Abstract
We present a method for the determination of triacylglycerol (TAG) profiles of oleaginous saltwater microalgae relevant for the production of biofuels, bioactive lipids, and high-value lipid-based chemical precursors. We describe a technique to remove chlorophyll using quick, simple solid phase extraction (SPE) and directly compare the intact TAG composition of four microalgae species (Phaeodactylum tricornutum, Nannochloropsis salina, Nannochloropsis oculata, and Tetraselmis suecica) using MALDI time-of-flight (TOF) mass spectrometry (MS), ESI linear ion trap-orbitrap (LTQ Orbitrap) MS, and 1H NMR spectroscopy. Direct MS analysis is particularly effective to compare the polyunsaturated fatty acid (PUFA) composition for triacylglycerols because oxidation can often degrade samples upon derivatization. Using these methods, we observed that T. suecica contains significant PUFA levels with respect to other microalgae. This method is applicable for high-throughput MS screening of microalgae TAG profiles and may aid in the commercial development of biofuels.
Keywords: lipids, biofuel, algae, MALDI-MS, SPE separation, nuclear magnetic resonance spectroscopy, PUFA
Microalgae have been identified as an attractive source for the production of biofuels in addition to bioactive lipids and high-value, lipid-based chemical precursors based on their ability to produce 20% or more of their mass as oil. Due to the conversion of triacylglycerols (TAG) to fatty acid alkyl esters for use as biodiesel (1, 2), current research has focused on TAG production and analysis to make biofuels from microalgae a more economically feasible option (3). Furthermore, the Clean Energy Act of 2007 mandates the production of one billion gallons of biodiesel by 2012. As the commercial use of alternative biofuels continues to gain attention, methods for analysis of the specific lipid composition in algae have become increasingly important. The methods and pursuit of algae-based fuels also have applications for bioactive lipids and high-value, lipid-based chemical precursors (4, 5), including polyunsaturated fatty acids (PUFA), carotenes, antioxidants, vitamins, and pigments, which have applications in the pharmaceutical, nutraceutical, and food industries. In all such applications, the methods of analysis, access to desired composition, total yield, and efficiency with which lipids can be converted to usable products will be the ultimate determinants of industrial success.
Previous analysis of algae lipid composition using GC/MS and MALDI-TOF mass spectrometry (MS) has shown a broad variety of lipid classes: glycerolipids, such as monogalactosyl-diacylglycerol (MGDG) and digalactosyl-diacylglycerol (DGDG); phospholipids, such as phosphatidylcholine (PC); and neutral lipids (triacylglycerols) (6–9). Many reports have focused on the analysis of polar lipids, but TAG analysis and composition have become increasingly important for biofuel applications. TAG analysis of mixed samples remains a challenge because of its lower ionization efficiency relative to polar lipids (such as phospholipids) (10). Analysis of neutral lipids in microalgae often relies on lipophilic fluorescence and colorimetric assays (11, 12); however, these assays do not provide compositional TAG characterization. Compositional analysis of microalgal TAG primarily involves the initial separation of extracts using either liquid chromatography or thin-layer chromatography followed by MS analysis or derivatization of extracts, such as transesterification, followed by analysis of fatty acid methyl esters (FAME) by GC/MS (13–15). The direct analysis of algal triacylglycerol using pyrolysis GC/MS has been reported as a promising alternative, but the purchase of specialty equipment and use of higher temperature ranges may limit the broad utility of this method (16, 17).
Herein, we describe the compositional analysis of intact triacylglycerol of oleaginous marine algae using MALDI-TOF MS. MALDI-TOF provides a direct method to determine the TAG composition of different saltwater microalgae strains or algae grown under different culturing conditions. Direct analysis of intact triacylglycerol is particularly useful to identify and compare PUFA components. ESI-LTQ-Orbitrap MS is also used to compare analyses and confirm levels of unsaturation. MALDI-TOF has been used for the rapid characterization of purified edible oils based on the lipid fingerprint (18–23), but the utility of this method with more complex algae lipid extracts has not been demonstrated. The method reported herein provides analysis of the TAG fingerprints of different algae species while circumventing the need to derivatize samples for fatty acid ester analysis or use specialized instruments. We also describe the use of 1H nuclear magnetic resonance (NMR) spectroscopy as a complementary method to monitor lipid extraction, the relative TAG purity, and the relative PUFA ratio based on integration and chemical shift of diagnostic TAG protons.
MATERIALS AND METHODS
Materials
Olive oil, used as a positive control for triacylglycerols, as well as 2,5-dihydroxybenzoic acid (DHB), was purchased from ACROS Organics. Solvents (ACS reagent-grade hexanes, methanol, acetic acid, diethyl ether, and chloroform), and acetate salts (LiOAc and NaOAc; >99% purity) were purchased from Sigma-Aldrich. SiliaPrep silica solid phase extraction (SPE) columns (Silicycle 500 mg, 3 ml capacity) were selected because they provided optimal separation of chlorophyll a from lipid samples. 3,4,5-Trichloropyridine was purchased from Alfa Aesar. Deuterated chloroform (99.8%) was purchased from Cambridge Isotope Laboratories.
Algae strains and culture conditions
Algae species were purchased from the University of Texas, Austin, TX (Phaeodactylum tricornutum, UTEX B 2089; and Nannochloropsis oculata, UTEX LB 2164), the Provasoli-Guillard Center for the Culture of Marine Phytoplankton at Bigelow Laboratory for Ocean Sciences, MA (Nannochloropsis salina, CCMP 537), and the Culture Collection of Algae and Protozoa, Scotland (Tetraselmis suecica, CCAP 6/4). P. tricornutum, N. salina, and T. suecica were cultured in medium per the supplier's recommendation. Cultures were aerated by a ThermoScientific MAXQ 2000 orbital shaker at a constant speed of 150 rpm at 22 ± 3°C on a 16:8 h light/dark cycle. N. oculata nitrogen-deficient comparison cultures were grown with stirring and constant bubbling of air on a 16:8 h light/dark cycle. Growth was monitored using absorbance with UV-Vis spectroscopy at 680 nm, and cell counts were performed using a hemocytometer. Algae cultures were harvested at day 10 of stationary phase. All experiments consisted of two or more biological replicates for each strain and culture condition. A culture of N. oculata was grown in nitrogen-deficient media that contained one-third the normal nitrogen content (with a final concentration of approximately 300 μM NaNO3), and it was analyzed at the same time as a control culture.
Lipid extraction
Nonpolar algal extracts were harvested from one-liter cultures in stationary phase, with cell counts greater than 1 × 106 cells. Cells were pelleted by centrifugation at 6,000 rpm for 20 min, and then lyophilized to dryness. For MS analysis, nonpolar lipids were obtained by hexane extraction, washed with water, and then concentrated by vacuum at room temperature (24, 25). For nitrogen-deficient culture comparison, nonpolar lipids were obtained using a modified Bligh Dyer method in which cells were sonicated for 30 s in chloroform; then 1:2 chloroform/methanol was added and cells were sonicated again for 30 s; and then extracts were washed with water and concentrated by vacuum at room temperature (26, 27). All extracts were stored in amber vials under argon in a −20°C freezer.
SPE separation
Prior to MS analysis, an additional separation step was performed on the nonpolar extract using SPE (28–30). Lipid samples were weighed prior to analysis. The SPE cartridge was primed with hexanes, and the nonpolar extract (up to 50 mg) was dissolved in 300 μl of hexanes before addition to the cartridge. An 80:20:1 mixture of hexanes/diethyl ether/acetic acid was used as mobile phase for TAG elution. A residual fraction containing polar lipids and chlorophyll was obtained by elution with acetone. Fractions were immediately dried and stored under argon in amber vials. Before using SPE for analysis of algae samples, oil standards were used to verify recovery and confirm that all TAG proportions were isolated in this method (see supplementary data).
MALDI-TOF MS analysis of algal lipid extracts
MADLI mass spectra were acquired on a 4700 MALDI-TOF-TOF (Applied Biosystems, Foster City, CA) with an internal MALDI source, a 355 nm pulsed Nd:YAG laser that was operated in positive-ion mode for analysis. The TOF was run in reflectron mode, and each run consisted of 2,500 shots with a focus mass of 850 Da and a scan range of m/z 400-1,100. Multiple acquisition scans were performed at different laser intensities to optimize conditions. Higher laser intensity was required to ionize the nonpolar TAG fraction whereas lower laser intensities (<12 μJ) were used to ionize the second SPE fraction that contained more polar constituents. Samples were diluted to 2 mg/ml in hexane, and then spotted in the fast-evaporation method in a 1:2 ratio with DHB as the matrix (31). To avoid glassing, spots were recrystallized on the stainless steel plate with 0.5 ml methanol, which allowed for optimal crystal formation. Data was normalized to base peak in all cases.
ESI-LTQ-Orbitrap analysis of algal lipid extracts
The ESI-LTQ-Orbitrap (ThermoFisher, San Jose, CA) was used for direct-injection electrospray analysis of the algal extracts. Extract samples for the LTQ-Orbitrap were diluted to 2 mg/ml in hexanes, and then diluted to 2 μM in acetone. Salt solutions of LiOAc and NaOAc (100 μM) in water were used to dope the acetone solution to a 20 nM concentration prior to injection; minimal salt solutions (1 μl) were used to prevent the oil from separating out of solution. The LTQ-Orbitrap was operated in positive-ion mode with an electrospray voltage of 5 kV. The sample was sprayed into the mass spectrometer with the sheath, auxiliary, and sweep gas set at 8, 0, and 0 units, respectively. Desolvation was further aided by an ion transfer tube temperature of 250°C. An isolation width of 2 amu was used with either 30% or 35% normalized collision energy (CID) for MSn experiments. All scan events were detected in the FT detector.
NMR spectroscopy of algal lipid extracts
1H NMR analysis was performed on crude lipid extracts to monitor TAG purity and determine relative compositional ratios based on integration. After SPE, 1H NMR analysis was also performed on purified lipid extracts to determine compositional ratios. Lipid extracts were dissolved in 1 ml of deuterated chloroform (CDCl3) with ∼5 mg of 3,4,5-trichloropyridine as an internal standard, and then 750 μl was transferred to a 5 mm NMR tube. 1H NMR spectra were acquired at 298 K on a 600 MHz NMR spectrometer (Varian, Palo Alto, CA) for 32 scans with 39k data points, a relaxation delay of 1 s, pulse angle of 45 degrees, and line broadening of 0.2 Hz. Samples were referenced to CDCl3 at 7.27 ppm and 3,4,5-trichloropyridine at 8.53 ppm (singlet, 2H). Figures were processed using MestReNova software (Mestrelab Research SL, Santiago de Compostela, Spain). Peak assignment was based on NMR spectroscopic studies of previously reported algal lipid extracts (32). Spectra are included in the supporting information.
RESULTS AND DISCUSSION
Three green algae species and one diatom strain were selected for analysis to compare TAG composition. Different algae species produce varying lipid compositions, which is important for their selection for fuel production (2). For this study, Phaeodactylum tricornutum (33), Nannochloropsis salina (34), Nannochloropsis oculata (35), and Tetraselmis suecica (36, 37) were selected due to their high lipid content and potential value for alternative fuel production (3). Prior to MS analysis, 1H NMR spectroscopy was used to verify the quantity and purity of the nonpolar algal extracts (32).
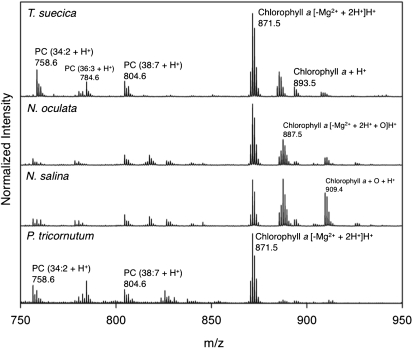
Preliminary MALDI-TOF analysis of the crude algal lipid extracts for all algae strains (Fig. 1) showed that signals from polar constituents, such as chlorophyll and phospholipids, dominated the spectrum and suppressed the TAG signals. Due to the preferential ionization of chlorophyll a and the fact that the signal overlaps with certain TAG species, even a small amount of chlorophyll a in the algae extracts could suppress TAG signals (6, 38, 39). While 1H NMR analysis indicated that <5% of chlorophyll was present in the sample (supplementary Fig. V) due to superior ionization, this was enough to completely suppress the desired TAG peaks in MS acquisition. MALDI-TOF analysis of the crude N. salina lipid extract showed that the dominant chlorophyll a signal was the species [chlorophyll a (−Mg2+ + 2H) + H]+ seen at m/z 871.5 (Fig. 1 and Fig. 2A). Oxidized chlorophyll a (chlorophyll a [−Mg2+ + 2H + O] + H+) was seen at a lower intensity at m/z 887.5, and intact chlorophyll a ([+Mg2+] + H+) was seen at lower intensity at m/z 893.5. Phosphatidylcholine (PC), which ionizes well and is known to suppress more nonpolar lipid signals, was also observed (10).
Fig. 1.
Comparison of the algae nonpolar lipid extracts (m/z 750-950) MALDI-TOF mass spectra from four algae species: T. suecica, N. oculata, N. salina, and P. tricornutum. Intensity has been normalized.
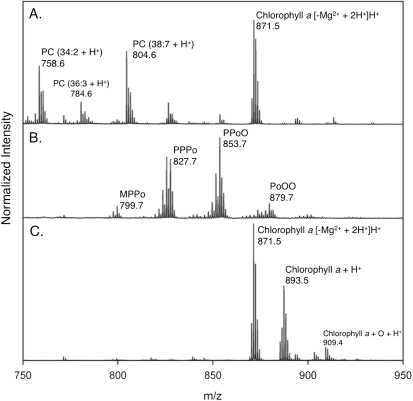
Fig. 2.
Comparison of the TAG region (m/z 750-950) in the MALDI-TOF mass spectra of lipid extracts from N. salina before and after SPE. (A) Crude lipid extract before SPE. (B) Triacylglycerols eluted from 80:20:1 hexanes/diethyl ether/acetic acid. All TAG ions are sodiated adducts. (C) Polar lipids and chlorophyll eluted in acetone. Fatty acids: M = C14:0, P = C16:0, Po = C16:1, and O = C18:1. PC, phosphatidylcholine.
SPE was implemented as an efficient pretreatment method for the lipid extract to reduce interfering signals and prevent competitive ionization. SPE methods, which can separate classes of constituents based on polarity, have found increasing use for separating biological samples (28–30, 40). SPE is an affordable and easy way to circumvent using an expensive liquid chromatography setup, and it is also suitable as a high-throughput technique for automation. Here, SPE elution and subsequent MALDI-TOF analysis of the nonpolar fraction showed the TAG components in the range from 800 Da and to ∼930 Da (Fig. 2B). The unsaturation of the triacylglycerol was visible as the peaks were separated by 2 Da. The identity of prominent TAG signals has been confirmed by MS/MS (supplementary Table II). Analysis of the second, more polar fraction (eluted with acetone) showed clear separation for any remaining constituents, including trace chlorophyll and polar lipids (Fig. 2C).
Both hexanes and chloroform/methanol extractions can be used for the extraction of nonpolar lipids (25, 27). Extraction using hexanes instead of chloroform/methanol provides a cleaner nonpolar extraction (10, 24, 25). We observed that hexane extraction reduced the level of chlorophyll present and that the hexane extraction could be directly loaded onto the SPE column without the need to remove and reconstitute the sample. Therefore, hexane extraction was generally more efficient for this method and allowed for more rapid MS analysis. It was noted that the chloroform/methanol sonication method had higher overall extraction weights; however, both MS and NMR analysis indicated a higher amount of chlorophyll and other polar constituents present in the chloroform/methanol extract compared with the hexane extract. For certain algae strains, such as N. oculata, higher chlorophyll levels were observed that overwhelmed smaller (500 mg) SPE columns when used at extract weights over 50 mg.
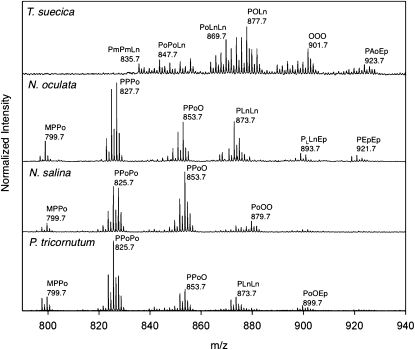
Using the optimized SPE procedure to remove residual polar components, we performed an analysis of the TAG profiles from four algal strains comparing both MALDI-TOF and ESI-LTQ-Orbitrap. This study represents the first analysis comparing the TAG profiles for several green marine microalgae using MALDI-TOF and ESI-Orbitrap (supplementary Table I). While oleaginous algae strains largely produced similar TAG populations, our study identified the unsaturation profiles and compositional differences observed among the triacylglycerols of these algae species. MALDI-TOF analysis is effective to compare the composition and patterns of unsaturation (Fig. 3). The identities of prominent TAG signals were confirmed by MS/MS (supplementary Table II). The observed triacylglycerols consisted primarily of unsaturated acyl side chains corresponding to 16 carbons and longer. P. tricornutum, N. oculata, and N. salina have more C16 chain fatty acids components and thus lower-weight TAG components. When comparing composition of diatom and green algae species, strong similarities were observed between the TAG profiles of P. tricornutum and N. salina, even though these two strains are phylogenetically distinct. However, in comparisons among green algae, T. suecica showed the highest degree of unsaturation and a greater quantity of longer acyl side chains. Also, T. suecica did not have triacylglycerols shorter than palmitic acid (C16:0), including a noticeable lack of myristic acid (C14:0). Tandem MS using both ESI-LTQ-Orbitrap and MALDI-TOF allowed us to correctly identify the specific fatty acid composition in TAG peaks (supplementary Table I). Some glycolipids, such as MGDG and DGDG, also eluted in the nonpolar fraction of SPE. Other lipids present, such as phosphatidylcholine, seen in the MALDI-TOF analysis of the crude extract, failed to be detected in either SPE fraction, which led us to believe that they were not eluting from the column under the conditions examined.
Fig. 3.
Comparison of the algae TAG footprints (m/z 750-1,000) in the MALDI-TOF mass spectra of nonpolar fraction of lipid extracts after SPE from four algae species: T. suecica, N. oculata, N. salina, and P. tricornutum. Intensity has been normalized to T. suecica. All TAG ions are sodiated adducts. Fatty acids: P = C16:0, Po = C16:1, PL = C16:3, Pm = C16:4, O = C18:1, Ln = C18:3, Ep = C20:5, and Ao = C20:4.
MALDI-TOF and ESI-LTQ-Orbitrap methods are particularly useful to identify and compare fatty acid side chains in the intact triacylglycerols (supplementary Table I). Lithium salts were primarily used in ESI analyses for comparison with sodium adducts produced in MALDI analyses. Initially, it was observed that ESI-Orbitrap produces ammonium adducts with lipids, which can prevent successful tandem MS. Thus, 100 mM LiOAc was added to the sample in microliter amounts to produce ions that were readily fragmented for tandem MS (41, 42). Our studies indicated that these cations increased the ionization ability of triacylglycerols; however, if the sample was not purified, salts increased the ionization of more polar molecular components in the crude samples. Most tandem MS was performed using ESI. Furthermore, tandem MS analysis is more time effective using ESI-Orbitrap due to the ability to optimize fragmentation in real time to produce the clearest MSn data needed.
On the basis of this study, MALDI-TOF is a preferred method for TAG analysis due to higher signal-to-noise ratio, ease of analysis, and the lack of carryover between samples. This method does not require a dedicated HPLC system, solvent pump, or specialized chromatography column. With ESI-LTQ-Orbitrap, residual triacylglycerols were detected for a significant time after use. Conversely, tandem MS analysis is more suited to the LTQ-Orbitrap because of the ability to adjust the analytical parameters, sample, and salt concentrations during analysis, as well as optimize the collision energy to produce the clearest MS2 data. Note that the MALDI-TOF response differed for each TAG analog, and thus, caution should be used when assigning relative amounts of the various triacylglycerols present.
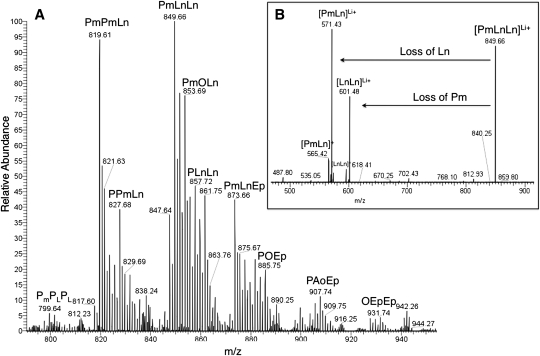
Whereas the different strains of algae produce similar TAG populations, the results showed some distinct differences among the triacylglycerols of the algae species examined, particularly with respect to the PUFA components. Algae species are known to possess PUFAs, although the extent to which they are present in triacylglycerols is less well documented in green saltwater microalgae (24, 35). The PUFA content of algal lipid extracts has been analyzed previously using derivative methods (43). Here, MALDI-TOF analysis identified C16, C18, and C20 fatty acid polyunsaturated components, reaching four degrees of unsaturation in C16, C18, and C20 fatty acids (Fig. 3 and supplementary Table I). The unsaturation levels of T. suecica were particularly notable, as the high levels of unsaturation observed were almost twice that of the other species. Analysis using ESI-LTQ-Orbitrap also confirmed the high level of unsaturation (Fig. 4A). Tandem MS confirmed the presence of PUFA (Fig. 4B). Previous analyses of P. tricornutum, T. suecica, and N. oculata lipid extracts have identified the 20:5 fatty acid components, but they have had variable detection of the 20:4 fatty acid component (36, 44, 45), which we observed in our studies. It is also notable that previous reports did not indicate the occurrence of the 16:3 or 16:4 fatty acid components, both of which were observed in our studies (36). It may be concluded that MALDI-TOF analysis can be used to identify TAG constituents, such as the C16:3, C16:4, and C18:4 components, that are not typically identified using GC/MS. This is either because these constituents occur in such small quantities, or because derivatization procedures required for GC/MS may allow or promote oxidation of these constituents. Although this method is useful to compare and identify constituents, it should not be used to assign relative amounts of the PUFA-containing triacylglycerols present because the MALDI-TOF response can differ for each TAG analog.
Fig. 4.
(A) Expansion of the m/z 750-1,000 region in the Orbitrap-ESI mass spectra for T. suecica with MS/MS verification. All species are lithiated adducts. (B) Inset represents tandem MS used to elucidate fatty acid composition of each TAG. Fatty Acids: P = C16:0, PL = C16:3, Pm = C16:4, O = C18:1, Ln = C18:3, Ep = C20:5, Ao = C20:4.
The method presented here was shown to be particularly useful to identify and contrast PUFA composition in the analysis of intact triacylglycerols to determine the ability of microalgae to produce a specific fatty acid composition. Both MS and 1H NMR spectroscopy results confirmed the ability to monitor the levels of unsaturation present in algae lipid extracts. Heavily polyunsaturated triacylglycerols are valuable precursors to lipid-based products, such as industrial lubricants and nutrition, whereas their use as fuel can be argued (5). Such PUFAs have reduced oxidative stability but superior operational temperature range, when it comes to biofuels. The desired fatty acid composition for biofuel purposes would consist of 8-16 carbons and one degree of unsaturation to promote oxidative stability and cold flow properties (46). The TAG profiles of most of the algae oils contained significant amounts of palmitic (C16:0) and oleic acid (C18:1), with the exception of T. suecica. These methods to determine the TAG profile are important for monitoring the compositional effect of various growth conditions directly on the algae oil profile, and they could also be useful to compare the effects of genetic modifications on metabolic pathways. Such analytical methods provide insight when endeavoring to achieve desired composition and produce viable quantities of important bioactive lipids and high-value, lipid-based chemical precursors.
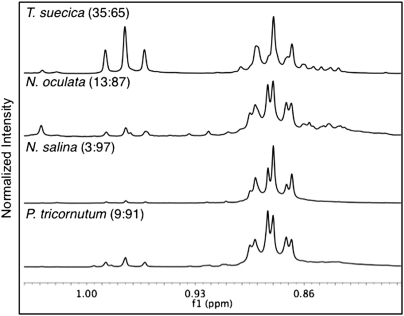
1H NMR analysis of both the initial and SPE-purified lipid extracts provides a complementary method to confirm the extent of unsaturation produced by different algae species for biofuel applications (Fig. 5 and supplementary Table VIII.). While NMR spectroscopy has been used to compare fatty acid profiles and identify unsaturated fatty acid components in various vegetable oils (47) and microalgae (32), this technique had not been applied previously to directly compare TAG composition among microalgae species and changes under various growth conditions. Two distinctive NMR signals can be used to compare the levels of unsaturation of algae TAG components: the signal at approximately 0.97 ppm represents the chemical shift of the methyl protons of ω3 PUFAs (i.e., C18:3) and the signal at 0.88 ppm represents the methyl protons of saturated and monounsaturated fatty acids. Comparing these diagnostic signals for TAG components confirmed that T. suecica exhibited a higher degree of unsaturation compared with microalgae, such as N. salina (supplementary Table VIII). Thus, NMR methods offer a valuable tool that complements MS analysis to identify PUFA components and assess the commercial applications of high-value, lipid-based chemicals (47).
Fig. 5.
Expansion of the 0.80-1.04 ppm region in the 1H NMR spectra comparing SPE-purified lipid extracts from four algae species: T. suecica, N. oculata, N. salina, and P. tricornutum. Region shows ratio of ω3-PUFA methyl protons (0.97 ppm) to other methyl protons (0.88 ppm). Intensity has been normalized. For full spectra, see supplementary Fig. V.
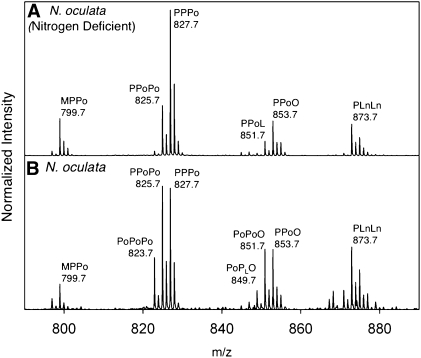
In addition to comparing different oleaginous algae strains, MALDI-TOF is effective in comparing the TAG profile of algae grown under nutrient deprivation. While nitrogen-deficient conditions are known to increase lipid production, there is limited information about the compositional changes that result from nutrient deprivation. MALDI-TOF had not been used previously to compare the TAG composition for algae grown under different conditions. Our MALDI-TOF analysis comparing the standard and nitrogen-deficient growth conditions showed TAG composition with an increase in C16 fatty acid components and also the shift to reduced levels of unsaturation in triacylglycerols (Fig. 6). 1H NMR analysis of SPE-purified nonpolar lipid extracts also showed a decreased level of PUFA-based triacylglycerols in N. oculata grown under nitrogen-deficient conditions (ratio in standard conditions = 13:87, and ratio in nitrogen-deficient conditions = 9:91; see supplementary Fig. VI) (48).
Fig. 6.
Expansion of the m/z 790-890 region in the MALDI-TOF mass spectra for lipid extracts from cultures of N. oculata comparing (A) nitrogen-deficient growth conditions with (B) standard growth conditions. Lipids were extracted using chloroform/methanol conditions.
In conclusion, combining a simple SPE step with MALDI-TOF MS provides a direct MS method to characterize intact nonpolar lipid samples from microalgae. SPE removes chlorophyll, making this method particularly useful for identifying microalgae TAG profiles with desirable fatty acid composition for biofuel purposes. The combination of MS and 1H NMR spectroscopy analysis provides the ability to monitor direct TAG compositional changes among species and growth conditions. Mass spectrometry confirmed that three algae, P. tricornutum, N. oculata, and N. salina, are suitable as biofuel feedstock, whereas T. suecica is more relevant for alternative lipid products where significant levels of PUFA are desired.
Supplementary Material
Acknowledgments
The authors thank Diana Wong and David Lam for their assistance with algae cultivation, algae harvesting, and helpful discussion, as well as Abel Silva, Skye Kelty, Jack Taylor, Kate Gibson, and the University of California Davis Young Scholars Program. The authors also acknowledge William Jewell for assistance with mass spectrometers.
Footnotes
Abbreviations:
- DGDG
- digalactosyl-diacylglycerol
- MGDG
- monogalactosyl-diacylglycerol
- SPE
- solid phase extraction
- TAG
- triacylglycerol
- TOF
- time of flight
This work was supported by Chevron Technology Ventures and the University of California. A.K.F. is recipient of a nontenured faculty award from 3M Corporation. L.A.A. is recipient of a graduate research fellowship from the National Science Foundation.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of seven figures and eight tables.
References
- 1.Chisti Y. 2007. Biodiesel from microalgae. Biotechnol. Adv. 25: 294–306 [DOI] [PubMed] [Google Scholar]
- 2.Hu Q., Sommerfeld M., Jarvis E., Ghirardi M., Posewitz M., Seibert M., Darzins A. 2008. Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J. 54: 621–639 [DOI] [PubMed] [Google Scholar]
- 3.Sheehan J., Dunahay T., Benemann J., Roessler P. 1998. A Look Back at the U.S. Department of Energy's Aquatic Species Program: Biodiesel from Algae. U. S. Department of Energy, National Renewable Energy Laboratory, Golden, Colorado [Google Scholar]
- 4.Mata T. M., Martins A. A., Caetano N. S. 2010. Microalgae for biodiesel production and other applications: a review. Renew. Sustain. Energy Rev. 14: 217–232 [Google Scholar]
- 5.Spolaore P., Joannis-Cassan C., Duran E., Isambert A. 2006. Commercial applications of microalgae. J. Biosci. Bioeng. 101: 87–96 [DOI] [PubMed] [Google Scholar]
- 6.Vieler A., Wilhelm C., Goss R., Süss R., Schiller J. 2007. The lipid composition of the unicellular green alga Chlamydomonas reinhardtii and the diatom Cyclotella meneghiniana investigated by MALDI-TOF MS and TLC. Chem. Phys. Lipids. 150: 143–155 [DOI] [PubMed] [Google Scholar]
- 7.Guschina I. A., Harwood J. L. 2006. Lipids and lipid metabolism in eukaryotic algae. Prog. Lipid Res. 45: 160–186 [DOI] [PubMed] [Google Scholar]
- 8.Khotimchenko S. V., Yakovleva I. M. 2005. Lipid composition of the red alga Tichocarpus crinitus exposed to different levels of photon irradiance. Phytochemistry. 66: 73–79 [DOI] [PubMed] [Google Scholar]
- 9.Ishida Y., Nakanishi O., Hirao S., Tsuge S., Urabe J., Sekino T., Nakanishi M., Kimoto T., Ohtani H. 2003. Direct analysis of lipids in single zooplankter individuals by matrix-assisted laser desorption/ionization mass spectrometry. Anal. Chem. 75: 4514–4518 [DOI] [PubMed] [Google Scholar]
- 10.Emerson B., Gidden J., Lay J. O., Durham B. 2010. A rapid separation technique for overcoming suppression of triacylglycerols by phosphatidylcholine using MALDI-TOF MS. J. Lipid Res. 51: 2428–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen W., Zhang C., Song L., Sommerfeld M., Hu Q. 2009. A high throughput Nile red method for quantitative measurement of neutral lipids in microalgae. J. Microbiol. Methods. 77: 41–47 [DOI] [PubMed] [Google Scholar]
- 12.Cheng Y-S., Zheng Y., VanderGheynst J. 2011. Rapid quantitative analysis of lipids using a colorimetric method in a microplate format. Lipids. 46: 95–103 [DOI] [PubMed] [Google Scholar]
- 13.Huerlimann R., de Nys R., Heimann K. 2010. Growth, lipid content, productivity, and fatty acid composition of tropical microalgae for scale-up production. Biotechnol. Bioeng. 107: 245–257 [DOI] [PubMed] [Google Scholar]
- 14.Sushchik N., Gladyshev M., Ivanova E., Kravchuk E. 2010. Seasonal distribution and fatty acid composition of littoral microalgae in the Yenisei River. J. Appl. Phycol. 22: 11–24 [Google Scholar]
- 15.Lepage G., Roy C. C. 1986. Direct transesterification of all classes of lipids in a one-step reaction. J. Lipid Res. 27: 114–120 [PubMed] [Google Scholar]
- 16.DeLuca S. J., Sarver E. W., Voorhees K. J. 1992. Direct analysis of bacterial glycerides by Curie-point pyrolysis–mass spectrometry. J. Anal. Appl. Pyrolysis. 23: 1–14 [DOI] [PubMed] [Google Scholar]
- 17.Miao X., Wu Q. 2004. High yield bio-oil production from fast pyrolysis by metabolic controlling of Chlorella protothecoides. J. Biotechnol. 110: 85–93 [DOI] [PubMed] [Google Scholar]
- 18.Fuchs B., Süss R., Schiller J. 2010. An update of MALDI-TOF mass spectrometry in lipid research. Prog. Lipid Res. 49: 450–475 [DOI] [PubMed] [Google Scholar]
- 19.Han X., Gross R. W. 2001. Quantitative analysis and molecular species fingerprinting of triacylglyceride molecular species directly from lipid extracts of biological samples by electrospray ionization tandem mass spectrometry. Anal. Biochem. 295: 88–100 [DOI] [PubMed] [Google Scholar]
- 20.Lay J. O., Liyanage R., Durham B., Brooks J. 2006. Rapid characterization of edible oils by direct matrix-assisted laser desorption/ionization time-of-flight mass spectrometry analysis using triacylglycerols. Rapid Commun. Mass Spectrom. 20: 952–958 [DOI] [PubMed] [Google Scholar]
- 21.Asbury G. R., Al-Saad K., Siems W. F., Hannan R. M., Hill H. H., Jr 1999. Analysis of triacylglycerols and whole oils by matrix-assisted laser desorption/ionization time of flight mass spectrometry. J. Am. Soc. Mass Spectrom. 10: 983–991 [Google Scholar]
- 22.Murphy R. C., Fiedler J., Hevko J. 2001. Analysis of nonvolatile lipids by mass spectrometry. Chem. Rev. 101: 479–526 [DOI] [PubMed] [Google Scholar]
- 23.Schiller J., Süss R., Arnhold J., Fuchs B., Lessig J., Müller M., Petkovic M., Spalteholz H., Zschörnig O., Arnold K. 2004. Matrix-assisted laser desorption and ionization time-of-flight (MALDI-TOF) mass spectrometry in lipid and phospholipid research. Prog. Lipid Res. 43: 449–488 [DOI] [PubMed] [Google Scholar]
- 24.Yu E. T., Zendejas F. J., Lane P. D., Gaucher S., Simmons B. A., Lane T. W. 2009. Triacylglycerol accumulation and profiling in the model diatoms Thalassiosira pseudonana and Phaeodactylum tricornutum (Baccilariophyceae) during starvation. J. Appl. Phycol. 21: 669–681 [Google Scholar]
- 25.Hara A., Radin N. S. 1978. Lipid extraction of tissues with a low-toxicity solvent. Anal. Biochem. 90: 420–426 [DOI] [PubMed] [Google Scholar]
- 26.Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917 [DOI] [PubMed] [Google Scholar]
- 27.Cooney M., Young G., Nagle N. 2009. Extraction of bio-oils from microalgae. Sep. Purif. Rev. 38: 291–325 [Google Scholar]
- 28.Kim H. Y., Salem, Jr N. 1990. Separation of lipid classes by solid phase extraction. J. Lipid Res. 31: 2285–2289 [PubMed] [Google Scholar]
- 29.Pié A., Giner A. 1966. Solvents for thin layer chromatography of blood serum lipids. Nature. 212: 402–403 [DOI] [PubMed] [Google Scholar]
- 30.Yongmanitchai W., Ward O. P. 1992. Separation of lipid classes from Phaeodactylum tricornutum using silica cartridges. Phytochemistry. 31: 3405–3408 [Google Scholar]
- 31.Nicola A. J., Gusev A. I., Proctor A., Jackson E. K., Hercules D. M. 1995. Application of the fast-evaporation sample preparation method for improving quantification of angiotensin II by matrix-assisted laser desorption/ionization. Rapid Commun. Mass Spectrom. 9: 1164–1171 [DOI] [PubMed] [Google Scholar]
- 32.Pollesello P., Toffanin R., Murano E., Paoletti S., Rizzo R., Kvam B. 1992. Lipid extracts from different algal species: 1H and 13C-NMR spectroscopic studies as a new tool to screen differences in the composition of fatty acids, sterols and carotenoids. J. Appl. Phycol. 4: 315–322 [Google Scholar]
- 33.Arao T., Kawaguchi A., Yamada M. 1987. Positional distribution of fatty acids in lipids of the marine diatom Phaeodactylum tricornutum. Phytochemistry. 26: 2573–2576 [Google Scholar]
- 34.Hu H., Gao K. 2003. Optimization of growth and fatty acid composition of a unicellular marine picoplankton, Nannochloropsis sp. , with enriched carbon sources. Biotechnol. Lett. 25: 421–425 [DOI] [PubMed] [Google Scholar]
- 35.Dunstan G., Volkman J., Barrett S., Garland C. 1993. Changes in the lipid composition and maximisation of the polyunsaturated fatty acid content of three microalgae grown in mass culture. J. Appl. Phycol. 5: 71–83 [Google Scholar]
- 36.Otero A., Fábregas J. 1997. Changes in the nutrient composition of Tetraselmis suecica cultured semicontinuously with different nutrient concentrations and renewal rates. Aquaculture. 159: 111–123 [Google Scholar]
- 37.Servel M-O., Claire C., Derrien A., Coiffard L., De Roeck-Holtzhauer Y. 1994. Fatty acid composition of some marine microalgae. Phytochemistry. 36: 691–693 [Google Scholar]
- 38.Suzuki T., Midonoya H., Shioi Y. 2009. Analysis of chlorophylls and their derivatives by matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry. Anal. Biochem. 390: 57–62 [DOI] [PubMed] [Google Scholar]
- 39.Van Breemen R. B., Canjura F. L., Schwartz S. J. 1991. Identification of chlorophyll derivatives by mass spectrometry. J. Agric. Food Chem. 39: 1452–1456 [Google Scholar]
- 40.Pinkart H. C., Devereux R., Chapman P. J. 1998. Rapid separation of microbial lipids using solid phase extraction columns. J. Microbiol. Methods. 34: 9–15 [Google Scholar]
- 41.Hsu F. F., Bohrer A., Turk J. 1998. Formation of lithiated adducts of glycerophosphocholine lipids facilitates their identification by electrospray ionization tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 9: 516–526 [DOI] [PubMed] [Google Scholar]
- 42.Hsu F-F., Turk J. 2010. Electrospray ionization multiple-stage linear ion-trap mass spectrometry for structural elucidation of triacylglycerols: assignment of fatty acyl groups on the glycerol backbone and location of double bonds. J. Am. Soc. Mass Spectrom. 21: 657–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robles M. A., Grima E. Molina, Giménez A., Ibañez González M. J. 1998. Downstream processing of algal polyunsaturated fatty acids. Biotechnol. Adv. 16: 517–580 [DOI] [PubMed] [Google Scholar]
- 44.Arao T., Sakaki T., Yamada M. 1994. Biosynthesis of polyunsaturated lipids in the diatom, Phaeodactylum tricornutum. Phytochemistry. 36: 629–635 [Google Scholar]
- 45.Yongmanltchal W., Ward O. 1992. Growth and eicosapentaenoic acid production by Phaeodactylum tricornutum in batch and continuous culture systems. J. Am. Oil Chem. Soc. 69: 584–590 [Google Scholar]
- 46.Knothe G. 2005. Dependence of biodiesel fuel properties on the structure of fatty acid alkyl esters. Fuel Process. Technol. 86: 1059–1070 [Google Scholar]
- 47.Annarao S., Sidhu O. P., Roy R., Tuli R., Khetrapal C. L. 2008. Lipid profiling of developing Jatropha curcas L. seeds using 1H NMR spectroscopy. Bioresour. Technol. 99: 9032–9035 [DOI] [PubMed] [Google Scholar]
- 48.Hodgson P., Henderson R., Sargent J., Leftley J. 1991. Patterns of variation in the lipid class and fatty acid composition of Nannochloropsis oculata (Eustigmatophyceae) during batch culture. I. The growth cycle. J. Appl. Phycol. 3: 169–181 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.