Abstract
The unicellular green alga Chlamydomonas has frequently been used as a eukaryotic model system to study intracellular phospholipid signaling pathways in response to environmental stresses. Earlier, we found that hypersalinity induced a rapid increase in the putative lipid second messenger, phosphatidic acid (PA), which was suggested to be generated via activation of a phospholipase D (PLD) pathway and the combined action of a phospholipase C/diacylglycerol kinase (PLC/DGK) pathway. Lysophosphatidic acid (LPA) was also increased and was suggested to reflect a phospholipase A2 (PLA2) activity based on pharmacological evidence. The question of PA's and LPA's origin is, however, more complicated, especially as both function as precursors in the biosynthesis of phospho- and galactolipids. To address this complexity, a combination of fatty acid-molecular species analysis and in vivo 32P-radiolabeling was performed. Evidence is provided that LPA is formed from a distinct pool of PA characterized by a high α-linolenic acid (18:3n-3) content. This molecular species was highly enriched in the polyphosphoinositide fraction, which is the substrate for PLC to form diacylglycerol. Together with differential 32P-radiolabeling studies and earlier PLD-transphosphatidylation and PLA2-inhibitor assays, the data were consistent with the hypothesis that the salt-induced LPA response is primarily generated through PLA2-mediated hydrolysis of DGK-generated PA and that PLD or de novo synthesis [via endoplasmic reticulum - or plastid-localized routes] is not a major contributor.
Keywords: diacylglycerol, lysophosphatidic acid, phospholipase A2, signal transduction Chlamydomonas, 32P-radiolabeling, α-linolenic acid, hypersalinity
The unicellular green alga Chlamydomonas has proven to be useful model organism to study phospholipid-signaling pathways in response to osmotic and salt stress (1, 2). Recently, particular interest has been focusing on the accumulation of phosphatidic acid (PA) as a lipid second messenger in plant and animal systems (3).
In Chlamydomonas moewusii, PA is a minor lipid, composing 0.67 mol% of the total phospholipid pool (4). In response to 150 mM NaCl, PA levels rapidly increase 3- to 4-fold within minutes of application (5). In a recent study, also lysophosphatidic acid (LPA) was shown to accumulate in C. moewusii under conditions of salt and nonionic hyperosmotic stress (6, 7). The response was dose-dependent within the range of 150 to 400 mM NaCl, reaching a maximum at 300 mM. Because of a pharmacological inhibitor, the LPA response was suggested to be generated via a phospholipase A2 (PLA2) (6).
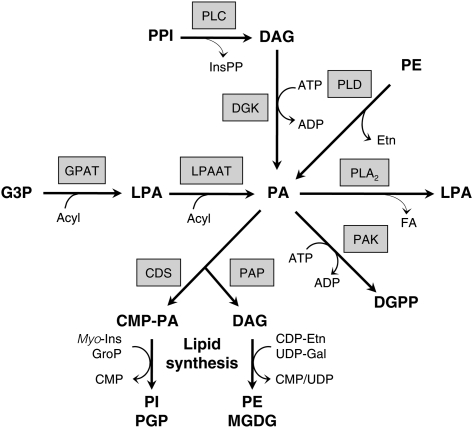
The origin of PA and LPA is, however, more complicated, because several pathways can be involved in their synthesis or breakdown. For example, the primary function of LPA and PA as precursors for de novo glycerolipid biosynthesis at the endoplasmic reticulum (ER) and plastidial membranes (Fig. 1) is often ignored. This pathway starts with two consecutive acylations of glycerol-3-phosphate (G3P) to generate LPA and PA, by the activities of G3P acyltransferase (GPAT) and LPA acyltransferase (LPAAT), respectively. For the synthesis of phosphatidylinositol (PI) and its phosphorylated derivatives, phosphatidylinositol 4-phosphate (PIP) and phosphatidylinositol 4,5-bisphosphate (PIP2) or phosphatidylglycerol (PG), PA is converted to cytidine monophosphate-PA (CMP-PA), which can react with myo-inositol or glycerolphosphate. Alternatively, for the synthesis of phosphatidylethanolamine (PE) or monogalactosyldiacylglycerol (MGDG), PA is dephosphorylated by PA phosphatase (PAP) to diacylglycerol (DAG), and acquires its headgroup via a reaction with CDP-ethanolamine (CDP-Etn) or UDP-galactose (UDP-Gal), respectively (Fig. 1).
Fig. 1.
Complexity of PA and LPA metabolism. PA is formed by successive acylations of G3P and LPA for de novo synthesis of glycerolipids, but it can also be formed through the stress-induced activities of PLC/DGK and PLD. For the synthesis of PI and PG(P), PA is converted to cytidine monophosphate-phosphatidic acid (CMP-PA) by CMP-PA synthase (CDS), and for the synthesis of PE and MGDG, it is dephosphorylated by PAP. Note that the PA/DAG substrates in the synthesis of PE and MGDG are different pools localized at the ER and plastidial envelope membrane, respectively. During salt stress, Chlamydomonas cells accumulate the alternative PA metabolites DGPP and LPA. The latter is suggested to originate from PA-hydrolyzing PLA2 activity.
While the PA formation by LPAAT may be prevalent under steady-state conditions, upon environmental stress, other pathways can be activated, generating distinct and transient PA responses that are likely to play a role in signaling (3, 8–14). These pathways include phospholipase D (PLD), which hydrolyzes structural phospholipids, like PE to generate PA, and DAG kinase (DGK), which produces PA by phosphorylation of DAG (4). Under stimulatory conditions, this DAG can be provided by PLC-mediated hydrolysis of polyphosphoinositides (PPI), i.e., PIP and PIP2 (3, 9, 15). Metabolically, DAG can also be generated through hydrolysis of phosphatidylcholine (PC) via nonspecific PLC, called NPC (16–18).
Here, the complexity of the salt stress-induced PA and LPA response in Chlamydomonas was studied in more detail to unravel their pathway of synthesis. Based on fatty acid molecular species analyses, in addition to in vivo 32P-labeling studies, we suggest that the salt stress-induced LPA response is produced via PLA2 hydrolysis of DGK-generated PA.
MATERIALS AND METHODS
Cell cultures
Chlamydomonas moewusii strain UTEX 10 (mating type minus) from the Culture Collection of Algae, University of Texas (Austin) was autotrophically grown as described before (19). Petri dishes containing cultured cells of ∼18 days old were flooded with 20 ml HMCK (10 mM HEPES, 1 mM MgCl2, 1 mM CaCl2, 1 mM KCl; pH 7.4), and after 16 h, suspensions of swimming gametes were harvested (4).
32P-labeling and lipid extraction
Cell suspensions were labeled with 32Pi,, and treated ± NaCl in the appropriate concentrations for the indicated times, and the lipids were extracted. To each 100 μl sample, 375 μl CHCl3/MeOH/HCl (50:100:1, by vol) was added to stop all reactions. After vigorous shaking, 375 μl CHCl3 and 100 μl 2 M HCl were added. The resultant upper phase was washed with 400 μl CHCl3/MeOH/1M HCl (3:48:47). Aliquots of concentrated lipid extracts were analyzed by TLC using silica gel 60 plates (Merck) in a solvent of CHCl3-MeOH-NH4OH-H2O (90:70:4:16, by vol) (20, 21). Radioactivity was quantified by phosphoimaging.
Fatty acid analysis
For fatty acid analyses, treatments were conducted in separation funnels containing 60 ml cells (density 2.0 × 107 cells/ml) to which 20 ml buffered NaCl solution (or only buffer) was added. Reactions were stopped by the addition of perchloric acid to a final concentration of 5% (w/v), and lipids were extracted by a previously described method with adjustment for the larger extraction volume (5).
PA and LPA were purified by column-adsorption chromatography on a 2 g silica column (Sep-Pak Plus). The lipid extract was dissolved in hexane and applied to the column. Elution solvents were (i) hexane-Et2O (99:1, 18 ml); (ii) hexane-Et2O (4:1, 15 ml); (iii) CHCl3 (10 ml); (iv) Me2CO-CHCl3 (2:1, 25 ml); (v) Me2CO-MeOH (29:1, 10 ml); (vi) Me2CO-MeOH (19:1, 30 ml); (vii) Me2CO-MeOH (2:1, 25 ml); and (viii) CHCl3-MeOH-H2O (1:2:0.8, 19 ml). The last eluate was collected in 10 ml tubes in four portions of 4.75 ml. To extract lipids from these aqueous eluates, 3.75 ml CHCl3 and 1 ml 0.9% (w/v) NaCl was added, and then tubes were vigorously shaken and centrifuged to separate two phases of which the lower one, containing LPA and PA, was dried down in a gyrovap at 50°C. The concentrated extracts were further purified by TLC as described. 32P-lipid markers served to calculate the recovery of each lipid after purification and for their localization on the TLC plate.
To generate fatty acid methyl esters (FAME), lipid spots were scraped from the TLC plates into 3% H2SO4 in MeOH. Known concentrations of heneicosanoic acid (21:0) methyl esters served as internal standard. Concentrated FAME extracts were analyzed by GC (Varian Chrompack, Bergen op Zoom, The Netherlands) using a 50 m WCOT fused silica column and FID with carrier gas N2 at 30 ml/min. Operating conditions were either 180°C isothermal or the temperature was programmed from 180°C to 220°C at 0.5°C/min with injector and detector temperatures at 250°C and 270°C, respectively.
In vitro PLA2 digestion of phospholipids
TLC-separated lipids were recovered from the silica and dissolved by sonication in 1 ml ethylether/MeOH (98:2, by vol). Five units of bee venom PLA2 was added in 100 μl 100 mM Tris HCl buffer (pH 8.9) containing 9.1 mM CaCl2. After 3 h incubation at 25°C with frequent shaking, reactions were stopped by adding 20 μl 0.5 M EDTA. Lipid products were extracted after evaporation of the ether phase by three consecutive extractions of the aqueous phase. Pooled extracts were dried, and the precipitate was resolved into CHCl3 for TLC purification and analysis. For the purpose of accurate quantitation of LPA in fatty acid analyses, a C17-LPA standard was prepared from the corresponding, commercially available di-C17-PA (Sigma) by in vitro PLA2 digestion.
Quantitation and statistics
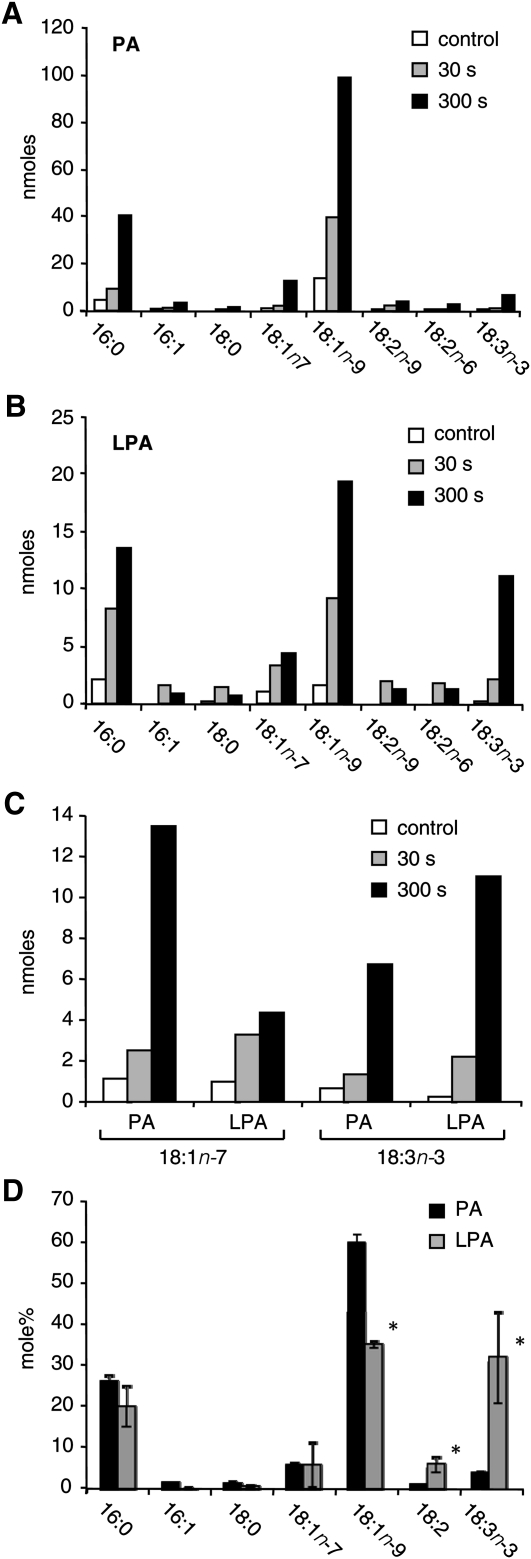
Calculation of the original lipid fatty acid masses was based on GC peak areas of algal FAMEs divided by the internal standard area, and subsequently corrected for variation in recovery, using 32P- and C17-lipid markers. Presented charts of fatty acid profiles show typical results, representative of three experiments showing similar trends. The data in Fig. 4D represent average values (n = 3). Statistical differences were determined using an unpaired t-test.
Fig. 4.
Comparison of PA (A) and LPA (B) fatty acid signatures after 30 s or 5 min of hyperosmotic stress. Lipids were isolated from Chlamydomonas after challenge with 300 mM NaCl for the respective times. Increments in 18:1n-7 and 18:3n-3, characteristic fatty acids of PPIs, were registered in both lipids after 30 s and 5 min (C). The fatty acid spectra of newly formed PA and LPA after 5 min of salt treatment (D) show LPA's significant enrichment in 18:3n-3 and 18:2, and a decrease in 18:1n-9 (mean ± SD, n = 3; *P < 0.003).
RESULTS
When Chlamydomonas cells were metabolically labeled with 32Pi and subsequently treated with 300 mM NaCl for only 5 min, several changes in the 32P-phospholipid pattern could be observed (Fig. 2), confirming earlier reports. Thus, typical 32P-increases were found in PA (by 1.5 to 14 times, depending on the prelabeling time), LPA (by 1.8 to 96 times, depending on the prelabeling time), diacylglycerol pyrophosphate (DGPP), PIP, PI(3,5)P2, and PI(4,5)P2 (2, 6, 22–25).
Fig. 2.
Effect of salt stress on 32P-phospholipid patterns of Chlamydomonas. Cells were labeled for 3 h and then treated for 5 min with buffer (Control) or 300 mM NaCl. Lipids were extracted, separated by TLC, and then visualized by PImaging. Results of a typical experiment are shown (n > 30).
The increase in 32P-PA was suggested to result from both a PLD pathway in which the structural phospholipid PE is hydrolyzed and the combined activities of the PLC/DGK pathway (5, 22). PLC hydrolyzes PI4P and PI(4,5)P2 into inositolpolyphosphates (InsPP) and DAG, and the latter can be converted to PA via DGK (Fig. 1). DGPP is the phosphorylated product of PA, which may represent an attenuation of PA as a signaling molecule, but it could also be a phospholipid signal itself (23, 26–28).
Like PA, the study of LPA is complicated by the multiplicity of pathways that synthesize it (Fig. 1). LPA is formed in the pathway of de novo PA synthesis, but it can also result from acyl hydrolysis of PA by a PLA2 activity. The first possibility implies that LPA is precursor to PA, whereas the latter implies that LPA is the product of PA. To study these product-precursor relationships, fatty acid analyses were performed, as distinct phospholipid classes and pools can have characteristic fatty-acid fingerprints that are inherited by their metabolites (5, 29, 30).
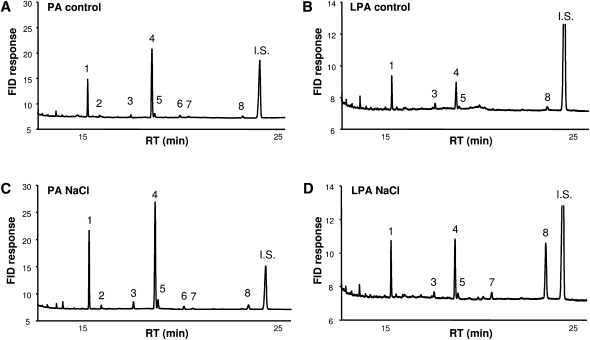
As shown in Fig. 3, salt stress induced major increases in 16:0 and 18:1n-9 in both LPA and PA pools. On the basis of quantitation of fatty acids, PA was increased by 4.2 ± 1.2 times (average ± SD, n = 4), and LPA by 7.2 ± 2.2 times (average ± SD, n = 4), the respective control levels. However, the LPA pool generated in response to salt also contained 18:3n-3 (α-linolenic acid, α-LA) (Fig. 3B, D, peak 8). Because this species was hardly detectable under steady-state conditions and increased in response to salt stress to become one of LPA's major constituents (Fig. 3B, D), these results strongly suggest an activation of a novel metabolic pathway rather than an upregulation of de novo synthesis. Subsequently, this possibility was investigated further.
Fig. 3.
Fatty acid composition analyses of PA and LPA in salt-stressed Chlamydomonas cells under control conditions (A, B) and treated with 300 mM NaCl for 5 min (C, D). FAMEs were made from purified phospholipids and analyzed by GC. Peak identification: 1, 16:0; 2, 16:1; 3, 18:0; 4, 18:1n-9; 5, 18:1n-7; 6, 18:2n-9; 7, 18:2n-6; 8, 18:3n-3 (α-LA); and IS, internal standard (21:0).
Interestingly, α-LA was not particularly abundant in the pool of PA under either condition (Fig. 3A, C). To explain this difference, it was hypothesized that the α-LA-enriched LPA was perhaps derived from a preceding, short-lived part of the PA increase with a similar α-LA enrichment in its fatty acid composition. To test this hypothesis, cells were treated for a shorter period (i.e., 30 s and 5 min) with 300 mM NaCl, and then the fatty acid compositions of LPA and PA quantitatively were analyzed (Fig. 4).
Within 30 s of NaCl stimulation, PA increased substantially and was characterized by the abundance of palmitic (16:0) and oleic acids (18:1), whereas α-LA, the hallmark of stimulated LPA, was relatively minor at both time points (Fig. 4A, B). Although this seems to argue against α-LA-enriched LPA resulting from a preceding phase of PA formation, it could be derived from a metabolically separated subpool of PA containing a relatively high α-LA content. Indeed, the increase in α-LA was found in the same order of magnitude in both lipids when plotted along the same axis (Fig. 4C). The PA increase could, therefore, underlie the significantly α-LA-enriched fatty acid spectrum of newly formed LPA (Fig. 4D). Simultaneously, the LPA increment displayed an enrichment in 18:2, whereas 18:1n-9 was diminished (Fig. 4D).
As salt stress-induced PA in Chlamydomonas is of mixed origin, it was hypothesized that LPA formation selectively drew upon one of the PA metabolic pools, explaining the disparate fatty acid compositions of PA and LPA. The LPA molecular species would then reflect the composition of either the PLD substrate, PE, or PLC's substrate, the PPIs. PE has been shown to contain mainly 18:1n-9 (92 mol%), and in the MS spectrum, the (18:1)2 species was predominant, accounting for over 95% of the total PE (5). Although a minor amount of α-LA (2 mol%) was present in PE (4), it probably is absent from the PA produced by PLD-mediated PE hydrolysis during salt stress (5). Although this suggested that PLD-derived PA could not lead to the increase in α-LA-containing LPA, it could still be precursor to the 18:1-containing LPA.
In contrast, PPIs contain both α-LA and 18:1n-7, a hallmark-fatty acid of PI and its derivatives (4). So their hydrolysis by PLC and subsequent phosphorylation of DAG could generate these molecular species of PA (Fig. 3A, C and Fig. 4) (5). If this PA pool could be a substrate to PLA2, it might account for the increase in α-LA and 18:1n-7 species of LPA.
PLA2 hydrolysis of PA from a mixed PLD- and PLC/DGK-derived origin could thus account for the production of the corresponding LPA species, provided that these fatty acids are esterified to the sn-1-OH position of the respective substrate lipids (i.e., PE and PPI). To test this hypothesis, a positional study of the fatty acids in PE and PI, which is similar to the PPIs (4, 5), was performed (Table 1). The lipids were purified from a total lipid extract, and then digested in vitro using PLA2 from bee venom to generate the corresponding lysophospholipids, which were purified on TLC and analyzed for their fatty acid content by GC.
TABLE 1.
Fatty acid composition of PE and PI and their lyso-derivatives acquired by in vitro PLA2-catalyzed digestion of C. moewusii lipids
| Lipid | 16:0 | 18:1n-9 | 18:1n-7 | 18:2n-6 | 18:3n-3 |
| Parenta | |||||
| PE | 5 ± 1 | 92 ± 4 | 0 | 1 | 2 |
| PI | 50 ± 4 | 14 ± 1 | 20 ± 2 | 9 ± 2 | 7 ± 1 |
| Daughterb | |||||
| LPE | 5 ± 1 | 92 ± 2 | 0 | 0 | 3 |
| LPI | 5 ± 1 | 19 ± 8 | 57 ± 10 | 6 ± 1 | 13 ± 2 |
Values are mol% average ± SD.
n = 4.
n = 2.
As shown in Table 1, the α-LA and 18:1n-7 content was not decreased in lyso-phosphatidylinositol (LPI) compared with its parent PI, suggesting that these fatty acids were predominantly linked to the sn-1 position, which is resistant to PLA2 digestion. These results are in agreement with the suggested pathway in Chlamydomonas involving PLA2-hydrolysis of PA derived from PPIs.
Some additional 32P-labeling experiments were performed that confirmed our hypothesis. This included a differential 32P-labeling technique (9, 21) that takes advantage of the extremely fast 32P-labeling of the cellular ATP pool when cells are only briefly incubated with 32P-Pi. Consequently, lipids that are direct products of ATP-dependent phosphorylation, such as DGK-generated PA, are quickly labeled, whereas PA that is derived from PLD activity is slowly labeled because of the relatively slow labeling kinetics of PE, which takes from hours to days (9, 12, 21). Salt-induced PA and LPA in Chlamydomonas were found to exhibit rapid labeling kinetics, similar to PPI, consistent with their formation through a DGK-PLA2 route (supplementary Fig. I). Conversely, a long-term labeling protocol designed to visualize the gradual label incorporation into structural lipids showed steady increases (up to 2 days) in 32P-PE, whereas salt-stimulated 32P-LPA declined, arguing against a major role for PLD in providing the PLA2 substrate (supplementary Fig. II).
DISCUSSION
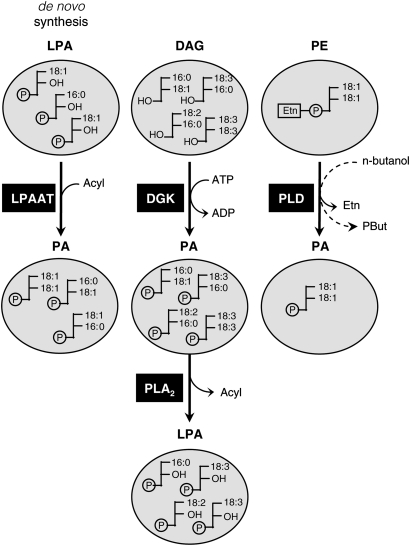
The rapid accumulation of PA (in minutes) seems to be an early hallmark of various plant stress responses (3, 9, 13, 31). Being the product of multiple pathways, however, the origin of this PA is complex. In Chlamydomonas cells, salt stress activates both PLD and DGK, generating a PA increment of mixed origin. In addition, 32P-LPA accumulated, which was proposed to be generated through PLA2 activity on PA (6). The present work used fatty acid analysis to provide evidence for a pathway connecting PLC/DGK signaling to the PA and LPA response. The suggested pathway was confirmed using differential radiolabeling techniques. A model is presented that maps different metabolic PA and LPA pools and their precursors and shows where the NaCl-induced LPA response fits in (Fig. 5).
Fig. 5.
Schematic representation of LPA and PA pools with distinct molecular species compositions, based on data from this and previous reports (5, 9). The fatty acids in de novo synthesized PA and LPA differ from the fatty acids in PA and LPA generated in the DGK and PLD signaling pathways. The accumulation of α-LA-enriched LPA in the salt stress response in Chlamydomonas is suggested to be derived from DGK-generated PA. In the presence of n-butanol, PLD can catalyze a transphosphatidylation reaction by which the artificial lipid PBut is formed, which is used as marker of PLD activity. The fatty acid composition of PBut matches that of the PLD substrate lipid pool (PE), and of PLD's normal product, PA.
LPA accumulates under salt stress, not via GPAT, but through PLA2 activity
The enzymes GPAT and LPAAT are responsible for de novo synthesis of LPA and PA, respectively (Fig. 1). This route prevails at basal conditions, but its activity might be induced under stress conditions, similar to observations of de novo synthesized DAG in hypo-osmotically stressed Dunaliella salina (32). However, the evidence presented indicates that the salt-induced LPA response in Chlamydomonas is not a consequence of stimulated GPAT activity. First, the α-LA-rich LPA under salt stress is clearly different from de novo synthesized LPA molecular species (Fig. 3B, D and Fig. 4B, D). Second, under basal conditions, 32P-radiolabeling of LPA occurred slowly, reaching equilibrium after days, whereas salt-stimulated LPA showed a contrary labeling trend (supplementary Fig. II). As a product of enhanced GPAT activity, LPA would display the slow equilibration kinetics of its precursors, which is not the case.
Not only did the results argue against de novo synthesis as the source of the LPA response, but several lines of evidence also suggested that it was derived from PA through PLA2 activity. Salt stress triggers a rapid increase in PA (Fig. 2), which has been shown previously to consist of different subpools (5, 22). LPA's peculiar fatty acid composition (Figs. 3 and 4) and its labeling kinetics (supplementary Figs. I and II) were consistent with its formation, through PLA2, from one of these PA subpools (discussed below). Accordingly, the increase in PA sets in a bit earlier than the LPA increase (supplementary Fig. I-A) (6). The results are also in agreement with a previous study, showing that pharmacological inhibition of PLA2 abrogated the LPA response (6). This treatment simultaneously increased the 32P-level of PA and its metabolite DGPP, again indicating that LPA, like DGPP, is derived from salt-induced PA.
Salt stress-induced PLA2 hydrolyses a subpool of PA that is generated via the PLC/DGK route
As summarized in Fig. 1 and represented in Fig. 5, several potential pathways could generate PA during salt stress. De novo synthesis of PA implicates LPA as precursor, whereas the PLC/DGK and PLD pathways may provide PA as substrate to PLA2, generating LPA as product. Previous studies have implicated the PLC/DGK and PLD pathways in the salt response in Chlamydomonas (5, 22). From fatty acid analyses of the artificial PLD product phosphatidylbutanol (PBut) (33), it was inferred that the enzyme hydrolyzed PE and that this did not produce α-LA-containing PA species (5), which is in agreement with our present conclusion that PLA2’s substrate is provided by another pathway (Fig. 5). NaCl-stimulated LPA species contain several fatty acids that are characteristic of PIP2, PIP, and PI: α-LA, 18:1n-7, and 18:2n-6 (4). Each is present at the sn-1 position of PI (Table 1), suggesting that sequential PIP and/or PIP2 hydrolysis by PLC and subsequent phosphorylation of DAG could provide the PA substrate to PLA2. This was further supported by our 32P-radiolabeling experiments. Under pulse-labeling conditions, which favor labeling of the products of DGK, salt induced rapid and large 32P-PA and 32P-LPA increments (supplementary Fig. I). Conversely, under conditions that favor label incorporation into PLD's substrate, 32P-LPA levels declined, likely following the decreased specific radioactivity of ATP (supplementary Fig. II). Interestingly, hyperosmotic stress in Arabidopsis seedlings induced the formation of DAG and PA species with a high 18:2n-6 and α-LA content, which was speculated to reflect a PLC/DGK pathway, drawing on a PPI pool with increased PUFA levels (33).
The finding that pharmacological inhibition of PLA2 not only decreased salt-induced LPA formation in Chlamydomonas but also simultaneously increased the levels of PA and DGPP (6) has interesting implications, as it suggests that PLA2 and PA kinase (PAK) compete for the same PA substrate pool. According to the present data, this is the PA pool generated through the PLC/DGK pathway. As DGK and PAK are predicted to be mainly localized at the plasma membrane (9, 34), PLA2 might be active there as well.
LPA and PLA2 signaling
In animal systems, LPA is an intercellular signaling molecule that can be produced by a highly PA-selective PLA2 (35), and it functions as ligand for G-protein coupled endothelial differentiation gene (EDG) receptors (36). In plants, however, these receptors are missing, and in a unicellular alga, it is not very likely that LPA is secreted and sensed by others. Nonetheless, other functions for PLA2 products have been described, and during the last few years, several advances in plant PLA2 signaling in higher plants have been made (37–40).
The PLA superfamily includes a broad range of enzymes. Although it is not always clear which substrate is used, both lysophospholipids and free fatty acids have been implicated in signaling (38, 40, 41). Literature on PA-specific enzymes is limited, however. From Arabidopsis, a putative PA-PLA1 gene (SRG1) has been cloned in which the knockout mutant displayed reduced gravitropic responses (42). In Arabidopsis, a novel PLA2, SOBER1, with no homology to any of the plant PLA families, was suggested to suppress the elicitor-induced induction of a hypersensitive response by reducing the accumulation of PA (43). However, based on in vitro activity of recombinant SOBER1, its substrate was speculated to be PC rather than PA. Other recent plant studies implicating a role for PLA2 includes auxin signaling (39), pH regulation (44, 45), plant defense (37, 43–46), stomatal opening (47), pollen development and germination (48), and vesicular transport (39, 48). For PA, putative protein targets have been identified in a proteomics screen using PA-affinity beads (49). It will be interesting to test LPA as a competitor in those assays, as some of the proteins may be LPA targets.
Salt stress responses in Chlamydomonas and other green algal systems
While a similar salt-induced LPA response seems to be lacking in higher plants, it has been reported in Dunaliella salina under hypersalinity stress (50). In this system also, 32P-lyso-PC increased and 32P-PA decreased. The distinct response may be related to the fact that D. salina lives in saline environments and is extremely salt tolerant. Phospholipid changes were only found when salt concentrations increased from 1.7 to 3.4 M NaCl, which may induce effects of NaCl toxicity, in addition to cell volume decrease, plasma membrane infolding, and shrinking of internal membranes. In the green alga Micrasterias denticulata, salt stress induced similar ultrastructural changes and a rapid (5 min) accumulation of reactive oxygen species, followed by symptoms of programmed cell death (51), which in higher plants has been associated with PA signaling (52, 53). With respect to osmoregulation, Chlamydomonas has a unique mechanism based on the function of its contractile vacuole, which has been implicated in the elimination of water in a low osmotic potential environment (54); however, its function in hyperosmotic stress has yet to be evaluated.
Possible functions of free fatty acids and their metabolites
In Chlamydomonas, the major molecular species of PI that have 18:3 are 18:3/16:0 and 18:3/18:3 (5). Upon PLA2 hydrolysis, they would generate free palmitic acid and α-LA. Free fatty acids and their metabolites may have functions in the regulation of enzymes, such as PLDδ, whose activity is enhanced in the presence of oleate and, to a lesser extent, α-LA (55). Moreover, α-LA can be metabolized to octadecanoids like jasmonic acid (JA), which acts as growth factor and modulator of stress resistance (56). Wounding-induced JA accumulation was shown to be generated by two chloroplast-localized galactolipases/PLAs, AtDAD1 and AtDGL1 (57).
In an evolutionary distant diatom, Thalassiosira rotula, wounding has been reported to trigger PLA2 activity, releasing C20 polyunsaturated fatty acids within minutes. These fatty acids were further metabolized to the defensive aldehydes 2,4-decadienal and 2,4,7-decatrienal (58). In Arabidopsis leaves, wounding leads to the formation of hexanal through a lipid-hydrolyzing activity, releasing α-LA as its precursor (59). Apart from the role of these aldehydes in toxic defense against grazers, they have been speculated to be chemical signals of unfavorable growth conditions, inducing programmed cell death within phytoplankton communities (1).
Biophysical aspects of PLA2-mediated LPA formation
Due to an intramolecular hydrogen bond between the headgroup of LPA and the sn-2-hydroxyl on the glycerol backbone, LPA carries more negative charge than PA (60). Moreover, at neutral pH, LPA has the shape of an inverted cone, whereas PA behaves as a cone shape, which could be even more pronounced if it contains PUFAs like α-LA (61). Hence, the interconversion of PA and LPA by PLA2 and LPAAT may affect local membrane charge and curvature depending on the membrane environment. Both effects may contribute to the function of the mammalian LPAATs C-terminal-binding protein/brefeldin A-ADP ribosylated substrate (CtBP/BARS) (62) and endophilin 1 (63) in vesicle formation. The response to hyperosmotic stress in Chlamydomonas may require membrane surface reduction by endocytosis to adapt to the decreased cell volume.
The question of where the different pools of PA and LPA characterized in this study (Fig. 5) are located in the cell will be important. Lipid biosensors for PA and LPA may yield valuable information about where and when PA and LPA are formed (64).
Supplementary Material
Acknowledgments
The authors thank Dr. Michel Haring (SILS, University of Amsterdam) for critical discussions and Dr. Harold Meijer (Wageningen University) for his earlier contribution in the 32P-labeling experiments.
Footnotes
Abbreviations:
- CDP-Etn
- CDP-ethanolamine
- CMP-PA
- cytidine monophosphate-PA
- DAG
- diacylglycerol
- DGK
- DAG kinase
- DGPP
- diacylglycerol pyrophosphate
- ER
- endoplasmic reticulum
- FAME
- fatty acid methyl ester
- G3P
- glycerol-3-phosphate
- GPAT
- G3P acyltransferase
- InsPP
- inositolpolyphosphate
- JA
- jasmonic acid
- α-LA
- α-linolenic acid
- LPA
- lysophosphatidic acid
- LPAAT
- LPA acyltransferase
- LPI
- lyso-phosphatidylinositol
- MGDG
- monogalactosyldiacylglycerol
- NPC
- nonspecific PLC
- PA
- phosphatidic acid
- PAK
- PA kinase
- PAP
- PA phosphatase
- PBut
- phosphatidylbutanol
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- PI
- phosphatidylinositol
- PIP
- phosphatidylinositol 4-phosphate
- PIP2
- phosphatidylinositol 4,5-bisphosphate
- PPI
- polyphosphoinositide
- PLA (A2/C/D)
- phospholipase A (A2/C/D)
- UDP-Gal
- UDP-galactose
This work was supported by Netherlands Organization for Scientific Research (NWO) Grants VIDI 864.05.001 and ECHO 700.56.007 and by European Cooperation in Science and Technology (COST) Grant FA0605.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two figures.
References
- 1.Guschina I. A., Harwood J. L. 2006. Lipids and lipid metabolism in eukaryotic algae. Prog. Lipid Res. 45: 160–186 [DOI] [PubMed] [Google Scholar]
- 2.Munnik T., Meijer H. J. 2001. Osmotic stress activates distinct lipid and MAPK signalling pathways in plants. FEBS Lett. 498: 172–178 [DOI] [PubMed] [Google Scholar]
- 3.Munnik T., Testerink C. 2009. Plant phospholipid signaling: “in a nutshell.” J. Lipid Res. 50: S260–S265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arisz S. A., van Himbergen J. A., Musgrave A., van den Ende H., Munnik T. 2000. Polar glycerolipids of Chlamydomonas moewusii. Phytochemistry. 53: 265–270 [DOI] [PubMed] [Google Scholar]
- 5.Arisz S. A., Valianpour F., van Gennip A. H., Munnik T. 2003. Substrate preference of stress-activated phospholipase D in Chlamydomonas and its contribution to PA formation. Plant J. 34: 595–604 [DOI] [PubMed] [Google Scholar]
- 6.Meijer H. J., Arisz S. A., Van Himbergen J. A., Musgrave A., Munnik T. 2001. Hyperosmotic stress rapidly generates lyso-phosphatidic acid in Chlamydomonas. Plant J. 25: 541–548 [DOI] [PubMed] [Google Scholar]
- 7.Meijer H. J., ter Riet B., van Himbergen J. A., Musgrave A., Munnik T. 2002. KCl activates phospholipase D at two different concentration ranges: distinguishing between hyperosmotic stress and membrane depolarization. Plant J. 31: 51–59 [DOI] [PubMed] [Google Scholar]
- 8.Arisz S. A., Munnik T. 2010. Diacylglycerol kinase. Lipid Signaling in Plants. Munnik T., editor Springer, New York: 107–114 [Google Scholar]
- 9.Arisz S. A., Testerink C., Munnik T. 2009. Plant PA signaling via diacylglycerol kinase. Biochim. Biophys. Acta. 1791: 869–875 [DOI] [PubMed] [Google Scholar]
- 10.Hong Y., Zhang W., Wang X. 2010. Phospholipase D and phosphatidic acid signalling in plant response to drought and salinity. Plant Cell Environ. 33: 627–635 [DOI] [PubMed] [Google Scholar]
- 11.Li M., Hong Y., Wang X. 2009. Phospholipase D- and phosphatidic acid-mediated signaling in plants. Biochim. Biophys. Acta. 1791: 927–935 [DOI] [PubMed] [Google Scholar]
- 12.Munnik T. 2001. Phosphatidic acid: an emerging plant lipid second messenger. Trends Plant Sci. 6: 227–233 [DOI] [PubMed] [Google Scholar]
- 13.Testerink C., Munnik T. 2005. Phosphatidic acid: a multifunctional stress signaling lipid in plants. Trends Plant Sci. 10: 368–375 [DOI] [PubMed] [Google Scholar]
- 14.Wang X. 2005. Regulatory functions of phospholipase D and phosphatidic acid in plant growth, development, and stress responses. Plant Physiol. 139: 566–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munnik T., Vermeer J. E. 2010. Osmotic stress-induced phosphoinositide and inositol phosphate signalling in plants. Plant Cell Environ. 33: 655–669 [DOI] [PubMed] [Google Scholar]
- 16.Gaude N., Nakamura Y., Scheible W. R., Ohta H., Dormann P. 2008. Phospholipase C5 (NPC5) is involved in galactolipid accumulation during phosphate limitation in leaves of Arabidopsis. Plant J. 56: 28–39 [DOI] [PubMed] [Google Scholar]
- 17.Nakamura Y., Koizumi R., Shui G., Shimojima M., Wenk M. R., Ito T., Ohta H. 2009. Arabidopsis lipins mediate eukaryotic pathway of lipid metabolism and cope critically with phosphate starvation. Proc. Natl. Acad. Sci. USA. 106: 20978–20983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peters C., Li M., Narasimhan R., Roth M., Welti R., Wang X. 2010. Non-specific phospholipase C NPC4 promotes responses to abscisic acid and tolerance to hyperosmotic stress in Arabidopsis. Plant Cell. 22: 2642–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schuring F., Smeenk J. W., Homan W. L., Musgrave A., Van den Ende H. 1987. Occurrence of O-methylated sugars in surface glycoconjugates in Chlamydomonas eugametos. Planta. 170: 322–327 [DOI] [PubMed] [Google Scholar]
- 20.Munnik T., Irvine R. F., Musgrave A. 1994. Rapid turnover of phosphatidylinositol 3-phosphate in the green alga Chlamydomonas eugametos: signs of a phosphatidylinositide 3-kinase signalling pathway in lower plants? Biochem. J. 298: 269–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munnik T., Van Himbergen J. A. J., Ter Riet B., Braun F-J., Irvine R. F., Van den Ende H., Musgrave A. R. 1998. Detailed analysis of the turnover of polyphosphoinositides and phosphatidic acid upon activation of phospholipases C and D in Chlamydomonas cells treated with non-permeabilizing concentrations of mastoparan. Planta. 207: 133–145 [Google Scholar]
- 22.Munnik T., Meijer H. J., Ter Riet B., Hirt H., Frank W., Bartels D., Musgrave A. 2000. Hyperosmotic stress stimulates phospholipase D activity and elevates the levels of phosphatidic acid and diacylglycerol pyrophosphate. Plant J. 22: 147–154 [DOI] [PubMed] [Google Scholar]
- 23.Munnik T., de Vrije T., Irvine R. F., Musgrave A. 1996. Identification of diacylglycerol pyrophosphate as a novel metabolic product of phosphatidic acid during G-protein activation in plants. J. Biol. Chem. 271: 15708–15715 [DOI] [PubMed] [Google Scholar]
- 24.Meijer H. J., Berrie C. P., Iurisci C., Divecha N., Musgrave A., Munnik T. 2001. Identification of a new polyphosphoinositide in plants, phosphatidylinositol 5-monophosphate (PtdIns5P), and its accumulation upon osmotic stress. Biochem. J. 360: 491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meijer H., Divecha N., Van den Ende H., Musgrave A., Munnik T. 1999. Hyperosmotic stress induces rapid synthesis of phosphatidyl-D-inositol 3,5-bisphosphate in plant cells. Planta. 208: 294–298 [Google Scholar]
- 26.van Schooten B., Testerink C., Munnik T. 2006. Signalling diacylglycerol pyrophosphate, a new phosphatidic acid metabolite. Biochim. Biophys. Acta. 1761: 151–159 [DOI] [PubMed] [Google Scholar]
- 27.Paradis S., Villasuso A. L., Aguayo S. S., Maldiney R., Habricot Y., Zalejski C., Machado E., Sotta B., Miginiac E., Jeannette E. 2011. Arabidopsis thaliana lipid phosphate phosphatase 2 is involved in abscisic acid signalling in leaves. Plant Physiol. Biochem. 49: 357–362 [DOI] [PubMed] [Google Scholar]
- 28.Racagni G., Villasuso A. L., Pasquare S. J., Giusto N. M., Machado E. 2008. Diacylglycerol pyrophosphate inhibits the alpha-amylase secretion stimulated by gibberellic acid in barley aleurone. Physiol. Plant. 134: 381–393 [DOI] [PubMed] [Google Scholar]
- 29.Wang X., Li W., Li M., Welti R. 2006. Profiling lipid changes in plant response to low temperatures. Physiol. Plant. 126: 90–96 [Google Scholar]
- 30.Welti R., Li W., Li M., Sang Y., Biesiada H., Zhou H. E., Rajashekar C. B., Williams T. D., Wang X. 2002. Profiling membrane lipids in plant stress responses. Role of phospholipase D alpha in freezing-induced lipid changes in Arabidopsis. J. Biol. Chem. 277: 31994–32002 [DOI] [PubMed] [Google Scholar]
- 31.Testerink C., Munnik T. 2011. Molecular, cellular, and physiological responses to phosphatidic acid formation in plants. J. Exp. Bot. 62: 2349–2361 [DOI] [PubMed] [Google Scholar]
- 32.Ha K. S., Thompson G. A., Jr 1992. Biphasic changes in the level and composition of Dunaliella salina plasma membrane diacylglycerols following hypoosmotic shock. Biochemistry. 31: 596–603 [DOI] [PubMed] [Google Scholar]
- 33.König S., Mosblech A., Heilmann I. 2007. Stress-inducible and constitutive phosphoinositide pools have distinctive fatty acid patterns in Arabidopsis thaliana. FASEB J. 21: 1958–1967 [DOI] [PubMed] [Google Scholar]
- 34.Wissing J. B., Behrbohm H. 1993. Phosphatidate kinase, a novel enzyme in phospholipid metabolism (purification, subcellular localization, and occurrence in the plant kingdom). Plant Physiol. 102: 1243–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snitko Y., Yoon E. T., Cho W. 1997. High specificity of human secretory class II phospholipase A2 for phosphatidic acid. Biochem. J. 321: 737–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noguchi K., Herr D., Mutoh T., Chun J. 2009. Lysophosphatidic acid (LPA) and its receptors. Curr. Opin. Pharmacol. 9: 15–23 [DOI] [PubMed] [Google Scholar]
- 37.Canonne J., Froidure-Nicolas S., Rivas S. 2011. Phospholipases in action during plant defense signaling. Plant Signal. Behav. 6: 13–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holk A., Rietz S., Zahn M., Quader H., Scherer G. F. 2002. Molecular identification of cytosolic, patatin-related phospholipases A from Arabidopsis with potential functions in plant signal transduction. Plant Physiol. 130: 90–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee O. R., Kim S. J., Kim H. J., Hong J. K., Ryu S. B., Lee S. H., Ganguly A., Cho H. T. 2010. Phospholipase A(2) is required for PIN-FORMED protein trafficking to the plasma membrane in the Arbidopsis root. Plant Cell. 22: 1812–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scherer G. F. E. 2010. Phospholipase A in plant signal transduction. Lipid Signaling in Plants. Munnik T., editor Springer, New York: 3–22 [Google Scholar]
- 41.Ryu S. B. 2004. Phospholipid-derived signaling mediated by phospholipase A in plants. Trends Plant Sci. 9: 229–235 [DOI] [PubMed] [Google Scholar]
- 42.Kato T., Morita M. T., Fukaki H., Yamauchi Y., Uehara M., Niihama M., Tasaka M. 2002. SGR2, a phospholipase-like protein, and ZIG/SGR4, a SNARE, are involved in the shoot gravitropism of Arabidosis. Plant Cell. 14: 33–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kirik A., Mudgett M. B. 2009. SOBER1 phospholipase activity suppresses phosphatidic acid accumlation and plant immunity in response to bacterial effector AvrBsT. Proc. Natl. Acad. Sci. USA. 106: 20532–20537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roos W., Viehweger K., Dordschbal B., Schumann B., Evers S., Steighardt J., Schwartze W. 2006. Intracellular pH signals in the induction of secondary pathways - the case of Eschscholzia californica. J. Plant Physiol. 163: 369–381 [DOI] [PubMed] [Google Scholar]
- 45.Viehweger K., Schwartze W., Schumann B., Lein W., Roos W. 2006. The Galpha protein controls a pH-dependent signal path to the induction of phytoalexin biosynthesis in Eschscholzia californica. Plant Cell. 18: 1510–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Froidure S., Canonne J., Daniel X., Jauneau A., Briere C., Roby D., Rivas S. 2010. AtsPLA2-alpha nuclear relocalization by the Arabidopsis transcription factor AtMYB30 leads to repression of the plant defense response. Proc. Natl. Acad. Sci. USA. 107: 15281–15286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seo J., Lee H. Y., Choi H., Choi Y., Lee Y., Kim Y. W., Ryu S. B., Lee Y. 2008. Phospholipase A2beta mediates light-induced stomatal opening in Arabidopsis. J. Exp. Bot. 59: 3587–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim H. J., Ok S. H., Bahn S. C., Jang J., Oh S. A., Park S. K., Twell D., Ryu S. B., Shin J. S. 2011. Endoplasmic reticulum- and Golgi-localized phospholipase A2 plays critical roles in Arabidopsis pollen development and germination. Plant Cell. 23: 94–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Testerink C., Dekker H. L., Lim Z. Y., Johns M. K., Holmes A. B., Koster C. G., Ktistakis N. T., Munnik T. 2004. Isolation and identification of phosphatidic acid targets from plants. Plant J. 39: 527–536 [DOI] [PubMed] [Google Scholar]
- 50.Einspahr K. J., Maeda M., Thompson G. A., Jr 1988. Concurrent changes in Dunaliella salina ultrastructure and membrane phospholipid metabolism after hyperosmotic shock. J. Cell Biol. 107: 529–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Affenzeller M. J., Darehshouri A., Andosch A., Lutz C., Lutz-Meindl U. 2009. Salt stress-induced cell death in the unicellular green alga Micrasterias denticulata. J. Exp. Bot. 60: 939–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yakimova E. T., Kapchina-Toteva V. M., Woltering E. J. 2007. Signal transduction events in aluminum-induced cell death in tomato suspension cells. J. Plant Physiol. 164: 702–708 [DOI] [PubMed] [Google Scholar]
- 53.Yamaguchi T., Tanabe S., Minami E., Shibuya N. 2004. Activation of phospholipase D induced by hydrogen peroxide in suspension-cultured rice cells. Plant Cell Physiol. 45: 1261–1270 [DOI] [PubMed] [Google Scholar]
- 54.Guillard R. R. L. 2007. A mutant of Chlamydomonas moewusii lacking contractile vacuoles. J. Eukar. Micr. 7: 262–268 [Google Scholar]
- 55.Wang C., Wang X. 2001. A novel phospholipase D of Arabidopsis that is activated by oleic acid and associated with the plasma membrane. Plant Physiol. 127: 1102–1112 [PMC free article] [PubMed] [Google Scholar]
- 56.Stratmann J. W. 2003. Long distance run in the wound response - jasmonic acid is pulling ahead. Trends Plant Sci. 8: 247–250 [DOI] [PubMed] [Google Scholar]
- 57.Hyun Y., Choi S., Hwang H. J., Yu J., Nam S. J., Ko J., Park J. Y., Seo Y. S., Kim E. Y., Ryu S. B., et al. 2008. Cooperation and functional diversification of two closely related galactolipase genes for jasmonate biosynthesis. Dev. Cell. 14: 183–192 [DOI] [PubMed] [Google Scholar]
- 58.Pohnert G. 2002. Phospholipase A2 activity triggers the wound-activated chemical defense in the diatom Thalassiosira rotula. Plant Physiol. 129: 103–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matsui K., Kurishita S., Hisamitsu A., Kajiwara T. 2000. A lipid-hydrolysing activity involved in hexenal formation. Biochem. Soc. Trans. 28: 857–860 [PubMed] [Google Scholar]
- 60.Kooijman E. E., Chupin V., Fuller N. L., Kozlov M. M., de Kruijff B., Burger K. N., Rand P. R. 2005. Spontaneous curvature of phosphatidic acid and lysophosphatidic acid. Biochemistry. 44: 2097–2102 [DOI] [PubMed] [Google Scholar]
- 61.Kooijman E. E., Chupin V., de Kruijff B., Burger K. N. 2003. Modulation of membrane curvature by phosphatidic acid and lysophosphatidic acid. Traffic. 4: 162–174 [DOI] [PubMed] [Google Scholar]
- 62.Weigert R., Silletta M. G., Spano S., Turacchio G., Cericola C., Colanzi A., Senatore S., Mancini R., Polishchuk E. V., Salmona M., et al. 1999. CtBP/BARS induces fission of Golgi membranes by acylating lysophosphatidic acid. Nature. 402: 429–433 [DOI] [PubMed] [Google Scholar]
- 63.Schmidt A., Wolde M., Thiele C., Fest W., Kratzin H., Podtelejnikov A. V., Witke W., Huttner W. B., Soling H. D. 1999. Endophilin I mediates synaptic vesicle formation by transfer of arachidonate to lysophosphatidic acid. Nature. 401: 133–141 [DOI] [PubMed] [Google Scholar]
- 64.Vermeer J. E., Munnik T. 2010. Imaging lipids in living plants. Lipid Signaling in Plants. Munnik T., editor Springer, New York: 185–201 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.