Abstract
The protein ubiquitin is an important post-translational modifier that regulates a wide variety of biological processes. In cells, ubiquitin is apportioned among distinct pools, which include a variety of free and conjugated species. Although maintenance of a dynamic and complex equilibrium among ubiquitin pools is crucial for cell survival, the tools necessary to quantify each cellular ubiquitin pool have been limited. We have developed a quantitative mass spectrometry approach to measure cellular concentrations of ubiquitin species using isotope-labeled protein standards and applied it to characterize ubiquitin pools in cells and tissues. Our method is convenient, adaptable and should be a valuable tool to facilitate our understanding of this important signaling molecule.
The protein ubiquitin is a highly conserved post-translational modifier that regulates a wide variety of biological processes in eukaryotes1,2. Most known ubiquitin signaling is initiated by conjugation of ubiquitin to protein substrates and terminated by hydrolysis of this linkage by deubiquitinating enzymes. In the cell, ubiquitin is dynamically apportioned among distinct pools which include ‘free’ (unconjugated) ubiquitin, ‘activated’ ubiquitin, which is conjugated via thioester linkages to enzyme-bound intermediates of the ubiquitin conjugation cascade, and peptide-linked conjugates to substrate proteins (Fig. 1a). The conjugate pool comprises monoubiquitin conjugates, in which one or more ubiquitin is attached to lysine residues on a substrate through an isopeptide linkage between the terminal carboxyl group of ubiquitin and the ε-amino group of the target lysine, and poly-ubiquitin conjugates, in which additional ubiquitin molecules are attached to the lysine residues in ubiquitin itself, producing substrate-linked polyubiquitin chains.
Figure 1.
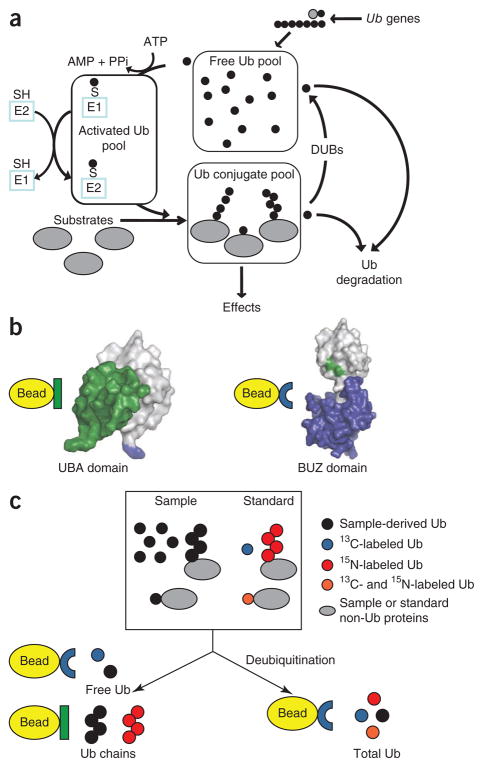
Ubiquitin pools and assay overview. (a) Ubiquitin (Ub) is present in eukaryotic cells as a mixture of free ubiquitin, monoubiquitinated substrates, polyubiquitin chains and activated species linked to enzymes by thioester bonds. DUBs, deubiquitinating enzymes. E1, ubiquitin activating enzyme. E2, ubiquitin transfer enzyme. SH, catalytic cysteine residues; S, catalytic cysteine residues conjugated via covalent thioester linkages to ubiquitin. (b) Cartoon representations of the interactions of the protein-affinity reagents used in the Ub-PSAQ assay with ubiquitin. The hP2 UBA domain binds the Ile44-centered hydrophobic patch on ubiquitin (patch is obscured on the left and is shown in green on the right) and preferentially binds ubiquitin chains without bias for specific ubiquitin-ubiquitin linkages8,23,24. The BUZ domain of IsoT binds the C-terminal Gly-Gly of free ubiquitin species (shown in blue on the left)13. (c) Schematic of the Ub-PSAQ assay. Isotope-labeled protein standards are added to lysates containing sample derived ubiquitin species. Half of the sample is treated with the deubiquitinating enzyme usp2cc, free ubiquitin species are bound to the BUZ affinity reagent, washed, eluted and then trypsinized before quantification of total sample–derived ubiquitin relative to the 13C-labeled free-ubiquitin protein standard by LC-ESI TOF MS. Free ubiquitin species in the untreated half of the sample are similarly captured by the BUZ affinity reagent and quantified by LC-ESI MS. Next, the untreated half of the sample is incubated with the hP2 UBA affinity reagent to capture polyubiquitin chains, which are quantified relative to the polyubiquitin chain standard.
Much of the complexity of cellular signaling by ubiquitin has been proposed to depend on its ability to form diverse covalent conjugates with other proteins1. For instance, attachment of Lys48-linked polyubiquitin chains to substrates is widely held to be the canonical signal for degradation by the proteasome, whereas Lys63-linked polyubiquitination of cell-surface receptors signals internalization by endocytosis, and monoubiquitination of histones regulates chromatin structure1,2. Additionally, free polyubiquitin chains that are not conjugated to substrates have emerging roles in cellular signaling3.
Levels of total cellular ubiquitin are regulated by transcriptional control at four different genetic loci and by post-translational mechanisms1. Disruption of total ubiquitin levels or the distribution of ubiquitin among different cellular pools is linked to a wide spectrum of diseases including cancer4 and neurodegeneration5. Given the importance of ubiquitin to most aspects of eukaryotic cell function and the recognition of the importance of maintaining ubiquitin homeostasis, the ability to accurately and precisely quantify ubiquitin pools in a cell or tissue is of paramount importance.
Most prior studies of ubiquitin pools have relied on antibodies to discriminate between free and conjugated ubiquitin species6,7. However, because traditional conjugate-selective antibodies react to a finite and indeterminate extent with free ubiquitin and antibodies to free ubiquitin react to an unknown extent with different types of conjugates, it has not been possible to accurately measure ubiquitin pools in biological specimens. Synthetic peptide absolute quantification (AQUA) mass spectrometry has enabled precise quantification of mono- and polyubiquitination of purified substrates and relative quantification of affinity-captured polyubiquitin species from cell and tissue lysates8–10. However, synthetic peptides cannot account for loss of protein that occurs through fractionation and processing procedures.
We developed a method that combines differential affinity chromatography and protein standard absolute quantification (PSAQ) mass spectrometry11 to enable the precise measurement of cellular molar concentrations of ubiquitin pool components. Our strategy, termed ubiquitin-PSAQ, uses stable isotope–labeled free ubiquitin and ubiquitin conjugates as recovery standards, which we ‘spiked’ into cell or tissue lysates and captured with affinity reagents that are selective for free ubiquitin or ubiquitin chains. Using this approach we determined the steady-state distribution of ubiquitin pools in tissue culture cells before and after induction of proteasome stress and in lysates of mouse and human brain. Our data indicate a surprising degree of heterogeneity in the distribution of ubiquitin among intracellular pools in different cell types and tissues and in the dynamic responses of these pools among different cell types. These findings constitute precise measurements of ubiquitin pool distribution and dynamics in a living system and have broad utility for enhancing our understanding of this important cellular signaling modifier.
RESULTS
Assay design
To measure cellular ubiquitin pools by quantitative mass spectrometry, we devised an approach (Ub-PSAQ) that combines the use of differential affinity chromatography (Fig. 1b) and isotope-labeled ubiquitin recovery standards (Fig. 1c and Supplementary Fig. 1). We prepared [13C]ubiquitin, [15N]ubiquitin and a doubly labeled, linear [13C,15N]ubiquitin-GFP fusion protein. We used the [13C,15N]ubiquitin-GFP as a monoubiquitin conjugate mimetic. We generated a polyubiquitin conjugate standard from the purified recombinant E3 enzyme Rsp5, autoubiquitinated with [15N]ubiquitin (Supplementary Fig. 1 and Supplementary Table 1).
To determine the cellular abundance of ubiquitin species, we lysed cells or tissues in the presence of 2% (w/v) sodium dodecyl sulfate (SDS), cleared the lysate by centrifugation and added a mixture of [13C]ubiquitin, Rsp5-[15N]polyubiquitin and [13C,15N]ubiquitin-GFP recovery standards. We divided samples into two equal portions then diluted them to reduce the SDS concentration to below 0.05%. One portion we used to determine total ubiquitin concentration after conversion of all ubiquitin conjugates to free ubiquitin with the catalytic domain of the deubiquitinating enzyme usp2cc12 (Fig. 1c). We isolated free ubiquitin species from this portion and from the portion not treated with deubiquitinating enzyme by exploiting the ability of the zinc-finger domain from isopeptidase T (BUZ) to bind specifically to the free C-terminal diglycine motif of ubiquitin13 (Fig. 1b). To confirm that our lysis conditions prevented deubiquitinating-enzyme activity, we monitored binding of the [15N,13C]ubiquitin-GFP standard by the BUZ affinity reagent in all samples and observed none when samples were lysed in the presence of 2% SDS and 5 mg ml−1 N-ethylmaleimide (NEM). We isolated polyubiquitin chains by affinity capture with the human PLIC2 ubiquitin-association domain (hP2 UBA) as previously described8 (Fig. 1b). To maximize conversion of conjugated ubiquitin to free ubiquitin, we used a molar excess of usp2cc to ubiquitin and monitored the extent of cleavage both as the loss of all ubiquitin chain linkages bound to hP2 UBA and as the increase of [15N,13C]ubiquitin binding to the BUZ domain (Supplementary Data 1). Next we washed and eluted affinity-captured material, digested it with trypsin and analyzed it by liquid chromatography–electrospray ionization time-of-flight mass spectrometry (LC-ESI TOF MS). We quantified sample-derived ubiquitin species using the ratio of ion intensities of the tracked endogenous peptides to labeled synthetic peptides (added during trypsinization to quantify material in the digest) and the protein standard–derived peptides (added to lysates to determine recovery) (Online Methods, Fig. 1c and Supplementary Data 1). To calculate the concentration of monoubiquitinated substrates, we subtracted amounts of measured free ubiquitin and polyubiquitin chains from measured total ubiquitin.
The addition of isotope-labeled protein standards to lysates and peptide standards to trypsin digestions enabled the determination of the recovery efficiency of ubiquitin conjugates throughout the isolation procedure, including capture by the affinity reagents. The extent of recovery depends on many parameters and is hindered by dilution and residual SDS, which is necessary to reduce deubiquitinating-enzyme activity. Typical recovery by the BUZ affinity reagent was typically 1–3%. Sample recovery using the hP2 UBA domain was typically 10–40%. Under the conditions used, we detected no binding of substrate-linked ubiquitin chains from our polyubiquitin standard binding to the BUZ domain, and the recovery of free ubiquitin with the hP2 UBA was ~0.03%, indicating that the bulk of polyubiquitin binding to hP2 UBA was due to avidity effects. To ensure accurate quantification of ubiquitin pools, we quantified the efficiency of recovery for each individual pulldown and carried this through into all calculations.
Assay performance and reproducibility
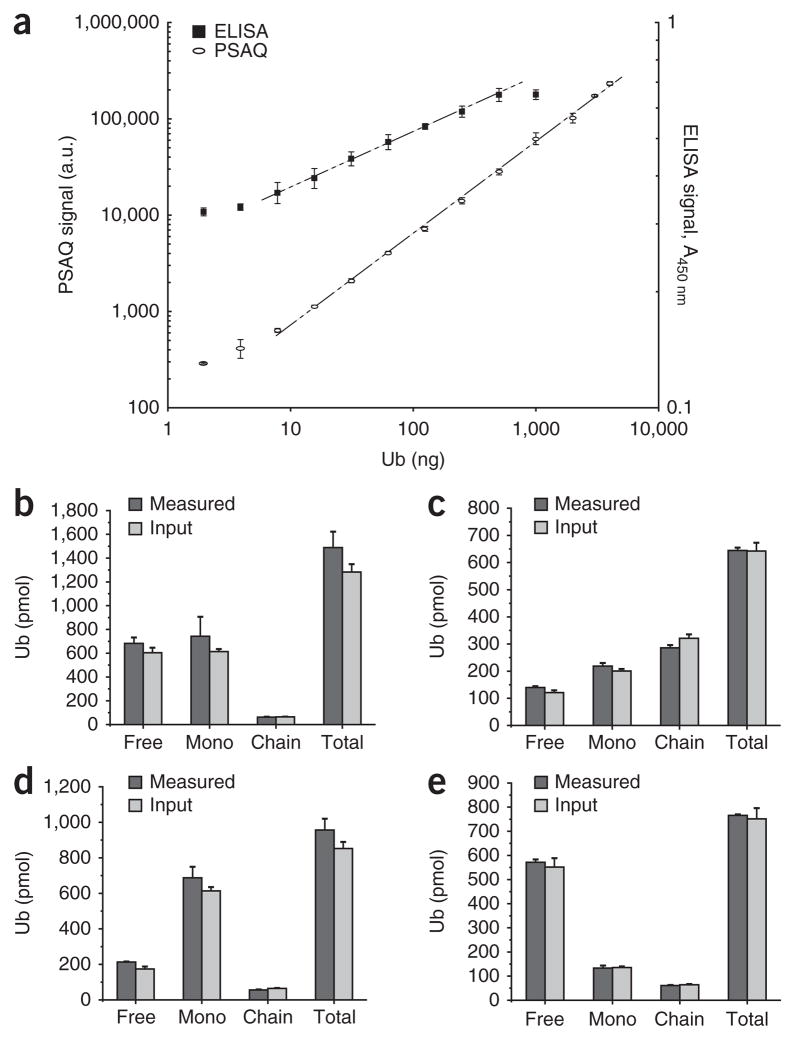
We assessed the feasibility of measuring total ubiquitin by BUZ-enriched Ub-PSAQ relative to free [13C]ubiquitin standard and compared the results to those obtained with an established indirect competitive ELISA14 (Fig. 2a). The sensitivity of Ub-PSAQ was comparable to that of the ELISA, with accurate and precise ubiquitin measurement down to ~10 ng (1.2 pmol). The PSAQ signal was linear over a range of three orders of magnitude of useful working concentrations. The percentage error associated with quantifying ubiquitin by AQUA mass spectrometry using ions from two equivalent linear tryptic peptides, TLSDYNIQK (amino acids 55–63; TLS) and ESTLHLVLR (amino acids 64–72; ESTL), was typically less than 20% (Supplementary Fig. 2). To ensure accurate quantification of ubiquitin, we averaged data derived from these two ions as previously described for synthetic peptide AQUA10.
Figure 2.
Assay validation. (a) Superimposed serial dilution curves of ubiquitin detection by BUZ-enriched PSAQ and ELISA comparing the linear dynamic range and the lower limit of detection. Mass spectrometry signal intensity is expressed in arbitrary units (a.u.) after normalization to the spiked 13C-labeled free-ubiquitin standard. A450 nm, absorbance at 450 nm. Data are represented as mean ± s.d. (n = 8 for ELISA and n = 3 for PSAQ). (b–e) Measurement of defined mixtures composed of unlabeled free ubiquitin, autoubiquitinated Rsp5 and ubiquitin-GFP mock monoubiquitinated substrate containing similar amounts of input free ubiquitin and ubiquitin-GFP substrate (b), higher levels of ubiquitin-GFP and ubiquitin chains (chain) than free ubiquitin (c), high levels of mock monoubiquitinated substrate (mono) (d) and higher levels of free ubiquitin (e). Error bars (b–e), ± s.d. (n = 3).
We first assessed the Ub-PSAQ method by measuring defined mixtures of unlabeled versions of our ubiquitin protein standards with varying ratios of free ubiquitin and conjugate species relative to isotope-labeled protein standards (Fig. 2b–e). These data indicate that Ub-PSAQ can accurately report the amounts of each ubiquitin species over an approximately fivefold range of relative abundance in the mixtures and that the relative abundance of each individual species does not impact the accurate measurement of the other species.
Ubiquitin pool distribution in cells and tissue
To determine total ubiquitin concentration in HEK293 cells by Ub-PSAQ, we treated HEK293 cell lysate with the deubiquitinating enzyme usp2cc, which releases free ubiquitin from all ubiquitin conjugates, and measured sample-derived ubiquitin relative to the 13C-labeled free ubiquitin standard after enrichment with the BUZ affinity reagent. In close agreement with previously reported values for the same cell line measured by ELISA14, we determined HEK293 ubiquitin total concentration to be 486.4 ± 42 pmol mg−1 total protein (±s.d.; n = 9), corresponding to a molar concentration of ~85 μM, ~8 × 107 molecules per cell or ~0.42% (weight ubiquitin/weight total protein).
We assessed the ubiquitin pool distribution in two cell lines, and in mouse and human brain tissues by Ub-PSAQ. Although HEK293 cells have about twofold more total ubiquitin than a mouse embryonic fibroblast (MEF) cell line, these cell lines have similar ubiquitin pool distributions with ~65% of ubiquitin present as monoubiquitinated substrates, ~23% as free ubiquitin and ~11% as polyubiquitin chains (Table 1). This distribution is consistent with the results of a prior study in which ubiquitin conjugates had been measured as a singular entity and found to reflect ~30–80% of cellular ubiquitin in various cell lines7. By contrast, our quantification of ubiquitin pools in mouse brain revealed that 59% of ubiquitin is present as free monomer, 35% as monoubiquitin on substrates and 5% as polyubiquitin chains (Table 1). In human frontal cortex, 82% of ubiquitin is free ubiquitin monomer, 13% is present as monoubiquitin on substrates and 4% is present as polyubiquitin chains (Table 1). We analyzed the major (Lys48, Lys63 and Lys11) ubiquitin-ubiquitin chain linkages in cell and tissue samples (Table 1). Although we could detect other polyubiquitin linkages (Lys6, Lys27, Lys29 and Lys33) in these samples, together they constitute less than 1% of total ubiquitin (data not shown) and were not included in this analysis. In sum, the steady-state distribution of ubiquitin among the different pools differs markedly between cell lines and brain tissue.
Table 1.
Quantification of ubiquitin pool components in cells and tissue
| Sample | Total Ub (pmol mg−1)a | Free Ub (pmol mg−1)a | Mono Ub (pmol mg−1)a | Ub chain (pmol mg−1)a | UbK48 isopeptide (pmol mg−1)a | UbK63 isopeptide (pmol mg−1)a | UbK11 isopeptide (pmol mg−1)a | Free Ub (%) | Mono (%) | Ub chain (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| HEK293 cell line | 499 ± 52 | 115 ± 26 | 341 ± 61 | 43 ± 14 | 36 ± 13 | 5.0 ± 1.7 | 2.0 ± 0.6 | 23 | 68 | 8.5 |
| MEF cell line | 262 ± 23 | 59 ± 6.5 | 167 ± 29 | 36 ± 6.3 | 27 ± 4.2 | 6.5 ± 1.6 | 3.3 ± 0.5 | 23 | 63 | 14 |
| Mouse brain | 121 ± 5.1 | 72 ± 5.6 | 43 ± 6.1 | 6.3 ± 0.5 | 3.0 ± 0.2 | 2.6 ± 0.3 | 0.7 ± 0.1 | 60 | 35 | 5 |
| Human frontal cortex | 160 ± 11 | 131 ± 5.5 | 22 ± 5.5 | 6.6 ± 0.4 | 3.6 ± 0.2 | 2.0 ± 0.2 | 1.0 ± 0.1 | 82 | 14 | 4 |
Ubiquitin (Ub) pools were measured by Ub-PSAQ from HEK293 and MEF cell lines, from whole mouse brain and from human frontal cortex.
Values are in picomoles ubiquitin species per milligram total protein and are shown as means ± s.d. (n = 3).
The precision and reproducibility of measured ubiquitin pool component concentrations were excellent in mouse brain lysate, with intra-assay (10 technical replicates measuring the same lysate within one assay with a single standard mixture) and interassay (4 technical replicates measuring the same lysate on different days with individually prepared standard mixtures) coefficients of variation less than 10% for total and free ubiquitin and less than 20% for ubiquitin chains and monoubiquitinated substrates (Table 1 and Supplementary Fig. 3).
Effect of acute proteasome inhibition on ubiquitin pools
To determine the distribution of ubiquitin pools before and after induction of proteasome stress, we measured ubiquitin pools in HEK293 and MEF cells after acute exposure to the proteasome inhibitor MG-132 (Fig. 3 and Supplementary Fig. 4). As expected, the major effect of MG-132 was to increase the abundance of ubiquitin chains, clearly evident from Ub-PSAQ and immunoblot analysis (Fig. 3 and Supplementary Fig. 4). In MEF cells, the sixfold increase in the abundance of polyubiquitin chains (Fig. 3b) at 12 h was accompanied by a twofold increase in free ubiquitin and monoubiquitinated substrates, leading to a robust increase in total ubiquitin, consistent with the previously reported transcriptional activation of ubiquitin gene expression by acute proteasome inhibition in MEFs15. In contrast, the increase in the abundance of polyubiquitin chains in HEK293 cells was not accompanied by an increase in total ubiquitin or other ubiquitin species, suggesting that the ubiquitin incorporated into chains in HEK293 cells treated with the proteasome inhibitor MG-132 may be derived principally from other cellular ubiquitin pools as opposed to de novo synthesis of additional ubiquitin. Indeed amounts of free ubiquitin and monoubiquitin on substrates were reduced (although not significantly P ≤ 0.05) in parallel with the increase in polyubiquitin chains (Fig. 3a). In agreement with prior work16,17, we observed a decrease in monoubiquitin conjugates linked to histones in MG-132–treated HEK293 cells (Fig. 3de, and Supplementary Figs. 5 and 6), likely reflecting a redistribution of ubiquitin pools during proteasomal stress.
Figure 3.
Effect of acute proteasome inhibition on ubiquitin pools. (a,b) Ub-PSAQ analysis of lysates from HEK293 (a) and MEF (b) cell lines treated with either DMSO (vehicle) or the proteasome inhibitor MG-132 (1 μM) over 12 h. Error bars, ± s.d. (n = 3). *P < 0.05; **P < 0.01; and ***P < 0.005 (unpaired t-test). MonoUb, monoubiquitin conjugates. (c) Representation of ubiquitin pool components in HEK293 and MEF cell lines. (d) Ubiquitin western blots with antibodies FK2 and A100 showing ubiquitin-immunoreactive material for HEK293 cells treated with either DMSO (vehicle) or MG-132 (1 μM) over 12 h. Bars on top right of blots denote high-molecular-weight ubiquitin conjugates (HMW Ub conjugates), and ubiquitin-modified histone H2A (UH) and free ubiquitin are indicated. (e) Distribution of monoubiquitinated substrates in cytosolic and histone-enriched fractions from HEK293 cells treated with either DMSO or MG-132 (10 μM) for 6 h.
Although Lys48-linked polyubiquitin chains are widely held to be the canonical signal for proteasome-mediated degradation, other polyubiquitin linkages have also been implicated in proteasomal targeting18,19. After 12 h of proteasome inhibition in the MEF cell line, we observed an approximately tenfold increase in Lys48-linked chains and a three- to fourfold increase in Lys63- and Lys11-linked chains (Supplementary Fig. 4). However, in HEK293 cells we observed a trend toward an approximately twofold increase in all polyubiquitin linkages by 12 h of treatment with MG-132, consistent with previous reports8,18,19 (Supplementary Fig. 4). The accumulation of Lys63- and Lys11-linked ubiquitin chains in addition to Lys48-linked chains could suggest that cells use multiple types of polyubiquitin chains as proteasome-targeting signals or reflect the existence of mixed-linkage polyubiquitin chains or of multiple chain types on individual proteins20,21. The differences in the kinetics of increase of the different polyubiquitin chain linkages, with a trend toward Lys48- and Lys11-linked polyubiquitin chains increasing at the 2 h time point, may suggest that Lys63-linked chains are unlikely to be a major proteasome-targeting signal. Overall, these data suggest that ubiquitin pool dynamics in response to proteasome stress differ greatly between these two cell types.
Ubiquitin pools in cytosolic and histone-enriched fractions
To understand the surprisingly high proportion of total ubiquitin present as monoubiquitin conjugates in cell lines (~65%), we used Ub-PSAQ to determine the fraction of monoubiquitin conjugates present in a cytosolic fraction and in a histone-enriched fraction prepared by acid extraction of a nuclear fraction from HEK293 cells (Supplementary Fig. 5). As expected, the histone fraction was highly enriched in monoubiquitin conjugates, with 91% of all ubiquitin in this fraction present as monoubiquitin-conjugated substrates (Fig. 4a), and accounting for ~23% of total cellular ubiquitin and ~34% of total cellular monoubiquitin conjugates (Fig. 4b). The remaining ~10% of ubiquitin associated with the histone fraction was approximately equally distributed between free ubiquitin and chain conjugates (Fig. 4c). The latter pool was somewhat enriched in Lys63 linkages, perhaps reflecting a high proportion of histone-associated DNA-repair enzymes, though we cannot speculate as to which substrates are modified by Lys63-linked ubiquitin chains.
Figure 4.
Ub-PSAQ analysis of ubiquitin pools in cytosolic and histone-enriched fractions from HEK293 cells. (a) Representation of ubiquitin pool components in cytosolic and histone-enriched fractions. (b) Distribution of ubiquitin pool components in cytosolic and histone-enriched fractions. Bar graphs show concentration of the indicated species per milligram of total protein in each fraction. Pie charts show the same data after normalization to show the distribution of each species. (c) Distribution of ubiquitin chain linkages in cytosolic and histone-enriched fractions. Bar graphs show the concentration of each species within each fraction, and pie charts show the distribution of each species between cytosolic and histone-enriched fractions. Error bars (b,c), means ± s.d. (n = 3).
DISCUSSION
The Ub-PSAQ method overcomes several problems commonly encountered in quantifying ubiquitin. First, the use of two affinity reagents that interact with ubiquitin through distinct surfaces allows the purification of specific ubiquitin pool components. Second, the use of usp2cc enables the conversion of all ubiquitin species to free ubiquitin and the accurate measurement of total ubiquitin12,14. Third, lysis of cells or tissues in the presence of 5 mg ml−1 NEM and 2% SDS prevents interconversion of ubiquitin species during lysis and sample processing. Finally, the method enables the analysis of sample- and protein standard–derived ubiquitin species directly after affinity capture without the need for SDS-PAGE steps that can confound quantification of complex analytes.
One potential limitation of the Ub-PSAQ method as implemented here is that it assumes that all ubiquitin-ubiquitin linkages bind with equal affinity to the hP2 UBA affinity reagent. Although data demonstrate that the UBA domains of ubiquilin-Plic family members bind ubiquitin Lys48 and ubiquitin Lys63 chains without linkage specificity22,23, not all linkages have been examined. Furthermore, it is possible that polyubiquitin chains measured binding to the hP2 UBA domain are underestimated owing to effects of chain length on binding affinity22,24 and to the effect of ubiquitin molecules at the end of chains, which do not contribute an isopeptide bond and are therefore not included in chain calculations. Prior work has demonstrated that the BUZ domain of IsoT binds ubiquitin via free C termini and that this reagent is unlikely to exhibit bias owing to ubiquitin-chain linkage type or length13.
The average length of cellular ubiquitin chains has, to our knowledge, not been reported. It will be important to generate a wider variety of isotope labeled ubiquitin protein standards and to extend the Ub-PSAQ method to obtain a more detailed picture of the abundance of ubiquitin species, including determining average chain lengths in cells.
Another potential limitation is that it may not be possible to measure all ubiquitin species simultaneously. For instance, although we observed binding of free, unanchored ubiquitin chains to the BUZ affinity reagent (Supplementary Data 1), their basal levels were low. Compounded with the low recovery by the BUZ domain under the conditions we used, the values for these species are too close to the noise to discern in the present work. Simple remedies, such as using larger samples for the BUZ domain capture should enable precise assessment of the concentration of free ubiquitin chains. Similarly, it should be possible to adapt the methods reported here to measure the abundance of thioester-linked ubiquitin in biological samples. We cannot currently estimate the abundance of thioester linked ubiquitin because the half-life of thioester-linked ubiquitin species at near-physiological pH is short relative to the time required for sample processing and affinity capture25. Consistent with this expectation, we observed no dithiothreitol-releasable ubiquitin binding to the BUZ domain in preliminary experiments (data not shown).
The general Ub-PSAQ method is versatile and could be readily extended by introducing new affinity reagents, such as chain-specific antibodies26, other ubiquitin binding domains or new standards (multiple types of ubiquitin chain standards with different linkages present as different isotope-labeled ubiquitin species, thioester linked ubiquitin species and free ubiquitin chains) to enable increasingly comprehensive quantification of specific species using the overall approach described.
Ubiquitin is important in many facets of biology because of its ability to reversibly form a diverse array of conjugates with other proteins and the ability of these conjugates to act as signals in a diverse set of biological processes1,2. Our work enables, to our knowledge for the first time, accurate and precise measurement of ubiquitin distribution among different cellular pools and will therefore be a valuable tool to facilitate our understanding of this important signaling molecule.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturemethods/.
Supplementary Material
Acknowledgments
We thank E. Bennett, M. Reese and M. Bowen for discussions and critical reading of the manuscript. This work was funded in part by a grant (NS 04842) from the US National Institute of Neurological Disorders and Stroke.
Footnotes
Supplementary information is available on the Nature Methods website.
AUTHOR CONTRIBUTIONS
S.E.K. and R.R.K. devised the Ub-PSAQ strategy with contributions from B.E.R. and T.A.S.; S.E.K. prepared all protein affinity reagents and standards, performed the experiments and analyzed the data with input from B.E.R. and T.A.S.; B.E.R. prepared all cellular samples; T.A.S. performed all mass spectrometry analyses; and R.S.T. contributed to mass spectrometry data analysis. S.E.K. and R.R.K. wrote the manuscript, and B.E.R. contributed to figure preparation. B.E.R., T.A.S. and R.S.T. contributed to editing. C.H.B. and H.S. contributed to conceptual and experimental design. All authors discussed the results and manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 3.Zeng W, et al. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458:438–444. doi: 10.1038/nature07960. [DOI] [PubMed] [Google Scholar]
- 5.Lee BH, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takada K, Hibi N, Tsukada Y, Shibasaki T, Ohkawa K. Ability of ubiquitin radioimmunoassay to discriminate between monoubiquitin and multi-ubiquitin chains. Biochim Biophys Acta. 1996;1290:282–288. doi: 10.1016/0304-4165(96)00032-3. [DOI] [PubMed] [Google Scholar]
- 7.Haas AL, Bright PM. The immunochemical detection and quantitation of intracellular ubiquitin-protein conjugates. J Biol Chem. 1985;260:12464–12473. [PubMed] [Google Scholar]
- 8.Bennett EJ, et al. Global changes to the ubiquitin system in Huntington’s disease. Nature. 2007;448:704–708. doi: 10.1038/nature06022. [DOI] [PubMed] [Google Scholar]
- 9.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci USA. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirkpatrick DS, et al. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- 11.Brun V, et al. Isotope-labeled protein standards: toward absolute quantitative proteomics. Mol Cell Proteomics. 2007;6:2139–2149. doi: 10.1074/mcp.M700163-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Catanzariti AM, Soboleva TA, Jans DA, Board PG, Baker RT. An efficient system for high-level expression and easy purification of authentic recombinant proteins. Protein Sci. 2004;13:1331–1339. doi: 10.1110/ps.04618904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reyes-Turcu FE, et al. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell. 2006;124:1197–1208. doi: 10.1016/j.cell.2006.02.038. [DOI] [PubMed] [Google Scholar]
- 14.Ryu KY, Baker RT, Kopito RR. Ubiquitin-specific protease 2 as a tool for quantification of total ubiquitin levels in biological specimens. Anal Biochem. 2006;353:153–155. doi: 10.1016/j.ab.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 15.Ryu KY, et al. The mouse polyubiquitin gene UbC is essential for fetal liver development, cell-cycle progression and stress tolerance. EMBO J. 2007;26:2693–2706. doi: 10.1038/sj.emboj.7601722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dantuma NP, Groothuis TA, Salomons FA, Neefjes J. A dynamic ubiquitin equilibrium couples proteasomal activity to chromatin remodeling. J Cell Biol. 2006;173:19–26. doi: 10.1083/jcb.200510071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mimnaugh EG, Chen HY, Davie JR, Celis JE, Neckers L. Rapid deubiquitination of nucleosomal histones in human tumor cells caused by proteasome inhibitors and stress response inducers: effects on replication, transcription, translation, and the cellular stress response. Biochemistry. 1997;36:14418–14429. doi: 10.1021/bi970998j. [DOI] [PubMed] [Google Scholar]
- 18.Xu P, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meierhofer D, Wang X, Huang L, Kaiser P. Quantitative analysis of global ubiquitination in HeLa cells by mass spectrometry. J Proteome Res. 2008;7:4566–4576. doi: 10.1021/pr800468j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phu L, et al. Improved quantitative mass spectrometry methods for characterizing complex ubiquitin signals. Mol Cell Proteomics. 2011;10:M110.003756. doi: 10.1074/mcp.M110.003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim HT, et al. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem. 2007;282:17375–17386. doi: 10.1074/jbc.M609659200. [DOI] [PubMed] [Google Scholar]
- 22.Raasi S, Varadan R, Fushman D, Pickart CM. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol. 2005;12:708–714. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- 23.Zhang D, Raasi S, Fushman D. Affinity makes the difference: nonselective interaction of the UBA domain of Ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains. J Mol Biol. 2008;377:162–180. doi: 10.1016/j.jmb.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raasi S, Orlov I, Fleming KG, Pickart CM. Binding of polyubiquitin chains to ubiquitin-associated (UBA) domains of HHR23A. J Mol Biol. 2004;341:1367–1379. doi: 10.1016/j.jmb.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 25.Song J, et al. Stability of thioester intermediates in ubiquitin-like modifications. Protein Sci. 2009;18:2492–2499. doi: 10.1002/pro.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newton K, et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008;134:668–678. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.