Abstract
Most cancers progress with the accumulation of genetic mutations with time and this is frequently associated with the acquisition of genomic instability in the form of whole chromosome changes, chromosomal rearrangements, gene amplifications or smaller changes at the nucleotide level. Whole chromosome instability (W-CIN), characterised by aneuploidy, is a major form of genomic instability observed in human cancers and several lines of evidence now support the argument that W-CIN is a promoter of tumourigenesis rather than being a passenger event. The primary mechanism proposed for evolution of CIN is abnormalities in mitosis/cytokinesis. However, mutations in genes directly involved in controlling mitosis/cytokinesis are rare in human cancers and so the mechanisms underpinning the evolution of CIN in cancers are not currently clear. On the other hand, mutations in RAS or BRAF are frequently found in human cancers, many of which demonstrate CIN, suggesting a possible link between deregulated signaling through the RAS/RAF/MEK/ERK pathway and CIN. In this review, we focus on a potential relationship between deregulated RAS/RAF signaling and CIN, and discuss possible mechanisms connecting the two.
Keywords: RAS, BRAF, CRAF, oncogenes, genome instability, aneuploidy, ERK, mouse models, cancer
1. Introduction
Genomic instability is a hallmark of cancer [1, 2], and chromosome instability (CIN) has been identified as a major form of genomic instability [3]. CIN is further categorized into either whole chromosome instability (W-CIN), characterized by numerical alterations of whole chromosomes, or structural chromosome instability (S-CIN), represented by chromosome rearrangements such as deletions, duplications and translocations [4]. While inappropriate DNA damage response/repair has been assumed to be the major cause of S-CIN, several different mechanisms have been proposed for W-CIN, including deregulated spindle assembly checkpoint (SAC) [5], alterations in kinetochore-microtubule (k-MT) dynamics [6], and defective chromosome cohesion [7], all of which facilitate, in principle, chromosome mis-segregation during mitosis.
The RAF/MEK/ERK MAP kinase pathway is one of the best-characterized signaling pathways downstream of the small GTPase RAS and comprises 3 RAF kinases (ARAF, BRAF, CRAF), two MEKs (MEK1 and MEK2) and two ERKs (ERK1 and ERK2) [8, 9]. In this pathway, RAF kinases phosphorylate MEK 1/2, which in turn phos-phorylate ERK 1/2 to transduce the signal flow. Ligand-stimulation of several types of cell surface receptors induces conversion of 3 RAS homologues (HRAS, NRAS, KRAS) to active forms leading to recruitment of the RAFs to the plasma membrane and activation of the downstream ERK MAP kinase pathway. It is well established that the ERK pathway promotes cell proliferation through G1/S cell cycle progression [8], and mutations in RAS/RAF/MEK/ERK pathway components are found in various types of human cancers [9-12].
The fact that CIN and RAS/RAF mutations are a frequent occurrence in human cancers suggests a possible link between them. However, such a link has not yet been clearly demonstrated in vivo, especially in human cancers. Below we elucidate growing evidence for a role of these oncogenes in driving CIN in the evolution of cancers.
2. Summary of RAS/RAF/MEK/ERK pathway mutations in human cancer
Although the presence of RAS mutations in human cancer samples were initially documented nearly 30 years ago [9, 10], the advent of high throughput DNA sequencing projects in recent years has consolidated the fact that other components of the RAS/RAF/MEK/ERK are also mutational targets in human cancer. These sequencing efforts have identified RAS mutations in ∼20-30% of human cancers, with KRAS mutations being far more common than either HRAS or NRAS mutations. BRAF is the next most commonly mutated gene in human cancers, at ∼7-8% [11, 12]. Mutations in CRAF, ARAF, MEK1 and ERK2 have been detected but are extremely rare while mutations affecting MEK2 and ERK1 have not been found so far. Although the vast majority of mutations are single point mutations, chromosomal translocations and rearrangements affecting BRAF or CRAF have been detected in some astrocytomas, melanomas, prostate and gastric cancer samples [13-16]. A summary of the current status of RAS/RAF/MEK/ERK pathway mutations in primary human cancer samples is provided in Table 1.
Table 1.
Summary of mutations of RAS/RAF/MEK/ERK pathway components detected in primary human cancer samples.
| Cancer Type | Organ | % Common point mutation | Rare point mutation (# of cases) | Gene fusion ( # of cases) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| KRAS | NRAS | HRAS | BRAF | CRAF | ARAF | MEK1 | BRAF | CRAF | ||
| Carcinoma | Oesophagus | 3% | 0% | 0% | 3% | ND | ND | ND | ND | ND |
| Stomach | 7% | 3% | 2% | 1% | 0% | 0% | 0% | AGTRAPBRAF(1) | ND | |
| Colorectal | 36% | 4% | 0% | 10% | 0% | G331C (1) T442T (1) | D67N (1) | N | ND | |
| Pancreas | 67% | 0% | 0% | 2% | 0% | 0% | ND | ND | ND | |
| Liver | 4% | 2% | 0% | 0% | N115S(1) | ND | ND | ND | ND | |
| Biliary Tract | 29% | 2% | 0% | 10% | 0% | ND | 0% | ND | ND | |
| Thyroid | 4% | 7% | 3% | 44% | ND | ND | ND | AKAP9-BRAF (4) | ND | |
| Lung | 16% | 1% | 0% | 2% | A319S (1) | Q383H (1) | K57N (3)Y240Y (1) | ND | ND | |
| Skin | 4% | 4% | 8% | 0% | 0% | ND | 0% | ND | ND | |
| Kidney | 1% | 1% | 0% | 0% | 0% | ND | 0% | ND | ND | |
| Urinary Tract | 4% | 0% | 8% | 1% | 0% | ND | 0% | ND | ND | |
| Prostate | 7% | 1% | 5% | 4% | 0% | 0% | 0% | SLC45A3-BRAF (1) | ESRP1-CRAF (2) | |
| Breast | 2% | 2% | 0% | 1% | 0% | 0% | 0% | ND | ND | |
| Ovary | 11% | 3% | 0% | 4% | S259A (1) | P561P (1) | 0% | ND | ND | |
| Endometrium | 15% | 1% | 0% | 4% | ND | ND | ND | ND | ND | |
| Cervix | 8% | 2% | 8% | 1% | 0% | ND | 0% | ND | ND | |
| Haemopoietic neoplasms | Lymphoid (Non-HCL) | 6% | 10% | 0% | 1% | 0% | 0% | 0% | ND | ND |
| HCL | ND | 0% | ND | 100% | ND | 0% | ND | ND | ND | |
| MM | 4% | 21% | 0% | 3% | 0% | 0% | 0% | ND | ND | |
| Myeloid (Non-LCH) | 4% | 10% | 0% | 1% | 0% | 0% | 0% | ND | ND | |
| LCH | ND | ND | ND | 57% | ND | 0% | ND | ND | ND | |
| CNS Tumours | 1% | 1% | 0% | 3% | 0% | 0% | 0% | KIAA154 9-BRAF (130)* | SRGAP3-CRAF (2)* | |
| Melanoma | 3% | 20% | 1% | 39% | 0% | ND | K57N (1) | ND | ND | |
ND: Not Determined; HCL: hairy cell leukemia, MM: multiple myeloma, LCH: Langerhans cell histiocytosis All data was obtained from the Catalogue Of Somatic Mutations In Cancer (COSMIC: http://www.sanger.ac.uk/genetics/CGP/cosmic/) except for mutations in HCL [114], MM [115] and LCH [116] that were described in the corresponding references and mutations in MEKl that were reported in references [117-120]. Only mutations detected in primary human cancer samples are included in the Table. A mutation in ERK2 has been detected in only one ovarian cancer sample (A143A) but, for convenience, is not included in this table. MEK2 and ERK1 mutations have not been reported as yet.
For CNS tumours, BRAF translocations were detected in 46% of pilocytic astrocytoma and CRAF translations in 4% of pilocytic astrocytoma
3. Signaling pathways downstream of RAS/RAF involved in mitosis
The ERK pathway is the best characterized signaling pathway downstream of the RAS/RAF cascade, promoting cell proliferation by facilitating G1/S cell cycle progression [8] or evoking premature cellular senescence when supra-physiological, robust activation is induced [17, 18]. In contrast to its well-established function in G1/S regulation, roles for the ERK pathway in mitosis are not entirely clear, especially in mammalian cells. The idea that ERK1/2 regulates mitotic progression comes primarily from studies on Xenopus egg extracts and oocytes and has been extensively reviewed elsewhere [19, 20]. In mammalian cells, although the activity of ERK1/2 has been reported to be enhanced during the G2/M transition [21, 22], the exact stage of the G2/M transition at which ERK1/2 is activated has been difficult to elucidate. A number of studies have shown that pharmacological and/or genetic inhibition of the ERK pathway delays the G2/M transition in mammalian cells [23-25]. However, long-term suppression of ERK1/2 activity in these studies makes it impossible to distinguish between direct effects of ERK1/2 and the ability of ERK1/2 to activate specific gene expression programmes required for progression through G2/M. A study utilizing live imaging and short-term MEK inhibition has demonstrated that direct ERK activity is dispensable for the G2/M transition, mitotic progression and spindle assembly in mammalian cells [26]. In contrast, depletion of ERK1/2 in human dermal keratinocytes, but not in fibroblasts, reportedly induced G2/M (or tetraploid G1) arrest through downregulation of cyclin B1 expression, suggesting that ERK-regulated gene expression programs, rather than direct ERK activity, could contribute to G2/M progression in a cell-type dependent manner [27]. Activated MEK1/2 and ERK1/2 have been reported to be detectable at centrosomes/spindle poles and kinetochores during mitosis, as assessed by immunofluorescence using phosphoantibodies [28-30]. However, there is little data to support functional roles at these locations and, furthermore, antibodies commonly used to detect phospho-MEK1/2 mitosis can cross-react with nucleophosmin, another nuclear phosphoprotein [31]. Another report has shown that ERK1/2 depletion and MEK inhibitor treatment failed to abrogate immunostaining of phosphorylated ERK at the mitotic apparatus [26], raising the possibility that the phosphorylated ERK in mitosis previously reported might be an immunological artifact. Overall, data indicating a role of the MEK/ERK pathway in G2/M progression needs to be carefully interpreted.
None of the above studies have addressed whether supra-physiological hyperactivation of the ERK pathway by oncogenic RAS/RAF affects mitotic progression. Indeed, when the ERK pathway is hyperactivated by ectopic expression of oncogenic RAS/RAF mutants, mitotic progression is reportedly perturbed [32-34]. In human melanoma cells, ectopic expression of V600E-BRAF, the most common BRAF mutant detected in human cancers, was shown to promote aberrant activation of the SAC through stabilization of the mitotic checkpoint protein MPS1 in a MEK/ERK-dependent manner [32], and this led to chromosome mis-segregation and aneuploidization [33]. In contrast, G12VHRAS conditionally expressed in rat thyroid cells has been reported to attenuate the SAC activation in a MT-depolymerized condition, but this phenotype did not require MEK/ERK activity [34]. Thus, the biological consequence of hyperactivation of the ERK pathway in mitosis seems context-dependent. Another study utilizing HeLa cells depleted for the Raf Kinase Inhibitory Protein (RKIP) in which CRAF, but not BRAF, was activated, also demonstrated that ERK hyperactivation through CRAF attenuates the SAC function in a MT-stabilized condition by inhibiting the chromosome passenger protein Aurora B [35]. However, this study relied on using the MEK inhibitor PD098059 at 10μM, which has been shown by others to be insufficient to inhibit ERK activation in HeLa cells even at 100μM [22], as well as partial inhibition by dominant-negative MEK, to prove the contribution of ERK activation in this response. Further studies using more potent and specific inhibitors will be needed to confirm the significance of CRAF-induced ERK hyperactivation in this context.
Whereas it is well established that BRAF signals primarily through the ERK pathway [36], CRAF has been reported to regulate other pathways independently of MEK/ERK activation [37]. These include ASK1-p38 MAP kinase, MST2-LATS-YAP1-p73, and ROKα-LIMK-COFILIN pathways, which may potentially contribute to mitotic progression through regulating the antephase checkpoint [38], mitotic cell death [39] and the SAC [40], and spindle positioning [41] and cytokinesis [42, 43] respectively. However, none of these pathways have been extensively investigated so far in the context of mitosis in the CRAF-deficient or -activated condition.
4. Aneuploidy and W-CIN
W-CIN is defined as a persistently high rate of losses/gains of whole chromosomes, and inevitably causes aneuploidy [3, 44]. However, single catastrophic events that transiently cause losses or gains of whole chromosomes can also result in aneuploidy without persistent instability, as seen in some types of congenital disorders. Even in cancers, chromosomally stable aneuploid tumors could develop through clonal evolution of aneuploid cells generated by such a single catastrophic event. Therefore, W-CIN must be evaluated by assessing the temporal dynamics of alterations in chromosome numbers or chromosome number variations at a single cell level, when only a single time point analysis is available. Single cell assays such as karyotyping and FISH are feasible in the latter case, but array CGH or DNA ploidy analysis using bulk populations on a single time point is not suitable for evaluating W-CIN, even if modal aneuploidy can be detected using these methodologies [44]. Of note, array CGH is a strong tool for detecting whole chromosome and segmental aneuploidy when clonally evolved, genetically homogenous cell populations are analyzed. Therefore, this method is useful for evaluating W-CIN when combined with single cell genomic DNA isolation or clonal subcultures.
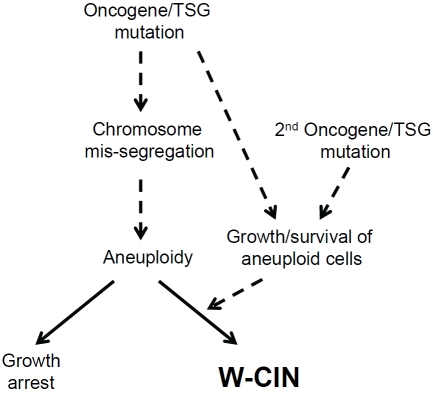
Recent studies using yeast and mouse models with stable aneuploidy have demonstrated that aneuploidy itself has a negative impact on cell proliferation [45, 46]. This was also found to be the case in aneuploid daughter cells developed from chromosomally stable, near-diploid human cancer cells following transient pharmacological perturbation of k-MT dynamics [47, 48]. In this case, proliferation of newly generated aneuploid cells was inhibited by p38 MAP kinase-mediated activation/stabilization of the tumor suppressor p53 [47]. Importantly, the cell line used in this study (HCT116) harbors the G13DKRAS oncogenic mutation, indicating that signals mediated by G13DKRAS are not sufficient for circumventing this inhibitory effect. In contrast, further aneuploidization of CIN cancer cells did not induce growth inhibition [48], indicating that this growth-inhibitory machinery is abrogated in CIN cancer cells. Thus, W-CIN can be characterized not only by an increased rate of chromosome mis-segregation, but also by cancellation of aneuploidy-induced growth inhibition (Figure 1). It is currently unknown whether abrogation of the p38/p53 pathway can entirely explain the escape from the aneuploidy-induced growth inhibition in CIN cancers, or whether other mechanisms are involved in this process. Although pharmacological MEK inhibition in the presence of functional p38/p53 has been reported to have no impact on aneuploidy-induced growth inhibition [47], it has yet to be clarified as to whether hyperactivation of the ERK pathway and/or MEK/ERK-independent CRAF effectors could be involved in escape from this growth-inhibitory machinery. Since the cell line used in this study harbors an oncogenic KRAS allele, it is important to elucidate whether oncogenic KRAS is still required for circumventing this growth inhibition when the p38/p53 pathway is disrupted.
Figure 1.
Evolution of Whole Chromosome Instability (W-CIN) in cancer. There are two distinct but intertwined characteristics that are required for evolution of CIN in cancer cells. The first is chromosomal heterogeneity that likely arises through chromosome mis-segregation and the second is compensatory mechanisms for avoiding aneuploidy-induced growth inhibition. These characteristics may be acquired either through mutation of a single oncogene/Tumour Suppressor Gene (TSG) or through sequential oncogene/TSG mutation.
5. The RAS/RAF cascade and W-CIN
5.1 Cell culture studies
Most of the early studies that evaluated the effect of oncogenic RAS on W-CIN were not ideal as they utilised indirect methodologies such as micronuclei formation [49-53]. However, a particularly elegant study by Woo and Poon utilised direct karyotyping combined with growth of cells under serum free conditions [54]. In this report, oncogenic G12VHRAS expressed in primary MEFs with a stable karyotype was shown to induce near-tetraploid aneuploidy within 24hrs of retro-viral expression [54]. This phenotype satisfies the criteria of W-CIN, and was further enhanced when G12VHRAS was expressed in p53-deficient MEFs. p53 deficiency itself did not cause significant alterations of the MEF karyotype under the same conditions [54]. These findings fit the two-step theory for W-CIN establishment; the first step involving the induction of chromosome mis -segregation as promoted by oncogenic RAS in this situation, and the second step mediated by p53 deficiency supporting the growth of the resulting aneuploid cells (Figure 1).
Consistent with this assumption, several previous studies investigating mitotic phenotypes induced by ectopic expression of mutant RAS point to a potential role for oncogenic RAS in chromosome mis-segregation [49-52]. A recent study using an isogenic pair of human cancer cells with or without an endogenous oncogenic KRAS allele demonstrated that an endogenous level of oncogenic KRAS evokes chromosome mis-segregation with mitotic stress phenotypes [55]. Although it is largely unknown how oncogenic RAS induces aberrant mitosis, it is notable that this study demonstrated synthetic lethality between oncogenic KRAS and inactivation of PLK1 [55], suggesting that oncogenic RAS could deregulate pathways functionally compensated by PLK1. PLK1 is involved in almost all aspects of mitotic progression, regulating a wide variety of downstream substrates [56], some of which, such as CDC25C, are also reportedly regulated by the ERK pathway [57]. Detailed analysis of such common targets between PLK1 and the RAS/RAF/ERK pathway could in the future provide clues as to how oncogenic RAS deregulates mitotic progression.
Another possible mechanistic link between oncogenic RAS and W-CIN has emerged from studies investigating the role of the RB/E2F pathway in mitotic progression [58-61]. The tumor suppressor RB, which is classically mapped as downstream of the RAS/RAF/ERK pathway via G1 cyclins and CDKs, has been shown to tran-scriptionally suppress the key SAC protein MAD2 through regulation of the transcription factor E2F [58], and to control centromeric localization of condensin and cohesin that are important in maintaining centromeric structure and function [59, 60]. RB depletion by gene targeting or shRNA caused MAD2 overexpression [58] and centromeric dysfunction [60], resulting in aberrant mitosis with chromosome mis-segregation and aneuploidy as confirmed by karyotyping and FISH. Although G1 cyclin/CDK activation by the RAS/RAF/ERK pathway could promote phosphorylation of RB and E2F-induced transcriptional activation of MAD2, it is currently unclear whether RB phosphorylation by upstream signals affects its centromeric function [59, 60]. Further studies elucidating the effect of post-translational modification of RB on chromosome integrity will be needed to consolidate the significance of the RB pathway in connecting the RAS/RAF cascade and W-CIN.
In addition to oncogenic RAS, a recent study utilizing human melanoma cells and primary melanocytes ectopically expressing V600EBRAF revealed that the most common cancer-associated BRAF mutant induces not only mitotic abnormalities but aneuploidization during a relatively short period as confirmed by karyotyping and FISH [33]. Given that BRAF primarily signals via the ERK pathway [36], it is most likely that the observed aneuploidization is attributable to V600EBRAF-induced ERK hyperactivation, though a causative role of the ERK pathway in the aneuploid phenotype was not directly proven in this study. Two of the melanoma cell lines used in this study for ectopic V600EBRAF expression are also known to harbor the Q61KNRAS mutation, suggesting that the aneuploid phenotype might be a consequence of the combinatorial effect of V600EBRAF and Q61KNRAS. In contrast, we have recently reported that primary splenic myeloid cells derived from Mx1-Cre;BrafLSL-V600E/+ mice, in which v600EBraf is expressed from its endogenous allele in hematopoietic cells, maintain a relatively stable karyotype in vivo and for two weeks in culture [62]. Similarly, splenocytes from Mx1Cre;KrasLSL-G12D/+ mice have also been reported to hold a stable diploid karyotype [63]. Thus, mouse hematopoietic cells could be relatively resistant to W-CIN induction by oncogenic RAS/RAF, compared to human melanocytes. It remains an open question as to what determines tissue type-specific sensitivity to W-CIN, if any. Protein expression levels of oncogenic RAS/RAF, downstream signal choice (PI3K, BRAF or CRAF) by oncogenic RAS, and compensatory mechanisms for aneuploidy-induced growth inhibition, may vary among tissue/tumor types and all of these could potentially affect development of the W-CIN phenotype.
5.2 Mouse models
5.2.1 RAS models
Since the first transgenic mouse model for oncogenic RAS was reported in 1987, numerous different models utilizing exogenous transgene expression under tissue-specific promoters or knock-in mutations of endogenous loci have been developed [9]. A few studies of these mouse models have focused on CIN and most of these suggest that the role of oncogenic RAS in W-CIN in vivo is cooperative with other genetic events, such as p53 mutation [64] or MAD2 overexpression [65], as summarized in Table 2. However, it is still somewhat vague as to whether oncogenic RAS, as a sole genetic event, could affect chromosome integrity in vivo.
Table 2.
RAS/RAF mouse models showing evidence of CIN. Although several RAS/RAF oncogenic mouse models have been reported in the past, the effects of oncogene expression on chromosome stability has only been examined in a few cases. The results of these are summarized in the table below.
| Mouse strain | Description of model | CIN phenotype | Reference |
|---|---|---|---|
| RAS models - transgenic | |||
| Villin-G12VKRAS | Expression of ectopic G12VKRAS under the control of the villin promoter and examination of effects in intestinal epithelium | None observed. 4/6 tumours analysed showed a DNA diploid profile resembling that of normal mucosa. | [66] |
| Eμ-G12DNRAS combined with p53 or Suv39h1 null alleles. | Expression of ectopic G12DNRAS under the control of the haemopoieticspecific promoter Eμ and examination of effects in lymphoid tissue. | Modest deviation from normal karyotype in Eμ-G12DNRAS lymphomas on wild-type and Suv39h1 null backgrounds. Tendency towards hyperdiploidy in Eμ-G12DNRAS;p53-null lymphomas | [67] |
| Doxycycline-inducible CCSP-rtTA transactivator with rtTA-responsive Mad2 and/or G12DKRAS transgenes | Inducible expression of ectopic Mad2 and/or G12DKRAS under the control of the Clara cell secretory protein promoter and examination of effects in lung epithelium | Higher levels of aneuploidy in G12DKRAS lung (13%) than normal (2%) but a further threefold increase in aneuploid cells (38%) in G12DKRAS plus Mad2-expressing lung. CGH analysis failed to detect any gross abnormalities. | [65] |
| RAS models – conditional knockins | |||
| LSL-G12DKRAS conditional knockin allele combined with Pdx1-Cre mouse, with or without the p53R172H/+ allele | Expression of endogenous G12DKRAS in progenitor cells of the mouse pancreas. | Aberrant karyotypes in the vast majority of pancreatic G12DKRAS; p53R172H/+ carcinoma cell lines but not from preinvasive ductal cells isolated from G12DKRAS pancreas. Abnormalities included W-CIN and S-CIN. | [64] |
| LSL-G12VKRAS/β-geo combined with CMV-Cre mouse | Expression of endogenous G12VKRAS in MEFs | Abnormal karyotypes observed in late passage MEFs involving W-CIN and S-CIN without evidence of p53 or p19ARF loss. | [70] |
| RAF models – conditional knockins | |||
| LSL-V600EBRAF combined with Mx1-Cre | Expression of endogenous V600EBRAF in haemopoietic cells and examination of effects in spleen | Stable karyotype in aberrantly growing V600EBRAF-expressing myeloid cells | [62, 81] |
| LSL-D594ABRAF combined with CMV-Cre | Expression of endogenous kinase inactive D594ABRAF constitutively and examination of effects in spleen and MEFs | W-CIN observed in aberrantly growing D594ABRAF-expressing Cd11b+ myeloid cells and MEFs without evidence of p53 or p19ARF loss. | [62] |
Using transgenic mouse models in which tissue-specific promoters drive oncogenic RAS, CIN and/or aneuploidy have been investigated in the resulting intestinal [66], lymphoid [67] and lung [65] tumors. When G12VKRAS was expressed under the control of the mouse villin promoter, more than 80% of aged mice developed intestinal tumors, ranging from adenomas with a moderate dysplasia to invasive adenocarcinomas [66]. However, none of these tumors exhibited aneuploidy when examined by flow cytometry, even in clonally-evolved advanced (invasive) adenocarcinomas [66]. Although flow cytometry is not sufficient for detecting cell-to-cell variability of chromosome numbers [44], these tumors also failed to display loss of heterozygosity (LOH) of the Apc locus. Since W-CIN has been suggested to promote tumorigenesis, at least in part through facilitating LOH of tumor suppressor loci [68], this study collectively implies that oncogenic KRAS in intestinal epithelial tumors may not induce W-CIN.
In contrast, T-cell lymphomas developed in Eμ-NRASG12D mice, in which mutant NRAS was driven by the immunoglobulin heavy (μ)-chain enhancer, have been reported to exhibit aberrant karyotypes involving translocations between chromosomes 6 and 15, and gains of chromosomes 10 and 17, indicative of S-CIN and W-CIN respectively [67]. In this model, some tumors displayed modal hyperdiploidy with significant variability of chromosome numbers even in the limited number of metaphases analyzed. Interestingly, this phenotype was further worsened on a p53-deficient background, suggesting the possible synergy between oncogenic NRAS and p53 deficiency in W-CIN induction.
Transgenic mice expressing ectopic G12DKRAS specifically in alveolar epithelial cells in a chemically inducible manner have been developed [65] and, in this model, G12DKRAS expression in alveolar epithelium resulted in the development of lung adenocarcinomas. These tumors exhibited higher levels of aneuploidy as detected by FISH than wild-type or MAD2-overexpressing lung epithelium, indicating that oncogenic KRAS might promote W-CIN more drastically than the deregulated SAC function caused by exogenous MAD2 expression. Intriguingly, when both G12DKRAS and exogenous MAD2 expression were induced, a strong synergistic effect on aneuploidy induction was observed. In this context, it is currently unknown as to whether oncogenic KRAS enhanced chromosome mis-segregation, or compensated for the growth inhibition of aneuploid cells generated by perturbation of MAD2 expression, or both.
Controlling RAS transgene expression within physiologically relevant levels is quite challenging in mouse models using the exogenous promoters described above. To overcome this problem, conditional knock-in mouse models, in which oncogenic KRAS is expressed from the endogenous locus after Cre recombinase (Cre)-mediated removal of a lox-stop-lox (LSL) element were developed by two independent research groups [69, 70]. The first model was designed to express G12DKRAS (hereafter referred to as G12D), and has been used as a model for lung [69], colon [71, 72], hematopoietic [63, 73] and pancreatic [64, 74] tumorigenesis. Notably, primary pancreatic ductal adenocarcinoma cells arising in the G12D model with Pdx1Cre on a p53R172H/+ background display a highly aberrant karyotype with characteristics of both W-CIN and S-CIN [64]. Again, the synergy between oncogenic KRAS and p53 deficiency regarding CIN induction was obvious. This was in contrast to preinvasive ductal cells with G12D alone, in which two thirds of metaphase spreads displayed a diploid karyotype in a similar way to normal (wild-type) pancreatic ductal cells, or the mice with p53 mutation alone which did not develop pancreatic tumors. However, in the future, detailed cytogenetic analyses of the preinvasive lesions and advanced tumors that arise in the context of G12D alone will be needed to elucidate any roles for G12DKRAS as a sole driver of CIN during pancreatic carcinogenesis. On the other hand, myeloproliferative neoplasms developed by crossing the G12D model to the Mx1Cre strain were reported to maintain a stable diploid karyotype [63]. Unfortunately, CIN phenotypes in colon and lung neoplasms arising in the G12D model have not been reported as yet.
The second knock-in KRAS model incorporated a bicistronic construct to express G12VKRAS as well as a marker gene β-geo (hereafter referred to as G12V/β-geo) [70]. Using this model, CIN was investigated in MEFs, in which G12VKRAS expression was induced by the CMVCre transgene. Although karyotype abnormalities were not evident in primary MEFs, tetraploidization and structural chromosome abnormalities were prominent at later passages, suggesting that in vitro culture stresses such as ROS production might contribute to the CIN phenotype. Since tetraploidization was commonly observed in both mutant and control MEFs at late passage, S-CIN rather than W-CIN seemed to be the predominant characteristic induced by G12VKRAS in this system. Surprisingly, in the G12V/β-geo model, no overt tumour formation was observed in adult epithelial tissues except for the lung when G12VKRAS expression was induced by CMVCre or 4OHT-induced CreERT activation, making it difficult to follow the effects of G12VKRAS on CIN during in vivo carcinogenesis.
A number of possible explanations for the discrepancy between the G12V/β-geo and G12D models have been previously proposed [72]. We also believe that the different Cre expression/activation systems used in these two studies may be a critical factor in determining the distinct phenotypes. In the G12D model, tissue-specific but constitutively expressed (activated) Cre strains were used to induce solid tumors in most cases, except for the induction of lung cancers (adenovirus-Cre) and skin papillomas (K14-CreERT). In contrast, the CMVCre strain used in the G12V/β-geo model is thought to possess relatively weak recombinase activity, based on the fact that it has been reported to induce only mosaic recombination for some loxP flanked lesions [75] and its expression in primary MEFs does not evoke growth inhibition [62] or karyotype abnormalities [70]. Constitutively expressed Cre causes continuous genome damage [76-78] and this could, potentially, cooperate with oncogenic RAS in inducing carcinogenesis or, more specifically, the CIN phenotype. Overall, it would seem to be important to test if the CIN phenotypes observed in the various oncogenic RAS models can be reproduced in the absence of Cre or with weak, transient Cre activation.
5.2.2 RAF models
Within a decade since human cancer-associated BRAF mutations were first reported [11], transgenic mouse models for the most common V600EBRAF mutant using either tissue-specific, chemically-inducible promoters [79, 80] or conditional knock-in strategies involving the LSL element [81-83], have already been developed by several independent research groups including our own. However, expression of V600EBRAF in alveolar [82]/intestinal [84] epithelial cells or skin melanocytes [85] using knockin models consistently promotes oncogene-induced senescence (OIS) and thus a time window for analyzing CIN in these models is limited to an initial proliferation phase that occurs before the OIS program is engaged. As mentioned earlier, we have recently taken advantage of our conditional V600EBRAF knock-in mouse model intercrossed with the poly I:C-inducible Mx1Cre strain [62] to investigate the role of V600EBRAF in CIN. In this model, fetal myeloproliferative neoplasia spontaneously develops within a short time after birth even without poly I:C injection [81]. Cre-induced genome damage is minimized in this situation by the fact that poly I:C-induced Cre activation is avoided and, furthermore, the aberrantly growing mye-loid cells in this strain do not exhibit obvious OIS responses. Interestingly, primary splenocytes derived from the MxlCre; BrafV600E/+ mice maintain a stable diploid karyotype in vivo as well as following culture [62], indicating that hyperactivation of the BRAF/ERK pathway in the myeloid lineage does not compromise chromosome stability. Similar analyses for clinically-relevant cell/tissue types such as melanocytes, thyroid, and colon will be needed to clarify whether V600E-BRAF is involved in W-CIN and, if so, whether tissue-specific differences exist.
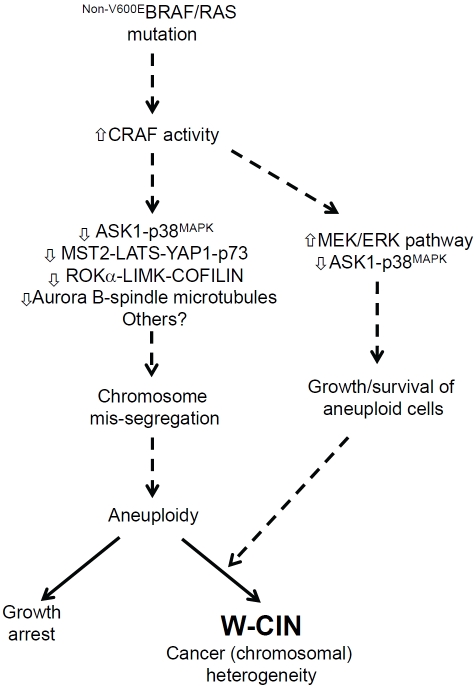
Although the V600EBRAF mutant is detected in more than 90% of cancers with BRAF mutations, a number of other residues in BRAF are mutated at a lower frequency [86]. Aspartate 594 is the fourth most common BRAF residue mutated in human cancer and, interestingly, mutants of this residue are kinase inactive. The mechanisms by which these mutants contribute to tumour development has been an unresolved question. To investigate this, we have generated a conditional knock-in mouse model for D594ABRAF and recently reported its characterization [62, 87]. We have found that constitutive, heterozygous expression of this mutant in mice promotes W-CIN/aneuploidization of primary MEFs and splenic myeloid cells, even in the absence of the Cre recombinase [62]. This mutant was also found to heterodimerise with CRAF [87], inducing its hyperactivation and activation of the downstream MEK/ERK pathway. The W-CIN phenotype of cultured splenocytes was rescued by chemical inhibition using the RAF inhibitor sorafenib and by intercrossing the D594ABraf mice with kinase-inactive D486ACraf mice. However, MEK inhibition using the U0126 was unable to rescue W-CIN but inhibited the growth of aneuploid cells. Even though sorafenib suppressed the MEK/ERK pathway to some degree, it was found to promote splenocyte growth, presumably because it allows the retention of diploid cells that have a growth advantage. The fact that hyperactivation of the MEK/ERK pathway by itself is not sufficient to induce the emergence of W-CIN is consistent with analysis of mouse splenocytes expressing V600EBRAF mentioned above that retain a diploid karyotype. Taken together, these data suggest a model in which CRAF transactivation by D594ABRAF functions through MEK-independent pathways to promote W-CIN and through MEK-dependent pathways to promote the growth of aneuploid cells (Figure 2). As such this model fits the previously proposed scenario (Figure 1) whereby CIN arises through cooperation between aberrant signaling pathways that, on the one hand, promote chromosome mis-segregation and, on the other, promote the growth of the resulting aneuploid cells.
Figure 2.
Role of CRAF in the evolution of Whole Chromosome Instability (W-CIN). Our investigations have provided evidence for a role of deregulated CRAF in the evolution of W-CIN [62, 87]. This finding came about through studies of impaired activity mutations of BRAF that are detected in human cancer and are involved in the cancer phenotype through their ability to heterodimerise and transactivate CRAF. It was found that deregulated CRAF was involved in driving the evolution of CIN downstream of mutant BRAF. However, the consequent deregulation of the MEK/ERK pathway was found not to be involved in the emergence of aneuploidy but, rather, was found to be required for the growth of aneuploid cells. This discovery fits the previously proposed dual model for the evolution of W-CIN (Figure 1). A number of MEK/ERK-independent effector pathways have been proposed previously for CRAF, as indicated, but which one(s) of these are involved in the two pathways leading to the evolution of W-CIN are not currently known. CRAF is a known effector of oncogenic RAS, and it will be important to assess in the future whether the emergence of W-CIN in RAS mutant tumours occurs through similar pathways. Our studies also ruled out a role for the most common BRAF mutant, V600EBRAF, as a single mutation in the evolution of W-CIN, which fits with the observation thatV600E-BRAF does not evidently signal through CRAF [86] and with the fact the V600EBRAF mutation is primarily linked with CIN-low CRCs [94]. However, V600EBRAF may function as a second oncogene to support growth/survival of aneuploid cells generated by the first oncogene/TSG in some types of cancers such as melanoma.
As mentioned above, CRAF has been suggested to activate a number of downstream signaling pathways in addition to the MEK/ERK pathway and an important next step will be to identify which of these CRAF effectors pathways is involved in chromosome mis-segregation and aneuploidy-induced growth. In preliminary investigations, Rok-a activity was found not to be altered in D594ABRAF-expressing cells synchronized at mitosis, suggesting the involvement of alternative pathways in the CIN response. In addition, it will be important to elucidate whether the same mechanisms operate in the context of other oncogenes acting upstream of CRAF, particularly RAS isoforms and Receptor Tyrosine Kinases (RTKs) that are mutated at a high frequency in human cancers. At least in human melanoma and mouse lung cancer, oncogenic RAS has been shown to be dependent on CRAF rather than BRAF [88, 89], raising the possibility that CRAF might be involved in W-CIN in these cancers.
5.3 Human colorectal cancer (CRC) studies
As mentioned earlier, the presence of aneuploid cells in human tumor samples does not necessarily mean W-CIN [44]. Because the evaluation of dynamic changes of karyotypes in culture is usually not feasible for primary human tumor samples, cell-to-cell variability of chromosome numbers as assessed by karyotyping or interphase FISH of primary tumour sections are the only realistic indicators of W-CIN. However, such examinations have not been routinely performed in clinical practice for solid cancers. Recently developed technologies including array CGH and SNP array are powerful tools for identifying aneuploidy, LOH and copy number variations when these abnormalities are homogenously distributed in the sample, but useless for evaluating on-going instability unless single cell or subclonal analysis is combined [44]. Thus, the major limitation in clinical studies for CIN is that the existence of aneuploidy has been used as a surrogate marker, but instability itself has not been directly measured. In this situation, it is important to validate the accuracy of static aneuploidy as a marker to estimate dynamic W-CIN, which might vary among tissues/tumor-types.
It has been well established that colon cancer lines with mismatch repair (MMR) deficiency maintain relatively stable, near-diploid karyotypes, but display microsatellite instability (MSI), another form of genomic instability [3]. On the other hand, MMR-proficient, microsatellite stable (MSS) colon cancer lines, which are usually aneuploid, have been reported to exhibit massively increased variability of chromosome number in clonal cultures [90], suggesting that static aneuploidy with the MSS phenotype in CRCs could faithfully reflect on-going W-CIN. Consistently, most near-diploid lines derived from colon cancers in the NCI-60 panel show minimal numerical heterogeneity in their karyotypes (which means they are chromosomally stable), whereas most aneuploid colon cancer lines in this panel display significant numerical heterogeneity (instability) [91]. In contrast, some ovarian, breast, and renal cancer lines display minimal numerical heterogeneity despite their aneuploid (hyper-diploid or near-triploid) karyotypes [91], suggesting that aneuploidy in these types of cancers does not specify W-CIN tumors. Thus, CRCs are ideal materials for correlation studies between BRAF/KRAS mutations and W-CIN (= aneuploidy = MSS) in humans.
KRAS mutations are detected in both MSI-high/CIN-negative and MSS/CIN+ CRCs, though the frequency is higher in MSS tumors [92-95]. Interestingly, while most mutations in MSS tumors (>80%) occur in codon 12 (G12) [92, 95], more than 50% of mutations in MSI-high tumors with hereditary MMR deficiency are in codon 13 (G13) [92], implying that there are potential differences in oncogenic functions between G12 and G13 mutants as well as in mutagenic mechanisms involved in the development of MSS/CIN+ and MSI-high/CIN-negative CRCs. KRAS mutations in MSS tumors are consistently detected throughout early-stage (Dukes' A) to advanced (Dukes' D) CRCs [93, 96], indicating that G12 mutations contribute to the establishment of early-stage MSS/CIN+ tumors [97, 98]. In MSI-high CRCs, however, the frequency of the mutations drastically increases during the progression from Dukes' A to Dukes' C [93], suggesting that the majority of KRAS mutations in CIN-negative tumors occur during a relatively late phase after MMR deficiency is established. These observations suggest a potential link between G12 mutations and CIN in the precancerous (adenoma) stage of MSS CRC development, whereas G13 mutations seem to facilitate later tumor progression without promoting CIN in MMR-deficient, MSI-high CRCs. Since more than two thirds of KRAS-mutated CRCs are reported not to harbor p53 mutation [99], oncogenic KRAS might be linked to CIN in MSS CRCs independently of p53 mutations.
On the other hand, V600EBRAF mutation is strongly associated with the CpG island methylator phenotype (CIMP) [98, 100, 101], which often involves promoter hypermethylation of MMR genes causing sporadic MSI-high (CIMP-high/CIN-negative) CRCs [102]. In addition, some CRCs with the V600EBRAF mutation are characterized by the unique MSS/CIMP-high phenotype, which shares clinicopathological features with sporadic MSI-high CRCs, including a predominant proximal localization, serrated and mucinous morphology with poor differentiation, and near-diploid ploidy with little evidence of CIN [94]. Thus, in a similar way to the mouse studies described above, the V600EBRAF mutation is primarily linked to the CIN-low/negative phenotype, at least in CRCs. In contrast, the kinase impaired D594BRAF mutations, are reportedly associated with CRCs with the MSS/CIMP-low phenotype [103] and have distinct gene expression profiles [104] that are more similar to MSS/CIN+ CRCs with KRAS mutations than CIMP-high/CIN-negative, V600EBRAF tumors. These data suggest a potential link between non-V600E (especially D594) BRAF mutations and CIN in CRCs. However, it will be important to analyse larger cohorts of CRCs to consolidate this link as well as to investigate the underlying mechanisms, and particularly to identify whether CRAF plays a key role in the emergence of CIN in this context.
6. Future perspectives
6.1 Resolving the complexity of the RAS/RAF pathway
Despite significant progress over the last few decades in cancer biology related to RAS and RAF oncogenes, the mechanistic basis of how RAS/RAF signals contribute to chromosome segregation is yet largely unknown. The involvement of multiple RAS/RAF genes and multiple distinct RAS/RAF mutations makes research in this field extremely complicated [9]. Each of the three RAS genes is mutated in a different spectrum of human cancers (Table 1) and mutations affect a number of different residues within each [10]. It will ultimately be necessary to elucidate the roles for gene/mutation combinations in clinically relevant contexts and, to this end, studies using knock-in mouse models for NRAS/HRAS mutations at the codons 12 and 61 have just started [72]. Another complexity is that the three different RAF homologues can be activated to different extents by oncogenic RAS and, given the data shown above that hyperactive BRAF and CRAF may cause different types of genomic instability, it will be important to know which RAF homologue(s) is activated by oncogenic RAS in each tumour type. At least in human melanoma cells with KRAS/NRAS Q61 mutations and mouse lung cells with the G12VKRAS mutation, ERK activation relies on CRAF but not on BRAF [88, 89], indicating that oncogenic RAS preferentially activates CRAF in these contexts. This type of analysis will be needed in other cancers with RAS mutations.
Similarly, although the majority of oncogenic BRAF mutations occur at the V600 residue, BRAF point mutations at non-V600 residues [11, 12, 86] as well as gene rearrangements generating gene fusions that contain the BRAF kinase domain [13-16] have been described in various types of human cancers. Given data indicating that V600E and non-V600E (including D594) mutations may be involved in distinct types of genomic instability [62, 103], a role for each distinct BRAF mutation or rearranged BRAF fusion protein in MSI and W-CIN/S-CIN must be individually investigated in relevant cell/tissue types, such as melanoma and thyroid cancers for V600E [11, 12, 105], and lung cancers for non-V600E mutations [106, 107]. In addition, although there are only a few reported cases of CRAF mutations in human cancer, chromosomal translocations generating fusion transcripts containing the kinase domain of CRAF have also been found in prostate cancer and pilocytic astrocytoma as with BRAF (Table 1) [13, 14]. Examination of the role of CRAF in inducing CIN in these various cancers will be an important next step.
6.2 Aneuploidy/CIN-targeting therapeutics
With respect to therapeutic development, a comprehensive understanding of the functional linkage between the RAS/RAF pathway and W-CIN may provide new therapeutic options for treating CIN-positive cancers. As described above, PLK1 and some other mitotic proteins have been identified in genome-wide RNAi synthetic lethal screen for oncogenic KRAS, and cell lines mutated for KRAS are hypersensitive to a PLK inhibitor and proteasome inhibitors [55]. Unfortunately, the cell lines (DLD-1 and HCT116) mainly used in this study were MMR-deficient, MSI-high/CIN-negative CRC cell lines, which leaves it an open question as to whether CIN cancers mutated for KRAS are also sensitive to PLK and/or proteasome inhibitors.
In CIN cancer cells, as indicated in Figure 1, there are two distinct but intertwined characteristics that might potentially be targeted by cancer-specific therapeutics. One is chromosomal (karyotypic) heterogeneity [108, 109] that likely drives clonal evolution of drug-resistant and/or metastatic subclones [110, 111], and the other is the compensative machinery against aneuploidy-induced growth inhibition [4, 112], which could be essential for the growth of CIN cancers but may be dispensable for normal diploid cells. A recent study using a conditional overexpressing G12DKRAS/MAD2 transgenic mouse model for lung cancer demonstrated that the enhanced CIN that occurred following co-overexpression of G12DKRAS and MAD2 increased tumour recurrence after abrogation of transgene expression [65]. It was argued that this increased relapse rate must be due to chromosomal heterogeneity established by preexisting CIN before abrogation of the transgenes. This study provides a cautionary note to current therapeutic interventions as it suggests that chromosomal heterogeneity induced by W-CIN promotes the emergence of therapy-resistant subclones lacking the primary driver oncogenes. Thus, late intervention against ongoing W-CIN in already established cancers might be ineffective in preventing cancer recurrence following treatment.
On the other hand, it has been proposed that imbalances in protein subunit stoichiometry caused by aneuploidy could evoke proteotoxic stress [2, 112] that attenuates cell proliferation and/or induces apoptotic cell death (aneuploidy -induced growth inhibition). CIN cancer cells are assumed to develop mechanisms such as p38/p53 inactivation to evade such stress responses [4, 47], but are still more sensitive to further stress loading by proteotoxic compounds like AICAR and 17-AAG than normal diploid or near-diploid MSI cancer cells [113]. Thus, the identification of pathways controlling aneuploidy -induced proteotoxicity/growth inhibition in CIN cancers will be a vital step towards the development of aneuploidy/CIN-targeting therapeutics. In this context, it will continue to be of great interest to determine whether deregulation of the RAS/RAF cascade in CIN cancers contributes to the surpassing of aneuploidy-induced proteotoxicity/growth inhibition, and if so, how pharmacological modulation of the RAS/RAF pathway affects these processes.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 4.Thompson SL, Compton DA. Chromosomes and cancer cells. Chromosome Res. 2011;193:433–444. doi: 10.1007/s10577-010-9179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schvartzman JM, Sotillo R, Benezra R. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat Rev Cancer. 2010;10:102–115. doi: 10.1038/nrc2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cimini D. Merotelic kinetochore orientation, aneuploidy, and cancer. Biochim Biophys Acta. 2008;1786:32–40. doi: 10.1016/j.bbcan.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Barber TD, McManus K, Yuen KW, Reis M, Parmigiani G, Shen D, Barrett I, Nouhi Y, Spencer F, Markowitz S, Velculescu VE, Kinzler KW, Vogelstein B, Lengauer C, Hieter P. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc Natl Acad Sci U S A. 2008;105:3443–3448. doi: 10.1073/pnas.0712384105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pearson G, Robinson F, Beers Gibson T, Xu BE, Karandikar M, Berman K, Cobb MH. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 9.Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;6:459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 10.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 11.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 12.Garnett MJ, Marais R. Guilty as charged: B-RAF is a human oncogene. Cancer Cell. 2004;6:313–319. doi: 10.1016/j.ccr.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 13.Palanisamy N, Ateeq B, Kalyana-Sundaram S, Pflueger D, Ramnarayanan K, Shankar S, Han B, Cao Q, Cao X, Suleman K, Kumar-Sinha C, Dhanasekaran SM, Chen YB, Esgueva R, Banerjee S, LaFargue CJ, Siddiqui J, Demichelis F, Moeller P, Bismar TA, Kuefer R, Fullen DR, Johnson TM, Greenson JK, Giordano TJ, Tan P, Tomlins SA, Varambally S, Rubin MA, Maher CA, Chinnaiyan AM. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat Med. 2010;16:793–798. doi: 10.1038/nm.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones DT, Kocialkowski S, Liu L, Pearson DM, Ichimura K, Collins VP. Oncogenic RAF1 rearrangement and a novel BRAF mutation as alternatives to KIAA1549:BRAF fusion in activating the MAPK pathway in pilocytic astrocytoma. Oncogene. 2009;28:2119–2123. doi: 10.1038/onc.2009.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones DT, Kocialkowski S, Liu L, Pearson DM, Bäcklund LM, Ichimura K, Collins VP. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008;68:8673–8677. doi: 10.1158/0008-5472.CAN-08-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciampi R, Knauf JA, Kerler R, Gandhi M, Zhu Z, Nikiforova MN, Rabes HM, Fagin JA, Nikiforov YE. Oncogenic AKAP9-BRAF fusion is a novel mechanism of MAPK pathway activation in thyroid cancer. J Clin Invest. 2005;115:94–101. doi: 10.1172/JCI23237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin AW, Barradas M, Stone JC, van Aelst L, Serrano M, Lowe SW. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev. 1998;19:3008–3019. doi: 10.1101/gad.12.19.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu J, Woods D, McMahon M, Bishop JM. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev. 1998;19:2997–3007. doi: 10.1101/gad.12.19.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torii S, Nakayama K, Yamamoto T, Nishida E. Regulatory mechanisms and function of ERK MAP kinases. J Biochem. 2004;136:557–561. doi: 10.1093/jb/mvh159. [DOI] [PubMed] [Google Scholar]
- 20.Bodart JF. Extracellular-regulated kinase-mitogen-activated protein kinase cascade: unsolved issues. J Cell Biochem. 2010;109:850–857. doi: 10.1002/jcb.22477. [DOI] [PubMed] [Google Scholar]
- 21.Tamemoto H, Kadowaki T, Tobe K, Ueki K, Izumi T, Chatani Y, Kohno M, Kasuga M, Yazaki Y, Akanuma Y. Biphasic activation of two mitogen-activated protein kinases during the cell cycle in mammalian cells. J Biol Chem. 1992;267:20293–20297. [PubMed] [Google Scholar]
- 22.Roberts EC, Shapiro PS, Nahreini TS, Pages G, Pouyssegur J, Ahn NG. Distinct cell cycle timing requirements for extracellular signal-regulated kinase and phosphoinositide 3-kinase signaling pathways in somatic cell mitosis. Mol Cell Biol. 2002;22:7226–7241. doi: 10.1128/MCB.22.20.7226-7241.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayne C, Tzivion G, Luo Z. Raf-1/MEK/MAPK pathway is necessary for the G2/M transition induced by nocodazole. J. Biol. Chem. 2000;275:31876–31882. doi: 10.1074/jbc.M002766200. [DOI] [PubMed] [Google Scholar]
- 24.Wright JH, Munar E, Jameson DR, Andreassen PR, Margolis RL, Seger R, Krebs EG. Mitogen-activated protein kinase kinase activity is required for the G(2)/M transition of the cell cycle in mammalian fibroblasts. Proc Natl Acad Sci USA. 1999;96:11335–11340. doi: 10.1073/pnas.96.20.11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu X, Yan S, Zhou T, Terada Y, Erikson RL. The MAP kinase pathway is required for entry into mitosis and cell survival. Oncogene. 2004;23:763–776. doi: 10.1038/sj.onc.1207188. [DOI] [PubMed] [Google Scholar]
- 26.Shinohara M, Mikhailov AV, Aguirre-Ghiso JA, Rieder CL. Extracellular signal-regulated kinase 1/2 activity is not required in mammalian cells during late G2 for timely entry into or exit from mitosis. Mol Biol Cell. 2006;17:5227–5240. doi: 10.1091/mbc.E06-04-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dumesic PA, Scholl FA, Barragan DI, Khavari PA. Erk1/2 MAP kinases are required for epidermal G2/M progression. J Cell Biol. 2009;185:409–422. doi: 10.1083/jcb.200804038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shapiro PS, Vaisberg E, Hunt AJ, Tolwinski NS, Whalen AM, Mclntosh JR, Ahn NG. Activation of the MKK/ERK pathway during somatic cell mitosis: direct interactions of active ERK with kinetochores and regulation of the mitotic 3F3/2 phosphoantigen. J Cell Biol. 1998;142:1533–1545. doi: 10.1083/jcb.142.6.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zecevic M, Catling AD, Eblen ST, Renzi L, Hittle JC, Yen TJ, Gorbsky GJ, Weber MJ. Active MAP kinase in mitosis: localization at kinetochores and association with the motor protein CENP-E. J Cell Biol. 1998;142:1547–1558. doi: 10.1083/jcb.142.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lou Y, Xie W, Zhang DF, Yao JH, Luo ZF, Wang YZ, Shi YY, Yao XB. NEK2 specifies the centrosomal location of ERK2. Biochem Biophys Res Commun. 2004;321:495–501. doi: 10.1016/j.bbrc.2004.06.171. [DOI] [PubMed] [Google Scholar]
- 31.Hayne C, Xiang X, Luo Z. MEK inhibition and phosphorylation of serine 4 on B23 are two coincident events in mitosis. Biochem Biophys Res Commun. 2004;321:675–80. doi: 10.1016/j.bbrc.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 32.Cui Y, Guadagno TM. B-Raf(V600E) signaling deregulates the mitotic spindle checkpoint through stabilizing Mps1 levels in melanoma cells. Oncogene. 2008;27:3122–3133. doi: 10.1038/sj.onc.1210972. [DOI] [PubMed] [Google Scholar]
- 33.Cui Y, Borysova MK, Johnson JO, Guadagno TM. Oncogenic B-Raf(V600E) induces spindle abnormalities, supernumerary centrosomes, and aneuploidy in human melanocytic cells. Cancer Res. 2010;70:675–684. doi: 10.1158/0008-5472.CAN-09-1491. [DOI] [PubMed] [Google Scholar]
- 34.Knauf JA, Ouyang B, Knudsen ES, Fukasawa K, Babcock G, Fagin JA. Oncogenic RAS induces accelerated transition through G2/M and promotes defects in the G2 DNA damage and mitotic spindle checkpoints. J Biol Chem. 2006;281:380–3809. doi: 10.1074/jbc.M511690200. [DOI] [PubMed] [Google Scholar]
- 35.Eves EM, Shapiro P, Naik K, Klein UR, Trakul N, Rosner MR. Raf kinase inhibitory protein regulates aurora B kinase and the spindle checkpoint. Mol Cell. 2006;23:561–674. doi: 10.1016/j.molcel.2006.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galabova-Kovacs G, Kolbus A, Matzen D, Meissl K, Piazzolla D, Rubiolo C, Steinitz K, Baccarini M. ERK and beyond: insights from B-Raf and Raf-1 conditional knockouts. Cell Cycle. 2006;5:1514–1518. doi: 10.4161/cc.5.14.2981. [DOI] [PubMed] [Google Scholar]
- 37.Niault TS, Baccarini M. Targets of Raf in tumorigenesis. Carcinogenesis. 2010;31:1165–1174. doi: 10.1093/carcin/bgp337. [DOI] [PubMed] [Google Scholar]
- 38.Mikhailov A, Shinohara M, Rieder CL. The p38-mediated stress-activated checkpoint. A rapid response system for delaying progression through antephase and entry into mitosis. Cell Cycle. 2005;41:57–62. doi: 10.4161/cc.4.1.1357. [DOI] [PubMed] [Google Scholar]
- 39.Niikura Y, Dixit A, Scott R, Perkins G, Kitagawa K. BUB1 mediation of caspase-independent mitotic death determines cell fate. J Cell Biol. 2007;178:283–296. doi: 10.1083/jcb.200702134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomasini R, Tsuchihara K, Tsuda C, Lau SK, Wilhelm M, Ruffini A, Tsao MS, lovanna JL, Jurisicova A, Melino G, Mak TW. TAp73 regulates the spindle assembly checkpoint by modulating BubR1 activity. Proc Natl Acad Sci U S A. 2009;106:797–802. doi: 10.1073/pnas.0812096106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaji N, Muramoto A, Mizuno K. LIM kinase-mediated cofilin phosphorylation during mitosis is required for precise spindle positioning. J Biol Chem. 2008;283:4983–4992. doi: 10.1074/jbc.M708644200. [DOI] [PubMed] [Google Scholar]
- 42.Kaji N, Ohashi K, Shuin M, Niwa R, Uemura T, Mizuno K. Cell cycle-associated changes in Slingshot phosphatase activity and roles in cytokinesis in animal cells. J Biol Chem. 2003;278:33450–33455. doi: 10.1074/jbc.M305802200. [DOI] [PubMed] [Google Scholar]
- 43.Gohla A, Birkenfeld J, Bokoch GM. Chronophin, a novel HAD-type serine protein phosphatase, regulates cofilin-dependent actin dynamics. Nat Cell Biol. 2005;7:21–29. doi: 10.1038/ncb1201. [DOI] [PubMed] [Google Scholar]
- 44.Geigl JB, Obenauf AC, Schwarzbraun T, Speicher MR. Defining ‘chromosomal instability’. Trends Genet. 2008;24:64–69. doi: 10.1016/j.tig.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 45.Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 46.Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol. 2010;188:369–381. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol. 2008;180:665–672. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saavedra HI, Knauf JA, Shirokawa JM, Wang J, Ouyang B, Elisei R, Stambrook PJ, Fagin JA. The RAS oncogene induces genomic instability in thyroid PCCL3 cells via the MAPK pathway. Oncogene. 2000;19:3948–3954. doi: 10.1038/sj.onc.1203723. [DOI] [PubMed] [Google Scholar]
- 50.Saavedra HI, Fukasawa K, Conn CW, Stambrook PJ. MAPK mediates RAS-induced chromosome instability. J Biol Chem. 1999;274:38083–38090. doi: 10.1074/jbc.274.53.38083. [DOI] [PubMed] [Google Scholar]
- 51.Hagag N, Diamond L, Palermo R, Lyubsky S. High expression of ras p21 correlates with increased rate of abnormal mitosis in NIH3T3 cells. Oncogene. 1990;5:1481–1489. [PubMed] [Google Scholar]
- 52.Denko N, Stringer J, Wani M, Stambrook P. Mitotic and post mitotic consequences of genomic instability induced by oncogenic Ha-ras. Somat Cell Mol Genet. 1995;21:241–253. doi: 10.1007/BF02255779. [DOI] [PubMed] [Google Scholar]
- 53.Abulaiti A, Fikaris AJ, Tsygankova OM, Meinkoth JL. Ras induces chromosome instability and abrogation of the DNA damage response. Cancer Res. 2006;66:10505–10512. doi: 10.1158/0008-5472.CAN-06-2351. [DOI] [PubMed] [Google Scholar]
- 54.Woo RA, Poon RY. Activated oncogenes promote and cooperate with chromosomal instability for neoplastic transformation. Genes Dev. 2004;18:1317–1330. doi: 10.1101/gad.1165204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, Wong KK, Elledge SJ. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barr FA, Silljé HH, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat Rev Mol Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- 57.Wang R, He G, Nelman-Gonzalez M, Ashorn CL, Gallick GE, Stukenberg PT, Kirschner MW, Kuang J. Regulation of Cdc25C by ERK-MAP kinases during the G2/M transition. Cell. 2007;128:1119–1132. doi: 10.1016/j.cell.2006.11.053. [DOI] [PubMed] [Google Scholar]
- 58.Hernando E, Nahlé Z, Juan G, Diaz-Rodriguez E, Alaminos M, Hemann M, Michel L, Mittal V, Gerald W, Benezra R, Lowe SW, Cordon-Cardo C. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 2004;430:797–802. doi: 10.1038/nature02820. [DOI] [PubMed] [Google Scholar]
- 59.Coschi CH, Martens AL, Ritchie K, Francis SM, Chakrabarti S, Berube NG, Dick FA. Mitotic chromosome condensation mediated by the retinoblastoma protein is tumor-suppressive. Genes Dev. 2010;24:1351–1363. doi: 10.1101/gad.1917610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Manning AL, Longworth MS, Dyson NJ. Loss of pRB causes centromere dysfunction and chromosomal instability. Genes Dev. 2010;24:1364–1376. doi: 10.1101/gad.1917310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Harn T, Foijer F, van Vugt M, Banerjee R, Yang F, Oostra A, Joenje H, te Riele H. Loss of Rb proteins causes genomic instability in the absence of mitogenic signaling. Genes Dev. 2010;24:1377–1388. doi: 10.1101/gad.580710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kamata T, Hussain J, Giblett S, Hayward R, Marais R, Pritchard C. BRAF inactivation drives aneuploidy by deregulating CRAF. Cancer Res. 2010;70:8475–8486. doi: 10.1158/0008-5472.CAN-10-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Braun BS, Tuveson DA, Kong N, Le DT, Kogan SC, Rozmus J, Le Beau MM, Jacks TE, Shannon KM. Somatic activation of oncogenic Kras in hematopoietic cells initiates a rapidly fatal myeloproliferative disorder. Proc Natl Acad Sci USA. 2004;101:597–602. doi: 10.1073/pnas.0307203101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, Tuveson DA. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 65.Sotillo R, Schvartzman JM, Socci ND, Benezra R. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature. 2010;464:436–440. doi: 10.1038/nature08803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Janssen KP, el-Marjou F, Pinto D, Sastre X, Rouillard D, Fouquet C, Soussi T, Louvard D, Robine S. Targeted expression of oncogenic K-ras in intestinal epithelium causes spontaneous tumorigenesis in mice. Gastroenterology. 2002;123:492–504. doi: 10.1053/gast.2002.34786. [DOI] [PubMed] [Google Scholar]
- 67.Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, Stein H, Dorken B, Jenuwein T, Schmitt CA. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436:660–665. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- 68.Baker DJ, Jin F, Jeganathan KB, van Deursen JM. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell. 2009;16:475–486. doi: 10.1016/j.ccr.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guerra C, Mijimolle N, Dhawahir A, Dubus P, Barradas M, Serrano M, Campuzano V, Barbacid M. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell. 2003;4:111–120. doi: 10.1016/s1535-6108(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 71.Tuveson DA, Shaw AT, Willis NA, Silver DP, Jackson EL, Chang S, Mercer KL, Grochow R, Hock H, Crowley D, Hingorani SR, Zaks T, King C, Jacobetz MA, Wang L, Bronson RT, Orkin SH, DePinho RA, Jacks T. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5:375–387. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- 72.Haigis KM, Kendall KR, Wang Y, Cheung A, Haigis MC, Glickman JN, Niwa-Kawakita M, Sweet-Cordero A, Sebolt-Leopold J, Shannon KM, Settleman J, Giovannini M, Jacks T. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008;40:600–608. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chan IT, Kutok JL, Williams IR, Cohen S, Kelly L, Shigematsu H, Johnson L, Akashi K, Tuveson DA, Jacks T, Gilliland DG. Conditional expression of oncogenic K-ras from its endogenous promoter induces a myeloproliferative disease. J Clin Invest. 2004;113:528–538. doi: 10.1172/JCI20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, Kawaguchi Y, Johann D, Liotta LA, Crawford HC, Putt ME, Jacks T, Wright CV, Hruban RH, Lowy AM, Tuveson DA. Pre-invasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 75.Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Loonstra A, Vooijs M, Beverloo HB, Allak BA, van Drunen E, Kanaar R, Berns A, Jonkers J. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc Natl Acad Sci U S A. 2001;98:9209–9214. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Silver DP, Livingston DM. Self-excising retro-viral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Mol Cell. 2001;8:233–843. doi: 10.1016/s1097-2765(01)00295-7. [DOI] [PubMed] [Google Scholar]
- 78.Higashi AY, Ikawa T, Muramatsu M, Economides AN, Niwa A, Okuda T, Murphy AJ, Rojas J, Heike T, Nakahata T, Kawamoto H, Kita T, Yanagita M. Direct hematological toxicity and illegitimate chromosomal recombination caused by the systemic activation of CreERT2. J Immunol. 2009;182:5633–5640. doi: 10.4049/jimmunol.0802413. [DOI] [PubMed] [Google Scholar]
- 79.Knauf JA, Ma X, Smith EP, Zhang L, Mitsutake N, Liao XH, Refetoff S, Nikiforov YE, Fagin JA. Targeted expression of BRAFV600E in thyroid cells of transgenic mice results in papillary thyroid cancers that undergo dedifferentiation. Cancer Res. 2005;65:4238–4245. doi: 10.1158/0008-5472.CAN-05-0047. [DOI] [PubMed] [Google Scholar]
- 80.Ji H, Wang Z, Perera SA, Li D, Liang MC, Zaghlul S, McNamara K, Chen L, Albert M, Sun Y, Al-Hashem R, Chirieac LR, Padera R, Bronson RT, Thomas RK, Garraway LA, Jänne PA, Johnson BE, Chin L, Wong KK. Mutations in BRAF and KRAS converge on activation of the mitogen-activated protein kinase pathway in lung cancer mouse models. Cancer Res. 2007;67:4933–4939. doi: 10.1158/0008-5472.CAN-06-4592. [DOI] [PubMed] [Google Scholar]
- 81.Mercer K, Giblett S, Green S, Lloyd D, DaRocha Dias S, Plumb M, Marais R, Pritchard C. Expression of endogenous oncogenic V600EB-raf induces proliferation and developmental defects in mice and transformation of primary fibroblasts. Cancer Res. 2005;65:11493–11500. doi: 10.1158/0008-5472.CAN-05-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dankort D, Filenova E, Collado M, Serrano M, Jones K, McMahon M. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes Dev. 2007;21:379–384. doi: 10.1101/gad.1516407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Urosevic J, Sauzeau V, Soto-Montenegro ML, Reig S, Desco M, Wright EM, Cañamero M, Mulero F, Ortega S, Bustelo XR, Barbacid M. Constitutive activation of B-Raf in the mouse germ line provides a model for human cardio-facio-cutaneous syndrome. Proc Natl Acad Sci USA. 2011;108:5015–5020. doi: 10.1073/pnas.1016933108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Carragher LA, Snell KR, Giblett SM, Aldridge VS, Patel B, Cook SJ, Winton DJ, Marais R, Pritchard CA. V600EBraf induces gastrointestinal crypt senescence and promotes tumour progression through enhanced CpG methylation of p16INK4a. EMBO Mol Med. 2010;2:458–471. doi: 10.1002/emmm.201000099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dhomen N, Reis-Filho JS, da Rocha Dias S, Hayward R, Savage K, Delmas V, Larue L, Pritchard C, Marais R. Oncogenic Braf induces melanocyte senescence and melanoma in mice. Cancer Cell. 2009;15:294–303. doi: 10.1016/j.ccr.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 86.Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, Jones CM, Marshall CJ, Springer CJ, Barford D, Marais R, Cancer Genome Project Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–67. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 87.Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, Hussain J, Reis-Filho JS, Springer CJ, Pritchard C, Marais R. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dumaz N, Hayward R, Martin J, Ogilvie L, Hedley D, Curtin JA, Bastian BC, Springer C, Marais R. In melanoma, RAS mutations are accompanied by switching signaling from BRAF to CRAF and disrupted cyclic AMP signaling. Cancer Res. 2006;66:9483–9491. doi: 10.1158/0008-5472.CAN-05-4227. [DOI] [PubMed] [Google Scholar]
- 89.Blasco RB, Francoz S, Santamaría D, Cañamero M, Dubus P, Charron J, Baccarini M, Barbacid M. c-Raf, but Not B-Raf, Is Essential for Development of K-Ras Oncogene-Driven Non-Small Cell Lung Carcinoma. Cancer Cell. 2011;19:652–663. doi: 10.1016/j.ccr.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 91.Roschke AV, Tonon G, Gehlhaus KS, McTyre N, Bussey KJ, Lababidi S, Scudiero DA, Weinstein JN, Kirsch IR. Karyotypic complexity of the NCI-60 drug-screening panel. Cancer Res. 2003;63:8634–8647. [PubMed] [Google Scholar]
- 92.Oliveira C, Westra JL, Arango D, Ollikainen M, Domingo E, Ferreira A, Velho S, Niessen R, Lagerstedt K, Alhopuro P, Laiho P, Veiga I, Teixeira MR, Ligtenberg M, Kleibeuker JH, Sijmons RH, Plukker JT, Imai K, Lage P, Hamelin R, Albuquerque C, Schwartz S, Jr, Lindblom A, Peltomaki P, Yamamoto H, Aaltonen LA, Seruca R, Hofstra RM. Distinct patterns of KRAS mutations in colorectal carcinomas according to germline mismatch repair defects and hMLH1 methylation status. Hum Mol Genet. 2004;13:2303–2311. doi: 10.1093/hmg/ddh238. [DOI] [PubMed] [Google Scholar]
- 93.Asaka S, Arai Y, Nishimura Y, Yamaguchi K, Ishikubo T, Yatsuoka T, Tanaka Y, Akagi K. Microsatellite instability-low colorectal cancer acquires a KRAS mutation during the progression from Dukes' A to Dukes' B. Carcinogenesis. 2009;30:494–499. doi: 10.1093/carcin/bgp017. [DOI] [PubMed] [Google Scholar]
- 94.Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50:113–130. doi: 10.1111/j.1365-2559.2006.02549.x. [DOI] [PubMed] [Google Scholar]
- 95.Andreyev HJ, Norman AR, Cunningham D, Oates JR, Clarke PA. Kirsten ras mutations in patients with colorectal cancer: the multicenter “RASCAL” study. J Natl Cancer Inst. 1998;90:675–684. doi: 10.1093/jnci/90.9.675. [DOI] [PubMed] [Google Scholar]
- 96.Oliveira C, Velho S, Moutinho C, Ferreira A, Preto A, Domingo E, Capelinha AF, Duval A, Hamelin R, Machado JC, Schwartz S, Jr, Carneiro F, Seruca R. KRAS and BRAF oncogenic mutations in MSS colorectal carcinoma progression. Oncogene. 2007;26:158–163. doi: 10.1038/sj.onc.1209758. [DOI] [PubMed] [Google Scholar]
- 97.O'Brien MJ, Yang S, Mack C, Xu H, Huang CS, Mulcahy E, Amorosino M, Farraye FA. Comparison of microsatellite instability, CpG island methylation phenotype, BRAF and KRAS status in serrated polyps and traditional adenomas indicates separate pathways to distinct colorectal carcinoma end points. Am J Surg Pathol. 2006;30:1491–1501. doi: 10.1097/01.pas.0000213313.36306.85. [DOI] [PubMed] [Google Scholar]
- 98.Velho S, Moutinho C, Cirnes L, Albuquerque C, Hamelin R, Schmitt F, Carneiro F, Oliveira C, Seruca R. BRAF, KRAS and PIK3CA mutations in colorectal serrated polyps and cancer: primary or secondary genetic events in colorectal carcinogenesis? BMC Cancer. 2008;8:255. doi: 10.1186/1471-2407-8-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Calistri D, Rengucci C, Seymour I, Leonardi E, Truini M, Malacarne D, Castagnola P, Giaretti W. KRAS, p53 and BRAF gene mutations and aneuploidy in sporadic colorectal cancer progression. Cell Oncol. 2006;28:161–166. doi: 10.1155/2006/465050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kambara T, Simms LA, Whitehall VL, Spring KJ, Wynter CV, Walsh MD, Barker MA, Arnold S, McGivern A, Matsubara N, Tanaka N, Higuchi T, Young J, Jass JR, Leggett BA. BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut. 2004;53:1137–1144. doi: 10.1136/gut.2003.037671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Samowitz WS, Albertsen H, Herrick J, Levin TR, Sweeney C, Murtaugh MA, Wolff RK, Slattery ML. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology. 2005;129:837–845. doi: 10.1053/j.gastro.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 102.Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, Koh H, Simms L, Barker M, Leggett B, Levine J, Kim M, French AJ, Thibodeau SN, Jass J, Haile R, Laird PW. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 103.Samowitz WS, Sweeney C, Herrick J, Albertsen H, Levin TR, Murtaugh MA, Wolff RK, Slattery ML. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res. 2005;65:6063–6069. doi: 10.1158/0008-5472.CAN-05-0404. [DOI] [PubMed] [Google Scholar]
- 104.Kim IJ, Kang HC, Jang SG, Kim K, Ahn SA, Yoon HJ, Yoon SN, Park JG. Oligonucleotide mi-croarray analysis of distinct gene expression patterns in colorectal cancer tissues harboring BRAF and K-ras mutations. Carcinogenesis. 2006;27:392–404. doi: 10.1093/carcin/bgi237. [DOI] [PubMed] [Google Scholar]
- 105.Ciampi R, Nikiforov YE. Alterations of the BRAF gene in thyroid tumors. Endocr Pathol. 2005;16:163–72. doi: 10.1385/ep:16:3:163. [DOI] [PubMed] [Google Scholar]
- 106.Schmid K, Oehl N, Wrba F, Pirker R, Pirker C, Filipits M. EGFR/KRAS/BRAF mutations in primary lung adenocarcinomas and corresponding locoregional lymph node metastases. Clin Cancer Res. 2009;15:4554–4560. doi: 10.1158/1078-0432.CCR-09-0089. [DOI] [PubMed] [Google Scholar]
- 107.Naoki K, Chen TH, Richards WG, Sugarbaker DJ, Meyerson M. Missense mutations of the BRAF gene in human lung adenocarcinoma. Cancer Res. 2002;62:7001–7003. [PubMed] [Google Scholar]
- 108.Park SY, Gonen M, Kim HJ, Michor F, Polyak K. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J Clin Invest. 2010;120:636–644. doi: 10.1172/JCI40724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Merlo LM, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nat Rev Cancer. 2006;6:924–935. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- 110.Duesberg P, Li R, Sachs R, Fabarius A, Upender MB, Hehlmann R. Cancer drug resistance: the central role of the karyotype. Drug Resist Updat. 2007;10:51–58. doi: 10.1016/j.drup.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 111.Schneider BL, Kulesz-Martin M. Destructive cycles: the role of genomic instability and adaptation in carcinogenesis. Carcinogenesis. 2004;25:2033–2044. doi: 10.1093/carcin/bgh204. [DOI] [PubMed] [Google Scholar]
- 112.Torres EM, Williams BR, Amon A. Aneuploidy: cells losing their balance. Genetics. 2008;179:737–746. doi: 10.1534/genetics.108.090878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tang YC, Williams BR, Siegel JJ, Amon A. Identification of aneuploidy-selective antiproliferation compounds. Cell. 2011;144:499–512. doi: 10.1016/j.cell.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tiacci E, Trifonov V, Schiavoni G, Holmes A, Kern W, Martelli MP, Pucciarini A, Bigerna B, Pacini R, Wells VA, Sportoletti P, Pettirossi V, Mannucci R, Elliott O, Liso A, Ambrosetti A, Pulsoni A, Forconi F, Trentin L, Semenzato G, Inghirami G, Capponi M, Di Raimondo F, Patti C, Arcaini L, Musto P, Pileri S, Haferlach C, Schnittger S, Pizzolo G, Foà R, Farinelli L, Haferlach T, Pasqualucci L, Rabadan R, Falini B. BRAF mutations in Hairy-Cell Leukemia. N Engl J Med. 2011;364:2305–2315. doi: 10.1056/NEJMoa1014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet JP, Ahmann GJ, Adli M, Anderson KC, Ardlie KG, Auclair D, Baker A, Bergsagel PL, Bernstein BE, Drier Y, Fonseca R, Gabriel SB, Hofmeister CC, Jagannath S, Jakubowiak AJ, Krishnan A, Levy J, Liefeld T, Lonial S, Mahan S, Mfuko B, Monti S, Perkins LM, Onofrio R, Pugh TJ, Rajkumar SV, Ramos AH, Siegel DS, Sivachenko A, Stewart AK, Trudel S, Vij R, Voet D, Winckler W, Zimmerman T, Carpten J, Trent J, Hahn WC, Garraway LA, Meyerson M, Lander ES, Getz G, Golub TR. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Badalian-Very G, Vergilio JA, Degar BA, Mac-Conaill LE, Brandner B, Calicchio ML, Kuo FC, Ligon AH, Stevenson KE, Kehoe SM, Garraway LA, Hahn WC, Meyerson M, Fleming MD, Rollins BJ. Recurrent BRAF mutations in Langerhans cell histiocytosis. Blood. 2010;116:1919–1923. doi: 10.1182/blood-2010-04-279083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Marks JL, Gong Y, Chitale D, Golas B, McLellan MD, Kasai Y, Ding L, Mardis ER, Wilson RK, Solit D, Levine R, Michel K, Thomas RK, Rusch VW, Ladanyi M, Pao W. Novel MEK1 mutation identified by mutational analysis of epidermal growth factor receptor signaling pathway genes in lung adenocarcinoma. Cancer Res. 2008;68:5524–5528. doi: 10.1158/0008-5472.CAN-08-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sasaki H, Hikosaka Y, Okuda K, Kawano O, Yukiue H, Yano M, Fujii Y. MEK1 gene mutation in Japanese lung adenocarcinoma patients. Mol Med Report. 2009;2:153–155. doi: 10.3892/mmr_00000076. [DOI] [PubMed] [Google Scholar]
- 119.Estep AL, Palmer C, McCormick F, Rauen KA. Mutation analysis of BRAF, MEK1 and MEK2 in 15 ovarian cancer cell lines: implications for therapy. PLoS One. 2007;2:e1279. doi: 10.1371/journal.pone.0001279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Murugan AK, Dong J, Xie J, Xing M. MEK1 mutations, but not ERK2 mutations, occur in melanomas and colon carcinomas, but none in thyroid carcinomas. Cell Cycle. 2009;8:2122–2124. doi: 10.4161/cc.8.13.8710. [DOI] [PubMed] [Google Scholar]