Abstract
An automated fluorescence microscopy assay using a nontoxic cholesterol binding protein, θ toxin domain 4, (D4), was developed in order to identify chemical compounds modifying intracellular cholesterol metabolism and distribution. Using this method, we screened a library of 1,056 compounds and identified 35 compounds that decreased D4 binding to the cell surface. Among them, 8 compounds were already reported to alter the biosynthesis or the intracellular distribution of cholesterol. The remaining 27 hit compounds were further analyzed biochemically and histochemically. Cell staining with another fluorescent cholesterol probe, filipin, revealed that 17 compounds accumulated cholesterol in the late endosomes. Five compounds decreased cholesterol biosynthesis, and two compounds inhibited the binding of D4 to the membrane. This visual screening method, based on the cholesterol-specific probe D4 in combination with biochemical analyses, is a cell-based, sensitive technique for identifying new chemical compounds and modifying cholesterol distribution and metabolism. Furthermore, it is suitable for high-throughput analysis for drug discovery.
Keywords: cholesterol binding toxin, θ toxin domain 4, filipin, cholesterol synthesis inhibitor, hydrophobic amines, late endosome, microscope-based screening, cell-based screening
Cholesterol is an essential constituent in the membranes of mammalian cells and has many important functions. It regulates membrane fluidity, increases membrane thickness, establishes the permeability barrier of the membranes (1), and modulates the activity of various membrane proteins (2). It is considered to promote the formation of cholesterol-sphingolipid-enriched microdomains, which are involved in signal transduction, trafficking, and virus infection (3). Cells tightly regulate cholesterol levels by a feedback pathway that controls the synthesis, esterification, and uptake of cholesterol (4). Its de novo synthesis occurs mainly in the endoplasmic reticulum (ER), and cells can also acquire exogenous cholesterol via LDL uptake. LDL particles carrying cholesteryl esters are internalized through the LDL receptor and are transported to the late endosomes, where cholesteryl esters are hydrolyzed to free cholesterol. Cholesterol is not uniformly distributed in cells, and its content varies among different organelles. The cholesterol level is low in the ER, but it increases through the Golgi apparatus, with the highest levels in the plasma membrane. Thus, cells properly maintain cellular cholesterol levels as well as intracellular cholesterol distribution (5–7).
The failure of cholesterol homeostasis is the cause of numerous diseases, including atherosclerosis and heart disease (8, 9). The compounds that affect cholesterol metabolism and transport, such as statins, have been employed as therapeutic agents for these disorders. Statins selectively inhibit HMG-CoA reductase, which is the primary regulatory enzyme in cholesterol biosynthesis (10, 11). These compounds are also used as tools in the field of cholesterol research.
Biochemical screening has been conducted to search for compounds that affect cholesterol metabolism and transport. Although quite useful, this method is time-consuming and is difficult to apply to high-throughput screening. Recently, Pipalia et al. (12) reported microscopy screening of a chemical library using a cholesterol binding fluorescent antibiotic, filipin. Filipin is a polyene antibiotic that binds to cholesterol and forms pores. Filipin has been employed in detecting cellular cholesterol distribution (13). The microscopy screening using filipin identified several compounds that decrease cholesterol accumulation in the late endosomes of Niemann-Pick C cells (12, 14). Because filipin penetrates the membrane, this antibiotic cannot be used to selectively label the plasma membrane.
In this study, we developed a fluorescence microscopy assay for detecting changes in the amount of cholesterol in the plasma membrane using a θ toxin derivative. θ Toxin is a cholesterol binding, pore-forming cytolysin produced by Clostridium perfringens (15, 16). It selectively binds to the cholesterol-enriched membrane, where the amount of cholesterol is more than 30 mol% (16, 17). This property enabled detection of even a slight decrease of cholesterol in the plasma membrane. θ Toxin domain 4 (D4) is the C-terminal fragment of the toxin. This domain is the smallest functional unit of the toxin, displaying the same cholesterol binding activity as the full-size protein (18). D4 binds to cholesterol with a high affinity (Kd ∼ 10−8), but does not exert cytotoxicity (18). Thus, in contrast to filipin, D4 can be used for the selective labeling of plasma membrane cholesterol in living cells.
Here, we screened 1,056 chemical compounds by cell surface labeling using fluorescent D4, and obtained 35 compounds that decrease D4 labeling in the plasma membrane. Among them, 8 compounds were already reported to alter the biosynthesis or the intracellular distribution of cholesterol. We examined the effects of the other 27 hit compounds on intracellular cholesterol distribution, using another cholesterol binding probe, filipin (13). Seventeen compounds accumulated cholesterol intracellularly. Two compounds inhibited the binding of D4 to the membrane. We further analyzed the effects of the remaining eight compounds on cholesterol synthesis and transport. Five compounds inhibited cholesterol biosynthesis, but none of them affected cholesterol transport from the ER to the plasma membrane. Thus, this new visual screening using cholesterol binding protein, in combination with biochemical analyses, successfully identified compounds based on their effects on cholesterol metabolism and distribution.
MATERIALS AND METHODS
Materials
The chemical library for screening was supplied by RIKEN Natural Products Depository (NPDepo). Egg phosphatidylcholine (PC) and brain SM were purchased from Avanti Polar Lipids (Alabaster, AL). [1-14C]acetic acid was purchased from American Radiolabeled Chemicals, Inc. (St. Louis, MO). Lipoprotein-deficient serum (LPDS), lovastatin, mevalonate, cholesterol, and stigmasterol were from Sigma (St. Louis, MO). Filipin was purchased from Polysciences, Inc. (Warrington, PA). Hoechst 33342 was from Nacalai Tesque, Inc. (Kyoto, Japan). 3-β-[2-(diethylamino)ethoxy]androst-5-en-17-one (U18666A) was purchased from BIOMOL (Plymouth Meeting, PA). Anti GM130 was from BD Bioscience (Franklin Lakes, NJ). Anti CD63 was from Cosmo Bio Co., Ltd. (Tokyo, Japan). His-enhanced green fluorescent protein (EGFP)-tagged θ toxin domain 4 (EGFP-D4) was expressed in Escherichia coli strain BL21 (DE3) and purified using HisTrap FF crude columns (GE Healthcare; England) (18).
Cells and cell culture
HeLa cells were cultured in DMEM supplemented with 10% FBS. Chinese hamster ovary (CHO) cells were cultured in Ham's F-12 supplemented with 10% FBS.
Preparation of multilamellar vesicles and small unilamellar vesicles
Vesicles were prepared by combining egg PC and cholesterol, or brain SM and cholesterol from chloroform stocks. The chloroform was evaporated under a stream of nitrogen gas, followed by drying under vacuum for 1 h. Then, PBS (pH 7.5, 55°C) was added to the dry lipid mixtures. The lipid mixture was vortexed to produce multilamellar vesicles (MLVs). Small unilamellar vesicles (SUVs) were prepared by sonication of MLVs using ASTRASON ULTRASONIC PROCESSOR XL (Misonix, Inc.; Farmingdale, NY).
EGFP-D4 binding to liposomes
EGFP-D4 binding to liposomes was analyzed as described (18) with the following modification. EGFP-D4 was incubated with MLVs in PBS (pH 7.5) for 30 min at room temperature. Then, the mixtures were centrifuged at 21,600 g for 10 min at 25°C. The pellets were subjected to SDS-PAGE followed by Coomassie Brilliant Blue or SYPRO Ruby (Invitrogen; Madison, WI) staining. For quantification of the protein, intensity of EGFP-D4 stained with SYPRO Ruby was measured using Typhoon 9410 (GE Healthcare; Piscataway, NJ) and analyzed by ImageQuant (Molecular Dynamics).
Filipin binding to liposomes
Filipin binding to liposomes was analyzed as described (19) with modification. Ten micromoles filipin was incubated with 100 μM SUVs in PBS (pH 7.5) for 30 min at room temperature. The ultraviolet (UV) absorption spectra were determined at 25°C using a V-650 spectrophotometer equipped with a thermostatic cell holder (Jasco; Tokyo, Japan). Absorbance at 320 nm and 356 nm was detected using Spectra Max M2 (Molecular Devices; Downingtown, PA).
D4 labeling
On day 0, cells were seeded in 96-well plates in medium with 10% FBS. On day 1, the medium was exchanged to medium with 10% LPDS containing various compounds. All plates were incubated with the compounds for 18 h at 37°C. On day 2, the cells were washed three times with HBSS, and then incubated with 5 μg/ml EGFP-D4 and Hoechst 33342 in binding buffer (0.1% BSA/HBSS) for 30 min at room temperature. The cells were washed and fixed with 3% paraformaldehyde in PBS (pH 7.5).
Fluorescence microscopy and image analysis
For screening, images were acquired using In Cell Analyzer 1000 (GE Healthcare, England) with a 20× objective. Images of cells stained with EGFP-D4 were analyzed using In Cell Analyzer image analysis software Developer. Three to five images per well were acquired. The sum of the fluorescence intensity in the microscopic field was divided by the cell number.
Filipin staining
Cells were washed and fixed with 3% paraformaldehyde in PBS (pH 7.5) followed by incubation with 50 μg/ml filipin for 30 min at room temperature.
Double labeling of cells with filipin and organelle markers
Cells were washed and fixed with 3% paraformaldehyde in PBS (pH 7.5). The cells were quenched with 50 mM NH4Cl and blocked with 0.1% BSA. After 1 h treatment with the first antibodies, cells were washed and labeled with Alexa Fluor 488-labeled second antibodies for 30 min. Filipin (50 μg/ml) was added both in primary and secondary antibody solutions. Images were acquired using In Cell Analyzer 1000 with a 20× objective or using a Zeiss LSM 510 confocal microscope equipped with C-Apochromat 63XW Korr (1.2 n.a.) objective.
Assay of de novo cholesterol synthesis
On day 0, cells were seeded in 12-well plates in DMEM containing 10% FBS. On day 1, the medium was replaced with DMEM containing 10% LPDS. On day 2, the cells were labeled with 0.5 μCi/ml [14C]acetic acid for 2 h in DMEM containing 10% LPDS in the presence of various compounds. Cells were then washed three times with PBS, and lipids were extracted and separated on high-performance thin-layer chromatography (HPTLC) using hexane-diethyl ether-acetic acid (80:20:2, v/v/v) as a solvent. Incorporation of [14C]acetic acid into the band corresponding to cholesterol was quantified by a BAS 5000 Bioimaging Analyzer (Fuji Film, Japan).
Assay of cholesterol transport
Cholesterol transport was analyzed as described (20, 21) with modification. On day 0, cells were seeded in 12-well plates in DMEM containing 10% FBS. On day 1, the medium was replaced with DMEM containing 10% LPDS. On day 2, the cells were labeled with 2 μCi/ml [14C]acetic acid for 15 min at 37°C in DMEM containing 10% LPDS in the presence of various compounds. The chase was performed in the presence of compounds in addition to 8 μM lovastatin and 100 μM mevalonate for 1 h at 37°C. During the last 10 min of the chase, the cells were incubated with 10 mM methyl-β-cyclodextrin. The medium was collected, and cells were then washed with PBS and scraped with 0.1% Triton X-100. Lipids were extracted from the cell suspension by the method of Bligh and Dyer (22). From the medium, lipids were extracted by shaking twice with three volumes of hexane-methanol (2:1). The lipids were separated and analyzed as described above. The arrival of cholesterol on the plasma membrane was defined as the relative portion of [14C]cholesterol radioactivity in the medium to the combined [14C]cholesterol radioactivity in the medium and the cellular fraction.
Quantification of cholesterol content by gas chromatography
Cells were treated with 12.5 μg/ml compounds for 18 h in 10% LPDS containing medium. Lipids were extracted from the cell suspension by the method of Bligh and Dyer (22). Stigmasterol (2 μg) was added as internal standard before the extraction. The lipid extracts were separated on HPTLC using hexane-diethyl ether-acetic acid (80:20:2, v/v/v) as a solvent. The bands corresponding to cholesterol, revealed under UV after spraying a nondestructive lipophilic dye, primuline solution, were collected and then extracted with a mixture of methanol-water-hexane (2:1:2). After centrifugation, the upper hexane phase was collected, dried under nitrogen, and then analyzed with a Shimadzu GC-14AH gas chromatograph (Shimadzu, Japan). An Econo-Cap EC-5 (Alltech Associates. Inc.) capillary column (30 m × 0.32 mm, 0.25 mm) was used with helium as carrier gas, and an oven temperature program from 290°C to 320°C at 2°C/min, held isothermal at 320°C for 10 min. The amount of cholesterol was normalized by the amount of protein, which was determined using the Bradford assay.
RESULTS AND DISCUSSION
Selection of cell type and domain 4 fragment of theta toxin to detect subtle alteration of cell surface cholesterol
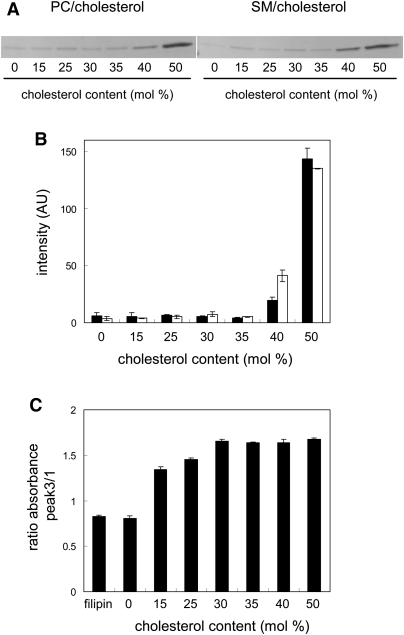
Recently, Maxfield and coworkers employed filipin for automated microscopy screening and discovered compounds that partially revert cholesterol accumulation in Niemann-Pick C cells (12, 14). Similar to filipin, θ toxin specifically binds cholesterol (16). However, the reported cholesterol binding properties of these reagents are different, in that θ toxin requires high concentration of cholesterol in membrane (16, 17). In the present study, we first compared the binding of filipin and domain 4 of θ toxin (hereafter D4) to cholesterol-containing liposomes. D4 is the C-terminal domain of θ toxin that has cholesterol binding activity, but does not exhibit cytotoxicity (18). Figure 1A shows the binding properties of D4. After incubation with egg PC or brain SM liposomes containing varying amounts of cholesterol, EGFP-D4 bound to the liposomes was recovered by centrifugation. The proteins in the pellet fractions were subjected to SDS-PAGE and stained with CBB (Fig. 1A) and SYPRO Ruby. Shown in Fig. 1B, intensity of SYPRO Ruby-stained EGFP-D4 in the bound fractions was quantitated. EGFP-D4 did not bind to PC or SM liposomes in the absence of cholesterol. The binding of EGFP-D4 to liposomes was not detected when the ratio of cholesterol to the phospholipids was 35% or less. The result indicates that D4 binding requires a high membrane concentration of cholesterol, as observed in the case of θ toxin (16, 17). We did not observe significant difference in the EGFP-D4 binding between PC/cholesterol liposomes and SM/cholesterol liposomes. These results suggest that the observed cholesterol dependence of D4 binding is not affected by other membrane lipids.
Fig.1.
Binding of EGFP-D4 and filipin to cholesterol-containing liposomes. A: EGFP-D4 was incubated with PC/cholesterol or SM/cholesterol liposomes (MLVs) for 30 min at room temperature. After centrifugation, the pellet fractions were subjected to SDS-PAGE followed by CBB staining. B: EGFP-D4 bound to PC/cholesterol (filled bar) and SM/cholesterol liposomes (open bar) was stained with SYPRO Ruby, and quantitated as described under MATERIALS AND METHODS. Data are the mean ± average deviation of two independent experiments. C: Filipin (10 μM) was incubated with 100 μM liposomes (SUVs) for 30 min at room temperature. Absorbance at 320 nm (Peak 3) and 356 nm (Peak 1) was determined as described under MATERIALS AND METHODS. Data represent the mean ± SD (n = 3).
Shown in Fig. 1C, is the analysis of the cholesterol dependence on the filipin binding to liposomes. Filipin has a characteristic UV absorption spectrum with four maxima. The ratio of absorbance of Peak 3, λ = 320 nm, to Peak 1, λ= 356 nm, was 0.83 ± 0.02 in aqueous solution (Fig. 1C). The binding of filipin to cholesterol causes a striking spectral change, and the ratio of absorbance of Peak 3 to Peak 1 becomes higher than 1 (19). When filipin was incubated with PC liposomes without cholesterol, the ratio of absorbance of Peak 3 to Peak 1 of filipin was 0.81 ± 0.03 (Fig. 1C), indicating that filipin did not bind to the liposomes. After incubation of filipin with liposomes containing cholesterol, the ratio of absorbance of Peak 3 to Peak 1 became higher than 1. Filipin bound to liposomes when cholesterol content was as low as 15%. The ratio of absorbance increased gradually when cholesterol content was increased from 15% to 30%. Then, the ratio became saturated at higher concentrations of cholesterol. These results indicate that filipin detects lower concentrations of cholesterol than EGFP-D4. However, EGFP-D4 is more sensitive than filipin to detect small alterations of cholesterol concentration (e.g., 40% to 35%) when membrane cholesterol is relatively high. Our recent study using super resolution fluorescence microscopy indicates that D4 stains cell surface lipid domains in living cells (23). These results suggest that D4 is suitable to label the cell surface and detect subtle alterations of plasma membrane cholesterol level induced by chemical compounds. Thus, we here employed D4 to perform automated fluorescence microscopy screening for compounds that decrease cholesterol in the plasma membrane.
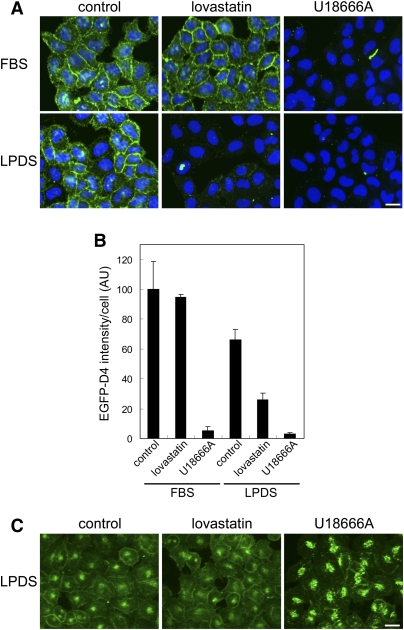
We then set up the experimental conditions for our cell-based screening using fluorescent D4. Figure 2A shows EGFP-D4 images of HeLa cells grown in DMEM containing FBS or LPDS. Living HeLa cells were first labeled with EGFP-D4, followed by fixation, and images were acquired using In Cell Analyzer 1000. In Fig. 2B, the fluorescence intensity of the microscopic field was divided by the number of nuclei using the In Cell Analyzer program, Developer. Cell surface was strongly labeled when cells were grown in the presence of FBS (control, FBS). The fluorescence intensity was not significantly altered when cells were treated with 8 μM lovastatin, an HMG-CoA reductase inhibitor, for 18 h in the presence of FBS before D4 labeling (lovastatin, FBS).
Fig. 2.
EGFP-D4 and filipin labeling of HeLa cells treated with lovastatin and U18666A. A: HeLa cells were treated with or without 8 μM lovastatin and 100 μM mevalonate, or 2 μg/ml U18666A in FBS- or LPDS-containing medium for 18 h. Cells were labeled with EGFP-D4 and Hoechst 33342 as described in MATERIALS AND METHODS. Images were acquired using In Cell Analyzer 1000 with a 20× objective. Bar = 20 μm. B: Fluorescence intensity of EGFP-D4 bound to cells was normalized by cell number as described in MATERIALS AND METHODS. Data represent the mean ± SD (n = 4). C: HeLa cells were treated with 8 μM lovastatin and 100 μM mevalonate, or 2 μg/ml U18666A in LPDS-containing medium for 18 h. The cells were labeled with filipin as described in MATERIALS AND METHODS. Images were acquired using In Cell Analyzer 1000 with a 20× objective. Bar = 20 μm.
When cells were grown in LPDS medium, the fluorescence intensity was decreased to 66.0 ± 21.6% compared with the cells in FBS medium (Fig. 2A control, LPDS and Fig. 2B). In contrast to its lack of effect in FBS medium, lovastatin in LPDS medium treatment significantly decreased the fluorescence (39.6 ± 19.4% compared with control in LPDS medium) (Fig. 2A lovastatin, LPDS and Fig. 2B). Cholesterol quantification by gas chromatography showed that its content of cells was only slightly decreased (93 ± 2.8% compared with LPDS control) under these conditions. These results indicate that EGFP-D4 detects subtle changes in cell surface cholesterol and can be used to screen compounds that inhibit de novo cholesterol synthesis. 3-β-[2-(diethylamino)ethoxy]androst-5-en-17-one (U18666A) is known to accumulate cholesterol in the late endosomes (24) and induces a Niemann-Pick type C-like phenotype (25, 26). U18666A treatment significantly reduced plasma membrane fluorescence after EGFP-D4 staining both in FBS and in LPDS medium, suggesting that the recycling through the late endosomes plays a major role in the maintenance of plasma membrane cholesterol level in HeLa cells.
Next, we examined the staining pattern of filipin under the same experimental conditions. Figure 2C shows HeLa cells grown in LPDS medium and stained with filipin in the presence and absence of inhibitors. Because filipin is toxic, cells were fixed before labeling. Filipin penetrated cells and labeled both the plasma membrane and the intracellular compartments. In contrast to EGFP-D4 labeling, lovastatin treatment did not significantly alter filipin labeling, suggesting that filipin is not a suitable probe to screen inhibitors of de novo cholesterol synthesis. U18666A enhanced the intracellular labeling of filipin, as already reported (24), and decreased fluorescence in the plasma membrane, again suggesting the importance of cholesterol traffic from the late endosomes in the maintenance of the plasma membrane cholesterol content.
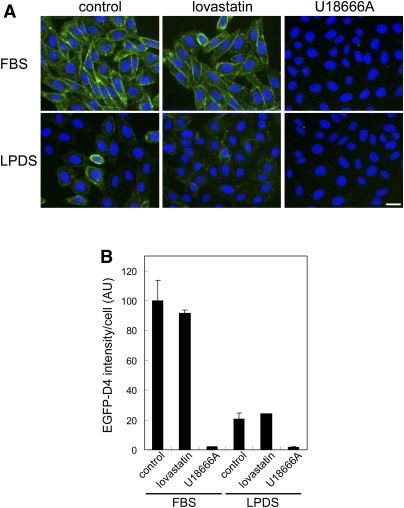
In Fig. 3, we observed EGFP-D4 labeling in Chinese hamster ovary (CHO) cells grown in the presence or the absence of lipoproteins. In contrast to HeLa cells, LPDS dramatically decreased the EGFP-D4 labeling in CHO cells (Fig. 3A control, LPDS and Fig. 3B), suggesting that the cellular cholesterol in CHO cells was largely dependent on exogenous lipoproteins. In support of this idea, lovastatin did not significantly alter EGFP-D4 labeling in cells grown either in FBS or in LPDS medium. Similar to its effect in HeLa cells, U18666A significantly decreased the cell surface fluorescence of EGFP-D4 in CHO cells cultured in both FBS and LPDS media.
Fig. 3.
EGFP-D4 labeling of CHO cells treated with lovastatin and U18666A. A: CHO cells were treated with or without 8 μM lovastatin and 100 μM mevalonate, or 2 μg/ml U18666A in FBS- or LPDS-containing medium for 18 h. Cells were labeled with EGFP-D4 and Hoechst 33342 as described in MATERIALS AND METHODS. Images were acquired using In Cell Analyzer 1000 with a 20× objective. Bar = 20 μm. B: Fluorescence intensity of EGFP-D4 bound to cells was normalized by cell number as described in MATERIALS AND METHODS. Data are the mean ± average deviation of two independent experiments.
On the basis of these preliminary assays, in the following screening, we employed HeLa cells grown in LPDS medium to screen inhibitors of de novo cholesterol synthesis as well as compounds that affect intracellular distribution of cholesterol.
Chemical library screening by cell labeling with EGFP-D4
Using EGFP-D4 labeling, we carried out screening of 1,056 compounds from RIKEN NPDepo library at a final concentration of 0.5, 5 and 50 μg/ml, searching for compounds that decrease the amount of cholesterol in the plasma membrane. Images of the cells stained with EGFP-D4 were acquired using In Cell Analyzer 1000. The EGFP-D4 fluorescence intensity of the microscopic field was divided by the number of nuclei using the In Cell Analyzer program. We selected compounds that decreased the EGFP-D4 fluorescence intensity of cells to approximately 70% of control. To find hit compounds, we also visually checked the EGFP-D4 images of cells, because the labeling pattern of the compound-treated cells was sometimes not homogeneous.
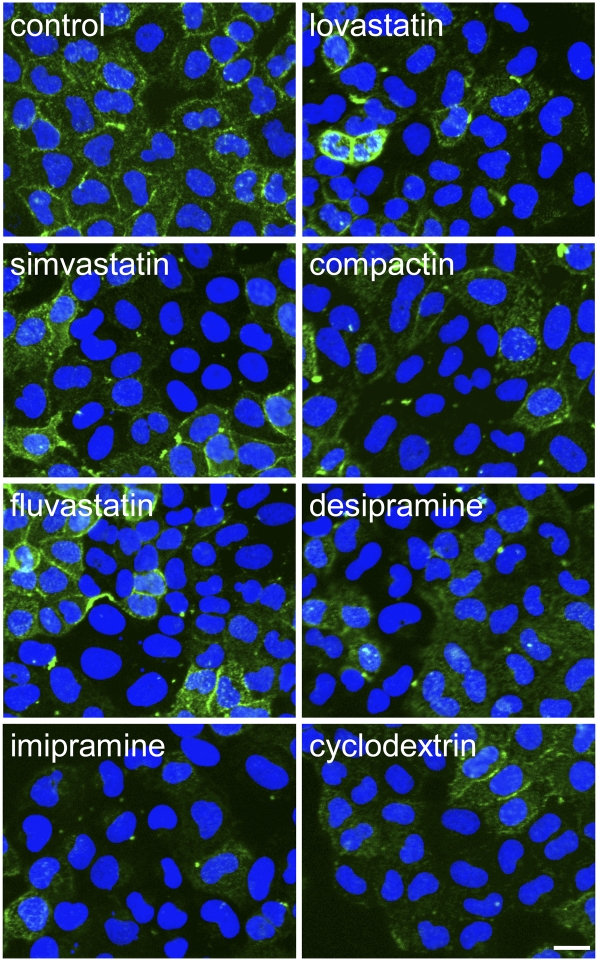
Through this screening, we identified compounds that reduced EGFP-D4 labeling in the plasma membrane. Among them, 8 compounds have already been reported to inhibit cholesterol biosynthesis or change the intracellular distribution of cholesterol. Figure 4 shows the EGFP-D4 labeling of cells treated with these compounds. Four compounds are known HMG-CoA reductase inhibitors, statins, such as lovastatin, compactin, simvastatin and fluvastatin (10). Cells treated with 0.5 μg/ml of these compounds showed lower EGFP-D4 fluorescence in the plasma membrane (Fig. 4). We observed that the cellular D4-EGFP labeling was not homogeneous.
Fig. 4.
EGFP-D4-labeling of HeLa cells treated with the compounds that are reported to inhibit cholesterol biosynthesis and intracellular cholesterol distribution. HeLa cells were treated without or with 0.5 μg/ml lovastatin, 0.5 μg/ml simvastatin, 0.5 μg/ml compactin, 0.5 μg/ml fluvastatin, 5 μg/ml desipramine, 5 μg/ml imipramine, or 50 μg/ml dimethyl-β-cyclodextrin in LPDS-containing medium for 18 h. Cells were labeled with EGFP-D4 and Hoechst 33342 as described in MATERIALS AND METHODS. Images were acquired using In Cell Analyzer 1000 with a 20× objective. Bar = 20 μm.
Among the hit compounds, three of the compounds are known to accumulate cholesterol in cells. Hydrophobic amines such as imipramine and desipramine have been reported to accumulate cholesterol in the late endosomes, like U18666A (27, 28). Treatment with 5 μg/ml imipramine and desipramine decreased the EGFP-D4 labeling of the cells (Fig. 4). Amiodarone also decreased the EGFP-D4 labeling of the cells (data not shown). Amiodarone is reported to accumulate cholesterol in the late endosomes (29). Cholesterol accumulation is also accompanied by phospholipid accumulation as the result of the inhibition of lysosomal phospholipase A2 and sphingomyelinase activities through a drug interaction with membrane lipids (29, 30).
Dimethyl-β-cyclodextrin is known to release cholesterol from the plasma membrane (31). Cells treated with this reagent at a concentration of 50 μg/ml displayed lower fluorescence after EGFP-D4 labeling (Fig. 4). The fact that known inhibitors of cholesterol metabolism were detected by this method indicates that our protocol is appropriate to identify compounds that affect cholesterol biosynthesis and its intracellular distribution.
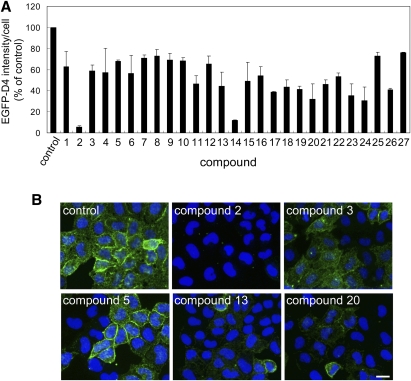
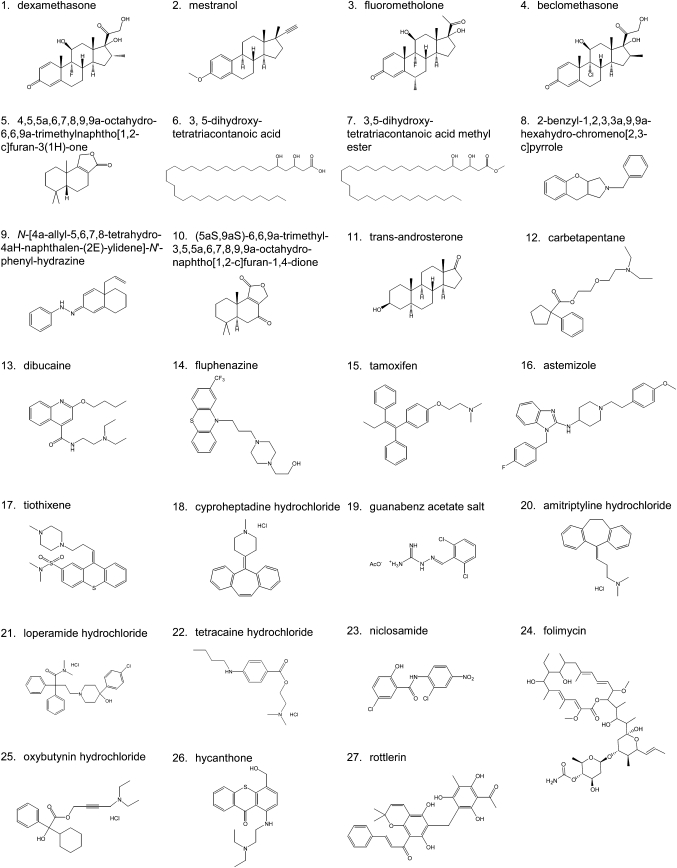
The other hit compounds were then rescreened at a final concentration of 5 μg/ml to confirm the reproducibility of the first screening. From the second screening, 27 compounds were selected that reproducibly decreased EGFP-D4 labeling in the plasma membrane. In Fig. 5A, the EGFP-D4 fluorescence intensity of the compound-treated cells is shown as a percentage of control. Figure 5B shows the screening plate images of the control and compound-treated wells. Compounds 2 (Fig. 5B) and 14 (data not shown) clearly abolished EGFP-D4 labeling in the plasma membrane. As observed previously with established inhibitors (see Fig. 4), most of the compounds resulted in a heterogeneous decrease of EGFP-D4 labeling (Fig. 5B, compounds 3, 5, 13, and 20). The chemical structures of the 27 hit compounds that decreased the EGFP-D4 labeling are displayed in Fig. 6.
Fig. 5.
EGFP-D4 labeling of cells treated with the hit compounds. HeLa cells were treated without or with 5 μg/ml compounds in medium with 10% LPDS for 18 h. Cells were labeled with EGFP-D4 and Hoechst 33342 as described in MATERIALS AND METHODS. A: Fluorescence intensity of EGFP-D4 per cell is shown as a percentage of control. Data are the mean ± average deviation of two independent experiments (compounds 2, 5, 7–11, 14–17, 19, 21, 22, and 25–27) or mean ± SD of three independent experiments (compounds 1, 3, 4, 6, 12, 13, 18, 20, 23, and 24). B: Images of EGFP-D4-labeled cells after treatment with hit compounds (compounds 2, 3, 5, 13, and 20). Images were acquired using In Cell Analyzer 1000 with a 20× objective. Bar = 20 μm.
Fig. 6.
Chemical structure of compounds that decreased EGFP-D4 labeling in the plasma membrane.
Effects of the hit compounds on intracellular cholesterol distribution
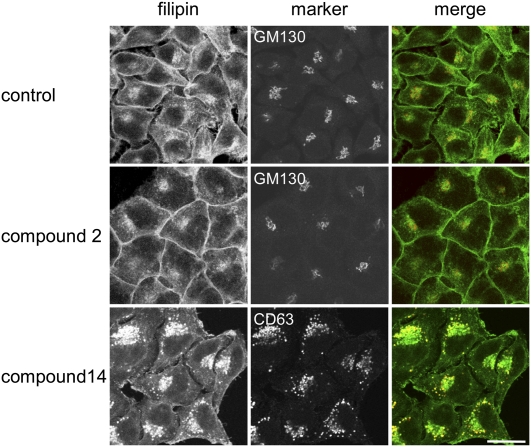
We then analyzed the possible molecular mechanism of the compound-induced alteration of cholesterol metabolism and distribution. First, we examined the possibility that the compounds accumulate cholesterol in intracellular organelles and decrease the amount of cholesterol in the plasma membrane. The 27 hit compounds were further analyzed by labeling cells with another cholesterol binding probe, filipin, to examine effects of the compounds on intracellular cholesterol distribution. Figure 7 shows the filipin labeling of the cells treated with or without hit compounds. In control cells, cholesterol was localized in the plasma membrane and the Golgi apparatus, the latter of which was stained with antibodies to GM130, a cis-Golgi matrix protein (32). The filipin fluorescence pattern of the cells treated with compound 2 was similar to that of the control cells. Compound 14 induced an increase of filipin intensity in the late endosomes that were labeled with antibodies to CD63, which is reported to accumulate in the late endosomes (33). Among the 27 hit compounds, 10 compounds (compounds 1–10) showed a filipin fluorescence pattern similar to that of the control cells, as was observed with compound 2 (see Fig. 7).
Fig. 7.
Filipin labeling of cells treated with hit compounds. HeLa cells were treated without or with 12.5 μg/ml hit compounds in 10% LPDS-containing medium for 18 h. The cells were labeled with filipin and anti GM130 antibodies or anti CD63 antibodies, as described in MATERIALS AND METHODS. Images were obtained under a Zeiss LSM510 confocal microscope with 63× objective. Bar = 20 μm.
Seventeen compounds (compounds 11–27) accumulated cholesterol in the late endosomes as displayed with compound 14( . Compound 11 was a steroid, trans-androsterone. Various steroids such as androstenedione and progesterone are reported to cause an accumulation of cholesterol in the late endosome/lysosomes and to inhibit cholesteryl ester formation (34, 35). This result suggests that trans-androsterone also inhibits cholesterol transport from the late endosomes. Folimycin (compound 24), also called concanamycin, is a specific inhibitor of vacuolar-type ATPase (36). Another vacuolar-type ATPase inhibitor, bafilomycin, is reported to accumulate cholesterol in the late endosomes (37, 38) and to act in the same way as the NPC mutation (39). It is suggested that the effects of folimycin on cholesterol accumulation in the late endosomes are attributed to its effect on vacuolar-type ATPase.
Most of compounds 11–27 can be categorized as hydrophobic amine. Hydrophobic amines such as imipramine are reported to accumulate cholesterol in the late endosomes/lysosomes (28). Amitriptyline (compound 20) is an antipsychotic drug. Certain antipsychotic drugs, including amitriptyline and imipramine, were shown to activate the sterol-regulatory element binding protein (SREBP) transcription factors (40, 41). Although the mechanism is unknown, the modulation of the SREBP pathway by amitriptyline may be related to the compound-induced accumulation of cholesterol in the late endosomes.
Thus, we can hypothesize that after cell treatment with these compounds, cholesterol might accumulate in the late endosomes/lysosomes owing to the inhibition of cholesterol exit from these organelles. As a result, the amount of cholesterol in the plasma membrane was reduced and EGFP-D4 labeling of the cell surface was decreased.
Effect of the incubation time of cells with compounds on EGFP-D4 labeling
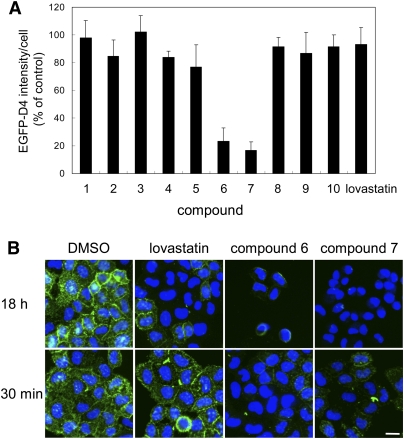
We next investigated the mechanisms of the inhibition of the EGFP-D4 labeling with compounds 1–10 that did not alter intracellular cholesterol distribution. To examine the possibility that compounds 1–10 directly inhibit D4 binding to cholesterol, we measured the effects of a short incubation time with these compounds. After cell treatment with compounds for 30 min, cells were labeled with EGFP-D4 and analyzed. Figure 8A shows the EGFP-D4 fluorescence intensity of cells treated with the compounds for 30 min, whereas Fig. 8B shows EGFP-D4 images of the cells treated with the compounds for 18 h or 30 min. Treatment of lovastatin for 30 min did not alter EGFP-D4 labeling, whereas 18 h treatment decreased the labeling (Fig. 8B). The EGFP-D4 labeling of cells was not changed even after cell treatment with lovastatin for 6 h (data not shown). The results indicate that longer incubation with lovastatin is required to decrease the amount of cholesterol in the plasma membrane. In contrast, compounds 6 and 7 drastically decreased EGFP-D4 labeling by treatment as short as 30 min, suggesting that these compounds are not involved in cholesterol metabolism. The structures of compounds 6 and 7 are similar (Fig. 6). Quantification of cholesterol by gas chromatography showed that the free cholesterol content of compound 6- and 7-treated cells was not significantly altered (data not shown). The results suggest that compounds 6 and 7 do not remove cholesterol from the plasma membrane. It is likely that compounds 6 and 7 inhibit the binding of EGFP-D4 to cholesterol. This result suggests that these compounds inhibited lipid-protein interaction. Alternatively, these compounds could alter the organization of lipid domains.
Fig. 8.
Effects of incubation time with the hit compounds on EGFP-D4 labeling of cells. A: HeLa cells were treated with 12.5 μg/ml hit compounds or 8 μM lovastatin in LPDS-containing medium for 30 min. Cells were labeled with EGFP-D4 and Hoechst 33342, and fluorescence intensity of EGFP-D4 bound to cell surface was normalized by cell number as described in MATERIALS AND METHODS. Data represent the mean ± SD (n = 4). B: HeLa cells were treated with DMSO, 8 μM lovastatin, 12.5 μg/ml compound 6, or compound 7 in 10% LPDS-containing medium for 18 h or 30 min. Cells were labeled with EGFP-D4 and Hoechst 33342. Images were acquired using In Cell Analyzer 1000 with a 20× objective. Bar = 20 μm.
Effects of the hit compounds on de novo cholesterol biosynthesis
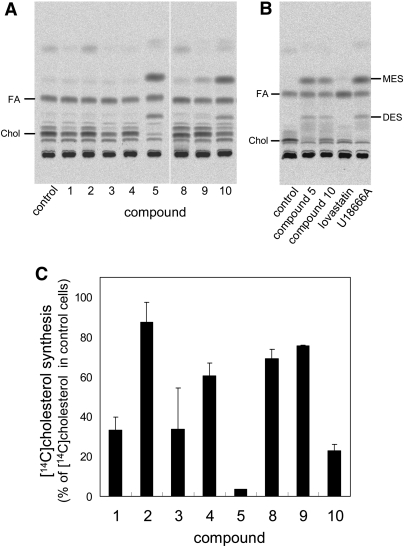
We then examined the effects of compounds 1–5 and 8–10 on cholesterol biosynthesis. In Fig. 9, cells were labeled with [14C]acetate for 2 h in the presence of the compounds, and de novo synthesized cholesterol was determined. The compounds induced distinct profiles of [14C]acetate-labeled lipids after TLC analysis (Fig. 9A). Quantification of the major bands corresponding to cholesterol revealed that compounds 1, 3, 5, and 10 exert potent inhibitory effects on cholesterol synthesis (Fig. 9C). Compound 4 moderately inhibited cholesterol synthesis (Fig. 9C). The chemical structures of the hit compounds are not similar to that of the HMG-CoA reductase inhibitors, statins. Compounds 1, 3, and 4 are steroids. Dexamethasone (compound 1) is reported to affect cholesterol synthesis in HeLa S3G cells, depending on the growth conditions (42, 43).
Fig. 9.
Effects of the hit compounds on de novo cholesterol biosynthesis. HeLa cells were cultured in the medium with LPDS for 18 h. Cells were then labeled with [14C]acetic acid for 2 h in the presence of 12.5 μg/ml compounds, 8 μM lovastatin or 2 μg/ml U18666A. Lipids were extracted and separated on HPTLC (A, B). The positions of 2,3-monoepoxysqualene (MES) and 2,3;22,23-diepoxysqualene (DES) were determined using nonradioactive standards (B). The incorporation of radioactivity into the band corresponding to cholesterol was measured as described in MATERIALS AND METHODS. Data are expressed as a percentage of control (C). Data are the mean ± average deviation of two independent experiments (compounds 5 and 8–10) or mean ± SD of three independent experiments (compounds 1–4).
The structures of compounds 5 and 10 are similar. Compounds 5 and 10 induced the formation of two additional lipids after [14C]acetate labeling, as shown in Fig. 9A. These bands were also observed after U18666A treatment but not after lovastatin treatment (Fig. 9B). In addition to its effect on the accumulation of cholesterol in late endosomes/lysosomes, U18666A is reported to inhibit oxidosqualene cyclase, an intermediate step in cholesterol synthesis (25). Inhibition of this enzyme led to the generation of 2,3-monoepoxysqualene and 2,3;22,23-diepoxysqualene, two substrates of oxydosqualene cyclase (38, 44, 45). Our results suggest that compounds 5 and 10 inhibit oxidosqualene cyclase. They also indicate that compounds 1, 3, 4, 5, and 10 decrease the amount of cholesterol in the plasma membranes by inhibition of cholesterol synthesis.
We also examined the effects of compounds 11–27, which accumulated cholesterol in the late endosomes, on cholesterol biosynthesis. Shown in supplementary Fig. I, cells were labeled with [14C]acetate for 2 h in the presence of the compounds, and labeled lipids were analyzed by HPTLC. Profiles of labeled lipids from cells treated with compounds 12–16, 22, and 25–26 were different from those of control cells. These profiles are similar to that of U18666A-treated cells. The results indicate that a number of compounds that accumulate cholesterol in the late endosomes also modify cholesterol biosynthesis. Indeed, tamoxifen (compound 15) is shown to be an effective inhibitor of the conversion of lanosterol to cholesterol, resulting in the accumulation of precursors of cholesterol, Δ8 cholesterol, zymosterol, and desmosterol in cells (46, 47).
Effects of the hit compounds on cholesterol transport from the ER to the plasma membrane
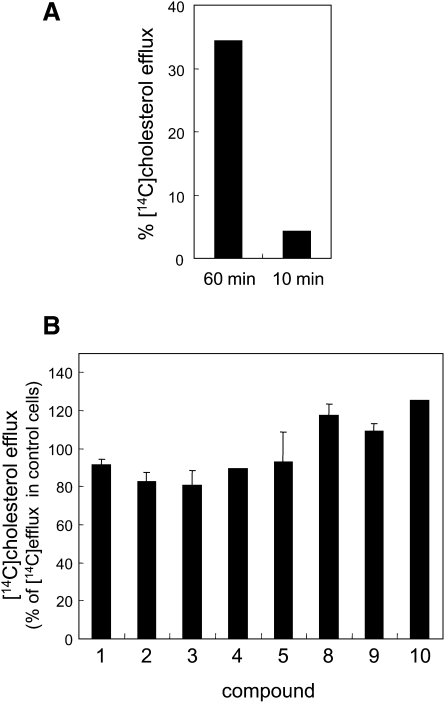
We next examined the effects of the hit compounds (compounds 1–5 and 8–10) on the cholesterol transport from the ER to the plasma membrane. Newly synthesized cholesterol was detected by pulse labeling the cells with [14C]acetate for 15 min in the presence of the compounds. We measured the appearance of newly synthesized [14C]cholesterol in a cell surface pool accessible for extraction with methyl-β-cyclodextrin after 1 h chase (20, 21). In control cells, 30–40% of the newly synthesized cholesterol was released into the medium (Fig. 10A). Newly synthesized cholesterol was not significantly detected in the medium when cells were chased for a period of 10 min after labeling with [14C]acetate for 15 min (Fig. 10A), whereas the amount of synthesized cholesterol displayed a level similar to that of the 1 h chase (data not shown). This result confirms that transport of newly synthesized cholesterol to the plasma membrane can be detected in this assay. In , the cholesterol transport observed in the presence of the hit compounds is shown as a percentage of the control. None of them inhibited cholesterol transport from the ER to the plasma membrane.
Fig. 10.
Effects of hit compounds on cholesterol transport. HeLa cells were cultured in medium with LPDS for 18 h. Cells were then labeled with [14C]acetic acid for 15 min in the presence of 12.5 μg/ml compounds, and chase was performed in the presence of 12.5 μg/ml compounds and 8 μM lovastatin for 1 h. In A, chase was also performed for 10 min. During the last 10 min of the chase, cells were incubated with 10 mM methyl-β-cyclodextrin. From medium and cells, lipids were extracted and separated on HPTLC. The arrival of cholesterol on the plasma membrane was determined as described in MATERIALS AND METHODS. Data are expressed as a percentage of control (B). Data are the mean ± average deviation of two independent experiments (compounds 1, 3, and 8–10) or mean ± SD of three independent experiments (compounds 2, 4, and 5).
CONCLUSION
In the present study, we employed EGFP-D4 as a sensitive cholesterol binding probe for automated fluorescence microscopy screening of compounds that modify intracellular cholesterol metabolism and distribution. The noteworthy advantages of EGFP-D4 over filipin, a commonly used cholesterol probe, are 1) it is nontoxic and thus can be used in living cells; and 2) it binds cholesterol only when cholesterol concentration in a membrane is very high. This makes it possible to detect small alterations of plasma membrane cholesterol.
In fact, known inhibitors of cholesterol metabolism and distribution were identified through this screening, strengthening that our method is useful to screen new inhibitors. In this study, we obtained 27 hit compounds that had not been reported to affect cholesterol metabolism and distribution.
In conclusion, our cell-based visual screening using EGFP-D4, in combination with biochemical and histochemical analyses, is a very sensitive and useful method to identify and classify compounds that affect cholesterol metabolism and intracellular distribution. Furthermore, it is suitable for high-throughput analysis using chemical and genetic libraries.
Supplementary Material
Acknowledgments
The authors thank the members of Kobayashi's laboratory for their support, critical reading of the manuscript, and discussion. We are grateful to Shizuya Yamashita of Osaka University for his advice.
Footnotes
Abbreviations:
- CHO
- Chinese hamster ovary
- D4
- θ toxin domain 4
- ER
- endoplasmic reticulum
- HPTLC
- high-performance thin-layer chromatography
- LPDS
- lipoprotein-deficient serum
- MLV
- multilamellar vesicle
- PC
- phosphatidylcholine
- SREBP
- sterol-regulatory element binding protein
- SUV
- small unilamellar vesicle
- UV
- ultraviolet
R.I. was supported by Hayashi Memorial Foundation for Female Natural Scientists. This work was supported by the Lipid Dynamics Program of RIKEN and Grant-in Aid for Scientific Research 22390018 (to T.K.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one figures.
REFERENCES
- 1.Simons K., Vaz W. L. 2004. Model systems, lipid rafts, and cell membranes. Annu. Rev. Biophys. Biomol. Struct. 33: 269–295 [DOI] [PubMed] [Google Scholar]
- 2.Epand R. M., Thomas A., Brasseur R., Epand R. F. 2010. Cholesterol interaction with proteins that partition into membrane domains: an overview. Subcell. Biochem. 51: 253–278 [DOI] [PubMed] [Google Scholar]
- 3.Simons K., Toomre D. 2000. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 1: 31–39 [DOI] [PubMed] [Google Scholar]
- 4.Goldstein J. L., DeBose-Boyd R. A., Brown M. S. 2006. Protein sensors for membrane sterols. Cell. 124: 35–46 [DOI] [PubMed] [Google Scholar]
- 5.Ikonen E. 2008. Cellular cholesterol trafficking and compartmentalization. Nat. Rev. Mol. Cell Biol. 9: 125–138 [DOI] [PubMed] [Google Scholar]
- 6.Maxfield F. R., van Meer G. 2010. Cholesterol, the central lipid of mammalian cells. Curr. Opin. Cell Biol. 22: 422–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mesmin B., Maxfield F. R. 2009. Intracellular sterol dynamics. Biochim. Biophys. Acta. 1791: 636–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniels T. F., Killinger K. M., Michal J. J., Wright R. W., Jr, Jiang Z. 2009. Lipoproteins, cholesterol homeostasis and cardiac health. Int. J. Biol. Sci. 5: 474–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gurr M. I., Harwood J. L., Frayn K. N. 2002. Lipid Biochemistry. 5th edition Blackwell Science, Ltd; Oxford, UK: 199–212 [Google Scholar]
- 10.Barrios-González J., Miranda R. U. 2010. Biotechnological production and applications of statins. Appl. Microbiol. Biotechnol. 85: 869–883 [DOI] [PubMed] [Google Scholar]
- 11.Endo A. 1992. The discovery and development of HMG-CoA reductase inhibitors. J. Lipid Res. 33: 1569–1582 [PubMed] [Google Scholar]
- 12.Pipalia N. H., Huang A., Ralph H., Rujoi M., Maxfield F. R. 2006. Automated microscopy screening for compounds that partially revert cholesterol accumulation in Niemann-Pick C cells. J. Lipid Res. 47: 284–301 [DOI] [PubMed] [Google Scholar]
- 13.Severs N. J. 1997. Cholesterol cytochemistry in cell biology and disease. Subcell. Biochem. 28: 477–505 [DOI] [PubMed] [Google Scholar]
- 14.Rujoi M., Pipalia N. H., Maxfield F. R. 2010. Cholesterol pathways affected by small molecules that decrease sterol levels in Niemann-Pick type C mutant cells. PLoS ONE. 5: e12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohno-Iwashita Y., Shimada Y., Hayashi M., Iwamoto M., Iwashita S., Inomata M. 2010. Cholesterol-binding toxins and anti-cholesterol antibodies as structural probes for cholesterol localization. Subcell. Biochem. 51: 597–621 [DOI] [PubMed] [Google Scholar]
- 16.Ohno-Iwashita Y., Shimada Y., Waheed A. A., Hayashi M., Inomata M., Nakamura M., Maruya M., Iwashita S. 2004. Perfringolysin O, a cholesterol-binding cytolysin, as a probe for lipid rafts. Anaerobe. 10: 125–134 [DOI] [PubMed] [Google Scholar]
- 17.Nelson L. D., Johnson A. E., London E. 2008. How interaction of perfringolysin O with membranes is controlled by sterol structure, lipid structure, and physiological low pH: insights into the origin of perfringolysin O-lipid raft interaction. J. Biol. Chem. 283: 4632–4642 [DOI] [PubMed] [Google Scholar]
- 18.Shimada Y., Maruya M., Iwashita S., Ohno-Iwashita Y. 2002. The C-terminal domain of perfringolysin O is an essential cholesterol-binding unit targeting to cholesterol-rich microdomains. Eur. J. Biochem. 269: 6195–6203 [DOI] [PubMed] [Google Scholar]
- 19.Norman A. W., Demel R. A., de Kruyff B., van Deenen L. L. 1972. Studies on the biological properties of polyene antibiotics. Evidence for the direct interaction of filipin with cholesterol. J. Biol. Chem. 247: 1918–1929 [PubMed] [Google Scholar]
- 20.Cruz J. C., Chang T. Y. 2000. Fate of endogenously synthesized cholesterol in Niemann-Pick type C1 cells. J. Biol. Chem. 275: 41309–41316 [DOI] [PubMed] [Google Scholar]
- 21.Heino S., Lusa S., Somerharju P., Ehnholm C., Olkkonen V. M., Ikonen E. 2000. Dissecting the role of the golgi complex and lipid rafts in biosynthetic transport of cholesterol to the cell surface. Proc. Natl. Acad. Sci. USA. 97: 8375–8380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917 [DOI] [PubMed] [Google Scholar]
- 23.Mizuno H., Abe M., Dedecker P., Makino A., Rocha S., Ohno-Iwashita Y., Hofkens J., Kobayashi T., Miyawaki A. 2011. Fluorescent probes for superresolution imaging of lipid domains on the plasma membrane. Chem. Sci. 2: 1548–1553 [Google Scholar]
- 24.Kobayashi T., Beuchat M. H., Lindsay M., Frias S., Palmiter R. D., Sakuraba H., Parton R. G., Gruenberg J. 1999. Late endosomal membranes rich in lysobisphosphatidic acid regulate cholesterol transport. Nat. Cell Biol. 1: 113–118 [DOI] [PubMed] [Google Scholar]
- 25.Cenedella R. J. 2009. Cholesterol synthesis inhibitor U18666A and the role of sterol metabolism and trafficking in numerous pathophysiological processes. Lipids. 44: 477–487 [DOI] [PubMed] [Google Scholar]
- 26.Liscum L., Faust J. R. 1989. The intracellular transport of low density lipoprotein-derived cholesterol is inhibited in Chinese hamster ovary cells cultured with 3-beta-[2-(diethylamino)ethoxy]androst-5-en-17-one. J. Biol. Chem. 264: 11796–11806 [PubMed] [Google Scholar]
- 27.Klingenstein R., Lober S., Kujala P., Godsave S., Leliveld S. R., Gmeiner P., Peters P. J., Korth C. 2006. Tricyclic antidepressants, quinacrine and a novel, synthetic chimera thereof clear prions by destabilizing detergent-resistant membrane compartments. J. Neurochem. 98: 748–759 [DOI] [PubMed] [Google Scholar]
- 28.Roff C. F., Goldin E., Comly M. E., Cooney A., Brown A., Vanier M. T., Miller S. P., Brady R. O., Pentchev P. G. 1991. Type C Niemann-Pick disease: use of hydrophobic amines to study defective cholesterol transport. Dev. Neurosci. 13: 315–319 [DOI] [PubMed] [Google Scholar]
- 29.Palmeri S., Battisti C., Malandrini A., Federico A. 1995. Amiodarone induced lipidosis similar to Niemann-Pick C disease. Biochemical and morphological study. Life Sci. 57: 1963–1971 [DOI] [PubMed] [Google Scholar]
- 30.Shayman J. A., Kelly R., Kollmeyer J., He Y., Abe A. 2010. Group XV phospholipase A, a lysosomal phospholipase A. Prog. Lipid Res. 50: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zidovetzki R., Levitan I. 2007. Use of cyclodextrins to manipulate plasma membrane cholesterol content: evidence, misconceptions and control strategies. Biochim. Biophys. Acta. 1768: 1311–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura N., Rabouille C., Watson R., Nilsson T., Hui N., Slusarewicz P., Kreis T. E., Warren G. 1995. Characterization of a cis-Golgi matrix protein, GM130. J. Cell Biol. 131: 1715–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kobayashi T., Vischer U. M., Rosnoblet C., Lebrand C., Lindsay M., Parton R. G., Kruithof E. K., Gruenberg J. 2000. The tetraspanin CD63/lamp3 cycles between endocytic and secretory compartments in human endothelial cells. Mol. Biol. Cell. 11: 1829–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aikawa K., Furuchi T., Fujimoto Y., Arai H., Inoue K. 1994. Structure-specific inhibition of lysosomal cholesterol transport in macrophages by various steroids. Biochim. Biophys. Acta. 1213: 127–134 [PubMed] [Google Scholar]
- 35.Butler J. D., Blanchette-Mackie J., Goldin E., O'Neill R. R., Carstea G., Roff C. F., Patterson M. C., Patel S., Comly M. E., Cooney A., et al. 1992. Progesterone blocks cholesterol translocation from lysosomes. J. Biol. Chem. 267: 23797–23805 [PubMed] [Google Scholar]
- 36.Dröse S., Bindseil K. U., Bowman E. J., Siebers A., Zeeck A., Altendorf K. 1993. Inhibitory effect of modified bafilomycins and concanamycins on P- and V-type adenosinetriphosphatases. Biochemistry. 32: 3902–3906 [DOI] [PubMed] [Google Scholar]
- 37.Furuchi T., Aikawa K., Arai H., Inoue K. 1993. Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, blocks lysosomal cholesterol trafficking in macrophages. J. Biol. Chem. 268: 27345–27348 [PubMed] [Google Scholar]
- 38.Issandou M., Guillard R., Boullay A. B., Linhart V., Lopez-Perez E. 2004. Up-regulation of low-density lipoprotein receptor in human hepatocytes is induced by sequestration of free cholesterol in the endosomal/lysosomal compartment. Biochem. Pharmacol. 67: 2281–2289 [DOI] [PubMed] [Google Scholar]
- 39.Lange Y., Ye J., Rigney M., Steck T. L. 2002. Dynamics of lysosomal cholesterol in Niemann-Pick type C and normal human fibroblasts. J. Lipid Res. 43: 198–204 [PubMed] [Google Scholar]
- 40.Raeder M. B., Ferno J., Glambek M., Stansberg C., Steen V. M. 2006. Antidepressant drugs activate SREBP and up-regulate cholesterol and fatty acid biosynthesis in human glial cells. Neurosci. Lett. 395: 185–190 [DOI] [PubMed] [Google Scholar]
- 41.Raeder M. B., Ferno J., Vik-Mo A. O., Steen V. M. 2006. SREBP activation by antipsychotic- and antidepressant-drugs in cultured human liver cells: relevance for metabolic side-effects? Mol. Cell. Biochem. 289: 167–173 [DOI] [PubMed] [Google Scholar]
- 42.Cavenee W. K., Johnston D., Melnykovych G. 1978. Regulation of cholesterol biosynthesis in HeLa S3G cells by serum lipoproteins: dexamethasone-mediated interference with suppression of 3-hydroxy-3-methylglutaryl coenzyme A reductase. Proc. Natl. Acad. Sci. USA. 75: 2103–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnston D., Matthews E. R., Melnykovych G. 1980. Glucocorticoid effects on lipid metabolism in HeLa cells: inhibition of cholesterol synthesis and increased sphingomyelin synthesis. Endocrinology. 107: 1482–1488 [DOI] [PubMed] [Google Scholar]
- 44.Mark M., Muller P., Maier R., Eisele B. 1996. Effects of a novel 2,3-oxidosqualene cyclase inhibitor on the regulation of cholesterol biosynthesis in HepG2 cells. J. Lipid Res. 37: 148–158 [PubMed] [Google Scholar]
- 45.Sexton R. C., Panini S. R., Azran F., Rudney H. 1983. Effects of 3 beta-[2-(diethylamino)ethoxy]androst-5-en-17-one on the synthesis of cholesterol and ubiquinone in rat intestinal epithelial cell cultures. Biochemistry. 22: 5687–5692 [DOI] [PubMed] [Google Scholar]
- 46.Holleran A. L., Lindenthal B., Aldaghlas T. A., Kelleher J. K. 1998. Effect of tamoxifen on cholesterol synthesis in HepG2 cells and cultured rat hepatocytes. Metabolism. 47: 1504–1513 [DOI] [PubMed] [Google Scholar]
- 47.Kedjouar B., de Medina P., Oulad-Abdelghani M., Payre B., Silvente-Poirot S., Favre G., Faye J. C., Poirot M. 2004. Molecular characterization of the microsomal tamoxifen binding site. J. Biol. Chem. 279: 34048–34061 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.