Abstract
Chronic alcohol consumption is associated with fatty liver disease in mammals. The object of this study was to gain an understanding of dysregulated lipid metabolism in alcohol-fed C57BL/6 mice using a targeted lipidomic approach. Liquid chromatography tandem mass spectrometry was used to analyze several lipid classes, including free fatty acids, fatty acyl-CoAs, fatty acid ethyl esters, sphingolipids, ceramides, and endocannabinoids, in plasma and liver samples from control and alcohol-fed mice. The interpretation of lipidomic data was augmented by gene expression analyses for important metabolic enzymes in the lipid pathways studied. Alcohol feeding was associated with i) increased hepatic free fatty acid levels and decreased fatty acyl-CoA levels associated with decreased mitochondrial fatty acid oxidation and decreased fatty acyl-CoA synthesis, respectively; ii) increased hepatic ceramide levels associated with higher levels of the precursor molecules sphingosine and sphinganine; and iii) increased hepatic levels of the endocannabinoid anandamide associated with decreased expression of its catabolic enzyme fatty acid amide hydrolase. The unique combination of lipidomic and gene expression analyses allows for a better mechanistic understanding of dysregulated lipid metabolism in the development of alcoholic fatty liver disease.
Keywords: liquid chromatography tandem mass spectrometry, LC/MS/MS, fatty acid, fatty acyl-CoA, sphingolipid, endocannabinoid, Lieber-DeCarli diet
Alcoholic fatty liver is generally the first presentation of alcoholic liver disease (ALD) and is thought to occur in ∼20% of alcoholics (1). Studies of hepatic lipid metabolism can provide mechanistic insights into the development of alcoholic fatty liver, as well as help in identifying potential biomarkers for progression to more severe disease. Lipid metabolism has been extensively studied in the liver of alcoholic humans and in animal models of ALD, implicating alcohol-associated alterations in several pathways, including fatty acid (FA) uptake, oxidation, and export (2, 3). However, until recently, technical limitations have prevented a survey of multiple lipid species in the same tissue, typically limiting studies to analyzing one lipid class or yielding only total levels of a specific lipid class. The development of high-sensitivity liquid chromatography tandem mass spectrometry (LC/MS/MS) allows for study of the so-called lipidome (i.e., the entire lipid content of tissues) in a specific model or disease of interest. Multiple lipid species can be assayed quantitatively in the same biological sample (4). Initial attempts to study the lipidome of alcohol-exposed liver have recently been reported (5–7). These studies have established the feasibility of using a lipidomic approach to characterize dysregulated lipid metabolism in response to alcohol. However, their impact was limited because only changes in lipid levels were reported. Indeed, Fernando et al. concluded that an evaluation of gene expression was required to provide a mechanistic understanding of their lipid data (5). In the present study, we have taken the next step by combining lipidomic analyses of alcohol-fed mice with a survey of gene expression, targeting key enzymes in the metabolic pathways for the lipid classes studied.
Our primary objective was to undertake a targeted lipidomic analysis of plasma and liver from control and alcohol-fed mice, and then to correlate observed changes in lipid levels with gene expression. Using specific internal standards, we were able to quantify levels of 62 defined lipid species by LC/MS/MS. Our analysis focused on three pathways: i) FA metabolism [measuring free fatty acids (FFA), fatty acyl CoAs (FA-CoA), and fatty acid ethyl esters (FAEE)]; ii) sphingolipid metabolism; and iii) the endocannabinoid system. These categories were chosen because of their importance in hepatic lipid metabolism, as well as their known associations with lipotoxicity and hepatic disease. Hepatic FFA and FA-CoA are essential precursors for many lipids in the liver. FAEEs are important in the context of ALD, as they are produced by the nonoxidative metabolism of alcohol. In addition to FA metabolites, we also focused on two major lipid-signaling families, sphingolipids (specifically ceramide and its precursors) and endocannabinoids [specifically anandamide (AEA) and 2-arachidonoylglycerol (2-AG)]. Ceramides are now recognized as important signaling molecules associated with control of cell survival and replication. Dysregulated sphingolipid and ceramide metabolism has been implicated in numerous diseases, including cancer, cardiovascular disease, and ALD (6, 8, 9). As discussed below, a link between endocannabinoid signaling and fatty liver disease has been identified, with one study also suggesting an involvement in ALD (10, 11).
We have integrated the results of our targeted lipidomic study with analyses of gene expression, providing an improved mechanistic understanding of alcohol-induced changes in hepatic lipid levels. Alcohol feeding was associated with pronounced increases in hepatic FFA levels, and conversely with a decrease in FA-CoA. Sphingolipid and ceramide levels were also increased in liver of alcohol-fed mice, as well as the endocannabinoid, AEA. Alcohol-induced changes in hepatic lipid levels are discussed in the context of our gene expression data.
MATERIALS AND METHODS
Animals and alcohol feeding
All animal experiments were carried out with the approval of the Institutional Animal Care and Use Committee of Columbia University according to criteria outlined in the Guide for the Care and Use of Laboratory Animals prepared by the National Academy of Sciences. Three- to four-month-old male C57BL/6 mice were used for all studies. Mice were maintained in an environmentally controlled facility with a 12 h light/dark cycle. All mice were maintained on a standard rodent chow diet until the beginning of the experiment, when they were randomized onto control or alcohol-containing liquid diets. Mice were fed alcohol using the Lieber-DeCarli liquid diet formulation (Bio-Serv, Frenchtown, NJ). This established alcohol-feeding paradigm employs nutritionally complete liquid diets, allowing mice fed the alcohol-containing diet to receive a defined volume of alcohol and control mice to receive an isocaloric control diet containing malto-dextrin in lieu of alcohol (12). All mice were housed singly to allow for measurement of diet consumption and to facilitate the pair feeding of control mice. We employed a run-in period to allow the mice receiving alcohol to acclimate to alcohol feeding. This period consisted of one week of control liquid diet, one week of 2.2% v/v alcohol, one week of 4.5% v/v alcohol, and two weeks of 6.7% v/v alcohol. Body weights were measured weekly. At the end of the experiment, mice were euthanized following a 4-5 h fasting period. Blood was drawn by intracardiac puncture, decanted into a tube containing 5 µl of 0.5 M EDTA, and then stored on ice. Plasma was separated from cells by centrifugation for 10 min at 12,000 rpm (Model 5145 D, Eppendorf AG, Hamburg, Germany). The plasma was then transferred into a clean tube and snap frozen in liquid N2. Liver was dissected, weighed, and immediately snap frozen in liquid N2. All tissues were stored at −80°C prior to analysis.
Biochemical analyses
All biochemical analyses were performed using kits and standard protocols, as recommended by the specific kit's manufacturer. Blood alcohol content (BAC) was measured in plasma using a NAD-Alcohol dehydrogenase reagent (Sigma-Aldrich, St Louis, MO). For analysis of BAC, blood was taken between midnight and 1 AM after one week of exposure to 6.7% alcohol. Alanine aminotransferase (ALT) was measured in plasma using an ALT-SL assay (Genzyme Diagnostics, Charlottetown, PE, Canada). Triglyceride measurements were made using a liquid stable triglyceride reagent (Thermo Fisher Scientific, Middleton, VA). Measurements for liver triglyceride content were taken from a solution of total lipids extracted from liver homogenates using a standard Folch extraction (13). Hepatic retinyl ester concentration was determined by reverse-phase HPLC, as previously described (14).
LC/MS/MS
A detailed description of the LC/MS/MS methodology is provided in the supplementary data. In brief, all lipid extractions were performed within one week of tissue collection. Levels of extracted lipids were measured on a Waters Xevo TQ MS ACQUITY UPLC system (Waters, Milford, MA). The identity of each lipid species was confirmed with internal standards.
RNA extraction, cDNA synthesis, and qPCR
RNA was extracted from liver samples using TRIzol (Invitrogen, Carlsbad, CA), according to the manufacturer's protocol. RNA cleanup and DNA digestion were performed on a Qiagen (Valencia, CA) RNeasy column. The concentration and quality of isolated RNA was determined using a NanoDrop1000 spectrophotometer (Thermo Fisher Scientific). One microgram of purified RNA was reverse-transcribed into cDNA using a high-capacity cDNA RT kit (Applied Biosystems, Carlsbad, CA). Quantitative PCR was performed using a LightCycler 480 (Roche Diagnostics, Indianapolis, IN) with SYBR green PCR master mix (Roche Diagnostics) under uniform reaction conditions. All primers were designed using LightCycler probe design software 2.0 (Roche Diagnostics). Where more than one transcript variant was found for a given gene, a region common to all variants was used for primer design. Supplementary Table I provides a complete list of genes studied and primer sequences. All qPCR data analysis was performed as described by Pfaffl (15). Two reference genes were used in these studies: 18S and cyclophilin A. Changes in expression of target genes relative to these reference genes were in good agreement; only data normalized to cyclophilin A expression are presented. Although our gene expression analysis is a strength of the study, we recognize that gene expression levels do not necessarily translate to changes in protein expression. Nevertheless, we feel our conclusions are supported by our lipidomic data, and they are discussed in the context of the supporting literature.
Statistical analyses and heat maps
The data presented below were obtained from two independent studies, which were identical in experimental design. The data from both experiments were in good agreement. Consequently, combined data from both experiments are presented here. All raw data were processed in Excel (Microsoft, Redmond, WA). Statistical analyses were performed using Prism 5 (GraphPad Software, La Jolla, CA). Data reported in the text are means ± standard deviations. Student's t-tests were performed to determine statistical differences between control and alcohol-fed mice. A P value < 0.05 was considered statistically significant. The multiple comparisons performed are a limitation in this study, leading to the possibility of a type 1 error. However, as noted above, it should be emphasized that two independent experiments were performed yielding confirmatory data, reinforcing the validity of our analysis. Heat maps were generated in Excel using conditional formatting; the heat maps represent the relative lipid level in alcohol-fed mice compared with control mice. Increasing intensity of green indicates a relative decrease in lipid level, whereas increasing intensity of red indicates an increased level. Black represents no change. White indicates that the specific lipid was assayed but found to be undetectable in the sample.
RESULTS
Induction of ALD in alcohol-fed mice
Following alcohol feeding, basic parameters associated with ALD were assessed in control and alcohol-fed mice (Table 1). No alcohol was detectable in the plasma of control mice, whereas alcohol-fed mice had an average BAC of 0.1 ± 0.05%, a level that is consistent with other alcohol-feeding studies in mice (16–18). When mice were euthanized, there were small but statistically nonsignificant decreases in body weight, coupled with nonsignificant increases in liver weight in alcohol-fed mice; this resulted in a significant increase in the liver:body weight ratio in this group. The total hepatic triglyceride content for alcohol-fed mice was significantly greater than for control mice, by an average of ∼15%. Similarly, alcohol-fed mice showed a significant elevation in the circulating level of triglycerides. Consistent with the previously described effect of alcohol on hepatic retinyl ester stores in humans, the hepatic retinyl ester concentration was lower in alcohol-fed mice relative to control mice (19). Plasma ALT levels tended to be greater in alcohol-fed mice than in control mice, albeit not statistically significantly. As expected, levels of the alcohol-inducible enzyme Cyp2e1 were significantly increased in alcohol-fed mice (supplementary Fig. I).
TABLE 1.
Physical and biochemical data
| Control (n = 8) | Alcohol (n = 11) | P | |
| Body weight (g) | 28.3 ± 2.2 | 26.5 ± 2.1 | ns |
| Liver weight (g) | 1.15 ± 0.14 | 1.20 ± 0.12 | ns |
| Liver:body weight ratio | 0.041 ± 0.003 | 0.045 ± 0.002 | <0.005 |
| Serum triglycerides (mg/dl) | 60.6 ± 18.8 | 106.7 ± 22.8 | <0.05 |
| Liver triglycerides (mg/g) | 45.6 ± 6.4 | 53.0 ± 1.7 | <0.05 |
| Retinyl esters (nmol/g) | 1,441.2 ± 657.3 | 697.8 ± 293.0 | <0.005 |
| Plasma ALT (U/l) | 167.1 ± 45.2 | 206.0 ± 54.0 | ns |
| Blood alcohol content (%) | – | 0.10 ± 0.05 | |
| Cyp2e1 expression (relative units) | 1.00 ± 0.98 | 12.21 ± 3.58 | <0.005 |
All data are means ± standard deviation; P values were determined by Student's t-test.
ns, no significant difference.
Liver and plasma samples obtained from control and alcohol-fed mice underwent targeted lipidomic analyses by LC/MS/MS. The relative changes in lipid levels in alcohol-fed mice versus control mice are described below (the absolute values for each lipid species analyzed in plasma and liver are presented in supplementary Tables II and III, respectively).
Fatty acid metabolites and gene expression analysis
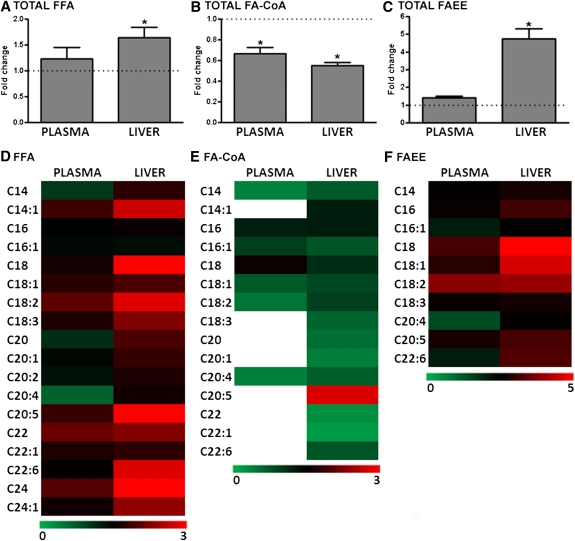
The relative changes in FFAs, FA-CoAs, and FAEEs levels were studied in alcohol-fed mice versus control mice (Fig. 1). Total FFAs in the plasma were not significantly increased by alcohol feeding, but a greater than 60% increase in hepatic FFA levels was observed (Fig. 1A). In contrast to the increase in total hepatic FFAs, alcohol-fed mice showed an approximately 50% lower level of total hepatic FA-CoA. Similarly, total FA-CoA levels in the plasma were also significantly reduced (Fig. 1B). Plasma levels of total FAEE trended up in alcohol-fed mice, but this did not reach statistical significance. However, for liver, there was an ∼5-fold increase in FAEE levels (Fig. 1C).
Fig. 1.
Relative levels of plasma and liver fatty acid metabolites in alcohol-fed mice. (A) The changes in plasma and hepatic total FFAs are shown for alcohol-fed mice (n = 11) relative to control mice (n = 8). In control mice, plasma total FFA levels are unchanged, whereas there is a significant increase in hepatic FFA levels for alcohol-fed mice. (B) Plasma and liver total FA-CoA levels are decreased in alcohol-fed mice. (C) Alcohol feeding is associated with a higher level of total FAEE in the liver; however there was no significant change in the plasma. Data are means ± SEM. Heat maps showing the relative change in plasma (left column) and hepatic lipids (right column) of different carbon-chain lengths are provided for FFA (D), FA-CoA (E), and FAEE (F). *P < 0.05 alcohol-fed versus control mice.
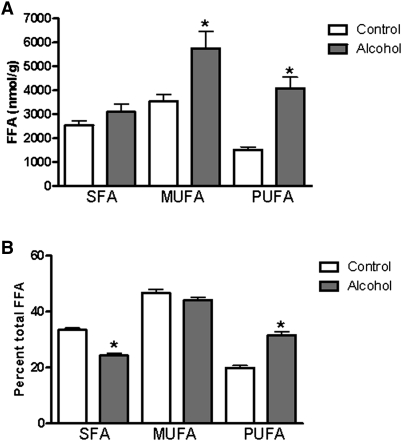
In addition to measuring total levels of these lipids, we analyzed the concentrations of individual fatty acyl species present. For plasma samples from alcohol-fed mice, statistically significant differences in FFA concentrations were observed only for the relatively unabundant myristoleic acid (C14:1) and docasanoic acid (22:0). There was a trend toward a decrease in plasma arachidonic acid (C20:4) levels in alcohol-fed mice, although this did not reach statistical significance owing to large standard deviations for these measures (control: 59.56 µM ± 66.59 versus alcohol: 26.85 µM ± 49.35). The majority of hepatic FFAs measured were found at higher levels in alcohol-fed mice (Fig. 1D). These increases were statistically significant for 11 of 18 FFAs analyzed. The greatest increases were seen with stearic acid (18:0) and eicosapentaenoic acid (EPA; 20:5), which were elevated 3- to 4-fold compared with control levels. When hepatic FFA levels were grouped according to the saturation of the FFAs, an interesting pattern emerged (Fig. 2). In absolute terms, the level of hepatic saturated FA (SFA) was not significantly changed by alcohol feeding; thus, the increase in total FFA arose from significant increases in monounsaturated FA (MUFA) and polyunsaturated FA (PUFA; Fig. 2A). In relative terms, the greatest increase was observed for PUFA, whereas MUFA was unchanged, and SFA was significantly decreased following alcohol feeding (Fig. 2B).
Fig. 2.
Increased unsaturated fatty acid levels in alcohol-fed mice. (A) Total hepatic saturated fatty acid (SFA), monounsaturated fatty acid (MUFA), and polyunsaturated fatty acid (PUFA) levels in control and alcohol-fed mice, demonstrating increased levels of MUFA and PUFA following alcohol feeding. (B) Relative levels of fatty acids in the liver, expressed as a percentage of total FFA. Following alcohol feeding, there was a relative decrease in SFA and a relative increase in PUFA. The relative amount of MUFA in the fatty acid pool was unchanged. Data are means ± SEM. *P < 0.05 alcohol-fed versus control mice.
With regard to the relative levels of individual FA-CoA in the plasma, there was a broad decrease in FA-CoAs, which is reflected in the significant decrease in total FA-CoA measured (Fig. 1E); however, statistically significant decreases for individual FA-CoA species were only observed for myristic acid (C14:0) and arachidonic acid (C20:4). In contrast to the increases in hepatic FFAs observed in alcohol-fed mice, FA-CoA levels were decreased across almost all carbon chain lengths (Fig. 1E). Of the 15 FA-CoA lipid species measured, 12 were significantly decreased.
The third group of FA metabolites analyzed was FAEEs (Fig. 1F). As noted above, we observed a nonsignificant increase in total plasma FAEE. When we analyzed individual FAEE species, we found a general trend toward increased levels, although these never reached significance. However, a large statistically significant increase in total hepatic FAEE was observed following alcohol feeding. Analysis of individual FAEE species revealed that all but one of the FAEEs measured had a greater mean hepatic concentration. This increase was significant in 4 of 10 FAEEs analyzed. Strikingly, hepatic levels of ethyl stearate (C18:0) were increased almost 20-fold in alcohol-fed mice. Indeed, large increases in 18-carbon FAEEs (C18:0, C18:1, C18:2) account for almost the entire absolute increase in hepatic total FAEEs.
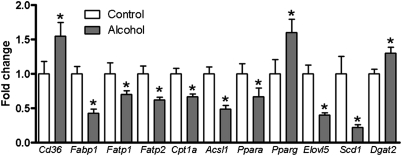
Hepatic gene expression profiles were determined by qPCR for the same control and alcohol-fed mice used for lipidomic analysis. To better understand the observed changes in FA metabolites, key enzymes and transcriptional regulators involved in hepatic FA metabolism were investigated. Genes that were significantly changed are presented in Fig. 3, and results of our gene expression analysis are provided in supplementary Table IV. Our analysis included genes involved in FA uptake (Cd36, Lpl, Hl, Fatp1, Fatp2, Fatp5, Fabp1); FA oxidation (Cpt1a); FA export (Apob); FA-CoA synthesis and oxidation (Acsl1, Acsl5, Acox1); de novo lipogenesis (Acc1, Fasn); desaturation and elongation (Scd1, Elovl5, Elovl6); triglyceride synthesis and hydrolysis (Dgat1, Dgat2, Tgh, Atgl, Hsl); and transcriptional regulation of FA metabolism (Ppara, Pparg, Srebp1c). With regard to FA uptake, we observed a significant increase in Cd36 expression and decreased expression of Fatp1, Fatp2, and Fabp1 in alcohol-fed mice. Expression of Apob, which encodes an apolipoprotein essential for export of lipid from liver into the circulation, was unchanged by alcohol feeding. Expression of Cpt1a, an important gene for FA oxidation, was significantly decreased following alcohol feeding. Similarly, Acsl1, which is associated with FA-CoA synthesis, was significantly downregulated, but Acsl5 expression was not significantly different. Acox1 catalyzes the oxidation of FA-CoA and is the rate-limiting step in peroxisomal FA oxidation (20, 21); its expression in alcohol-fed mice was unchanged compared with control mice. We studied three transcriptional regulators associated with lipid metabolism in the liver (Ppara, Pparg, Srebp1c). Signaling through PPARα is typically associated with increased FA oxidation. This gene was downregulated in our alcohol-fed mice. Conversely, PPARγ, which promotes lipogenesis, was significantly increased. Genes in the de novo lipogenesis pathway are regulated in part by Srebp1c, but we observed no change in the expression of this gene or genes it regulates (Acc1, Fasn). Scd1 encodes an enzyme that catalyzes the conversion of saturated FA to monounsaturated FA. This gene was significantly downregulated in alcohol-fed mice. The elongase Elovl5 also exhibited decreased expression following alcohol feeding. The final group of genes analyzed was those involved in triglyceride synthesis and hydrolysis. In this group, we observed a significant increase only in Dgat2 expression. The expression of all other genes tested was unchanged following alcohol feeding.
Fig. 3.
Alcohol feeding is associated with altered expression of genes involved in FA metabolism. Relative mRNA expression of several genes associated with FA metabolism is shown. Data are means ± SEM. *P < 0.05 alcohol-fed versus control mice.
Sphingolipid levels and gene expression analysis
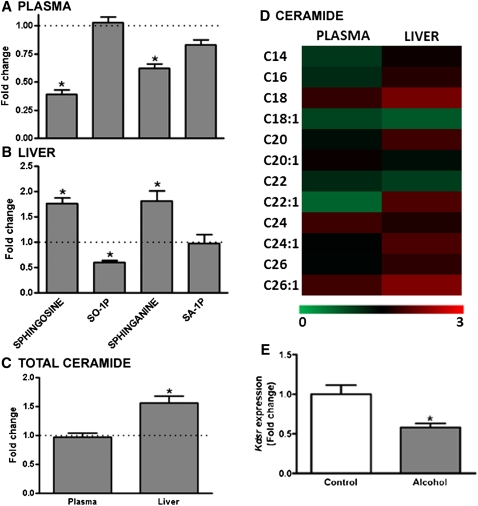
Sphingosine, sphingosine-1-phosphate (SO-1P), sphinganine, sphinganine-1-phosphate (SA-1P), and ceramide concentrations were measured in plasma and liver of control and alcohol-fed mice (Fig. 4). Alcohol exposure was associated with a significant decrease in circulating levels of sphingosine and sphinganine, although the phosphorylated forms of these sphingolipids were unchanged (Fig. 4A). In the liver, the opposite phenomenon was observed: sphingosine and sphinganine were both increased in alcohol-fed mice, whereas there was a ∼50% decrease in SO-1P and SA-1P was unchanged (Fig. 4B). Sphingosine and sphinganine, in combination with FAs of varying chain length, are precursors for the generation of ceramide. Differences in the fatty acyl compositions of plasma and liver ceramides are presented in Fig. 4D. In plasma, the total level of ceramide was equivalent in alcohol-fed and control mice (Fig. 4C). For the 12 different ceramides analyzed, significant decreases were observed in N-oleoyl ceramide (C18:1) and N-docasenoyl ceramide (C22:1), although these are low-abundance lipids in the plasma ceramide pool. There was an almost 2-fold increase in total ceramide levels in the liver of alcohol-fed mice (Fig. 4C). With regard to specific ceramides, of the 12 analyzed, 2 were significantly decreased and 2 increased. However, these were all relatively low-abundance ceramides. The increase in total ceramide apparently reflects the combined effect of small nonsignificant increases in the most abundant ceramides: N-palmitoyl ceramide (C16:0), N-tetracosanoyl ceramide (C24:0), and N-tetracosenoyl ceramide (C24:1), which, when totaled, result in a statistically significant increase in total ceramides.
Fig. 4.
Relative levels of sphingolipids in the plasma and liver of alcohol-fed mice. (A) Plasma levels of sphingosine and sphinganine were significantly lower in alcohol-fed mice (n = 11) relative to control mice (n = 8), with no change in SO-1P or SA-1P observed. (B) Liver levels of sphingosine and sphinganine were higher in alcohol-fed mice, whereas SO-1P was decreased, and SA-1P was unchanged. (C) Total ceramide levels were unchanged in the plasma of alcohol-fed mice; however there was a significant increase in total ceramide in the liver. (D) A heat map showing relative changes in individual ceramide molecules with different fatty acyl chain lengths is shown for plasma (left column) and liver (right column). (E) Kdsr expression is decreased in the liver of alcohol-fed mice. All data are means ± SEM. *P < 0.05 alcohol-fed versus control mice.
Sphingolipid and ceramide metabolism is complex and is only now beginning to be understood (22). Our analysis of gene expression in this pathway focused on specific genes associated with synthesis of sphinganine (Sptlc1, Sptlc2, Kdsr); phosphorylation and dephosphorylation of sphinganine and sphingosine (Spk1, Spk2, Spp1, Spp2); SO-1P and SA-1P catabolism (Sgpl1); and ceramide synthesis (Cers2, Smpd1). Of the eight genes analyzed, only one significant change was observed (supplementary Table V). Specifically, expression of Kdsr in alcohol-fed mice was reduced to approximately 60% of control mice (Fig. 4E).
Endocannabinoid levels and gene expression analysis
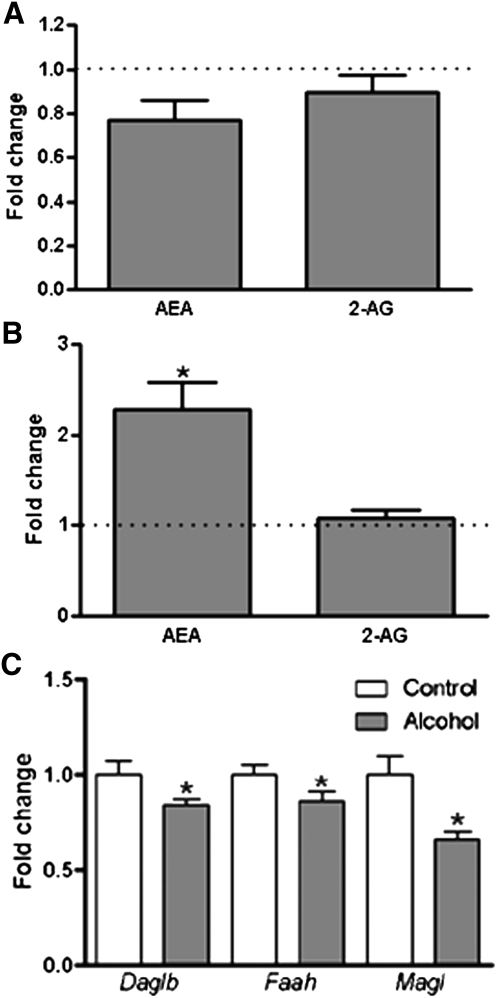
The levels of the endocannabinoids AEA and 2-AG were measured in plasma and liver of control and alcohol-fed mice (Fig. 5). We observed no significant difference in plasma level of AEA or 2-AG following alcohol exposure (Fig. 5A). In liver, the 2-AG level was also unchanged, although there was a more than 2-fold increase in AEA observed in alcohol-fed mice (Fig. 5B).
Fig. 5.
Relative plasma and hepatic levels of AEA and 2-AG in alcohol-fed mice (n = 11) versus control mice (n = 8). (A) Plasma levels of AEA and 2-AG in alcohol-fed mice relative to control mice are shown; there was no effect of alcohol feeding observed. (B) Liver levels of AEA were significantly increased in the alcohol-fed mice relative to control mice; however there was no change in 2-AG observed. (C) The expressions of Daglb, Faah, and Magl were significantly deceased in liver of alcohol-fed mice. Data are means ± SEM. *P < 0.05 alcohol-fed versus control mice.
The expression of genes representing key factors in the endocannabinoid system, which were significantly changed by alcohol feeding, are shown in Fig. 5C (a complete list of gene expression data is reported in supplementary Table VI) (23). Synthesis and breakdown of AEA was studied by examining the expression of Nape-pld and Faah, respectively. Synthesis (Dagla, Daglb) and breakdown (Magl) of 2-AG and cannabinoid receptor expression (Cb1 and Cb2) were also assessed. We observed a significant decrease in the expression of Faah, Daglb, and Magl in alcohol-fed mice. Expression levels for the other genes in this pathway were unchanged.
DISCUSSION
We have undertaken a targeted lipidomic analysis of plasma and liver from control and alcohol-fed mice. The study's goal was to gain an understanding of the development of fatty liver in this model of ALD. Several recent studies have used lipidomic analyses in the context of ALD, and as discussed below, many of their reported changes are in agreement with our study (5–7). We have extended the interpretation of these studies by performing a systematic analysis of gene expression in alcohol-fed mice, targeting genes within the lipid pathways studied. This unified approach allows us to interpret changes in lipid levels and gene expression together, providing a mechanistic understanding of the development of alcoholic fatty liver in mice.
Decreased FA oxidation and decreased FA-CoA synthesis is observed in liver of alcohol-fed mice
The first lipid class focused upon consisted of key FA metabolites, specifically FFAs, FA-CoAs, and FAEEs. In broad terms, our data revealed striking trends for these lipids. There was a significant increase in total FFA and FAEE levels in liver of alcohol-fed mice, whereas total FA-CoA concentration was decreased. These findings can be understood in the context of our gene expression data. Alcohol increased expression of the FA transporter Cd36, which we take to reflect increased FA uptake from the circulation by liver of alcohol-fed mice, similar to the recent report by Ge et al. (24). Expression levels of Fatp1 and Fatp2 were decreased following alcohol exposure. FATP1 and FATP2 can transport FA across plasma membranes, and both are functionally coupled with FA-CoA synthesis. Thus, decreased expression of these genes likely accounts for decreased FA-CoA levels in liver of alcohol-fed mice (25, 26). Decreased expression of Fabp1 in the liver of alcohol-fed mice was also observed. Studies of Fabp1 knockout mice reveal a requirement for this gene in hepatic FA oxidation (27, 28). Decreased Fabp1 expression is therefore consistent with increased FFA levels in alcohol-fed mice, arising from decreased mitochondrial FA oxidation. Decreased expression of other genes associated with mitochondrial FA oxidation was also observed. Specifically, expression of Cpt1a and Acsl1 was significantly reduced. Interestingly, peroxisomal FA oxidation was unaffected in alcohol-fed mice, as reflected by an absence of change in the expression of Acox1, which encodes the enzyme catalyzing the rate-determining step in this pathway (20, 21). No alcohol-induced changes in genes associated with de novo lipogenesis (Srebp1c, Acc1, Fasn) were observed. Taken together, our data establish that the increased level of FFA in the liver of alcohol-fed mice resulted from both increased FFA uptake from the circulation and decreased mitochondrial FA oxidation. On the basis of the observed decreases in genes associated with FA-CoA synthesis, we also conclude that the decreased FA-CoA levels are the result of decreased FA-CoA synthesis. Both of these differences are likely associated with decreased PPARα signaling, the expression of which was decreased in alcohol-fed mice, as several genes in both pathways are controlled by this transcription factor (29).
In keeping with alcohol's established effect on liver, our alcohol-fed mice showed enlarged liver and modest, but statistically significant, increased hepatic triglyceride content. We consider our mice to be in the early stages of developing alcoholic fatty liver disease. We would expect prolonged alcohol feeding to be associated with more pronounced steatosis that is consistent with the literature (30, 31). Excessive hepatic triglyceride accumulation is associated with signaling through the transcription factor PPARγ (32). Expression of Pparg was increased in liver of alcohol-fed mice, consistent with the increased triglyceride content observed. With respect to the expression of genes directly involved in hepatic triglyceride synthesis, we only observed an increase in Dgat2. Moreover, expression levels of genes involved in triglyceride hydrolysis were unchanged (Tgh, Atgl, Hsl). Investigations involving longer periods of alcohol feeding will be needed to determine the significance of decreased FA oxidation and FA-CoA synthesis as early contributing factors to ALD.
A question arises regarding the decreased FA-CoA levels observed in alcohol-fed mice together with elevated triglyceride levels. It seems counterintuitive that increased triglyceride accumulation would occur, given that FA-CoAs are an obligatory intermediate in triglyceride synthesis. However, liver-specific knockout of Acsl1 decreases hepatic FA-CoA levels, without protecting mice from diet-induced steatosis (33). This result suggested that there are metabolically distinct FA-CoA pools within hepatocytes partitioned for different pathways, including one for triglyceride synthesis and one for FA oxidation. In this respect, Acsl5 is thought to catalyze the synthesis of FA-CoA in the triglyceride synthesis pathway (34, 35). In alcohol-fed mice, Acsl1 expression was significantly reduced compared with controls, whereas Acsl5 was unchanged. Although this remains to be definitively demonstrated, our data suggest that the FA-CoA pool needed for triglyceride synthesis is preserved in alcohol-fed mice, whereas the pool associated with FA oxidation is depleted.
FAEEs are nonoxidative metabolites of alcohol that are believed to be hepatotoxic (36). Their presence in multiple tissues has led to considerable research aimed at establishing FAEEs as biomarkers for chronic alcohol abuse (37). FAEEs have also been associated with disease development in the alcoholic liver, heart, and pancreas (36, 38, 39). The molecular identities of enzymes that synthesize and degrade FAEEs are currently not established (40, 41). It is thought that multiple hepatic microsomal carboxyesterases (CES) are able to catalyze the synthesis of FAEE; but because of this uncertainty, we did not exhaustively study expression of CES that might be involved in FAEE synthesis (41). Our results, however, identify ethyl oleate (C18:1) as the most abundant FAEE in the plasma and liver of alcohol-fed mice. This finding is consistent with work done in other animal models and humans (7, 42, 43). Accumulation of FAEE in the liver of alcohol-fed mice may contribute to the development of ALD, and although methods have been developed to detect these toxic lipids, their metabolism and the basis for their cytotoxic effects remain to be elucidated.
The high sensitivity of LC/MS/MS allowed quantification of individual lipid species levels in plasma and liver, allowing analysis of FA metabolites with different carbon-chain lengths. In this regard, we observed significant increases in PUFAs, EPA (C20:5), and docasahexaenoic acid (DHA; C22:6) in the liver of alcohol-fed mice. These two n-3 PUFAs, which are abundant in fish oils, are proposed to have beneficial effects, particularly in the cardiovascular system (44). However, extensive literature also links consumption of PUFAs with more severe ALD in humans and rodents (45–47). Our observation of elevated endogenous levels of these two PUFAs in the liver of alcohol-fed mice implies that elevations in EPA and DHA may contribute to the development of ALD.
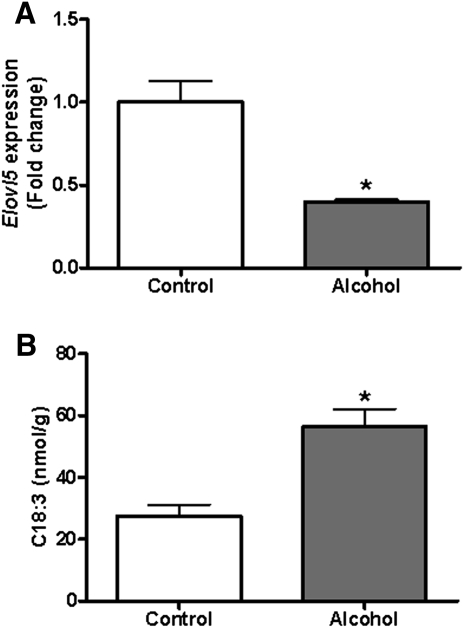
The combined analysis of hepatic gene expression patterns and lipid levels provides insights into hepatic FFAs and their effects on gene expression. For instance, we observed a significant decrease in Elovl5 expression in the liver of alcohol-fed mice. In vivo, ELVOL5 catalyzes elongation of specific long-chain FAs, including C18:3 to C20:3. It has previously been shown that Elovl5 knockout mice accumulate C18:3 FFA in their liver because they cannot elongate it to C20:3 (48). Similarly, we found a decrease in hepatic Elvol5 gene expression coupled with increased C18:3 FFA following alcohol feeding (Fig. 6). In the context of ALD, this observation is significant because Elovl5 knockout mice also develop fatty liver, although further study will be needed to establish whether this mechanism contributes to ALD (43). Also of interest was a significant decrease in hepatic Scd1 expression in alcohol-fed mice. SCD1 introduces a double bond into saturated fatty acyl chains ranging in length from C12:0 to C19:0, yielding a monounsaturated product (49). Given that Scd1 is a known target of PPARα, the observed decrease in Ppara expression correlates with decreased Scd1 expression (45). Another likely contributing factor to decreased Scd1 expression is the fat composition of the Lieber-DeCarli diet. The FA profile of the Lieber-DeCarli liquid diet is rich in unsaturated FAs. Alcohol-fed mice take up more FFA into their liver (24), resulting in higher absolute and relative levels of hepatic unsaturated FA (Fig. 2). It is established that rat primary hepatocytes downregulate Scd1 expression when cultured with unsaturated FAs of varying chain lengths and that rats fed high amounts of unsaturated FAs also show decreased Scd1 expression (50, 51). Interestingly, PUFAs are the most potent inhibitors of Scd1 expression, and it is this subtype of FFA that is most increased in the liver of alcohol-fed mice. Thus, increased levels of unsaturated FFA in the liver of alcohol-fed mice, coupled with decreased Ppara expression, drives down expression of Scd1.
Fig. 6.
Decreased Elovl5 expression is associated with increased hepatic C18:3 FFA. (A) Elovl5 gene expression is decreased to ∼40% of control in the liver of alcohol-fed mice. (B) The Elovl5 substrate C18:3 is significantly increased in liver of alcohol-fed mice. Data are means ± SEM. *P < 0.05 alcohol-fed versus control mice.
Hepatic sphingolipid levels are increased following alcohol feeding
Hepatic levels of sphingosine, sphinganine, and total ceramides were significantly increased in alcohol-fed mice, whereas levels of SO-1P were decreased. The observed elevations in hepatic ceramides are in agreement with recently published data (6, 9). Given that sphingosine and sphinganine are both substrates for ceramide synthesis, their increased abundance in the liver correlates well with the increased ceramide levels. Unfortunately, our gene expression analysis did not provide more insight into our lipidomic data. The only significant change observed was a decrease in Kdsr expression in the liver of alcohol-fed mice. The Kdsr gene product, 3-ketodihydrosphingosine reductase, is the enzyme that synthesizes sphinganine (22). Thus, we would have expected to observe a decrease in sphinganine levels, not an increase. It is possible that elevated levels of hepatic sphinganine led to a downregulation of Kdsr via a negative feedback loop. While our gene expression data could not definitively explain the increased hepatic levels of sphingosine and sphinganine, the concomitant decrease in plasma levels of these lipids suggests that their hepatic concentration is modulated by flux between the liver and circulation, a process that may be dysregulated by alcohol exposure.
The pathophysiologic significance of increased sphingolipid levels in the liver of alcohol-fed mice requires further study. We have confirmed that hepatic ceramide levels are increased following alcohol exposure, and we are the first to report that levels of the ceramide precursors, sphingosine and sphinganine, are also elevated. Reported increases in ceramide levels from different models of nonalcoholic fatty liver have lead to suggestions that ceramides are important in fatty liver disease development, although the mechanistic link between hepatic steatosis and ceramide is unclear (52–55). Increased hepatic ceramide levels have been reported following alcohol exposure in vivo and in H4IIEC3 cells. These increases were associated with increased activity of acidic sphingomyelinase (Smpd1), which uses sphingomyelin as a substrate for ceramide synthesis. There was, however, no corresponding decrease in sphingomyelin reported (6, 9, 56). In our studies, we found no change in the mRNA level of Smpd1 in alcohol-fed mice.
Hepatic anandamide is increased in alcohol-fed mice
We observed a significant increase in AEA levels in the liver of alcohol-fed mice, whereas levels of 2-AG were unchanged. Plasma levels of both lipids were also unaffected by alcohol feeding. The increased hepatic AEA levels were associated with decreased expression of Faah, which encodes an enzyme responsible for AEA breakdown (23). This observation leads us to conclude that the increased hepatic level of AEA was the result of decreased degradation. A substantial body of work links endocannabinoid signaling and hepatic steatosis (10, 57). A previous study exploring endocannabinoid signaling in alcohol-fed mice reported an increase in hepatic 2-AG levels, but not in AEA levels (11). We found no change in the level of 2-AG in the liver of alcohol-fed mice. Nevertheless, a report by Jeong et al. suggests that an eventual disruption of this tightly regulated 2-AG concentration may be significant in ALD (11). Differences in the diets employed or the duration of alcohol exposure might account for the discrepancy between studies. These authors also reported that the increase in 2-AG occurred specifically in hepatic stellate cells; our study was performed in whole-liver homogenates and may therefore have lacked the resolution to detect this change. Further research will be required to resolve this issue and to determine the relative importance of AEA and 2-AG in ALD.
SUMMARY
Our data supports the concept that decreased mitochondrial FA oxidation is one of the contributing factors in alcoholic fatty liver disease. Alcohol feeding led to elevated hepatic FFA levels, coupled with decreased expression of genes associated with FA oxidation. A central mechanism underlying this observation is decreased PPARα signaling (58, 59). As far as we are aware, we are the first to report broad decreases in FA-CoA levels in the liver of alcohol-fed mice that are associated with decreased expression of FA-CoA-synthesizing genes. Consistent with previous reports, we observed increased ceramide levels in alcohol-fed mice (6, 9). Our data established that this is associated with increased levels of the precursor metabolites sphingosine and sphinganine. Dysregulated ceramide homeostasis is proposed to be an important factor in nonalcoholic fatty liver disease. Our research also indicates an important role in ALD; this will be an important question for future research to resolve. Lastly, we demonstrated an increased concentration of the endocannabinoid AEA in the liver of alcohol-fed mice. Our data conclusively establish that the metabolic pathways for each of the three lipid classes studied are markedly dysregulated following alcohol consumption and that this very likely contributes to the development of ALD.
Supplementary Material
Footnotes
Abbreviations:
- AEA
- anandamide
- 2-AG
- 2-arachidonoylglycerol
- ALD
- alcoholic liver disease
- ALT
- alanine aminotransferase
- BAC
- blood alcohol content
- DHA
- docasahexaenoic acid
- EPA
- eicosapentaenoic acid
- FA-CoA
- fatty acyl-CoA
- FAEE
- fatty acid ethyl ester
- MRM
- multiple reaction monitoring
- PPAR
- peroxisome proliferator-activated receptor
- SA-1P
- sphinganine-1-phosphate
- SIR
- selected ion recording
- SO-1P
- sphingosine-1-phosphate
- TEA
- triethylamine
- UPLC
- ultra performance liquid chromatography
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one figure and six tables.
This work was supported by National Institutes of Health Grants RC2 AA-019413, R01 DK-068437, R01 DK-079221, and UL1 RR-024156. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health. This work was also supported by a Clinical and Translational Science Award from the Irving Institute, Columbia University.
REFERENCES
- 1.Mann R. E., Smart R. G., Govoni R. 2003. The epidemiology of alcoholic liver disease. Alcohol Res. Health. 27: 209–219 [PMC free article] [PubMed] [Google Scholar]
- 2.Lakshman M. R. 2004. Some novel insights into the pathogenesis of alcoholic steatosis. Alcohol. 34: 45–48 [DOI] [PubMed] [Google Scholar]
- 3.Sozio M. S., Liangpunsakul S., Crabb D. 2010. The role of lipid metabolism in the pathogenesis of alcoholic and nonalcoholic hepatic steatosis. Semin. Liver Dis. 30: 378–390 [DOI] [PubMed] [Google Scholar]
- 4.Oresic M. 2009. Metabolomics, a novel tool for studies of nutrition, metabolism and lipid dysfunction. Nutr. Metab. Cardiovasc. Dis. 19: 816–824 [DOI] [PubMed] [Google Scholar]
- 5.Fernando H., Kondraganti S., Bhopale K. K., Volk D. E., Neerathilingam M., Kaphalia B. S., Luxon B. A., Boor P. J., Shakeel Ansari G. A. 2010. 1H and 31P NMR lipidome of ethanol-induced fatty liver. Alcohol. Clin. Exp. Res. 34: 1937–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao Z., Yu M., Crabb D., Xu Y., Liangpunsakul S. 2011. Ethanol-induced alterations in fatty acid-related lipids in serum and tissues in mice. Alcohol. Clin. Exp. Res. 35: 229–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loftus N., Barnes A., Ashton S., Michopoulos F., Theodoridis G., Wilson I., Ji C., Kaplowitz N. 2011. Metabolomic investigation of liver profiles of nonpolar metabolites obtained from alcohol-dosed rats and mice using high mass accuracy MSn analysis. J. Proteome Res. 10: 705–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng W., Kollmeyer J., Symolon H., Momin A., Munter E., Wang E., Kelly S., Allegood J. C., Liu Y., Peng Q., et al. 2006. Ceramides and other bioactive sphingolipid backbones in health and disease: lipidomic analysis, metabolism and roles in membrane structure, dynamics, signaling, and autophagy. Biochim. Biophys. Acta. 1758: 1864–1884 [DOI] [PubMed] [Google Scholar]
- 9.Liangpunsakul S., Sozio M. S., Shin E., Zhao Z., Xu Y., Ross R. A., Zeng Y., Crabb D. W. 2010. Inhibitory effect of ethanol on AMPK phosphorylation is mediated in part through elevated ceramide levels. Am. J. Physiol. Gastrointest. Liver Physiol. 298: G1004–G1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mallat A., Lotersztajn S. 2010. Endocannabinoids and their role in fatty liver disease. Dig. Dis. 28: 261–266 [DOI] [PubMed] [Google Scholar]
- 11.Jeong W. I., Osei-Hyiaman D., Park O., Liu J., Bátkai S., Mukhopadhyay P. 2008. Paracrine activation of hepatic CB1 receptors by stellate-cell derived endocannabinoids mediates alcoholic fatty liver. Cell Metab. 7: 227–235 [DOI] [PubMed] [Google Scholar]
- 12.DeCarli L. M., Lieber C. S. 1967. Fatty liver in the rat after prolonged intake of ethanol with a nutritionally adequate new liquid diet. J. Nutr. 91: 331–336 [DOI] [PubMed] [Google Scholar]
- 13.Folch J., Lees M., Sloane-Stanley G. H. 1957. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226: 497–509 [PubMed] [Google Scholar]
- 14.O'Byrne S. M., Wongsiriroj N., Libien J., Vogel S., Goldberg I. J., Baehr W., Palczewski K., Blaner W. S. 2005. Retinoid absorption and storage is impaired in mice lacking lecithin:retinol acyltransferase (LRAT). J. Biol. Chem. 280: 35647–35657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfaffl M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mankes R. F., Battles A. H., LeFevre R., van der Hoeven T., Glick S. D. 1992. Preferential alcoholic embryopathy: effects of liquid diets. Lab. Anim. Sci. 42: 561–566 [PubMed] [Google Scholar]
- 17.Partridge C. R., Sampson H. W., Forough R. 1999. Long-term alcohol consumption increases matrix-metalloproteinase-2 activity in rat aorta. Life Sci. 65: 1395–1402 [DOI] [PubMed] [Google Scholar]
- 18.Kane M. A., Folias A. E., Wang C., Napoli J. L. 2010. Ethanol elevates physiological all-trans-retinoic acid levels in select loci through altering retinoid metabolism in multiple loci: a potential mechanism of ethanol toxicity. FASEB J. 24: 823–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leo M. A., Lieber C. S. 1982. Hepatic vitamin A depletion in alcoholic liver injury. N. Engl. J. Med. 307: 597–601 [DOI] [PubMed] [Google Scholar]
- 20.Reddy J. K., Goel S. K., Nemali M. R., Carrino J. J., Laffler T. G., Reddy M. K. 1986. Transcription regulation of peroxisomal fatty acyl-CoA oxidase and enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase in rat liver by peroxisome proliferators. Proc. Natl. Acad. Sci. USA. 83: 1747–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varanasi U., Chu R., Huang Q., Castellon R., Yeldandi A. V., Reddy J. K. 1996. Identification of a peroxisome proliferator-responsive element upstream of the human peroxisomal fatty acyl coenzyme A oxidase gene. J. Biol. Chem. 271: 2147–2155 [DOI] [PubMed] [Google Scholar]
- 22.Bartke N., Hannun Y. A. 2009. Bioactive sphingolipids: metabolism and function. J. Lipid Res. 50: S91–S96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Marzo V. 2008. Endocannabinoids: synthesis and degradation. Rev. Physiol. Biochem. Pharmacol. 160: 1–24 [DOI] [PubMed] [Google Scholar]
- 24.Ge F., Zhou S., Hu C., Lobdell H., 4th, Berk P. D. 2010. Insulin- and leptin-regulated fatty acid uptake plays a key causal role in hepatic steatosis in mice with intact leptin signaling but not in ob/ob or db/db mice. Am. J. Physiol. Gastrointest. Liver Physiol. 299: G855–G866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coe N. R., Smith A. J., Frohnert B. I., Watkins P. A., Bernlohr D. A. 1999. The fatty acid transport protein (FATP1) is a very long chain acyl-CoA synthetase. J. Biol. Chem. 274: 36300–36304 [DOI] [PubMed] [Google Scholar]
- 26.Falcon A., Doege H., Fluitt A., Tsang B., Watson N., Kay M. A., Stahl A. 2010. FATP2 is a hepatic fatty acid transporter and peroxisomal very long-chain acyl-CoA synthetase. Am. J. Physiol. Endocrinol. Metab. 299: E384–E393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erol E., Kumar L. S., Cline G. W., Shulman G. I., Kelly D. P., Binas B. 2004. Liver fatty acid binding protein is required for high rates of hepatic fatty acid oxidation but not for the action of PPARalpha in fasting mice. FASEB J. 18: 347–349 [DOI] [PubMed] [Google Scholar]
- 28.Atshaves B. P., Martin G. G., Hostetler H. A., McIntosh A. L., Kier A. B., Schroeder F. 2010. Liver fatty acid-binding protein and obesity. J. Nutr. Biochem. 21: 1015–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gavrilova O., Haluzik M., Matsusue K., Cutson J. J., Johnson L., Dietz K. R., Nicol C. J., Vinson C., Gonzalez F. J., Reitman M. L. 2003. Liver peroxisome proliferator-activated receptor gamma contributes to hepatic steatosis, triglyceride clearance, and regulation of body fat mass. J. Biol. Chem. 278: 34268–34276 [DOI] [PubMed] [Google Scholar]
- 30.Pritchard M. T., McMullen M. R., Stavitsky A. B., Cohen J. I., Lin F., Medof M. E., Nagy L. E. 2007. Differential contributions of C3, C5, and decay-accelerating factor to ethanol-induced fatty liver in mice. Gastroenterology. 132: 1117–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng Z., Borea P. A., Varani K., Wilder T., Yee H., Chiriboga L., Blackburn M. R., Azzena G., Resta G., Cronstein B. N. 2009. Adenosine signaling contributes to ethanol-induced fatty liver in mice. J. Clin. Invest. 119: 582–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berger J., Moller D. E. 2002. The mechanisms of action of PPARs. Annu. Rev. Med. 53: 409–435 [DOI] [PubMed] [Google Scholar]
- 33.Li L. O., Ellis J. M., Paich H. A., Wang S., Gong N., Altshuller G., Thresher R. J. 2009. Liver-specific loss of long chain acyl-CoA synthetase-1 decreases triacylglycerol synthesis and beta-oxidation and alters phospholipid fatty acid composition. J. Biol. Chem. 284: 27816–27826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mashek D. G., McKenzie M. A., Van Horn C. G., Coleman R. A. 2006. Rat long chain acyl-CoA synthetase 5 increases fatty acid uptake and partitioning to cellular triacylglycerol in McArdle-RH7777 cells. J. Biol. Chem. 281: 945–950 [DOI] [PubMed] [Google Scholar]
- 35.Bu S. Y., Mashek D. G. 2010. Hepatic long-chain acyl-CoA synthetase 5 mediates fatty acid channeling between anabolic and catabolic pathways. J. Lipid Res. 51: 3270–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu H., Cai P., Clemens D. L., Jerrells T. R., Ansari G. A., Kaphalia B. S. 2006. Metabolic basis of ethanol-induced cytotoxicity in recombinant HepG2 cells: role of nonoxidative metabolism. Toxicol. Appl. Pharmacol. 216: 238–247 [DOI] [PubMed] [Google Scholar]
- 37.Kaphalia B. S., Cai P., Khan M. F., Okorodudu A. O., Ansari G. A. 2004. Fatty acid ethyl esters: markers of alcohol abuse and alcoholism. Alcohol. 34: 151–158 [DOI] [PubMed] [Google Scholar]
- 38.Beckemeier M. E., Bora P. S. 1998. Fatty acid ethyl esters: potentially toxic products of myocardial ethanol metabolism. J. Mol. Cell. Cardiol. 30: 2487–2494 [DOI] [PubMed] [Google Scholar]
- 39.Wu H., Bhopale K. K., Ansari G. A., Kaphalia B. S. 2008. Ethanol-induced cytotoxicity in rat pancreatic acinar AR42J cells: role of fatty acid ethyl esters. Alcohol Alcohol. 43: 1–8 [DOI] [PubMed] [Google Scholar]
- 40.Diczfalusy M. A., Björkhem I., Einarsson C., Hillebrant C. G., Alexson S. E. 2001. Characterization of enzymes involved in formation of ethyl esters of long-chain fatty acids in humans. J. Lipid Res. 42: 1025–1032 [PubMed] [Google Scholar]
- 41.Kaphalia B. S., Ansari G. A. 2001. Purification and characterization of rat hepatic microsomal low molecular weight fatty acid ethyl ester synthase and its relationship to carboxylesterases. J. Biochem. Mol. Toxicol. 15: 165–171 [DOI] [PubMed] [Google Scholar]
- 42.Dan L., Laposata M. 1997. Ethyl palmitate and ethyl oleate are the predominant fatty acid ethyl esters in the blood after ethanol ingestion and their synthesis is differentially influenced by the extracellular concentrations of their corresponding fatty acids. Alcohol. Clin. Exp. Res. 21: 286–292 [PubMed] [Google Scholar]
- 43.Laposata M., Hasaba A., Best C. A., Yoerger D. M., McQuillan B. M., Salem R. O., Refaai M. A., Soderberg B. L. 2002. Fatty acid ethyl esters: recent observations. Prostaglandins Leukot. Essent. Fatty Acids. 67: 193–196 [DOI] [PubMed] [Google Scholar]
- 44.Russo G. L. 2009. Dietary n-6 and n-3 polyunsaturated fatty acids: from biochemistry to clinical implications in cardiovascular prevention. Biochem. Pharmacol. 77: 937–946 [DOI] [PubMed] [Google Scholar]
- 45.Tsukamoto H., Cheng S., Blaner W. S. 1996. Effects of dietary polyunsaturated fat on ethanol-induced Ito cell activation. Am. J. Physiol. 270: G581–G586 [DOI] [PubMed] [Google Scholar]
- 46.Nanji A. A. 2004. Role of different dietary fatty acids in the pathogenesis of experimental alcoholic liver disease. Alcohol. 34: 21–25 [DOI] [PubMed] [Google Scholar]
- 47.Bridges F. S. 2009. Relationship between dietary beef, fat, and pork and alcoholic cirrhosis. Int. J. Environ. Res. Public Health. 6: 2417–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moon Y. A., Hammer R. E., Horton J. D. 2009. Deletion of ELOVL5 leads to fatty liver through activation of SREBP-1c in mice. J. Lipid Res. 50: 412–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Enoch H. G., Catala A., Strittmatter P. 1976. Mechanism of rat liver microsomal stearyl-CoA desaturase. Studies of the substrate specificity, enzyme-substrate interactions, and the function of lipid. J. Biol. Chem. 251: 5095–5103 [PubMed] [Google Scholar]
- 50.Miller C. W., Ntambi J. M. 1996. Peroxisome proliferators induce mouse liver stearoyl-CoA desaturase 1 gene expression. Proc. Natl. Acad. Sci. USA. 93: 9443–9448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Landschulz K. T., Jump D. B., MacDougald O. A., Lane M. D. 1994. Transcriptional control of the stearoyl-CoA desaturase-1 gene by polyunsaturated fatty acids. Biochem. Biophys. Res. Commun. 200: 763–768 [DOI] [PubMed] [Google Scholar]
- 52.Alkhouri N., Dixon L. J., Feldstein A. E. 2009. Lipotoxicity in nonalcoholic fatty liver disease: not all lipids are created equal. Expert Rev. Gastroenterol. Hepatol. 3: 445–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang G., Badeanlou L., Bielawski J., Roberts A. J., Hannun Y. A., Samad F. 2009. Central role of ceramide biosynthesis in body weight regulation, energy metabolism, and the metabolic syndrome. Am. J. Physiol. Endocrinol. Metab. 297: E211–E224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chocian G., Chabowski A., Zendzian-Piotrowska M., Harasim E., Łukaszuk B., Górski J. 2010. High fat diet induces ceramide and sphingomyelin formation in rat's liver nuclei. Mol. Cell. Biochem. 340: 125–131 [DOI] [PubMed] [Google Scholar]
- 55.Summers S. A. 2010. Sphingolipids and insulin resistance: the five Ws. Curr. Opin. Lipidol. 21: 128–135 [DOI] [PubMed] [Google Scholar]
- 56.Liu J. J., Wang J. Y., Hertervig E., Cheng Y., Nilsson A., Duan R. D. 2000. Activation of neutral sphingomyelinase participates in ethanol-induced apoptosis in Hep G2 cells. Alcohol Alcohol. 35: 569–573 [DOI] [PubMed] [Google Scholar]
- 57.Tam J., Liu J., Mukhopadhyay B., Cinar R., Godlewski G., Kunos G. 2011. Endocannabinoids in liver disease. Hepatology. 53: 346–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moriya T., Naito H., Ito Y., Nakajima T. 2009. “Hypothesis of seven balances”: molecular mechanisms behind alcoholic liver diseases and association with PPARalpha. J. Occup. Health. 51: 391–403 [DOI] [PubMed] [Google Scholar]
- 59.Gyamfi M. A., Wan Y. J. 2010. Pathogenesis of alcoholic liver disease: the role of nuclear receptors. Exp. Biol. Med. (Maywood). 235: 547–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.