Abstract
To elucidate the lutein biosynthesis pathway in the lutein-producing alga, Chlorella protothecoides CS-41, the ζ-carotene desaturase gene (zds) was isolated from Chlorella protothecoides using the approach of rapid amplification of cDNA ends. The full-length cDNA sequence was 2031 bp and contained 1755 bp putative open reading frame which encodes a 584 amino acid deduced polypeptide whose computed molecular weight was 63.7 kDa. Sequence homology research indicated that the nucleotide and putative protein had sequence identities of 72.5% and 69.5% with those of the green alga Chlamydomonas reinhardtii, respectively. Phylogenetic analysis demonstrated that the ZDS from C. protothecoides CS-41 had a closer relationship with those of chlorophyta and higher plants than with those of other species. In addition, we also found that the zds gene expression was upregulated in response to light.
1. Introduction
Green algae in the genus Chlorella are widely distributed in freshwater environments. They are typically small, nonmotile, unicellular organisms with a single chloroplast and a rigid cell wall [1]. Chlorella protothecoides CS-41 is a strain that produces a high yield of lutein, which triggered our interest to investigate it.
Carotenoids are widely distributed in the world. They are a structurally diverse class of isoprenoids which are synthesized by all photosynthetic organisms and many nonphotosynthetic bacteria and fungi. They play a critical role in human nutrition and health [2]. In addition, they protect the photosynthetic apparatus in plants [3] from photo-oxidation. So far, more than 700 types of carotenoids have been found from natural sources [4]. For their extensive commercial and industrial uses, carotenoids especially lutein and astaxanthin are in high demand around the world.
Currently, for lutein production, the most widely used source is French marigold (Tagetes patula). However, mass plantation of marigold occupies a large land area and is easily influenced by season and climate. Previous research in our laboratory showed that Chlorella protothecoides CS-41 cultivated heterotrophically can produce a considerable amount of lutein [5]. By optimizing their cultivation conditions and the composition of the medium, one can improve the production of lutein for a higher yield [6, 7]. However, there are no reports about enhancing lutein production by genetic modification, although the genetic engineering technology has become more and more popular in the area of molecular biology. Therefore, the purpose of this research was to characterize the genes involved in lutein biosynthesis, with the expectation of increasing the lutein content in Chlorella protothecoides CS-41 by genetic engineering in the near future.
The carotenoid biosynthesis pathway in Chlorella protothecoides CS-41 is similar to that of higher plants, which is a complicated secondary metabolic system [8, 9] and is highly conserved in all carotenogenic organisms [10] (Figure 1). The process starts with the condensation of two geranylgeranyl diphosphate molecules to form phytoene. Phytoene is converted into colored ζ-carotene in a two-step desaturation reaction by phytoene desaturase. Further desaturation and cyclization reactions lead to the biosynthesis of α- and β-carotene, which are subsequently converted into xanthophylls. ZDS is a key enzyme in carotenogenesis serving downstream of ζ-carotene in the carotenoid biosynthetic pathway. In order to have a fuller understanding of the carotenogenic pathway of Chlorella protothecoides, we decided to isolate the cDNA of the zds gene and characterize this essential enzyme as well as other key enzymes such as PSY, PDS, and LCYE using a series of bioinformatics tools and functional analysis methods.
Figure 1.
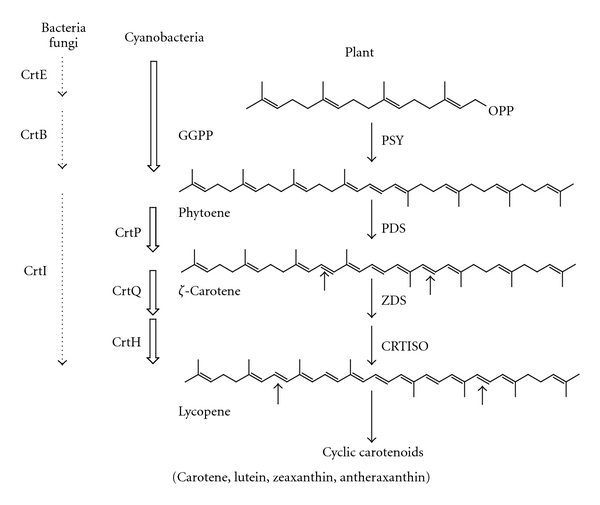
Putative biosynthesis pathway of carotenoids. The names of enzymes are abbreviated as follows: CrtB, bacterial phytoene synthase; CrtI, bacterial phytoene desaturase; PSY, phytoene synthase; PDS/CrtP, phytoene desaturase; ZDS/CrtQ, ζ-carotene desaturase; CRTISO/CrtH, carotenoid isomerase.
2. Materials and Methods
2.1. Microbial Strains and Culture Conditions
The microalgal strain used in this study was Chlorella protothecoides CS-41, obtained from the CSIRO Marine Laboratories (Hobart, Australia). They were grown in modified Basal medium [11], which contained 10 g L−1 glucose, at 28°C and shaken at 180 rpm. The cultures were later collected at the log or late log phase of growth.
Escherichia coli strain DH5α was used as the host for the multiplication of plasmids.
2.2. Genomic DNA and RNA Isolation
Genomic DNA was extracted using a modified cetyltrimethylammonium bromide (CTAB) method [12]. The total RNA was isolated from 15 mL of C. protothecoides CS-41 cells grown to the late log phase using RNAprep Plant Kit (Tiangen Biotech, China). DNA contamination was removed with DNaseI (Tiangen Biotech, China) according to the manufacturer's instruction. The quantity of the total DNA and RNA was determined using a Nanodrop1000 spectrophotometer (Thermo scientific, USA).
2.3. Cloning of Full-Length zds cDNA and Genomic DNA
Degenerate primers were designed for the amplification of the partial cDNA from C. protothecoides CS-41. The primers were derived from the highly conserved nucleotide and amino acid sequences reported for the zds gene from four kinds of algae: Volvox carteri (XM_002954182), Chlorella NC_64A (gm_36_00005), Chlamydomonas reinhardtii (XM_001700734), and Dunaliella salina (HM754265); and four kinds of higher plants: Arabidopsis thaliana (NM_202493), Solanum lycopersicum (EF650012), Carica papaya (FJ812088), and Zea mays (EU974617).
One-step reverse transcriptase-polymerase chain reaction (RT-PCR) with the primers ZDSF and ZDSR (Table 1) was performed to amplify a portion of the zds cDNA. RT-PCR was performed using an RNA PCR Kit (AMV) ver.3.0 (TaKaRa, Dalian, China) according to the manufacturer's protocol for cDNA synthesis, followed by PCR amplification (1 cycle of 94°C, 5 min; 20 cycles of 94°C, 30 s, 65–50°C, 30 s (decrease 0.5°C per cycle), and 72°C, 2 min; 5 cycles of 94°C, 30 s, 44°C, 30 s, and 72°C, 2 min; 1 cycle of 72°C, 10 min). The PCR product was purified with an AxyPrep PCR column (Axygen Biosciences, USA) to remove excess primers. Then the product was ligated into pMD-18T vector (TaKaRa, Dalian, China) according to the manufacturer's instructions and chemically transformed into E. coli strain DH5α competent cells. Transformants were selected on LB-agar plates containing 100 mg/L ampicillin. Positive clones were selected and sequenced in both directions using M13 sequencing primers.
Table 1.
PCR primers and targeted fragments of the zds gene in C. protothecoides CS-41.
| Primers | Primer sequence (5′-3′) | Product size (bp) |
|---|---|---|
| Core fragment | 413 | |
| ZDSF | TCTTTGGCTGCTATTTCAACCTG | |
| ZDSR | ATCGTCACAGTTCAGGAAGC | |
| 5′-RACE | 691 | |
| ZRO1 | TTCAGGAAGCCCAGAGCGTAG | |
| ZRI1 | TTGCCGCCGAGGAAGAAGC | |
| 3′-RACE | 1390 | |
| ZFO1 | ACCTGCTGCTCAAGGACCAC | |
| ZFI1 | AGACGTGCGTGAGCTGGACTTC | |
| ZFI2 | CCGATTGCTGACTACATCACTGCCA | |
| ZDS-ORF | 1755 for cDNA; 4587 for DNA | |
| ZDSF1 | ATGCAGGCATCAGCGGTGGCC | |
| ZDSR1 | TCACGCAGCCACCGCCTGCT | |
| qRT-PCR | 171 | |
| ZDSRTF | TGGGCACCTCTCCCGTTG | |
| ZDSRTR | AGTTCAGGAAGCCCAGAGCG | |
| Chlorella β-actin | 166 | |
| c-actinF | TGATGAAGATCCTCACAGAGC | |
| c-actinR | TCTGGCAGTTCAAAGGTCTT |
Sets of specific primers were synthesized based on the sequence of putative insert for 5′ and 3′ rapid amplification of cDNA ends (RACE) [13]. ZRO1 and ZRI1 were used for 5′ RACE of zds gene, ZFO1 and ZFI1, ZFO1 and ZFI2 (Table 1) for 3′RACE of zds gene. RACE was performed using the 5′-Full RACE Kit and 3′-Full RACE Core Set Ver.2.0 (TaKaRa, Dalian, China) according to the manufacturer's protocol. The RACE products were gel purified and sequenced as previously described. One pair of specific primer, ZDSF1 and ZDSR1 (Table 1), was designed from the sequences of the 5′ and 3′RACE fragments to amplify a full-length zds cDNA and its corresponding gene.
2.4. Sequence Alignment and Phylogenetic Analysis
Comparative and bioinformatics analysis of the nucleotide sequences and deduced amino acid sequences were carried out online at the websites (http://www.ncbi.nlm.nih.gov/) and (http://cn.expasy.org/). The nucleotide sequence, deduced amino acid sequence, and open reading frame [14] were analyzed, and the sequence comparison was conducted through a database search using BLAST programs. The phylogenetic analysis of zds gene from other plant species was aligned with Clustal X program version 1.83 using default parameters [15] and manual adjustments when necessary. Phylogenetic trees were constructed using MEGA (Molecular Evolutionary Genetics Analysis) version 4.0 [16]. The neighbor-joining (NJ) [17] method was used to construct the tree. In the NJ method, the P distance was used to analyze the amino acid sequences. A total of 1000 repetitions were performed using the bootstrap method to determine the reliability of each node of the tree.
2.5. zds Expression Analysis in Response to Light
To analyze the light regulation pattern of the zds gene in C. protothecoides, the algal cells from late log phase were cultivated in the dark for more than 2 days, then collected by centrifugation at 5 500 g for 15 min under dark conditions. The pellet was resuspended in fresh medium without glucose, and then subjected to light treatment under a light intensity of 120 μmol m−2 s−1 for different induction times (0 h, 0.5 h, 1 h, 2 h, and 4 h). Each treatment was carried out in three replicates.
QPCR experiments were performed on an iQ Cycler (Bio-Rad, Watford, UK) with the specific primers (Table 1) and the SYBR ExScript RT-PCR kit (TaKaRa, Dalian, China). For each reaction, 1 μL of cDNA was added as template, 12.5 μL of 2 × SYBR Premix Ex Taq, 0.25 μM of forward primer, and 0.25 μM of reverse primer, and nuclease-free water were added to a final volume of 25 μL. The thermal cycling conditions were 3 min at 95°C for activation of the hot start Taq polymerase and 40 cycles of 95°C, 15 s, 60°C, 15 s, and 72°C, 30 s. Melting curve analysis following each qPCR was performed to assess product specificity. Each qPCR measurement was carried out independently at least three times, and the mean value was used for quantification. The 2−ΔΔCT method was used to analyze the relative changes in gene expression, and the expression of β-actin gene was used as a normalized control, and the expression of the untreated samples was used as a negative control.
3. Results and Discussion
3.1. Cloning of C. protothecoides CS-41 zds Gene
Carotenoids comprise a diverse group of multifunctional, taxonomically widespread, and biotechnologically important pigments. Their biosynthetic pathway is a model system for understanding the evolution of secondary metabolism. ZDS is an essential carotenogenic enzyme that carries out the conversion from ζ-carotene to lycopene in eukaryotes. So far, nuclear genome sequences of several eukaryotic algae, including C. reinhardtii [18, 19] and Chlorella NC_64A [1, 20] have been completed, but the characteristics of ZDS in these eukaryotic algae have not yet been released by the authors. As a result of the incomplete molecular investigation of carotenogenesis of eukaryotic algae, the carotenogenic enzymes of C. protothecoides CS-41 have not been isolated and characterized thoroughly, and the carotenogenic pathway of C. protothecoides CS-41 is still unclear. Nonetheless, several DNA sequences of carotenogenic enzymes of C. protothecoides CS-41 have been determined, such as the psy, pds, and lcye genes. In this study, the cDNA of zds was isolated using the RACE-PCR approach.
TD-PCR with primers ZDSF and ZDSR (Table 1) generated a predicted 413-bp fragment (Figure 2, lane 1). BLAST analysis showed that the nucleotide sequence of this fragment shared about 76% identity with that of C. reinhardtii, which suggested that the sequence of the fragment was derived from a putative ζ-carotene desaturase.
Figure 2.
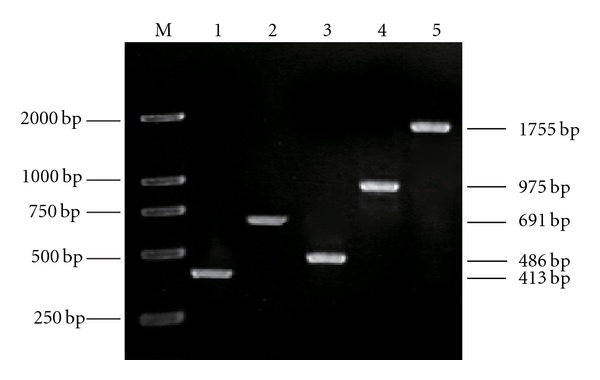
The PCR products of the zds gene of C. protothecoides CS-41. Lane 1: The core fragment of zds; lane 2: 5′-RACE fragment of zds; lanes 3 and 4: 3′-RACE fragments of zds; lane 5: the ORF fragment of zds; M: DL 2000.
With this sequence information, specific primers were designed for 5′ and 3′RACE of the related gene. A 691-bp fragment (Figure 2, lane 2) was generated with the primers ZRO1/ZRI1; a 486-bp fragment (Figure 2, lane 3) generated at the first cycle of 3′RACE with the primers ZFO1/ZFI1; a 975-bp fragment (Figure 2, lane 4) at the second cycle of 3′RACE with ZFO1/ZFI2. They were confirmed by sequencing to be the 5′ and 3′ regions of a phytoene desaturase gene, respectively. RT-PCR with the primer pair, ZDSF1/ZDSR1, generated a 1755-bp fragment (Figure 2, lane 5), which was identified as a full-length zds cDNA (submitted to GenBank, accession no. GU269622).
To characterize the corresponding gene of the zds cDNA, genomic PCR was performed. A 4587-bp fragment (submitted to GenBank, accession No. GU269623) was generated and sequenced. Analysis of the obtained nucleotide sequence revealed that the product was the zds gene. After analysis using online software (http://www.ncbi.nlm.nih.gov/IEB/Research/Ostell/Spidey/), this gene was found to consist of fourteen exons and thirteen introns (Figure 3).
Figure 3.
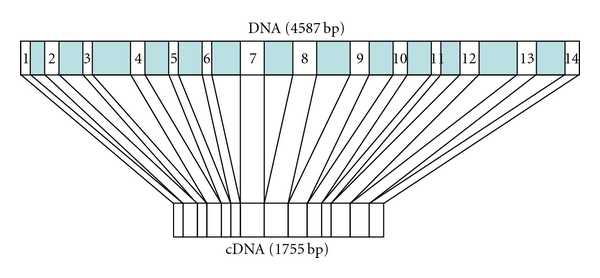
Diagram of exons and introns of zds gene in C. protothecoides CS-41. Ten exons were (1) 1st bp to 112nd bp; (2) 217th bp to 332nd bp; (3) 509th bp to 586th bp; (4) 893rd bp to 1001st bp; (5) 1194th bp to 1285th bp; (6) 1500th bp to 1598th bp; (7) 1812nd bp to 1995th bp; (8) 2243rd bp to 2438th bp; (9) 2707th bp to 2851st bp; (10) 3070th bp to 3171st bp; (11) 3366th bp to 3427th bp; (12) 3589th bp to 3748th bp; (13) 4060th bp to 4238th bp; and (14) 4467th bp to 4587th bp.
3.2. Characterization of the Deduced ZDS Protein
By using the Compute pI/Mw tool at the website http://web.expasy.org/compute_pi/, the open reading frame of zds cDNA encodes a protein of 584 amino acids, which has a calculated molecular mass of 63.67 kDa and an isoelectric point of 8.68. The protein contained 64 strongly basic amino acids (Lys and Arg), 58 strongly acidic amino acids (Asp and Glu), 219 hydrophobic amino acids (Ala, Ile, Leu, Phe, Trp and Val), and 145 polar amino acids (Asn, Cys, Gln, Ser, Thr and Tyr).
A sequence homology search revealed that the putative protein had high homology with ZDS sequences of other Chlorella species, such as 64.8% identities and 69.1% positives with that from Chlorella NC_64A. ZDS was also highly similar to that from C. reinhardtii (69.5% identities, 75.4% positives) and D. salina (57.4% identities, 67.0% positives), further confirming that ZDS belongs to the algal ZDS family. The deduced ZDS similar to the PDS protein contained a conserved dinucleotide-binding site (GXGX2GX3AX2LX3GX6EX5GG) at the N-terminus and a carotenoid-binding domain (AGDYTKQRYLASMEGATFSG) at the C-terminus [21] (Figure 4).
Figure 4.
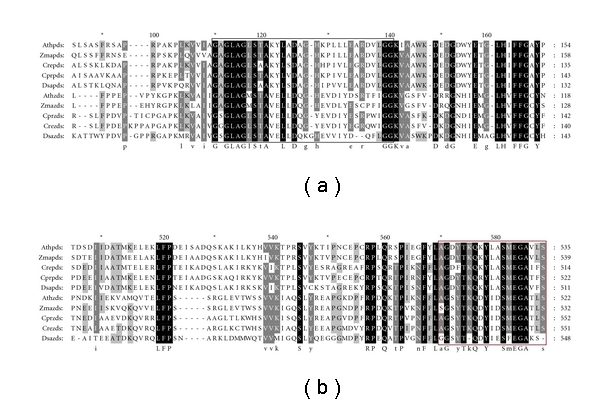
Sequence alignment of deduced PDS and ZDS with those from other organisms. Species designations are A. thaliana (Ath), Z. mays (Zma), C. reinhardtii (Cre), C. protothecoides (Cpr), and D. salina (Dsa). Amino acids in black boxes indicate identical residues, while gray boxes indicate conserved residues. The blank frame shows the consensus motif of dinucleotide-binding domain. The red frame indicates the postulated carotenoid-binding domain.
BLASTP analysis suggested that this protein contains a FAD binding domain belonging to the Pyridine nucleotide-disulphide oxidoreductase superfamily that is involved in oxidative metabolism of a variety of hydrocarbons [21]. Prediction of a transmembrane region was performed using the TMHMM server version 2.0, which suggested that the C. protothecoides ZDS was not a potential transmembrane protein. The SignalP-HMM analysis results suggested that there is a signal peptide in this polypeptide with the maximum cleavage site probability of 0.307 between positions 16 and 17 (Figure 5), which implies that it belongs to the outside protein. The function of this protein in C. protothecoides CS-41 needs further investigation.
Figure 5.
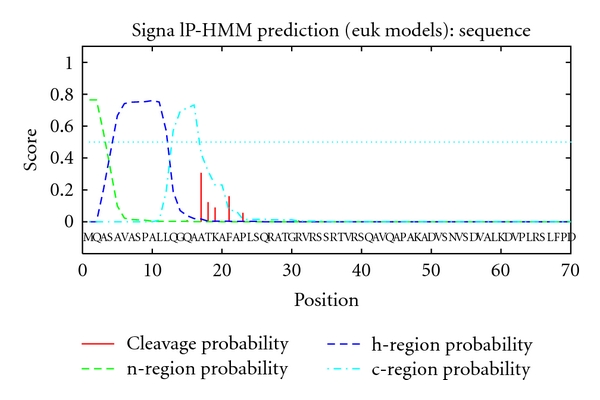
Analysis of the signal peptide of the ZDS from C. protothecoides CS-41.
3.3. Molecular Evolution Analysis
The zds gene was first cloned from C. protothecoides CS-41, prompting an investigation of their evolutionary position in the phylogenetic tree of various genes. Using MEGA 4.0, phylogenetic analysis of 20 ZDS protein sequences demonstrated that four clusters were displayed on the phylogenetic tree as follows: higher plants, chlorophyta, cyanobacteria, and bacteria. As displayed in the phylogenetic tree, the ZDS from C. protothecoides had a closer relationship with those of chlorophyta and higher plants than with those of cyanobacteria (Figure 6). This suggests that ZDS was derived from a common ancestor gene from bacteria via cyanobacteria to plants. This is consistent with the previous conclusion from the phylogenetic analysis that PSY and PDS from C. protothecoides are more closely related to those from other algae and higher plants.
Figure 6.
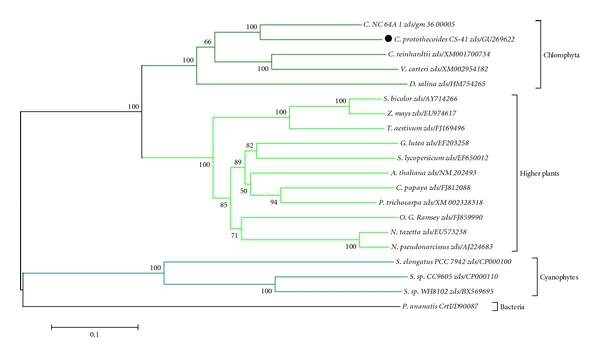
Phylogenic tree of the zds sequences from various species. The phylogeny was derived using Neighbor-joining analysis. Bootstrap values >50% are indicated near the base of each branch (1000 replicates). Horizontal branch lengths represent relative evolutionary distances with the scale bar corresponding to 0.1 amino acid substitutions per site.
3.4. Gene Expression Response to Light
Light, as a particularly important environmental factor for all photosynthetic organisms, is essential for photosynthesis, and also involved in the regulation of many aspects of plant growth and development [22]. Carotenoids are ubiquitous and essential components of photosynthetic tissues in unicellular microalgae because they participate in the light harvesting process and prevent photooxidative damage of the photosynthetic apparatus [23]. Hence, investigating gene expression in the biosynthetic pathway of carotenoids is very important. As a key enzyme in the pathway of carotenoids, the transcription level of the zds gene was investigated after treatment under light by using real-time PCR. The result showed that the expression of zds gene increased to 5.3 times than that in the dark, which indicated that the expression of zds gene was upregulated in response to light (Figure 7). In addition to zds gene, we also investigated the expression of the psy, pds, and lcye genes in the unicellular green alga C. protothecoides (data not shown). Our results revealed that all of these genes in the biosynthesis pathway showed upregulation in response to light.
Figure 7.
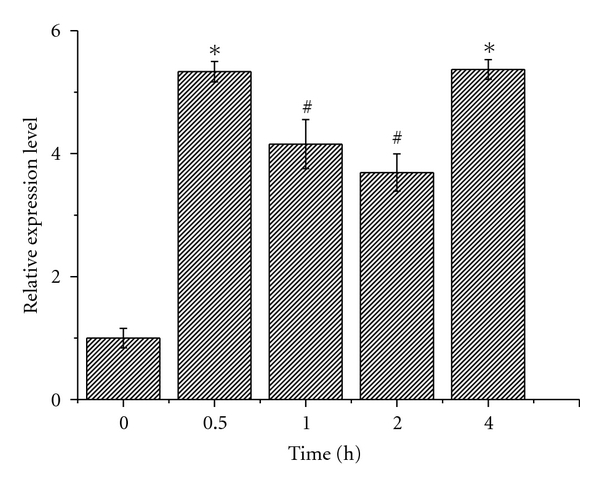
Gene expression levels determined from cells exposed to light for different lengths of time in C. protothecoides CS-41. Data (mean + SEM) are combined from three independent experiments. *indicated that gene expression levels were highly significantly different from that at 0 h (P < 0.01); #showed that gene expression levels were significantly different from that at 0 h (P < 0.05).
Since metabolically engineered microorganisms [24] and transgenic plants [25–28] are widely used in carotenoid production and carotenoid breeding to meet the high global demand, large carotenogenic gene pool should be provided first from the targeted engineered microorganisms for carotenoid production. The carotenoid biosynthesis pathway in higher plants and C. reinhardtii has been described in great detail using genetic, biochemical, and molecular data; however, this is the first research on the carotenoid pathway of a lutein-producing microalga, Chlorella protothecoides CS-41. Once these genes of key enzymes have been successfully isolated and characterized including the zds gene, it will lead to the elucidation of the carotenogenic pathway of C. protothecoides as well as the evolution of secondary metabolism.
Acknowledgments
This work was supported by National Natural Science Foundation of China (Grant nos. 31000779 and U1031003) and National 863 program of China (Grant no. 2006AA02Z226), and Shanghai Jiao Tong University Innovation Fund for Postgraduates. The authors also thank Professor James T. Staley at University of Washington for the critical review of this paper.
References
- 1.Eckardt NA. The Chlorella genome: big surprises from a small package. Plant Cell. 2010;22(9):p. 2924. doi: 10.1105/tpc.110.220911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krinsky NI, Johnson EJ. Carotenoid actions and their relation to health and disease. Molecular Aspects of Medicine. 2005;26(6):459–516. doi: 10.1016/j.mam.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Hirschberg J. Carotenoid biosynthesis in flowering plants. Current Opinion in Plant Biology. 2001;4(3):210–218. doi: 10.1016/s1369-5266(00)00163-1. [DOI] [PubMed] [Google Scholar]
- 4.Ladislav F, Vere P, Karel S, Karel V. Reliability of carotenoid analyses: a review. Current Analytical Chemistry. 2005;1(1):93–102. [Google Scholar]
- 5.Shi XM, Jiang Y, Chen F. High-yield production of lutein by the green microalga Chlorella protothecoides in heterotrophic fed-batch culture. Biotechnology Progress. 2002;18(4):723–727. doi: 10.1021/bp0101987. [DOI] [PubMed] [Google Scholar]
- 6.Shi XM, Liu HJ, Zhang XW, Chen F. Production of biomass and lutein by Chlorella protothecoides at various glucose concentrations in heterotrophic cultures. Process Biochemistry. 1999;34(4):341–347. [Google Scholar]
- 7.Shi X, Wu Z, Chen F. Kinetic modeling of lutein production by heterotrophic Chlorella at various pH and temperatures. Molecular Nutrition and Food Research. 2006;50(8):763–768. doi: 10.1002/mnfr.200600037. [DOI] [PubMed] [Google Scholar]
- 8.Liang C, Zhao F, Wei W, Wen Z, Qin S. Carotenoid biosynthesis in cyanobacteria: structural and evolutionary scenarios based on comparative genomics. International Journal of Biological Sciences. 2006;1(4):197–207. doi: 10.7150/ijbs.2.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang F, Jiang JG, Chen Q. Progress on molecular breeding and metabolic engineering of biosynthesis pathways of C(30), C(35), C(40), C(45), C(50) carotenoids. Biotechnology Advances. 2007;25(3):211–222. doi: 10.1016/j.biotechadv.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 10.Sieiro C, Poza M, de Miguel T, Villa TG. Genetic basis of microbial carotenogenesis. International Microbiology. 2003;6(1):11–16. doi: 10.1007/s10123-003-0097-0. [DOI] [PubMed] [Google Scholar]
- 11.Shi XM, Chen F, Yuan JP, Chen H. Heterotrophic production of lutein by selected Chlorella strains. Journal of Applied Phycology. 1997;9(5):445–450. [Google Scholar]
- 12.Stewart CN, Jr., Via LE. A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. BioTechniques. 1993;14(5):748–750. [PubMed] [Google Scholar]
- 13.Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proceedings of the National Academy of Sciences of the United States of America. 1988;85(23):8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peers G, Truong TB, Ostendorf E, et al. An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature. 2009;462(7272):518–521. doi: 10.1038/nature08587. [DOI] [PubMed] [Google Scholar]
- 15.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 17.Zhang W, Sun Z. Random local neighbor joining: a new method for reconstructing phylogenetic trees. Molecular Phylogenetics and Evolution. 2008;47(1):117–128. doi: 10.1016/j.ympev.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 18.Wheeler GL, Miranda-Saavedra D, Barton GJ. Genome analysis of the unicellular green alga Chlamydomonas reinhardtii indicates an ancient evolutionary origin for key pattern recognition and cell-signaling protein families. Genetics. 2008;179(1):193–197. doi: 10.1534/genetics.107.085936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godman J, Balk J. Genome analysis of Chlamydomonas reinhardtii reveals the existence of multiple, compartmentalized iron-sulfur protein assembly machineries of different evolutionary origins. Genetics. 2008;179(1):59–68. doi: 10.1534/genetics.107.086033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blanc G, Duncan G, Agarkova I, et al. The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell. 2010;22(9):2943–2955. doi: 10.1105/tpc.110.076406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cunningham FX, Jr., Sun Z, Chamovitz D, Hirschberg J, Gantt E. Molecular structure and enzymatic function of lycopene cyclase from the cyanobacterium Synechococcus sp strain PCC7942. Plant Cell. 1994;6(8):1107–1121. doi: 10.1105/tpc.6.8.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neff MM, Fankhauser C, Chory J. Eight: an indicator of time and place. Genes and Development. 2000;14(3):257–271. [PubMed] [Google Scholar]
- 23.Frank HA, Cogdell RJ. Carotenoids in photosynthesis. Photochemistry and Photobiology. 1996;63(3):257–264. doi: 10.1111/j.1751-1097.1996.tb03022.x. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt-Dannert C, Umeno D, Arnold FH. Molecular breeding of carotenoid biosynthetic pathways. Nature Biotechnology. 2000;18(7):750–753. doi: 10.1038/77319. [DOI] [PubMed] [Google Scholar]
- 25.Paine JA, Shipton CA, Chaggar S, et al. Improving the nutritional value of golden rice through increased pro-vitamin A content. Nature Biotechnology. 2005;23(4):482–487. doi: 10.1038/nbt1082. [DOI] [PubMed] [Google Scholar]
- 26.Aluru M, Xu Y, Guo R, et al. Generation of transgenic maize with enhanced provitamin A content. Journal of Experimental Botany. 2008;59(13):3551–3562. doi: 10.1093/jxb/ern212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bai C, Twyman RM, Farré G, et al. A golden era-pro-vitamin A enhancement in diverse crops. In Vitro Cellular and Developmental Biology—Plant. 2011;47(2):205–221. [Google Scholar]
- 28.Ducreux LJM, Morris WL, Hedley PE, et al. Metabolic engineering of high carotenoid potato tubers containing enhanced levels of β-carotene and lutein. Journal of Experimental Botany. 2005;56(409):81–89. doi: 10.1093/jxb/eri016. [DOI] [PubMed] [Google Scholar]


