Summary
Transport and biosynthesis of folate and its derivatives is frequently controlled by the tetrahydrofolate (THF) riboswitch in Firmicutes. We have solved the crystal structure of the THF riboswitch aptamer in complex with folinic acid, a THF analog. Uniquely, this structure reveals two molecules of folinic acid binding to a single structured domain. These two sites interact with ligand in a similar fashion, primarily through recognition of the reduced pterin moiety. 7-deazaguanine, a soluble analog of guanine, binds the riboswitch with nearly the same affinity as its natural effector. However, 7-deazaguanine effects transcriptional termination to a substantially lesser degree than folinic acid, suggesting that the cellular guanine pool does not act upon the THF riboswitch. Under physiological conditions the ligands display strong cooperative binding, with one of the two sites playing a greater role in eliciting the regulatory response, which suggests that the second site may play another functional role.
Introduction
Folic acid is an essential micronutrient that plays a central role in one carbon metabolism. In its reduced form, tetrahydrofolate (THF, Figure 1A (1)), it is a carrier of one carbon units in the form of methyl, methylene or formyl groups that are utilized in methionine, thymidine, and purine biosynthesis, respectively (Bermingham and Derrick, 2002). Its deficiency is directly linked to broad range of human health issues including birth defects, cardiovascular disease, and increased cancer risk (Suh et al., 2001). As an important nutrient, efforts are being devoted to improving biofortification of folate in bacteria and plants to combat a significant health problem in developing counties (Bekaert et al., 2008; Sybesma et al., 2004; Zhu et al., 2005).
Figure 1.
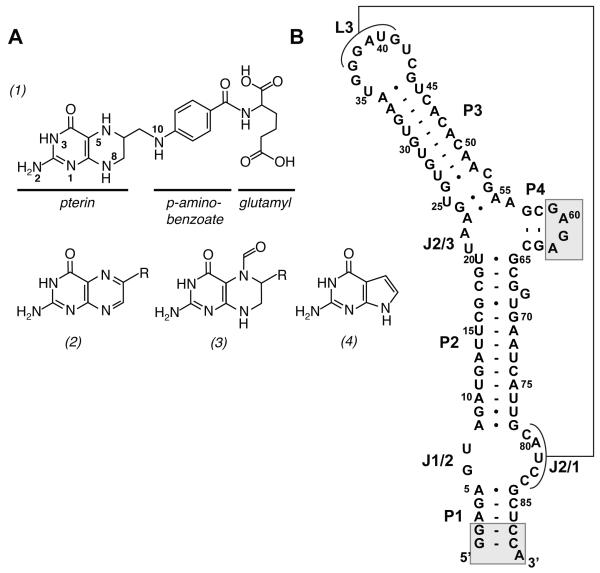
(A) Chemical structures of tetrahydrofolate (1), folate (2), folinic acid (3), and 7-deazaguanine (4). The R group of compounds (2) and (3) correspond to the para-aminobenzoate/glutamyl moieties in THF. (B) Secondary structure of the aptamer domain of the Streptococcus mutans folT riboswitch. The regions shaded in gray were changed from the wild type sequence to facilitate transcription and crystallizability.
Folate biosynthesis is also a significant target for antimicrobial therapeutics (Bermingham and Derrick, 2002). For example, one of the most longstanding and successful classes of drugs, the sulfonamides, targets the attachment of the para-aminobenzoate (pABA) moiety to 7,8-dihydropterin. As these and other folate biosynthetic reactions are absent in animals, it makes this pathway a proven and important avenue for the design of new antibacterial agents (Suh et al., 2001). For both bioengineering and drug discovery applications, exploiting the regulation of bacterial folic acid synthesis and transport are potentially important avenues.
In bacteria, particularly the Firmicutes and Fusobacteria, many central metabolic pathways are regulated at the RNA level. One of the most pervasive forms of regulation by RNA is the riboswitch, an element located in the 5′-leader sequence that directly binds a metabolite, which determines the state of a downstream secondary structural switch (Roth and Breaker, 2009). These regulatory elements direct expression at either the translational or transcriptional level. For example, small molecule binding to a transcriptional repressor element fates the mRNA to form a classic rho-independent transcriptional terminator that causes RNA polymerase to abort synthesis (Roth and Breaker, 2009). Knowledge of this regulation has enabled engineering of bacteria for enhanced production of inosine and thiamin by altering the regulation by riboswitches associated with the regulation of the biosynthesis of these compounds (Asahara et al., 2010; Tokui et al., 2011).
Recently, a new class of riboswitches that bind THF was discovered (Ames et al., 2010). Found primarily in Firmicutes, the THF riboswitch is found associated with mRNAs encoding genes that regulate folate transport (folT) or biosynthetic (folC and folE) genes. The THF receptor (aptamer domain) of this riboswitch comprises a ~100 nucleotide element predicted to form four helices and contain a three-way junction and a pseudoknot (Figure 1B) (Ames et al., 2010). Nucleotides conserved in >97% of identified sequences are distributed around the three-way junction and pseudoknot. This RNA specifically binds reduced forms of folate, including THF and DHF, as well as 5- and 10-modified forms of THF while rejecting folic acid (Figure 1A (2)). Since the biologically active form of folate is THF and its derivatives, this suggests that the riboswitch is only monitoring the active fraction of the total intracellular folate pool and turning off folate transport and/or biosynthesis in response. Furthermore, the RNA is able to bind tetrahydrobiopterin with approximately the same affinity as THF, suggesting that the riboswitch does not significantly interact with the p-aminobenzoate or glutamyl moieties (Ames et al., 2010).
To reveal the molecular basis for recognition of THF and regulation by this riboswitch, we determined its three-dimensional structure in the presence of the THF analog, folinic acid (Figure 1A, (3)). The RNA folds into a commonplace architecture involving side-by-side placement of two sets of coaxially stacked helices organized by a three-way junction and distal pseudoknot interaction. The reduced pterin moiety of folinic acid mediates most of its interaction with the RNA. Surprisingly, two molecules of folinic acid are observed in the structure, with specific binding sites located in the junction and the pseudoknot. At physiological magnesium concentrations we observe strongly cooperative binding of the two ligands. However, only one of these sites is necessary to elicit a strong regulatory response, suggesting that the second site may fulfill another role in regulation such as promoting efficient co-transcriptional folding of the RNA.
Results
Structure of the THF riboswitch
To obtain crystals of the aptamer domain of the THF riboswitch, five sequences were surveyed for initial crystallization. One of the sequences, a THF riboswitch that controls the folT gene in Streptococcus mutans (Smu) UA159 (Ames et al., 2010) yielded crystals in a sparse matrix and was subsequently modified to improve the crystal quality. Two sets of changes were made to this sequence: two base pairs of the first helix (P1) were changed to G-C to promote transcription and P4 was changed to a tetraloop to facilitate crystallization (Figure 1B, gray boxes). This sequence, which binds with slightly lower affinity than the wild type aptamer (KD,app = 18 ± 1 μM vs. 7.9 ± 0.1 μM, respectively) reproducibly yielded diffraction quality crystals. Further improvements in crystal quality were made by using the THF derivative folinic acid (FA, Figure 1A, (3)), which contains a formyl modification at the 5-position. This compound binds the riboswitch with the same affinity as THF (KD,app = 23 ± 7 μM) consistent with the observation that the RNA does not discriminate between various one-carbon carrier forms of THF (Ames et al., 2010). Cocrystallization in the presence of 1 mM iridium hexammine (Keel et al., 2007) yielded a derivative used to calculate an experimental electron density map containing very clear RNA features. The final model, refined against data extending to 2.27 Å, has good statistics and geometry (Table 1).
Table 1.
Crystallographic Statistics
| Crystal | Smu wild type | Smu (U25C) |
|---|---|---|
| Data Collection | ||
| Space Group | P 21 21 21 | P 21 21 21 |
| Cell dimensions | ||
| a, b, c (Å) | 27.6, 68.2, 157.6 | 27.5, 68.6, 158.1 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 52.5-2.27 (2.33-2.27) | 62.9-1.95 (2.01-1.95) |
| Rp.i.m. | 0.038 (0.493) | 0.025 (0.431) |
| Mn (I/σI) | 14.3 (2.0) | 20.3 (2.5) |
| Completeness (%) | 96.4 (98.9) | 98.4 (97.1) |
| Redundancy | 4.1 (3.6) | 3.5 (3.7) |
| Wavelength (Å) | 1.1051 | 1.1051 |
| Phasing | ||
| Number of heavy atoms | 5 | 5 |
| Figure of merit (before DM) | 0.34 | 0.36 |
| Refinement | ||
| Resolution (Å) | 28.6-2.27 | 62.9-1.96 |
| No. Reflections | 23076 | 22038 |
| Rwork/Rfree | 23.4/27.2 | 22.8/24.4 |
| No. Atoms | ||
| RNA | 1913 | 1913 |
| Ion | 49 | 35 |
| Ligand | 50 | 22 |
| Water | 40 | 103 |
| B-factors | ||
| RNA | 33.6 | 39.5 |
| Ion | 40.8 | 93.4 |
| Ligand | 31.9 | 41.0 |
| Water | 29.0 | 35.4 |
| RMS deviations | ||
| Bond lengths (Å) | 0.002 | 0.002 |
| Bond angles (°) | 0.513 | 0.502 |
| PDB accession code | 3SD1 | 3SD3 |
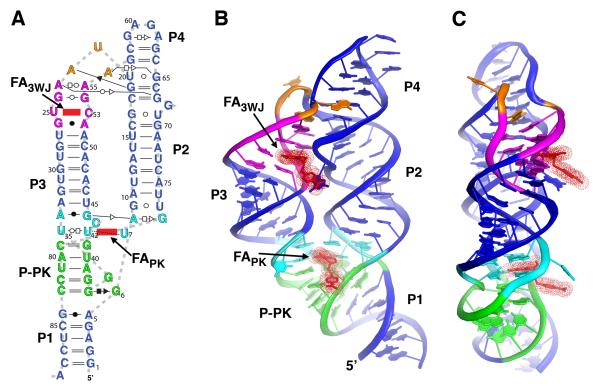
The overall architecture of the THF riboswitch consists of two sets of coaxially stacked helices positioned in a side-by-side arrangement via formation of a pseudoknot (Figure 2). The first set of coaxially stacked helices comprises helices P1, J2/1-L3, and P3, while the second is formed by P2 and P4. The placement of these coaxial stacks is enforced by a three-way junction (3WJ) motif between P2, P3 and P4, and a pseudoknot between L3 and J2/1. The overall topology of the 3WJ is similar to global architecture of the thiamine pyrophosphate and cyclic-di-GMP riboswitches (Kulshina et al., 2009; Serganov et al., 2006; Smith et al., 2009; Thore et al., 2006), all of which belong to the “family B” 3WJ category according to a catagorization by Westhof (Lescoute and Westhof, 2006). Like many three way junction motifs, the organization of the helices is enforced by a distal tertiary interaction (de la Pena et al., 2009). In the THF riboswitch it is a pseudoknot formed through a series of five Watson-Crick pairs between L3 and J2/1 (P-PK, Figure 2A). The architecture of this pseudoknot is that of an HL-type (helix-loop) pseudoknot in which the principal two helices, P1 and P-PK, are coaxially stacked (Han and Byun, 2003).
Figure 2.
Global structure of the aptamer domain of the THF riboswitch. (A) Secondary structure of the RNA used in the structure determination, redrawn to reflect the tertiary structure and include non-canonical base pairs, denoted using the Leontis-Westhof notation (Leontis and Westhof, 2001). The two molecules of folinic acid (FAPK and FA3WJ) observed in the crystal structure are denoted in red. (B) Cartoon representation of the global architecture of the THF aptamer. Coloring of the RNA is consistent with panel (a). (C) 90° rotation of the perspective shown in panel (b); note the placement of both THF binding sites on the same face of the RNA. See also Figure S1.
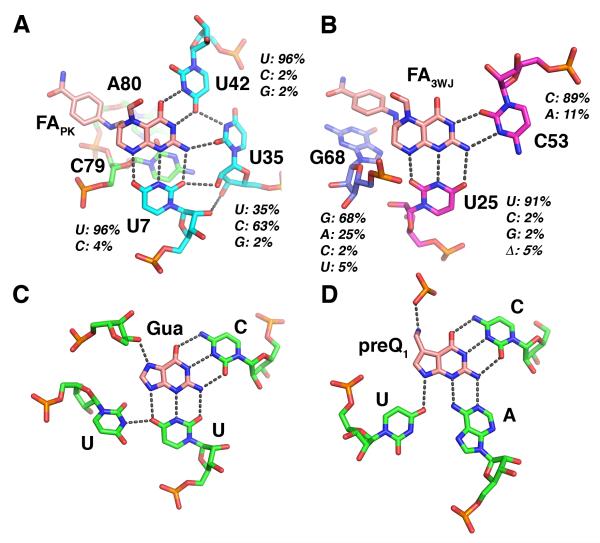
Unlike any other RNA aptamer characterized to date, natural or in vitro selected, our structure revealed that the THF riboswitch contains two ligand binding sites presented within a single structured domain. This is in contrast to the dual aptamer domains found in the glycine riboswitch in which each of two structured domains binds a single ligand (Mandal et al., 2004). The first binding site (FAPK) is adjacent to the pseudoknot on the minor groove face of the site of coaxial stacking between P-PK and P3 (Figure 2B). Recognition of the reduced pterin moiety is mediated by three uridine residues: U35 and U42 in L3, and U7 in J1/2. The N2/N3 edge of the pterin (numbering shown in Figure 1A) forms hydrogen bonds with the two uridines in L3 (Figure 3A). The N1/N8 of the pterin interacts with the Watson-Crick face of U7. While the first ring stacks with A8, the second ring, which is in the reduced form and thus not planar, displays little stacking with the RNA. The pABA moiety interacts with the RNA through stacking of the benzene ring with backbone atoms in A80 of J2/1. No clear electron density in the final 2Fo-Fc map or an omit map (Figure S1) could be associated with the glutamate moiety. It is likely that it does not interact with the RNA and thus disordered, consistent with previous observations (Ames et al., 2010). This likely reflects the fact that folate is polyglutamylated in many bacteria to promote intracelluar retention (Shane, 1989). Tetrahydrobiopterin, which lacks both the pABA and glutamyl moieties, binds with only ~4-fold lower affinity than THF (Ames et al., 2010), indicating that the majority of binding energy is obtained through interactions mediated by the pterin moiety.
Figure 3.
Recognition of folinic acid by the THF riboswitch. (A) Interactions between the RNA and the ligand in site PK. Hydrogen bonding interactions are denoted by gray dashes. For each nucleotide in direct contact with folinic acid, the degree of conservation observed across 57 sequences (Ames et al., 2010) is given as a percent identity of each nucleotide type. (B) Interactions between RNA and ligand in site 3WJ. (C) Recognition of guanine by the guanine riboswitch (from PDB ID 1Y27 (Serganov et al., 2004)). (D) Recognition of preQ1 by the preQ1 riboswitch (from PDB ID 2KFC (Klein et al., 2009)). See also Figures S1 and S5, and Table S1.
The second ligand (FA3WJ) is bound to a site in the minor groove in the P3 adjacent to the three-way junction in a fashion similar to the FAPK site (Figure 3B). The N2/N3 edge of the pterin moiety of FA3WJ forms a reverse Watson-Crick type pair with C53, while the N1/N8 edge forms three-hydrogen bonding interactions with U25. Like the FAPK site, the first ring of the pterin moiety shows stacking interactions with adjacent nucleobases (G26 and G54), but the second ring does not significantly stack with the RNA. The pABA moiety interacts with this site through stacking interactions with a bulged guanosine (G68). Also, similar to FAPK, no electron density supporting specific positioning of the glutamate moiety in the second site was observed.
There are two likely reasons for this RNA’s ability to discriminate between reduced forms of folate (THF and DHF) and the fully oxidized form in both sites. First is the formation of a hydrogen bonding interaction between N8 of the ligand and U7 or U25. This can only occur in the reduced forms of folate, dihydrofolate (DHF) and THF, which carry a hydrogen at this position (Figure 1A). Another possible means of discrimination is the different positioning of the pABA/glutamyl moiety in folate and THF. Reduction of the second ring of the pterin moiety breaks the planarity of the ring, resulting in a repositioning of the pABA/glutamyl moieties. Superimposition of folate onto folinic acid via their pterin rings in each of the two sites revealed no potential steric clashes between folate and the RNA in either site (data not shown). However, in folate, the pABA moiety can no longer stack upon A80 in the pseudoknot site or G68 in the junction site.
Binding of guanine to the THF riboswitch
In consideration that the RNA primarily interacts with the pterin moiety of folinic acid, this riboswitch might be able to interact with planar compounds with a similar constellation of hydrogen bond donors and acceptors. The most obvious candidate is guanine, which can easily be modeled into both THF sites and satisfy all of the hydrogen bonding and stacking interactions as the reduced pterin moiety. The limited solubility of guanine—100 μM at saturation—became a technical problem for some of the experiments used in this study. However, in both sites, the N7 of guanine would likely not interact with the RNA, and thus the soluble analog 7-deazaguanine (7DG, Figure 1A (4)) was considered as a reasonable proxy. To confirm that 7-deazaguanine is a suitable analog for guanine, we measured their affinity for the wild type THF aptamer and observed that they were nearly identical.
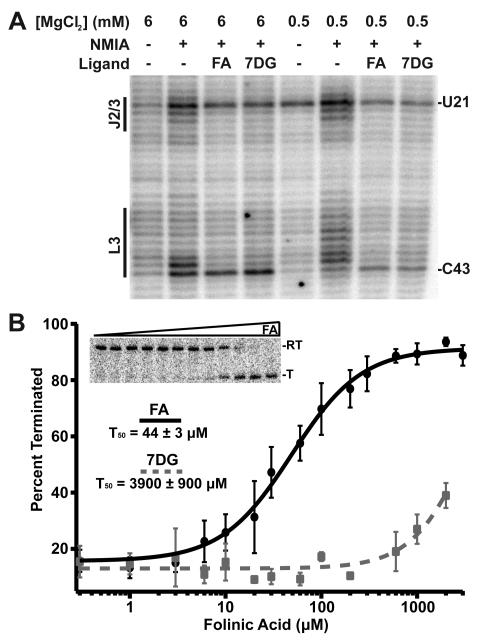
To determine whether THF and guanine bind to the Smu THF riboswitch in similar fashions, we probed its solution structure with N-methyisatoic acid (NMIA), a chemical reagent that selectively reacts with conformationally dynamic regions of RNA (Wilkinson et al., 2006). Probing the RNA in the presence of folinic acid and 7-deazaguanine demonstrated that the latter is capable of inducing a nearly identical pattern of reactivity changes as folinic acid (Figure 4A). Importantly, we observe a protection pattern that is consistent with 7-deazaguanine occupying both sites. The only significant difference is G68, which stacks with the pABA moiety in the crystal structure. While folinic acid protects this nucleotide from NMIA modification, in the presence of 7-deazaguanine, which lacks an analogous chemical moiety, G68 remains reactive.
Figure 4.
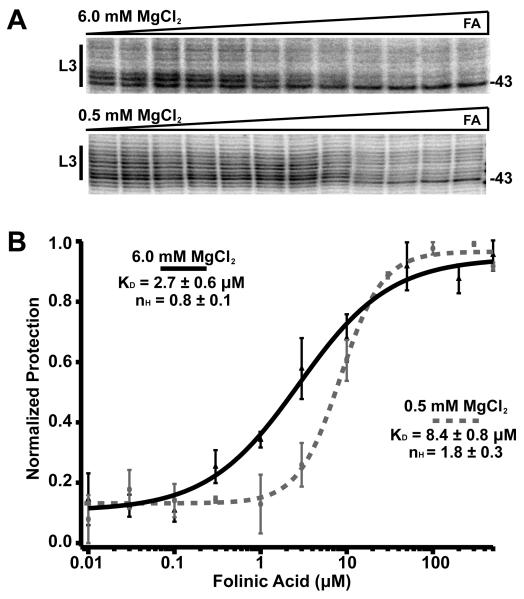
Binding of a guanine derivative to the THF riboswitch. (A) NMIA chemical probing of the THF riboswitch at two different magnesium conditions in the presence of either folinic acid or 7-deazaguanine. Note the nearly identical protection patterns induced by the two ligands at both the pseudoknot site (L3, FAPK) and the three-way junction (J2/3, FA3WJ). (B) Fitting of representative data from transcription assay titration of folinic acid and 7-deazaguanine to the chimeric switch to the Hill equation. Each data point represents the average of three independent experiments with error bars associated with the standard deviation. See also Figures S2, S3 and S4.
This result was further validated using isothermal titration calorimetry (ITC) (Table 2). Under high magnesium conditions, 7-deazaguanine displays nearly equal binding affinity as folinic acid. Both compounds also display similar enthalpic and entropic contributions to binding, further reinforcing that THF and guanine can interact with this riboswitch in nearly identical fashions. At a more physiological magnesium concentration (0.5 mM for the purposes of this study), the affinity of the RNA for 7-deazaguanine drops by approximately 2-fold relative to folinic acid.
Table 2.
Thermodynamic parameters for ligand binding to the THF riboswitch at 25 °C.
| RNA | Ligand | MgCl2 [mM] |
KD (μM) |
ΔH, (kcal mol−1) |
TΔS (kcal mol−1) |
n1 |
|---|---|---|---|---|---|---|
| wt | folinic acid | 6 | 18 ± 1 | 15 ± 1 | 8.0 ± 0.2 | 2.1 ± 0.1 |
| wt | folinic acid | 0.5 | 22 ± 2 | −15 ± 1 | −9.1 ± 0.1 | 2.0 ± 0.1 |
| wt | 7-deazaG | 6 | 24 ± 1 | −20 ± 1 | −14 ± 1 | 2.0 ±0.1 |
| wt | 7-deazaG | 2.5 | 26 ± 2 | −19 ± 1 | −13 ± 1 | 2.0 ± 0.1 |
| wt | 7-deazaG | 0.5 | 38 ± 1 | −30 ± 1 | −24 ± 1 | 2.1 ± 0.1 |
| U7C | folinic acid | 6 | 23 ± 2 | −20 ± 1 | −13 ± 1 | 1.1 ± 0.1 |
| U25C | folinic acid | 6 | 18 ± 1 | −11 ± 1 | −4.1 ± 0.2 | 1.3 ± 0.1 |
| U7C/U25C | folinic acid | 6 | n.d.2 | n.d. | n.d. | n.d. |
| U7C | folinic acid | 0.5 | 140 ± 7 | −30 ± 1 | −25 ± 1 | 1.1 ± 0.1 |
| U25C | folinic acid | 0.5 | 340 ± 10 | −19 ± 1 | −14 ± 1 | 1.2 ± 0.1 |
| U7C/U25C | folinic acid | 0.5 | n.d. | n.d. | n.d. | n.d. |
n is a fitted parameter that reflects the number of ligand binding sites.
n.d. no detectable binding.
To determine whether guanine is also capable of acting as an effector of the THF riboswitch, we employed an in vitro transcription assay that has been previously used to study riboswitch function (Artsimovitch and Henkin, 2009; Dann et al., 2007). The Smu THF riboswitch, unfortunately, does not contain an easily recognizable expression platform that would indicate whether it regulates at the transcriptional or translational level. Thus, we tested a riboswitch found in the human gut metagenome (BABG01026644.1/586-688) (Ames et al., 2010) that clearly contains a stem-loop structure that could act as a classic rho-independent transcriptional terminator. Unfortunately, we did not observe any folinic acid dependent switching of this RNA under various conditions that yields robust regulatory responses with other riboswitches in single-turnover transcription assays.
In the course of our studies of various riboswitches, we have observed that activities of the aptamer and expression platform are separable to the extent that they can be “mixed-and-matched” (Stoddard and Batey, 2006). For example, we can create a chimera in which aptamer domains from various riboswitches can be spliced into the expression platform of the B. subtilis metE SAM-I riboswitch to yield synthetic riboswitches regulated by the effector of the transplanted aptamer (Ceres et al, manuscript in preparation). The T50 (the concentration of ligand that elicits a half-maximal regulatory response) of these chimeras are principally dictated by the aptamer domain; the yitJ SAM-I riboswitch aptamer regulates at nearly identical S-adenosylmethionine concentrations in the context of the B. subtilis metE and lysC expression platforms as the wild type riboswitch (Ceres et al, manuscript in preparation). This chimeric approach has previously been employed to dissect functional domains of bacterial small noncoding RNAs (Papenfort et al., 2010). We therefore designed a chimeric riboswitch that would allow us to study THF-dependent regulation (Figures S2, S3). Nucleotides 5-84 (Figure 1B) of the aptamer domain of the wild type Smu THF riboswitch was attached to the expression platform of the B. subtilis metE expression platform, such that the secondary structural switch in the expression platform was not altered.
Regulation of the Smu folT/Bsu metE chimeric riboswitch showed strong folinic acid dependent transcriptional termination (Figure 4B). Importantly, this chimera shows no response to SAM (Figure S4), indicating that the Smu aptamer controls the effector specificity of the riboswitch. Fitting the data to the Hill binding equation yields a T50 = 44 ± 3 μM, consistent with the previous binding data. Titration of this chimera with 7-deazaguanine yielded an observed T50 > 3500 μM (Figure 4B). A partial titration of the riboswitch with guanine up to 60 μM showed no termination, consistent with 7-deazaguanine. While the transcription conditions do not exactly match the expected in vivo magnesium or NTP concentrations and uses E. coli RNAP, these data indicate that guanine is a substantially less effective regulator of the S. mutans riboswitch than THF despite it binding with nearly identical affinity.
Cooperative ligand binding to the THF riboswitch
An unusual aspect of this structure is the presence of two bound ligands. A formal possibility is that one of these sites is fortuitously occupied by THF and does not represent a biologically relevant site. Examination of the published phylogeny of the THF riboswitches reveals that each of the nucleotides that directly contacts the pterin moiety of each ligand are amongst the most highly conserved in the RNA (Figure 2A, B). These data suggest that in fact both ligand bindings sites are conserved in the THF family, and not an artifact of crystallization at high THF concentrations or an unusual sequence.
To explore the relationship between two sites, we made a set of point mutations that ablate each site. These mutants disrupt folinic acid binding by changing the uridine that interacts with the N1/N8 edge of pterin with cytosine: U7C (FAPK site), U25C (FA3WJ site), and U7C/U25C (both sites). Based upon the structure, each mutation would reduce the potential number of hydrogen bonds with the ligand by two, severely destabilizing the interaction. Each of these mutants was probed using NMIA to verify that the secondary and tertiary structure of the RNA was not altered and that the unmodified site retains folinic acid binding activity (data not shown). As a further validation, we obtained a crystal structure of the U25C mutant bound to a single folinic acid (Table 1), and observed a nearly identical global structure with the wild type complex (least squares r.m.s.d. = 0.71 Å). The only significant difference is that the G24•G54 pair immediately above the 3WJ site rearranges such that the Watson-Crick face of G54 hydrogen bonds with a non-bridging phosphate oxygen of A23 (Figure S5). While site PK was occupied by a well-ordered ligand, site 3WJ does not contain a ligand as no electron density was observed adjacent to C25 in a 2Fo-Fc map (Figure S5). This demonstrates that this mutant completely abrogates binding at site 3WJ. Notably, crystals of the U7C mutant were not obtained, suggesting that folinic acid binding to this site may be critical for proper structure formation under the crystallization conditions.
The affinity and stoichiometry of each RNA for THF was determined using ITC (Table 2). At high magnesium concentration (6 mM), the wild type aptamer shows an apparent affinity (KD) of ~20 μM for THF and a stoichiometry of 2:1 THF:RNA. While the apparent affinity is lower than that observed for the Alkaliphilus metalliredigens folT riboswitch (70 nM) (Ames et al., 2010), it is not outside the range of observed dissociation constants for riboswitch-ligand interactions (Ames and Breaker, 2011; Sudarsan et al., 2003). Each of the two single point mutants displayed similar affinities at 6 mM MgCl2, but each binds a single ligand (Table 2), indicating that these mutations ablate THF binding at the corresponding site. These data further reveal that each site binds ligand independently--there is no cooperative binding. To further validate this finding, we measured the binding of folinic acid for the Smu THF aptamer using NMIA chemical probing at 6 mM magnesium (Figure 5A). Fitting the folinic acid dependent protections at each binding site to a multisite binding model yielded a Hill coefficient ~1, clearly indicating independent binding (Figure 5B).
Figure 5.
Cooperative binding of folinic acid to the THF riboswitch. (A) Bands corresponding to nucleotides 36 – 43 in a NMIA probing experiment in which the RNA was bound by various concentrations of FA ranging from 0.1 – 500 μM at two different magnesium conditions. (B) Plot of data averaged from three individual titrations fit to a Hill equation. The two curves represent the fit to data taken in 6 mM (solid) and 0.5 mM magnesium (dashed).
At a lower, more physiological magnesium concentration (0.5 mM) the affinity of the wild type RNA for folinic acid remains essentially unchanged (Table 2). However, for both single-site mutants, the apparent affinity for folinic acid dropped by 5-10 fold. The lowered values for each individual site indicate that abolishing ligand binding to one site is deleterious to binding to the other site under physiological magnesium concentrations. One explanation for this result is that at physiological concentrations of monovalent and divalent cations, binding to the two sites is cooperative. However, these data indicate that the single point mutants are not entirely non-perturbing to the other site, preventing our quantifying the energetic contribution of cooperativity to ligand binding. Instead, we performed NMIA footprinting of the wild type RNA at 0.5 mM magnesium chloride and quantified folinic acid dependent protections in both sites using a Hill equation (Weiss, 1997). This analysis yielded a fitted Hill coefficient (nH) of 1.8 (Figure 5B), indicative of strong cooperative binding between the two sites. Together, with the above data, this suggests that at low magnesium the local binding sites are not fully structured, despite a nearly identical NMIA reactivity pattern at 6 mM Mg2+.
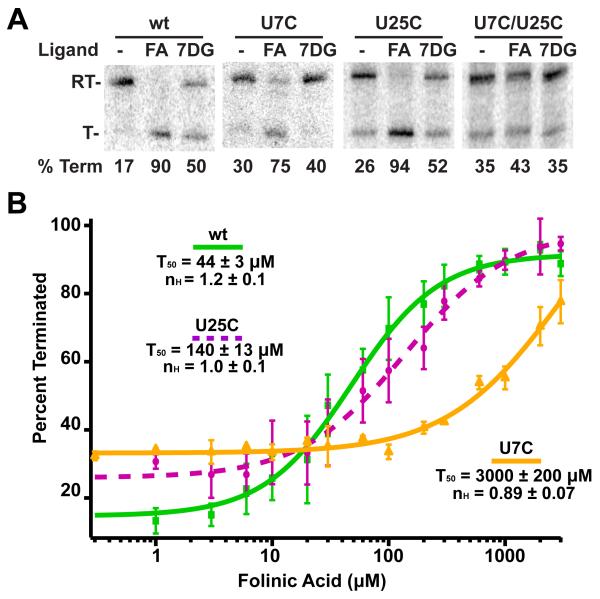
Regulation by riboswitches is often obligatorily cotranscriptional such that it is a “kinetic controlled” process in which the ligand concentration required to elicit a response is inconsistent with affinity measurements of the isolated aptamer (Wickiser et al., 2005a; Wickiser et al., 2005b). Thus, we measured the ability of each of the mutants to regulate transcription in the context of the Smu folT/Bsu metE chimeric switch (Figure 6A). The U25C mutant that knocks out the three-way junction showed a ~3-fold drop in its T50 compared to the wild type chimera, indicating that site PK is capable of regulation without a functional 3WJ site. This RNA shows no cooperativity in its ligand response curve, unlike the wild type RNA that shows weak apparent cooperativity (nH= 1.2 ± 0.1) (Figure 6B). This value is consistent with a moderate Hill coefficient of binding (nH= 1.4 ± 0.2) measured at a magnesium concentration equivalent to that in the transcription assay (2.5 mM). Conversely, the U7C mutant showed very low regulatory activity (Figure 6B, yellow curve), suggesting that site PK is most important to THF riboswitch function. The strong disparity between the two sites ability to regulate expression despite their similar affinities for ligand may indicate that they have separate roles in the function of the riboswitch.
Figure 6.
Single turnover transcription assays of THF riboswitch mutants. (A) Transcription assays of the wild type (wt), U7C (PK mutant), U25C (3WJ mutant), and double mutants. Each riboswitch was transcribed in the absence of ligand (- lanes), 200 μM folinic acid (FA lanes), or 7-deazaguanine (7DG lanes). RT denotes the read through transcription product and T denotes the terminated product. The percent of transcript terminated is given below each lane. These values represent the average of at least three independent replications of the assay. (B) Data from titration of the wild type (wt), U7C, and U25C THF riboswitches with folinic acid and fitting the each data set to a Hill equation. See also Figures S2 and S3.
Discussion
The THF riboswitch is unique amongst known riboswitches and aptamers in that it contains two ligand binding sites within a single structured domain. While each site is in the context of a different structural motif, a multihelix junction and a pseudoknot, the modes of THF recognition are strikingly similar. This is reflected by the similar affinities of each site for folinic acid and 7-deazaguanine. In contrast, single round transcriptional assays reveal that the pseudoknot site is substantially more important for eliciting a regulatory response. These data may suggest that the ancestral THF riboswitch could have functioned with a single site in the pseudoknot with the second site evolving subsequently. Of the 57 sequences identified as candidate THF riboswitches by Ames et al. (Ames et al., 2010), it is notable that there is slightly less conservation in site 3WJ than in site PK (Figure 3 and Table S1), suggesting that single site THF riboswitches may still be biologically relevant. Most of these potential single site variants have multiple changes in the altered site, indicating that they contain only a single functional binding site for THF.
Interestingly, the recognition of the pterin moiety by the THF riboswitch is in many ways analogous to nucleobase recognition by other natural and in vitro selected RNA aptamers. For example, the purine riboswitch forms a base triple with guanine similar to the base triples observed in both THF sites (Figure 3C) (Batey et al., 2004; Noeske et al., 2005; Serganov et al., 2004). Within the purine riboswitch family, the pyrimidine that hydrogen bonds to the Watson-Crick face of the nucleobase dictates the specificity for guanine versus adenine, while the pyrimidine that interacts with the N3/N9 face (equivalent of the N1/N8 edge of the pterin ring) face determines the specificity for guanine versus deoxyguanosine (Edwards and Batey, 2009). A similar mode of recognition is used by the preQ1 riboswitch to recognize guanine derivatives (Figure 3D), where hydrogen bonding interactions between the Watson-Crick and N3/N9 faces of the ligand are the primary means of interaction (Kang et al., 2009; Klein et al., 2009; Spitale et al., 2009). Recognition of theophylline (1,3-dimethylxanthine) by an in vitro selected aptamer is also achieved using two pyrimidines that form a base triple with the ligand (Zimmermann et al., 1997). These examples of convergence upon a similar solution for binding nucleobases suggests that it may be possible to evolve simple binding sites in numerous structural contexts.
A consequence of recognition of primarily the pterin moiety of THF is that the riboswitch also binds guanine, suggesting the possibility that it is regulated by both metabolites. In glucose-fed rapidly growing E. coli, the intracellular concentration of guanine has been measured to be ~200 μM (Bennett et al., 2009). While a similar measurement for THF in E. coli has not been performed, the KM for serine hydroxymethyltransferase, an enzyme that transfers a one carbon unit from serine to THF, is ~25 μM (Schirch et al., 1985), which serves as a good estimate for the minimal intracellular concentration of THF (Bennett et al., 2009). Additionally, estimates of the folate pools in B. subtilis and Lactococcus are in the 10-50 μM range (Sybesma et al., 2003; Zhu et al., 2005). Given these estimates and the observation that the T50 of folinic acid is ~100-fold lower than 7-deazaguanine, it is likely that the S. mutans riboswitch is almost entirely responsive to the total reduced folate pool in the cell rather than guanine. However, another THF riboswitch from A. metalliredigens binds THF with ~100-fold greater affinity (70 nM) (Ames et al., 2010), which may translate into a much lower T50 for THF and guanine such that both could regulate under physiological conditions. Given that GTP is the starting substrate in the synthesis of THF, as well as THF being involved in two separate steps of GTP biosynthesis, it may not be surprising that a riboswitch may need to sense both reduced folate and guanine pools in vivo. This idea is reinforced by the observation that the PurR repressor, which uses hypoxanthine and guanine as corepressors, regulates the expression of the serine hydroxyltransferase (glyA) in E. coli (Steiert et al., 1992).
The inability of 7-deazaguanine to effect transcriptional termination by the S. mutans THF aptamer despite a similar equilibrium dissociation constant (KD) to folinic acid underscores a kinetic relationship between ligand binding and regulation. In regulation at the transcriptional level, readout of the secondary structural switch by RNA polymerase occurs within seconds of the aptamer domain acquiring the ability to interrogate the cellular environment (Wickiser et al., 2005b). The rate at which the aptamer reaches equilibrium is governed by the relaxation equation τ−1 = kon[ligand] + koff where τ, the time constant, is the time required for 1-e−1 (63.2%) of the population to reach the final steady state equilibrium. If genetic readout occurs prior to the aptamer fully reaching equilibrium, then the kinetic rates determine the concentration of ligand required to achieve a regulatory response, resulting in regulation at ligand concentrations in excess of that expected by the measured KD. While we have not measured association and dissociation rates for folinic acid and 7-deazaguanine, the latter may have a significantly longer time constant. This results in substantially higher concentrations to effect transcriptional termination by 7-deazaguanine as compared to folinic acid (Figure 4B). This observation is relevant to the design of antimicrobials that target riboswitches (Blount and Breaker, 2006); compounds that bind with an affinity comparable to the natural ligand may not be capable of genetic regulation.
There are several reasons why this riboswitch might have evolved multiple sites to facilitate regulation. The first is that it senses the total THF/guanine intracellular pool. In vitro transcription data demonstrates that by itself guanine is a poor effector of regulation, but that may be the result of poor binding to the FAPK site. Instead, guanine might act primarily at the FA3WJ site to assist in folding of the aptamer (see below). A second, and more likely, explanation for two ligand binding sites is to enable a more digital response to THF via cooperative binding. Under physiological magnesium conditions we observe strong cooperative binding (nH = 1.8) between the two ligands using NMIA chemical probing (Figure 4). This is a similar value for the Hill coefficient measured for the glycine riboswitch, an RNA that uses two tandem aptamers and achieves cooperative binding through ligand-dependent formation of interdomain interactions (Butler et al., 2011; Mandal et al., 2004). Unfortunately, we were not able to perform transcription assays under these conditions; the lowest concentration of magnesium at which reliable data could be acquired was 2.5 mM. At this condition, we observe only moderate (nH = 1.4, Figure 5B) cooperative binding between sites as well as in the transcription assay (nH = 1.2, Figure 6B). Nonetheless, the close agreement of these values indicate that transcriptional regulation under physiological conditions could be facilitated through cooperative binding of ligand at the two sites.
Cooperative binding of the ligands may also be required to promote rapid and efficient folding of the aptamer. Many riboswitches act co-transcriptionally and therefore have a short temporal window in which to act prior to the escape of RNA polymerase into the coding region. Thus, the RNA must be able to efficiently and rapidly fold and interrogate the cellular environment for the presence of the effector in order to control gene expression. During transcription, the three-way junction would be synthesized first and start to fold. An RNA spanning nucleotides 19-90 of the A. metalliredigens RNA (equivalent of 9-78 of the Smu riboswitch) is capable of binding ligand, albeit at a reduced affinity (Ames et al., 2010). Thus, binding of ligand to site 3WJ could facilitate organization of the three-way junction to place L3 in proximity of J2/1 to form the pseudoknot and site PK. In this model, site 3WJ is not essential for regulatory activity, as shown by the transcription assays, but rather for rapid folding of the RNA and formation of the regulatory site in the pseudoknot. In most other riboswitches, folding of the RNA occurs using motifs that do not require an additional ligand. For example, studies of the B. subtilis xpt guanine and pbuE adenine riboswitches have demonstrated that a distal loop-loop interaction facilitates the folding of a three-way junction element that hosts the ligand binding site (Greenleaf et al., 2008; Lemay et al., 2006; Stoddard et al., 2008). It is therefore possible that the THF riboswitch initially evolved the site in the pseudoknot as a means of establishing a regulatory site and subsequently a second site to facilitate rapid and efficient folding of the RNA via the three way junction. This is consistent with our measurement of binding at physiological magnesium concentrations where it is likely that binding of THF within the three-way junction facilitates formation of the pseudoknot site to achieve cooperativity.
Methods
RNA transcription and purification
A DNA template for transcription was created using PCR amplification of overlapping oligonucleotides. The template was transcribed in vitro using T7 RNA polymerase and purified using previously described methods (Reyes et al., 2009). Briefly, transcribed products were purified using a denaturing polyacrylamide gel (8 M urea, 12% 29:1 acrylamide:bisacrylamide) . Gel fragments containing RNA were soaked in 0.5X T.E. (10 mM Tris, pH 8.0, 1 mM EDTA) buffer to elute. Buffer exchange and concentration was performed using Amicon Ultra 10,000 molecular weight cutoff centrifugal concentrators. The final concentration was calculated from the absorbance at 260 nm and the molar extinction coefficient calculated from the summation of the contributions from the individual bases.
RNA crystallization, structure solution and refinement
RNA was crystallized set up in a hanging drop/vapor diffusion method. 1 μL of RNA ligand mix (500 μM RNA, 1 mM folinic acid) was added to 2 μl of 6% 2,4-methyl-pentanediol, 10 mM spermine, 40 mM Na-cacodylate, pH 7.0, 80 mM NaCl and 1 mM DTT was incubated over 500 μl of 35% MPD. Drops were allowed to equilibrate at 30° C for 2-4 days, during which time rod-like crystals formed. To prepare a suitable derivative, 1 mM iridium hexammine was added to drop solution prior to cryoprotection. To cryoprotect, 10% MPD was added to the crystallization solution, allowed to equilibrate for 5-10 minutes and the crystals were frozen in liquid nitrogen.
Complete datasets of a wild type RNA and a U7C mutant were collected at the Advanced Light Source (5.0.2). The data was indexed, integrated, and scaled with Xia2(Winter, 2010). Significant anomalous signal throughout the entire resolution range allowed for location of iridium heavy atom sites, calculation of SAD phases, and density modification via Phenix (Adams et al., 2002). The experimental density map was of sufficient quality to allow for manual building of the entire RNA in Coot (Emsley and Cowtan, 2004). The RNA was refined, initially against the experimental phases and as a last step against the model phases using phenix.refine. The refinement protocol included restrained coordinate refinement, combined individual and TLS atomic displacement parameters for the entire RNA, and occupancies for the ligand and heavy atoms. The ligand was placed at the last step of refinement with the assistance of 2Fo-Fc, Fo-Fc, and experimental phasing maps. The crystallographic, phasing, and refinement statistics are presented in Table 1. The 2Fo-Fc map, and simulated annealing maps omitting the ligands are shown in Figure S2. The model, structure factors, and experimental phases were deposited to the Protein Data Bank (PDB 3SD1 and 3SD3 for wild type and the U7C mutant, respectively).
Isothermal titration calorimetry (ITC)
The affinity of the RNA for a specific ligand of interest was performed using previously described methods (Gilbert and Batey, 2009; Gilbert et al., 2007) with a MicroCal ITC200 (GE Healthcare). RNA samples were dialyzed overnight at 4 °C in buffer containing 10 mM HEPES pH 8.0, 100 mM NaCl, 10 mM KCl, 1 mM DTT, and either 0.5 or 6 mM MgCl2. The ligand was brought up from powder in matching dialysis buffer. 270 μL of 200-500 μM RNA was added to the sample cell and the injector contained 40 μl of 1-5 mM ligand. The amount of the titrant and titrate were such that the c-value for each experiment was between 10 and 50 to ensure accurate analysis of the resulting titration. Each titration was performed at 25° C. Raw data was integrated and fit using Origin ITC software (Microcal Software Inc.).
Chemical probing with N-methylisatoic acid (NMIA)
RNA was prepared as described previously (Stoddard et al., 2008; Stoddard et al., 2010). Structure cassettes flanking the 5′ and 3′ ends of the RNA was added to facilitate reverse transcription (Wilkinson et al., 2006). NMIA modification was performed using the established protocols (Mortimer and Weeks, 2007; Stoddard et al., 2008). RNA in 0.5X T.E. buffer was incubated for 2 min at 90 °C then immediately placed on ice for 10 min. 8 μL of stock solution contained 900 nM RNA, 1X folding buffer (3.3X folding buffer: 333 mM K-HEPES pH 8.0, 333 mM NaCl, and 20 mM MgCl2 for a final of 6 mM or 1.65 mM MgCl2 for a final of 0.5 mM) was added to thin walled PCR tubes. 1 μL ligand was dissolved in a solution of 1 mM DTT to make a final concentration of 500 μM. For titrations, 1 μL ligand was added of appropriate concentration. For ligand free samples, 1 μL of 1 mM DTT was added. To probe RNA, 1 μl of 130 mM in DMSO was added to each aliquot. For control reactions, 1 μl of DMSO was added. Samples were incubated for 5 half-lives of NMIA at 25 °C. Compensating amounts of MgCl2 was added back to samples containing 0.5 mM MgCl2 to insure condition matching for reverse transcription.
Samples were reverse transcribed by adding 3 μL of 32P 5′-end labeled DNA oligomer to the solution. Reactions were incubated at 65 °C for 5 min 35 °C for 10 min and held at 54 °C. Samples were then supplemented with 7 μL of enzyme mix (250 mM KCl, 167 mM Tris-HCl pH 8.3, 16.7 mM DTT, 1.67 mM each dNTP, and 0.33 units of Superscript III reverse transcriptase (Invitrogen)). Each reaction was incubated at 54 °C for 10 min. Reaction was stopped by adding 1 μL of 4 M NaOH and incubated at 90 °C for 5 min. Incubation continued for an additional 5 min after the addition of 29 μL acid stop mix. DNA was separated using a 12% denaturing polyacrylamide gel, the gel dried and a Typhoon PhosphorImager (Molecular Dynamics) was used to visualize results. Bands were aligned using SAFA(Das et al., 2005) and quantified using SAFA or Image J. Data was fit to the Hill equation, Θ = a + (b - a)([ligand]n/([ligand]n + KDn) where a and b are the lower and upper baselines and n is the Hill coefficient, using Igor Pro (Wavemetrics).
Transcription template construction for in vitro transcription assays
DNA templates used for transcription assays were constructed by PCR amplification of overlapping oligonucleotides using standard methods. The THF aptamer domain (nucleotides 5 to 84) was spliced into the expression platform of B. subtilis metE SAM-I riboswitch (Figure S2). This riboswitch has been previously shown to regulate transcription by E. coli RNA polymerase (RNAP) in an S-adenosylmethionine (SAM) dependent fashion (Artsimovitch and Henkin, 2009). The chimera was constructed in two pieces. The 5′ piece consisted of a T7A1 promoter followed by the native 5′-metE sequence (including most of P1), with the THF aptamer in place of the natural aptamer domain. The 3′ portion was composed of the metE expression platform, which was PCR amplified from B. subtilis genomic DNA. Recombinant PCR was used to fuse these two portions. Mutant templates were obtained by altering the desired nucleotides in the DNA oligonucleotides; the sequence of the Smu folT/Bsu metE chimera is given in Figure S3.
In vitro transcription assay
Single-turnover transcription reactions were performed essentially as described in Dann et al. (Dann et al., 2007). Briefly, 100 ng of DNA were incubated at 37 °C for 15 minutes in 25 μL of 2X transcription buffer (140 mM Tris-HCl, pH 8.0, 140 mM NaCl, 0.2 mM EDTA, 28 mM β-mercaptoethanol and 70 μg/mL BSA), 5 μL of 25 mM MgCl2, 1 μCi of α-32P-ATP and 0.5 units of E. coli RNA polymerase σ70 holoenzyme obtained from Epicentre Biotechnologies (Madison, WI) to a final volume of 35 μL. The reactions were initiated with the addition of 15 μL of NTP mix (NTPs in equal molar ratios of 82.5 μM), 0.2 mg/mL heparin and the desired concentration of ligand. After incubation at 37 °C for 15 minutes, the reactions were quenched with equal volume of 8 M urea and 2 minutes incubation at 65 °C. The terminated and read through transcription products were separated using 6% denaturing polyacrylamide gel, dried and exposed overnight. Quantification of bands was performed with ImageQuant software (Molecular Dynamics) and the data fit to a Hill equation.
Supplementary Material
Highlights.
Crystal structure of the tetrahydrofolate (THF) riboswitch reveals two ligand sites
Binding of the two ligands is highly cooperative at physiological magnesium
Guanine binds to the THF riboswitch but does not effect regulation
One THF binding site is more effective in regulating gene expression
Acknowledgments
We would like to thank Jennifer Pfingsten, Quentin Vicens and members of the Batey lab for critical review of this manuscript. The work was funded through grants to R.T.B. from the National Institutes of Health (GM083953 and GM073850) and J.J.T. was funded through a Creative Training in Molecular Biology grant (NIH 5-T32-GM07135).
Footnotes
Accession codes Protein Data Bank: Coordinates and structure factors for THF riboswitch have been deposited with accession number 3SD1 and 3SD3
Author contributions J.J.T. conducted crystallization trials, collected diffraction data, assisted in solving the crystal structure, and performed all ITC and chemical probing experiments. F.E.R. collected diffraction data, solved the phase problem, and built and refined the THF aptamer structures. P.C. designed and implemented the chimeric riboswitch transcription assay and analyzed the resulting data. All authors contributed to the design and implementation of experiments and writing of the manuscript.
Competing Financial Interests R.T.B. is a paid consultant of BioRelix, a company dedicated to developing antibiotic therapeutics targeted against riboswitches.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Ames TD, Breaker RR. Bacterial aptamers that selectively bind glutamine. RNA Biol. 2011;8 doi: 10.4161/rna.8.1.13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames TD, Rodionov DA, Weinberg Z, Breaker RR. A eubacterial riboswitch class that senses the coenzyme tetrahydrofolate. Chem Biol. 2010;17:681–685. doi: 10.1016/j.chembiol.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I, Henkin TM. In vitro approaches to analysis of transcription termination. Methods. 2009;47:37–43. doi: 10.1016/j.ymeth.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahara T, Mori Y, Zakataeva NP, Livshits VA, Yoshida K, Matsuno K. Accumulation of gene-targeted Bacillus subtilis mutations that enhance fermentative inosine production. Appl Microbiol Biotechnol. 2010;87:2195–2207. doi: 10.1007/s00253-010-2646-8. [DOI] [PubMed] [Google Scholar]
- Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- Bekaert S, Storozhenko S, Mehrshahi P, Bennett MJ, Lambert W, Gregory JF, 3rd, Schubert K, Hugenholtz J, Van Der Straeten D, Hanson AD. Folate biofortification in food plants. Trends Plant Sci. 2008;13:28–35. doi: 10.1016/j.tplants.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham A, Derrick JP. The folic acid biosynthesis pathway in bacteria: evaluation of potential for antibacterial drug discovery. Bioessays. 2002;24:637–648. doi: 10.1002/bies.10114. [DOI] [PubMed] [Google Scholar]
- Blount KF, Breaker RR. Riboswitches as antibacterial drug targets. Nat Biotechnol. 2006;24:1558–1564. doi: 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]
- Butler EB, Xiong Y, Wang J, Strobel SA. Structural basis of cooperative ligand binding by the glycine riboswitch. Chem Biol. 2011;18:293–298. doi: 10.1016/j.chembiol.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dann CE, 3rd, Wakeman CA, Sieling CL, Baker SC, Irnov I, Winkler WC. Structure and mechanism of a metal-sensing regulatory RNA. Cell. 2007;130:878–892. doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Pena M, Dufour D, Gallego J. Three-way RNA junctions with remote tertiary contacts: a recurrent and highly versatile fold. RNA. 2009;15:1949–1964. doi: 10.1261/rna.1889509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AL, Batey RT. A structural basis for the recognition of 2′-deoxyguanosine by the purine riboswitch. J Mol Biol. 2009;385:938–948. doi: 10.1016/j.jmb.2008.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Gilbert SD, Batey RT. Monitoring RNA-ligand interactions using isothermal titration calorimetry. Methods Mol Biol. 2009;540:97–114. doi: 10.1007/978-1-59745-558-9_8. [DOI] [PubMed] [Google Scholar]
- Gilbert SD, Love CE, Edwards AL, Batey RT. Mutational analysis of the purine riboswitch aptamer domain. Biochemistry. 2007;46:13297–13309. doi: 10.1021/bi700410g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenleaf WJ, Frieda KL, Foster DA, Woodside MT, Block SM. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Byun Y. PSEUDOVIEWER2: Visualization of RNA pseudoknots of any type. Nucleic Acids Res. 2003;31:3432–3440. doi: 10.1093/nar/gkg539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M, Peterson R, Feigon J. Structural Insights into riboswitch control of the biosynthesis of queuosine, a modified nucleotide found in the anticodon of tRNA. Mol Cell. 2009;33:784–790. doi: 10.1016/j.molcel.2009.02.019. [DOI] [PubMed] [Google Scholar]
- Keel AY, Rambo RP, Batey RT, Kieft JS. A general strategy to solve the phase problem in RNA crystallography. Structure. 2007;15:761–772. doi: 10.1016/j.str.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DJ, Edwards TE, Ferre-D’Amare AR. Cocrystal structure of a class I preQ1 riboswitch reveals a pseudoknot recognizing an essential hypermodified nucleobase. Nat Struct Mol Biol. 2009;16:343–344. doi: 10.1038/nsmb.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulshina N, Baird NJ, Ferre-D’Amare AR. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nat Struct Mol Biol. 2009;16:1212–1217. doi: 10.1038/nsmb.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay JF, Penedo JC, Tremblay R, Lilley DM, Lafontaine DA. Folding of the adenine riboswitch. Chem Biol. 2006;13:857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. RNA. 2006;12:83–93. doi: 10.1261/rna.2208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J Am Chem Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- Noeske J, Richter C, Grundl MA, Nasiri HR, Schwalbe H, Wohnert J. An intermolecular base triple as the basis of ligand specificity and affinity in the guanine- and adenine-sensing riboswitch RNAs. Proc Natl Acad Sci U S A. 2005;102:1372–1377. doi: 10.1073/pnas.0406347102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Bouvier M, Mika F, Sharma CM, Vogel J. Evidence for an autonomous 5′ target recognition domain in an Hfq-associated small RNA. Proc Natl Acad Sci U S A. 2010;107:20435–20440. doi: 10.1073/pnas.1009784107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes FE, Garst AD, Batey RT. Strategies in RNA crystallography. Methods Enzymol. 2009;469:119–139. doi: 10.1016/S0076-6879(09)69006-6. [DOI] [PubMed] [Google Scholar]
- Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem. 2009;78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirch V, Hopkins S, Villar E, Angelaccio S. Serine hydroxymethyltransferase from Escherichia coli: purification and properties. J Bacteriol. 1985;163:1–7. doi: 10.1128/jb.163.1.1-7.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shane B. Folylpolyglutamate synthesis and role in the regulation of one-carbon metabolism. Vitam Horm. 1989;45:263–335. doi: 10.1016/s0083-6729(08)60397-0. [DOI] [PubMed] [Google Scholar]
- Smith KD, Lipchock SV, Ames TD, Wang J, Breaker RR, Strobel SA. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat Struct Mol Biol. 2009;16:1218–1223. doi: 10.1038/nsmb.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitale RC, Torelli AT, Krucinska J, Bandarian V, Wedekind JE. The structural basis for recognition of the PreQ0 metabolite by an unusually small riboswitch aptamer domain. J Biol Chem. 2009;284:11012–11016. doi: 10.1074/jbc.C900024200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiert JG, Kubu C, Stauffer GV. The PurR binding site in the glyA promoter region of Escherichia coli. FEMS Microbiol Lett. 1992;78:299–304. doi: 10.1016/0378-1097(92)90044-o. [DOI] [PubMed] [Google Scholar]
- Stoddard CD, Batey RT. Mix-and-match riboswitches. ACS Chem Biol. 2006;1:751–754. doi: 10.1021/cb600458w. [DOI] [PubMed] [Google Scholar]
- Stoddard CD, Gilbert SD, Batey RT. Ligand-dependent folding of the three-way junction in the purine riboswitch. RNA. 2008;14:675–684. doi: 10.1261/rna.736908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard CD, Montange RK, Hennelly SP, Rambo RP, Sanbonmatsu KY, Batey RT. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure. 2010;18:787–797. doi: 10.1016/j.str.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 2003;17:2688–2697. doi: 10.1101/gad.1140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh JR, Herbig AK, Stover PJ. New perspectives on folate catabolism. Annu Rev Nutr. 2001;21:255–282. doi: 10.1146/annurev.nutr.21.1.255. [DOI] [PubMed] [Google Scholar]
- Sybesma W, Burgess C, Starrenburg M, van Sinderen D, Hugenholtz J. Multivitamin production in Lactococcus lactis using metabolic engineering. Metab Eng. 2004;6:109–115. doi: 10.1016/j.ymben.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Sybesma W, Starrenburg M, Tijsseling L, Hoefnagel MH, Hugenholtz J. Effects of cultivation conditions on folate production by lactic acid bacteria. Appl Environ Microbiol. 2003;69:4542–4548. doi: 10.1128/AEM.69.8.4542-4548.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thore S, Leibundgut M, Ban N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- Tokui M, Kubodera T, Gomi K, Yamashita N, Nishimura A. Construction of a thiamine pyrophosphate high-producing strain of Aspergillus oryzae by overexpression of three genes involved in thiamine biosynthesis. J Biosci Bioeng. 2011 doi: 10.1016/j.jbiosc.2010.12.011. [DOI] [PubMed] [Google Scholar]
- Weiss JN. The Hill equation revisited: uses and misuses. Faseb J. 1997;11:835–841. [PubMed] [Google Scholar]
- Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005a;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell. 2005b;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- Winter G. xia2: an expert system for macromolecular crystallography data reduction. Journal of Applied Crystallography. 2010;43:186–190. [Google Scholar]
- Zhu T, Pan Z, Domagalski N, Koepsel R, Ataai MM, Domach MM. Engineering of Bacillus subtilis for enhanced total synthesis of folic acid. Appl Environ Microbiol. 2005;71:7122–7129. doi: 10.1128/AEM.71.11.7122-7129.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann GR, Jenison RD, Wick CL, Simorre JP, Pardi A. Interlocking structural motifs mediate molecular discrimination by a theophylline-binding RNA. Nat Struct Biol. 1997;4:644–649. doi: 10.1038/nsb0897-644. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.