 in the living cells. The dominant PLA1 activity over PLA2 activity, as well as the preference of 2-acyl-lyso PC in the lyso PC O-acylation, suggested the involvement of A-C1 in the remodeling at the sn-1 position of glycerophospholipids in a CoA-independent manner. As for tissue distribution, mRNA of AC-1 was highly expressed in testes and skeletal muscles of human, rat, and mouse. In addition, its moderate expression was seen in heart and brain. Our results were in agreement with previous reports that A-C1 was predominantly expressed in skeletal muscle, heart, brain, and bone marrow of mouse (16), and skeletal muscle, testis, heart, and brain of human (24). Such a tissue distribution of A-C1 distinguishable from those of other members of the HRASLS subfamily suggests a unique physiological role of this protein.
The present study and our previous studies (17–20) revealed that all proteins of the HRASLS subfamily possess phospholipid-metabolizing activities. Similarity among their enzymatic properties is in good agreement with high homology among their primary structures (Fig. 1). The histidine and cysteine residues corresponding to the catalytic dyad of LRAT are completely conserved throughout the five proteins. Because subtle structural differences of the members should explain different availabilities of acyl acceptor substrates, further investigation will be required to elucidate the structure-function relationship. Contribution of the catalytic activities to their tumor-suppressive activities currently remains unclear. The tumor-suppressing activity was mostly implicated in Ras-transformed cells (11, 13, 16, 33), and the mutants of TIG3 addressed to the conserved asparagine and cysteine residues failed to induce the apoptosis of HtTA cervical cancer cells, which was caused by the wild-type (34). Because this cysteine residue functions as the catalytic center, it is possible that the phospholipid-metabolizing activity of the tumor suppressors regulates the function of Ras by altering the membrane structures of microdomains where Ras is specifically localized (35).
All the HRASLS subfamily members contain the sequence NCEHFV (amino acids 118–123 in the case of A-C1) (Fig. 1). Human FAM84A and FAM84B show homology to LRAT and the HRASLS subfamily members (Fig. 6A). Although both of the proteins contain a sequence similar to the sequence NCEHFV, serine is substituted for the cysteine residue. The lack of PLA1/2 activity in FAM84B may be related to this substitution. As shown in the phylogenetic tree (Fig. 6A), the distinct evolution of LRAT, FAM84B, and the HRASLS subfamily members appears to explain the difference in their catalytic properties. However, we cannot rule out a possibility that FAM84B has another enzyme activity.
To date, various names have been used for each member of the HRASLS subfamily (Table 2). According to the nomenclature proposed by the HUGO Gene Nomenclature Committee, HRASLS1-5s are assigned to genes for A-C1, HRASLS2, H-rev107, TIG3, and iNAT, respectively. Considering that all these proteins possess PLA1/2 and acyltransferase activities, here we propose to term the products of HRASLS1-5 genes as phospholipase A/acyltransferase (PLA/AT)-1 to -5, respectively (Table 2).
TABLE 2.
A new nomenclature for proteins belonging to the HRASLS subfamily
| Gene name | Former names | New name |
| HRASLS1 | A-C1, HRASLS | PLA/AT-1 |
| HRASLS2 | HRASLS2 | PLA/AT-2 |
| HRASLS3 | H-rev107, H-REV107-1, HRSL3, AdPLA | PLA/AT-3 |
| HRASLS4 | TIG3, RIG1, RARRES3 | PLA/AT-4 |
| HRASLS5 | iNAT, HRLP5 | PLA/AT-5 |
In conclusion, we characterized for the first time the tumor suppressor protein A-C1 as a phospholipid-metabolizing enzyme. Considering that five human members of the HRASLS subfamily, including A-C1, share similar catalytic properties, these proteins appear to form a novel class of enzymes showing PLA1/2 and acyltransferase activities.
Acknowledgments
The authors are grateful to Ms. Akiko Yamamoto for technical assistance, and acknowledge technical assistance from the Division of Research Instrument and Equipment and the Division of Radioisotope Research, Kagawa University.
Footnotes
Abbreviations:
- HRASLS
- HRAS-like suppressor
- iNAT
- Ca2+-independent N-acyltransferase
- LRAT
- lecithin retinol acyltransferase
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PLA/AT
- phospholipase A/acyltransferase
This study was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (T.U., K.T.), and the Japan Society for the Promotion of Science (X-H.J., N.U.), Kagawa University Faculty of Medicine Specially Promoted Research Fund 2010 (T.U.), and grants from the Ichiro Kanehara Foundation (T.U.), the Sumitomo Foundation (T.U.), and the Suzuken Memorial Foundation (K.T.).
REFERENCES
- 1.Kudo I., Murakami M. 2002. Phospholipase A2 enzymes. Prostaglandins Other Lipid Mediat. 68–69: 3–58 [DOI] [PubMed] [Google Scholar]
- 2.Schaloske R. H., Dennis E. A. 2006. The phospholipase A2 superfamily and its group numbering system. Biochim. Biophys. Acta. 1761: 1246–1259 [DOI] [PubMed] [Google Scholar]
- 3.Shindou H., Shimizu T. 2009. Acyl-CoA:lysophospholipid acyltransferases. J. Biol. Chem. 284: 1–5 [DOI] [PubMed] [Google Scholar]
- 4.Aoki J., Inoue A., Makide K., Saiki N., Arai H. 2007. Structure and function of extracellular phospholipase A1 belonging to the pancreatic lipase gene family. Biochimie. 89: 197–204 [DOI] [PubMed] [Google Scholar]
- 5.Shindou H., Hishikawa D., Harayama T., Yuki K., Shimizu T. 2009. Recent progress on acyl CoA:lysophospholipid acyltransferase research. J. Lipid Res. 50 (Suppl.): 46–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shimizu T. 2009. Lipid mediators in health and disease: enzymes and receptors as therapeutic targets for the regulation of immunity and inflammation. Annu. Rev. Pharmacol. Toxicol. 49: 123–150 [DOI] [PubMed] [Google Scholar]
- 7.Choi J. W., Herr D. R., Noguchi K., Yung Y. C., Lee C. W., Mutoh T., Lin M. E., Teo S. T., Park K. E., Mosley A. N., et al. 2010. LPA receptors: subtypes and biological actions. Annu. Rev. Pharmacol. Toxicol. 50: 157–186 [DOI] [PubMed] [Google Scholar]
- 8.Rando R. R. 2002. Membrane-bound lecithin-retinol acyltransferase. Biochem. Biophys. Res. Commun. 292: 1243–1250 [DOI] [PubMed] [Google Scholar]
- 9.Blaner W. S., O'Byme S. M., Wongsiriroj N., Kluwe J., D'Ambrosio D. M., Jiang H., Schwabe R. F., Hillman E. M., Piantedosi R., Libien J. 2009. Hepatic stellate cell lipid droplets: a specialized lipid droplet for retinoid storage. Biochim. Biophys. Acta. 1791: 467–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anantharaman V., Aravind L. 2003. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 4: R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shyu R-Y., Hsieh Y-C., Tsai F-M., Wu C-C., Jiang S-Y. 2008. Cloning and functional characterization of the HRASLS2 gene. Amino Acids. 35: 129–137 [DOI] [PubMed] [Google Scholar]
- 12.Hajnal A., Klemenz R., Schäfer R. 1994. Subtraction cloning of H-rev107, a gene specifically expressed in H-ras resistant fibroblasts. Oncogene. 9: 479–490 [PubMed] [Google Scholar]
- 13.Sers C., Emmenegger U., Husmann K., Bucher K., Andres A. C., Schäfer R. 1997. Growth-inhibitory activity and downregulation of the class II tumor-suppressor gene H-rev107 in tumor cell lines and experimental tumors. J. Cell Biol. 136: 935–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiSepio D., Ghosn C., Eckert R. L., Deucher A., Robinson N., Duvic M., Chandraratna R. A., Nagpal S. 1998. Identification and characterization of a retinoid-induced class II tumor suppressor/growth regulatory gene. Proc. Natl. Acad. Sci. USA. 95: 14811–14815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang S-L., Shyu R-Y., Yeh M-Y., Jiang S-Y. 2000. Cloning and characterization of a novel retinoid-inducible gene 1 (RIG1) deriving from human gastric cancer cells. Mol. Cell. Endocrinol. 159: 15–24 [DOI] [PubMed] [Google Scholar]
- 16.Akiyama H., Hiraki Y., Noda M., Shigeno C., Ito H., Nakamura T. 1999. Molecular cloning and biological activity of a novel Ha-Ras suppressor gene predominantly expressed in skeletal muscle, heart, brain, and bone marrow by differential display using clonal mouse EC cells, ATDC5. J. Biol. Chem. 274: 32192–32197 [DOI] [PubMed] [Google Scholar]
- 17.Jin X-H., Okamoto Y., Morishita J., Tsuboi K., Tonai T., Ueda N. 2007. Discovery and characterization of a Ca2+-independent phosphatidylethanolamine N-acyltransferase generating the anadamide precursor and its congeners. J. Biol. Chem. 282: 3614–3623 [DOI] [PubMed] [Google Scholar]
- 18.Uyama T., Morishita J., Jin X-H., Okamoto Y., Tsuboi K., Ueda N. 2009. The tumor suppressor gene H-rev107 functions as a novel Ca2+-independent cytosolic phospholipase A1/2 of the thiol hydrolase type. J. Lipid Res. 50: 685–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uyama T., Jin X-H., Tsuboi K., Tonai T., Ueda N. 2009. Characterization of the human tumor suppressors TIG3 and HRASLS2 as phospholipid-metabolizing enzymes. Biochim. Biophys. Acta. 1791: 1114–1124 [DOI] [PubMed] [Google Scholar]
- 20.Jin X-H., Uyama T., Tsuboi K., Tonai T., Ueda N. 2009. cDNA cloning and characterization of human and mouse Ca2+-independent phosphatidylethanolamine N-acyltransferases. Biochim. Biophys. Acta. 1791: 32–38 [DOI] [PubMed] [Google Scholar]
- 21.Ueda N., Tsuboi K., Uyama T. 2010. N-acylethanolamine metabolism with special reference to N-acylethanolamine-hydrolyzing acid amidase (NAAA). Prog. Lipid Res. 49: 299–315 [DOI] [PubMed] [Google Scholar]
- 22.Ueda N., Tsuboi K., Uyama T. 2010. Enzymological studies on the biosynthesis of N-acylethanolamines. Biochim. Biophys. Acta. 1801: 1274–1285 [DOI] [PubMed] [Google Scholar]
- 23.Duncan R. E., Sarkadi-Nagy E., Jaworski K., Ahmadian M., Sul H. S. 2008. Identification and functional characterization of adipose-specific phospholipase A2 (AdPLA). J. Biol. Chem. 283: 25428–25436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito H., Akiyama H., Shigeno C., Nakamura T. 2001. Isolation, characterization, and chromosome mapping of a human A-C1 Ha-Ras suppressor gene (HRASLS). Cytogenet. Cell Genet. 93: 36–39 [DOI] [PubMed] [Google Scholar]
- 25.Kaneda A., Kaminishi M., Yanagihara K., Sugimura T., Ushijima T. 2002. Identification of silencing of nine genes in human gastric cancers. Cancer Res. 62: 6645–6650 [PubMed] [Google Scholar]
- 26.Jahng W. J., Xue L., Rando R. R. 2003. Lecithin retinol acyltransferase is a founder member of a novel family of enzymes. Biochemistry. 42: 12805–12812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmid P. C., Reddy P. V., Natarajan V., Schmid H. H. O. 1983. Metabolism of N-acylethanolamine phospholipids by a mammalian phosphodiesterase of the phospholipase D type. J. Biol. Chem. 258: 9302–9306 [PubMed] [Google Scholar]
- 28.Arai H., Inoue K., Natori Y., Banno Y., Nozawa Y., Nojima S. 1985. Intracellular phospholipase activities of Tetrahymena pyriformis. J. Biochem. 97: 1525–1532 [DOI] [PubMed] [Google Scholar]
- 29.Bradford M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- 30.Yamano Y., Asano A., Ohyama K., Ohta M., Nishio R., Morishima I. 2008. Expression of the Ha-Ras suppressor family member 5 gene in the maturing rat testis. Biosci. Biotechnol. Biochem. 72: 1360–1363 [DOI] [PubMed] [Google Scholar]
- 31.Kobayashi T., Masaki T., Sugiyama M., Atomi Y., Furukawa Y., Nakamura Y. 2006. A gene encoding a family with sequence similarity 84, member A (FAM84A) enhanced migration of human colon cancer cells. Int. J. Oncol. 29: 341–347 [PubMed] [Google Scholar]
- 32.Huang X-P., Rong T-H., Wang J-Y., Tang Y-Q., Li B-J., Xu D-R., Zhao M-Q., Zhang L-J., Fang Y., Su X-D., et al. 2006. Negative implication of C-MYC as an amplification target in esophageal cancer. Cancer Genet. Cytogenet. 165: 20–24 [DOI] [PubMed] [Google Scholar]
- 33.Tsai F-M., Shyu R-Y., Jiang S-Y. 2006. RIG1 inhibits the Ras/mitogen-activated protein kinase pathway by suppressing the activation of Ras. Cell. Signal. 18: 349–358 [DOI] [PubMed] [Google Scholar]
- 34.Tsai F-M., Shyu R-Y., Lin S-C., Wu C-C., Jiang S-Y. 2009. Induction of apoptosis by the retinoid inducible growth regulator RIG1 depends on the NC motif in HtTA cervical cancer cells. BMC Cell Biol. 10: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henis Y. I., Hancock J. F., Prior I. A. 2009. Ras acylation, compartmentalization and signaling nanoclusters. Mol. Membr. Biol. 26: 80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]