SUMMARY
Background
The eukaryotic cell cycle begins with a burst of Cdk phosphorylation. In budding yeast, several Cdk substrates are preferentially phosphorylated at the G1/S transition rather than later in the cell cycle when Cdk activity levels are high. These early Cdk substrates include signaling proteins in the pheromone response pathway. Two such proteins, Ste5 and Ste20, are phosphorylated only when Cdk is associated with the G1/S cyclins Cln1 and Cln2, and not G1, S, or M cyclins. The basis of this cyclin specificity is unknown.
Results
Here we show that Ste5 and Ste20 have recognition sequences, or “docking” sites, for the G1/S cyclins. These docking sites, which are distinct from Clb5/cyclin A-binding “RxL” motifs, bind preferentially to Cln2. They strongly enhance Cln2-driven phosphorylation of each substrate in vivo, and function largely independent of position and distance to the Cdk sites. We exploited this functional independence to re-wire a Cdk regulatory circuit in a way that changes the target of Cdk inhibition in the pheromone response pathway. Furthermore, we uncover functionally active Cln2 docking motifs in several other Cdk substrates. The docking motifs drive cyclin-specific phosphorylation, and the cyclin preference can be switched by using a distinct motif.
Conclusions
Our findings indicate that some Cdk substrates are intrinsically capable of being phosphorylated by several different cyclin-Cdk forms, but they are inefficiently phosphorylated in vivo without a cyclin-specific docking site. Docking interactions may play a prevalent but previously unappreciated role in driving phosphorylation of select Cdk substrates preferentially at the G1/S transition.
INTRODUCTION
Cyclin-dependent kinases (Cdks) are central regulatory enzymes of the eukaryotic cell cycle [1]. In most eukaryotes, different Cdk forms are specialized for driving distinct events in the cell cycle. In metazoans, these forms can differ in both the cyclin subunit and the Cdk enzyme. In simpler eukaryotes such as yeasts, a single Cdk enzyme associates with multiple different cyclins, which impart different functional properties to the cyclin-Cdk complex. The ways in which the cyclin affects these properties are understood partly though still incompletely. For example, cyclins can affect the specific activity of the Cdk enzyme, the interaction with particular substrates, or the targeting to distinct subcellular locations [2, 3].
In the budding yeast Saccharomyces cerevisiae, a single Cdk protein (Cdc28) associates with nine different cyclins [2, 4]. Six B-type cyclins (Clb1–Clb6) drive DNA synthesis and mitosis (S and M phases), whereas the transition from G1 to S phase is driven by the G1 cyclin Cln3 and the G1/S cyclins Cln1 and Cln2. While there is functional overlap, these various cyclins are clearly specialized for optimum performance of discrete tasks. Even the three semi-redundant Cln proteins show functional distinctions, as cln3Δ and clnΔ cln2Δ cells each display unique phenotypic defects [5–8]. Interestingly, there are several Cdk substrates whose phosphorylation peaks during maximum expression of Cln1/2 [9–12], and it is not clear why later cyclins are less effective; e.g., despite the fact that the M-phase cyclin Clb2 confers especially strong Cdk activity [13], some substrates are phosphorylated more readily by Cln2-Cdc28 than by Clb2-Cdc28. To date, no molecular mechanism explains why any particular substrate is preferentially phosphorylated in a Cln1/2-specific manner.
Cln1/2-specific substrates include proteins in the pheromone response pathway. This signaling pathway triggers a G1 phase arrest that synchronizes cells prior to mating [14, 15]. In cells that have already begun the cell cycle, this pathway is transiently inactivated so that cell division can conclude [16, 17]. Three proteins in this pathway are phosphorylated at the G1/S transition: Far1 (a Cdk inhibitor protein), Ste20 (a PAK-family kinase), and Ste5 (a MAP kinase cascade scaffold protein) [10–12, 18]. While the role of Ste20 phosphorylation remains unknown [19], Cdk phosphorylation of Far1 and Ste5 inactivates the cell cycle arrest and signal transduction functions of the pheromone pathway, respectively [12, 18, 20]. Notably, Ste20 and Ste5 are Cln1/2-specific substrates [10–12], but the mechanistic basis of this specificity is unknown. S-phase cyclins such as mammalian cyclin A and yeast Clb5 have a “hydrophobic patch” that allows them to recognize specific “RxL” motifs in some substrates [21–23]. Some related examples exist for other cyclins [24, 25], but not for the yeast G1 or G1/S cyclins. Moreover, as with many Cdk substrates, Ste20 and Ste5 are each phosphorylated at a large number of sites, and this can be important for proper regulation [12].
In this study we probe the molecular basis of cyclin specificity during G1/S Cdk phosphorylation in vivo. We find that both Ste5 and Ste20 have specific recognition sequences, or “docking” sites, that interact preferentially with the cyclin Cln2. In each protein, the docking sites promote efficient phosphorylation of Cdk sites, and they do so in a cyclin-specific manner. Furthermore, these docking sites function largely irrespective of distance or orientation relative to the phosphorylation sites, and they are interchangeable between substrates. We identify functionally similar behavior for motifs in several other Cdk substrates, suggesting that these docking sites may represent the first of many previously unrecognized recognition sequences for the G1 or G1/S cyclins in yeast.
RESULTS
Cyclin-specific phosphorylation of Ste5 and Ste20
Pheromone responsiveness rises and falls in opposition to the periodic fluctuations in Cln1/2 levels [16, 17]. Similarly, both Ste20 and Ste5 are phosphorylated in a periodic pattern dependent on Cln1/2 cyclins [10–12]. To investigate the basis of this cyclin specificity, we asked if it is an intrinsic feature of the Cdk phosphorylation sites themselves or if it involves other structural or functional features of each substrate protein. Ste5 and Ste20 are large, multi-domain proteins (>900 residues) that can be roughly divided into N-terminal regulatory/localization regions and C-terminal catalytic/signaling regions (Figure 1A). In each case the Cdk sites are concentrated in the N-terminal regions. Using N-terminal fragments of each protein (Ste51–370, Ste51–337, Ste2080–590) as substrates, we found that cyclin specificity was maintained. First, their phosphorylation in synchronous cultures peaked at the G1/S boundary (bud emergence; Figure 1B) rather than in M-phase (when levels of Clb2 were maximal). Second, when using the inducible GAL1 promoter (PGAL1) to drive expression of various cyclins, phosphorylation of the Ste5 and Ste20 fragments was observed only with Cln1 and Cln2, and not with Cln3, Clb5, or Clb2 (Figure 1C). (Although unequal levels of cyclins could contribute to these differences, Clb2 was able to drive phosphorylation of the Cdk substrate Swe1 [Figure 1C] [26], and later results will show activity for both Clb5 and Clb2 in similar assays.) Thus, stage-specific and cyclin-specific phosphorylation of Ste5 and Ste20 does not require their signaling functions, and the determinants of specificity must lie within their N-terminal fragments.
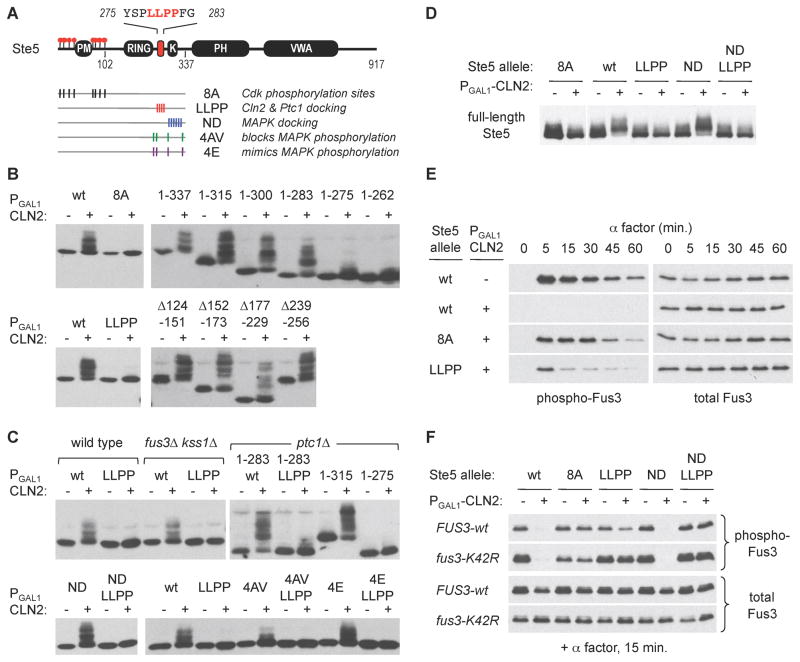
Figure 1. Cyclin specific phosphorylation of Ste5 and Ste20 regulatory domains.
(A) Domain structures of Ste5 and Ste20. Red circles indicate Cdk sites [12, 19]; in Ste20, only the 13 confirmed sites (of 23 possible) are shown
(B) Phosphorylation of Ste5 and Ste20 fragments in synchronous cdc15-2 cultures, after release from M phase arrest. Full-length Ste20 is shown for comparison; its phosphorylation behavior was described previously [10, 11]. Cell cycle progression was monitored by anti-Clb2 immunoblot and by budding
(C) HA-tagged Ste5 or V5-tagged Ste20 fragments, expressed from native promoters, were monitored after galactose-induced expression of GST-tagged cyclins. Reduced electrophoretic mobility signifies phosphorylation, as confirmed by phosphatase treatment (data not shown). For comparison, V5-tagged Swe1 demonstrates activity for Clb2. The relative expression levels for GST-cyclins were highly reproducible; one representative example is shown. A variant of Cln3 lacking ten Cdk sites (Cln310A) was used to increase its stability. Results were similar in sic1Δ cells (data not shown), in which the Clb-Cdk inhibitor Sic1 is absent.
A distal recognition sequence promotes Cdk phosphorylation of the Ste5 N-terminus
We used the Ste51–337 fragment to study the requirements for phosphorylation by Cln2-Cdc28. As expected, phosphorylation required the same 8 Cdk sites shown previously to regulate full-length Ste5 (Figure 2B) [12]. Nevertheless, these sites were not sufficient to ensure efficient phosphorylation. By making further truncations, we discovered that phosphorylation required a short stretch of sequence (276–283) far away from the Cdk sites (Figure 2B). Similarly, alanine replacement of four residues within this region (LLPP) also disrupted phosphorylation (Figure 2B). In contrast, internal deletions showed that several other large segments of the Ste5 N-terminus were dispensable (Figure 2B). We conclude that efficient Cln2-Cdc28 phosphorylation of Ste5 requires specific sequences that are separate from the phosphorylation sites themselves. For now we tentatively refer to this required region as a Cln2 docking site.
Figure 2. A distal docking motif promotes Cln2-Cdc28 phosphorylation of Ste5.
(A) Locations of key Ste5 features and mutations. Residues 275–283 contain the putative Cln2 docking motif required for efficient phosphorylation of the N-terminal Cdk sites. Mutations used in later panels are indicated; see Figure S1A for details
(B) Phosphorylation of the Ste5 N-terminus (Ste51–337) requires both the Cdk sites and a distal LLPP motif between residues 275 and 283. Phosphorylation was triggered by galactose-induced expression of a PGAL1-CLN2 construct (+) or a vector control (−)
(C) The role of the LLPP motif is independent of MAPKs (fus3Δ kss1Δ), the phosphatase Ptc1 (ptc1Δ), and the MAPK binding site (ND mutant). Results show the Ste51–337 fragment except as indicated otherwise. The fus3Δ kss1Δ strain (PPY1173) was tested in parallel with a congenic wild-type strain (PPY640). LLPP function also does not require the MAPK phosphorylation sites, but non-phosphorylatable mutations at these sites (4AV) mildly reduce the extent of Cln2-Cdc28 phosphorylation. Also see Figure S1B,C
(D) Cln2-driven phosphorylation of full-length Ste5 (V5-tagged) requires the LLPP motif. For the 8A lanes, a longer exposure (of the same blot) is shown to compensate for imperfect loading
(E) Mutation of the LLPP motif disrupts the ability of Cln2 to inhibit pheromone signaling. Pheromone-induced phosphorylation of Fus3 was monitored in ste5Δ fus3Δ kss1Δ strains (± PGAL1-CLN2) harboring STE5 variants and wild-type FUS3 on plasmids
(F) The LLPP motif mediates regulation by Cln2 in the absence of Fus3-Ste5 binding (Ste5 ND mutant) and Fus3 kinase activity (fus3-K42R mutant). Strains (as in panel D) harbored plasmids with the indicated forms of STE5 and FUS3.
This putative Cln2 docking site lies within a larger inter-domain region of Ste5 with several notable features (Figure S1A), including: (i) a MAPK binding site; (ii) four MAPK phosphorylation sites, which possibly could also act as Cdk sites due to similar target sequences (i.e., SP/TP); and (iii) a binding site for the phosphatase Ptc1 [27, 28]. None of these features seemed crucial for docking site function. MAPK binding played no evident role, as the results were unaffected by deleting MAPK genes (fus3Δ kss1Δ) or by mutating the MAPK binding site (ND mutant) (Figure 2C,S1B). At the MAPK phosphorylation sites, phospho-mimetic mutations (4E) had no effect (Figure 2C), but non-phosphorylatable mutations (4AV) caused a mild reduction; thus, phosphorylation at one or more of these sites (e.g., by a MAPK or Cdk) may enhance further Cdk phosphorylation elsewhere, or the mutations may mildly disrupt recognition of the docking motif. (All four MAPK sites cannot be required, because the shorter Ste51–283 fragment retains only two sites yet remains a good substrate; Figure 2B,C). Finally, although the required LLPP motif overlaps a binding site for Ptc1 [28], it promoted Cdk phosphorylation identically in ptc1Δ cells (Figure 2C, S1C). Altogether, these data show that recognition of the putative Cln2 docking site can be separated from MAPK and Ptc1 binding, though it may be affected by either the sequence or phosphorylation status of adjacent SP/TP sites. Furthermore, as with the N-terminal fragments, the LLPP motif was also required for Cdk phosphorylation of full-length Ste5 (Figure 2D).
Next, we tested if the Cln2 docking site is required for Cdk inhibition of Ste5 signaling [12]. Indeed, as with mutation of the Cdk sites (8A), mutation of the LLPP motif restored the ability of pheromone to activate the MAPK Fus3 in PGAL1-CLN2 cells (Figure 2E). However, this restored activation was unusually transient (Figure 2E), and the transcriptional response was only partially restored (Figure S1D). This complex behavior likely reflects the additional role of the LLPP region in binding to Ptc1, which antagonizes the ability of Fus3 to bind Ste5 and down-regulate signaling [28]. That is, although the LLPP mutation disrupts Cln2 docking, it will simultaneously lead to excessive binding and inhibition by Fus3, which may have a compensatory effect. To eliminate contributions from Fus3, we used the Ste5 ND mutation to disrupt Fus3 binding [27], or a kinase-inactive form of Fus3 (K42R). In these contexts, the LLPP mutation clearly blocked the inhibitory effect of Cln2 (Figures 2F, S1D). Therefore, the LLPP motif can mediate Cdk inhibition in the absence of both Fus3 binding and kinase activity, whereas excess Fus3 activity may obscure this role. Although the overlap between Cln2 and Ptc1 sites creates an added layer of complexity, overall these results establish that Cdk regulation of Ste5 involves both Cdk phosphorylation sites and a separate Cln2 docking motif.
Cyclin-specific binding sites in Ste20 and Ste5
Efforts to test if Ste5 binds Cln2 were hampered by technical issues (e.g., non-specific precipitation), but binding of Ste20 to Cln2 is readily detectable in cell extracts [19, 29]. Therefore, we searched for the responsible sequences in Ste20 using GST-Ste20 and Cln2-myc13 fusions. An N-terminal fragment (Ste201–333) was sufficient to bind Cln2, and this required residues 73–119 (Figure 3B). The C-terminal end of this fragment, which harbors a membrane-binding “basic-rich” (BR) domain, was not required but it did enhance binding (Figure S2B); hence, further analyses kept the C-terminus fixed at position 333. The required region (73–119) includes a conserved block of ~20 residues (Figure S2C), and mutating consecutive sets of residues in this region revealed that an eight-residue stretch (SLDDPIQF) was critical for binding to Cln2 (Figure 3Ci). We then asked if the role of this Ste20 motif could be replaced with a small fragment from Ste5 that harbors its putative Cln2 docking site. Indeed, the Ste5 site mediated Cln2 binding, and this required the same LLPP sequence that promotes Ste5 phosphorylation (Figure 3Cii). Furthermore, each sequence preferentially bound to Cln2 over other cyclins (Figure 3D), and weakly to Cln1. Collectively, these experiments identify docking sites in both Ste20 and Ste5 that can discriminate among different cyclins to mediate specific binding. The two sites are not highly similar but each contains the motif LxxPΦxΦ (where Φ is hydrophobic), raising the possibility that a degenerate pattern of hydrophobic side chains forms the Cln2 recognition motif.
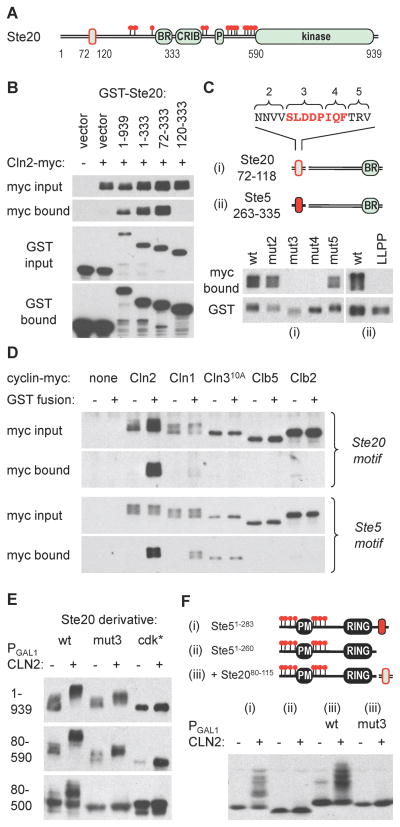
Figure 3. Cyclin-specific binding by docking motifs in Ste20 and Ste5.
(A) The diagram indicates endpoints used for mapping the Cln2-binding region in Ste20, which is outlined in red
(B) Cells co-expressing GST fusions to Ste20 fragments and Cln2-myc13 were lysed, and complexes were recovered using glutathione sepharose. Input (5%) and bound proteins were analyzed by anti-myc and anti-GST blots
(C) Starting with a Ste2072–333 fragment, alanine substitutions were made at blocks of residues in the 72–118 region (see Figure S2C; numbering starts at 2 because additional flanking mutations were used in other assays). Cln2 binding was tested as in panel B. Separately, the required Ste20 region was replaced by a Ste5263–335 fragment (ii), in both wt and LLPP mutant forms, to test the ability of this Ste5 sequence to mediate Cln2 binding
(D) Ste20 and Ste5 docking sites show cyclin-specific binding. GST alone (−) or GST fusions (+) were used to co-precipitate myc13-tagged cyclins (expressed from the CYC1 promoter) in yeast lysates. The GST fusions were to Ste201–333 (Ste20 motif) or to the Ste5263–335-Ste20120–333 chimera used in panel Cii (Ste5 motif). Cln310A showed varying levels of non-specific precipitation but no reproducible binding to either GST fusion
(E) Cln2-induced phosphorylation was assayed for V5-tagged forms of full-length Ste20 (1–939) or N-terminal fragments (80–590, 80–500), with or without mutations in the docking site (mut3) or the 13 confirmed Cdk sites (cdk*)
(F) The Cln2 docking site from Ste20 can drive phosphorylation of a heterologous substrate. Phosphorylation was analyzed using Ste51–283 (i), Ste51–260 (ii), and wt or mut3 versions of the Ste20 docking site (residues 80–115) fused to Ste51–260 (iii).
The Cln2 docking site in Ste20 enhances phosphorylation
Ste20 contains 23 potential Cdk sites, of which at least 13 are used in vivo [19]. To test if Cln2 docking affects Ste20 phosphorylation, we compared mutations in either the docking site (mut3) or the 13 confirmed Cdk sites (cdk*), using full-length Ste20 and several N-terminal fragments (Figure 3E). In full-length Ste20 (1–939), the docking site mutation reduced the magnitude of the Cln2-induced mobility shift, but did not eliminate it (i.e., unlike the cdk* mutation). This partial phenotype could signify less efficient use of all sites, or a specific defect at particular sites due to their position or sequence context; in this regard, it is noteworthy that 9 of the 13 confirmed Cdk sites are “minimal” sites (S/T-P) whereas only 4 are “consensus” sites (S/T-P-x-K/R). The docking site mutation again caused a partial phenotype for the largest N-terminal fragment (80–590), but it caused a strong disruptive phenotype for smaller fragments such as 80–500 (Figure 3E) and 80–333 (data not shown). Though we did not parse these differences further, we note that the two smaller fragments retain only one consensus Cdk site, raising the possibility that docking is especially important for phosphorylating minimal sites. As a separate test of the activity of the Ste20 docking motif, we asked if it could substitute for the analogous site in Ste5. Indeed, the Ste20 motif restored strong phosphorylation to the Ste51–260 fragment, and this activity was blocked by mutations that disrupt Cln2 binding (Figure 3Fiii). Thus, the Ste20 docking motif can stimulate Cln2-Cdc28 phosphorylation of native sites in Ste20 or sites in a heterologous substrate.
Re-wiring a Cdk regulatory circuit via a Ste20Ste5PM chimera
Cdk phosphorylation of Ste5 serves to inhibit pheromone response at the G1/S transition [12]. We wished to explore whether the responsible regulatory mechanism could be moved to other proteins, and whether such synthetic approaches [30, 31] could help further probe the factors controlling Cdk regulation. Therefore, we attempted to transfer the regulatory effects of Cdk phosphorylation from Ste5 onto Ste20. Our rationale stems from the fact that each protein requires a short membrane-binding motif for its plasma membrane localization and signaling activity [32, 33], and the function of the Ste5 motif, termed the PM domain, is inhibited by phosphorylation at adjacent Cdk sites [12]. Thus, we replaced the membrane-binding (BR) motif in Ste20 with Ste5 fragments that contain both the PM domain and its flanking Cdk sites (Figure 4A), and then asked if Ste20 now could be inhibited by Cdk phosphorylation.
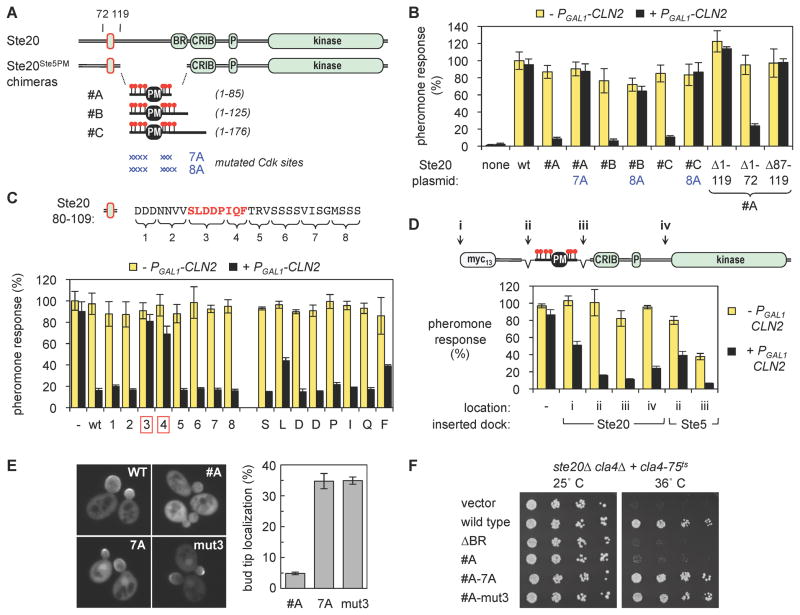
Figure 4. Re-wiring a Cdk regulatory circuit with a Ste20Ste5PM chimera.
(A) The Ste20Ste5PM chimeras. Ste20 residues 124–311, containing the membrane-binding BR domain, were replaced with three fragments from Ste5 that include its membrane-binding PM domain plus 7 or 8 flanking Cdk sites
(B) Pheromone signaling by Ste20Ste5PM chimeras is inhibited by Cln2. Because Cln2-Cdk normally inhibits signaling via Ste5, these tests used cells with a non-phosphorylatable Ste5 variant (ste20Δ STE5-8A). Wild-type Ste20 (wt) or Ste20Ste5PM chimeras (from panel A) were introduced on plasmids, and pheromone response was measured using a transcriptional reporter (FUS1-lacZ). Signaling by all three chimeras (#A, #B, #C) was inhibited by Cln2, but this was blocked by mutations in the Cdk phosphorylation sites (7A, 8A). Deletions of the Ste20 N-terminus, made in the #A chimera, show that residues 87–119 are required for regulation by Cln2. Bars, mean ± SD (n = 3)
(C) Sequences required for regulation by Cln2 were analyzed using a chimera similar to #A, containing only Ste20 residues 80–109 upstream of Ste51–85 (see Figure S3C). Alanine mutations replaced eight blocks of residues (left) or individual residues in the SLDDPIQF motif (right). These were compared to an intact sequence (wt) and a chimera that lacks the sequence entirely (−). Signaling was assayed as in panel B. Bars, mean ± SD (n = 3)
(D) Docking sites from Ste20 (80–115) or Ste5 (257–330) were inserted at different positions (i–iv) into a variant of chimera #A that lacks residues 87–119 (see panel B). Insertions at position ii also removed residues 1–86. Signaling was assayed as above. Bars, mean ± SD (n = 3)
(E) Bud tip localization of Ste20Ste5PM chimera in cycling cells is inhibited via the Cdk sites (7A) and the Cln2 docking site (mut3). Strain BY4741 harbored GFP-Ste20 plasmids. Representative images show unfixed cells (left); localization was quantified after formaldehyde fixation (right). Bars, mean ± SD (n = 3 experiments; >150 cells per allele per experiment)
(F) The growth function of Ste20 is inhibited in the Ste20Ste5PM chimera, if Cdk and docking sites are intact. Serial (1/5x) dilutions of strain KBY211 harboring the indicated Ste20 plasmids were incubated at 25 or 36 C for 4 days. As a control, the ΔBR allele removes the BR domain in full-length Ste20 [33].
The Ste5 PM domain was a highly effective substitute for the Ste20 BR domain (Figure 4B), irrespective of the precise size of the transferred Ste5 fragment. As hypothesized, the function of these Ste20Ste5PM chimeras in the pheromone response pathway was inhibited by expression of Cln2 (Figure 4B). (Because Cln2 ordinarily inhibits pheromone response via Ste5, these assays used cells harboring a Cdk-resistant derivative, Ste5-8A.) Importantly, this inhibition specifically required intact Cdk sites flanking the PM domain (Figure 4B), despite the presence of other Cdk sites elsewhere in Ste20. Moreover, the inhibition was cyclin-specific and was not accompanied by any reduction in protein levels (Figures S4A,B). Thus, these experiments show that the Cdk regulatory target in the pheromone pathway can be switched to a different protein, and that the Cdk-inhibited domain from Ste5 constitutes a portable regulatory module. Furthermore, inhibition of the Ste20Ste5PM chimera required the region of Ste20 (residues 87–119) that contains its Cln2 docking site (Figure 4B, right). This finding establishes two further points: (i) the mere presence of Cdk sites in the transferred Ste5 fragment is insufficient to make it an effective target of Cdk down-regulation; and (ii) the Cln2 docking site in Ste20 can control functional regulation via Cdk phosphorylation.
We used the Ste20Ste5PM chimera to dissect which docking site residues were required for regulatory activity. This function was fully contained in a 30-residue region (80–109) spanning the conserved stretch noted earlier (Figure S3C), and it required the same 8-residue motif involved in Cln2 binding (SLDDPIQF) (Figure 4C, left). Notably, no single residue in this motif was absolutely required, though partial phenotypes were seen for mutations at the L, P, and F residues (Figure 4C, right). Thus, Cln2 docking may involve contacts distributed throughout the motif. We also found that docking site function is highly flexible, as it remained active when placed on either side of the PM domain and up to ~280 residues from the nearest Cdk site (Figure 4D). Moreover, the docking site from Ste5 could also function in the chimera (Figure 4D, right). Finally, additional tests showed that the Cdk regulation conferred upon the Ste20Ste5PM chimera is not limited to pheromone response or to over-expressed Cln2. Specifically, Ste20 performs additional functions in cycling cells, and its ability to localize to growing bud tips, or to sustain viability in the absence of the related PAK Cla4, requires plasma membrane contact [33–35]. These abilities were inhibited in the Ste20Ste5PM chimera in a manner that required both docking and Cdk sites (Figure 4E, F). Hence, the chimera converts Ste20 from a multi-functional kinase into a form that is restricted to functioning in non-cycling cells. Altogether, the results clearly reveal the feasibility of creating new Cdk regulatory circuits via a combination of Cln2 docking and Cdk phosphorylation sites, and they establish the Ste20Ste5PM chimera as a functionally flexible platform with which to assay docking site activity.
Functional Cln2 docking sites in other Cdk substrates
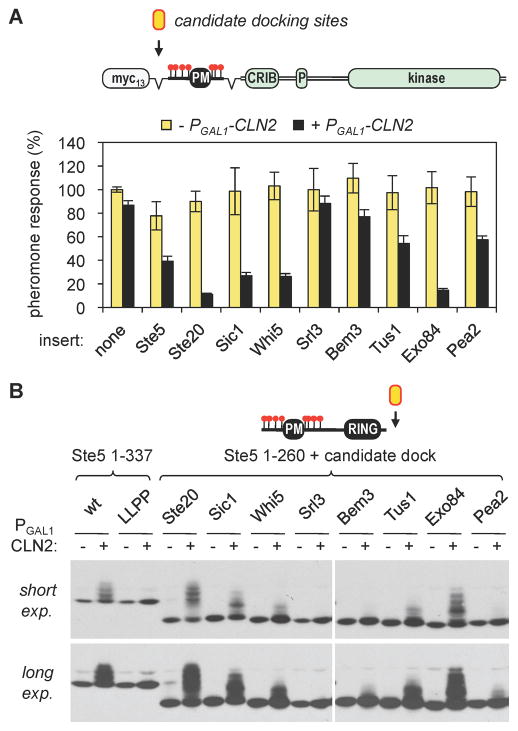
To assess whether the Cln2 docking behavior seen with Ste5 and Ste20 might be more widespread, we searched for similar sites in other Cdk substrates. Matches to the LxxPΦxΦ motif or the core Ste5 site (LLPP) were too numerous for a proteome-wide analysis. Nevertheless, we scanned the sequences of known Cln1/2-Cdc28 targets and Cln2 binding partners, plus Cdc28 substrates found in large-scale screens [4, 29, 36, 37], for Leu/Pro-rich sequence motifs that are conserved among fungal orthologs and that lie outside of known or predicted structural domains. While not comprehensive, as an initial test case we chose seven such sequences (from Sic1, Whi5, Srl3, Bem3, Tus1, Exo84, and Pea2; see Figure S4), including one recently implicated in Cln2-Cdc28 phosphorylation of Sic1 [38]. We then tested these sequences for their ability to functionally substitute for the Ste5/Ste20 docking sites, using two experimental settings: (i) Cln2 inhibition of signaling by the Ste20Ste5PM chimera; and (ii) Cln2-driven phosphorylation of the Ste5 N-terminus.
Remarkably, the majority of these sequences had measurable activity. In the signaling assays (Figure 5A), they mediated Cln2 inhibition to varying degrees, from strong (Sic1, Whi5, Exo84) to moderate (Tus1, Pea2) to weak (Bem3); only the Srl3 sequence was ineffective. Importantly, their inhibitory effects were a specific response to Cln2 expression, and no non-specific effects were seen. The three most potent sequences contain exact matches to the Ste5 LLPP motif (Figure S4B), though it is notable that each was in fact more potent than the Ste5 sequence in these assays (Figure 5A); thus, additional context features likely influence the efficacy of this motif. Similarly, the Pea2 sequence closely resembles the motif in Ste20, and yet was less potent. The phosphorylation assays yielded similar overall results, with a range of activity (Figures 5B and S4C). The relative activities in the two assays were generally correlated, but there were some discrepancies; for example, the Whi5 and Pea2 sequences were each weaker in the phosphorylation assay than the sequences with which they showed comparable potency in the signaling assay. Together, these results clearly establish the utility of each assay for their ability to rapidly evaluate multiple candidate Cln2 docking sites, and to compare efficacy in parallel. Overall, given the large fraction of sequences that were effective, the findings suggest that Cln2 recognition sites of various strengths may be quite prevalent among Cln-Cdk targets.
Figure 5. Identification of additional candidate Cln2 docking sites.
(A) Candidate Cln2 docking sites from seven Cdk substrates (see Figure S4) were inserted into a Ste20Ste5PM chimera lacking its endogenous docking site. These derivatives were compared to chimeras containing the Ste5 or Ste20 docking sites inserted at the same position, or no docking site (none). Cln2 inhibition of pheromone response was assayed as in Figure 4. Bars, mean ± SD (n = 5)
(B) The same candidate docking sites used in panel A were inserted at the end of a Ste5 fragment (Ste51–260) that lacks its endogenous docking site, and Cln2-driven phosphorylation was assayed. See Figure S4C for additional repetitions.
The phosphorylation role of docking sites is cyclin-specific
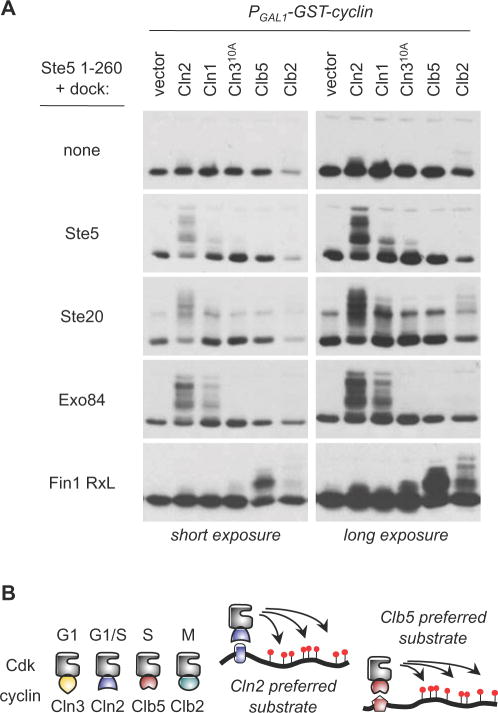
Finally, we tested whether the Cln2 docking sites enhance phosphorylation by all forms of cyclin-Cdk or only by specific forms. We used the Ste51–260 fragment as a starting substrate, and monitored its phosphorylation by inducing expression of different cyclins in asynchronous cells (Figure 6A). In the absence of any docking site, none of the cyclins drove appreciable phosphorylation. When Cln2 docking sites from Ste5, Ste20, or Exo84 were appended to this substrate, phosphorylation was enhanced in a manner that was clearly cyclin-specific. Cln2 was generally most effective, followed by Cln1. (With the Ste20 site, Clb2 could drive some weak phosphorylation detectable in longer exposures.) As a further test, we fused the same substrate to a different type of docking site: namely, a short sequence containing an “RxL” motif from Fin1, a Cdk substrate that is normally preferred by Clb5 [13]. This motif switched the cyclin preference and caused phosphorylation to be driven strongly by Clb5, plus moderately by Clb2 (Figure 6A, bottom). (It is noteworthy that the band patterns were not identical, suggesting a different extent or distribution of phosphorylation events.) These results clearly indicate that Cdk sites in the Ste5 N-terminus are intrinsically capable of being phosphorylated by several different cyclin-Cdk forms, but they are not effectively phosphorylated in vivo in the absence of cyclin-specific docking sites, and hence this provides specificity to the Cdk phosphorylation.
Figure 6. Docking sites drive cyclin-specific phosphorylation.
(A) Phosphorylation of a single substrate (Ste51–260) was monitored with and without the addition of the indicated docking sites, in cells expressing different PGAL1-induced cyclins. Docking sites from Ste5, Ste20, and Exo84 preferentially enhance phosphorylation by Cln1/2-Cdc28, whereas an RxL-containing fragment from Fin1 converts the substrate into one that is preferred by Clb5 and Clb2
(B) Schematic comparison of different cyclin-Cdk complexes, with a general model for how cyclin-specific docking interactions can selectively enhance substrate phosphorylation by individual forms of cyclin-Cdk.
DISCUSSION
In this study we identified docking sites for the yeast G1/S cyclins in several Cdk substrates. These sites bind preferentially to Cln2 and enhance phosphorylation of Cdk substrates in a cyclin-specific manner in vivo. They are also functionally modular, in that they can promote phosphorylation at a variety of distances and positions relative to the Cdk sites, and the cyclin specificity of phosphorylation can be switched by exchanging docking sites. We exploited this functional modularity to re-wire a Cdk regulatory circuit so as to change the target of Cln2-Cdc28 regulation in the pheromone response pathway, and to identify candidate Cln2 docking sites in several other Cdk substrates. The relative ease with which these other sites were found suggests that there may be numerous examples of such sites for the G1 or G1/S cyclins in yeast. Indeed, recent studies from another group show that the Sic1 sequence which we used to drive Cln2-Cdc28 phosphorylation of heterologous substrates does in fact promote phosphorylation of Sic1 itself and can also act as a competitive inhibitor of other Cln2-Cdc28 substrates [38]. Collectively, these findings suggest that docking interactions play a prevalent but previously unappreciated role in driving phosphorylation of G1/S Cdk substrates.
The use of separate docking and phosphorylation sites offers functional and regulatory flexibility. At one extreme, it can allow different kinases to phosphorylate the same sites, as we observed by replacing a Cln2 docking site with an RxL motif favored by B-type cyclins. It may also allow the sequence requirements at phosphorylation sites to be relaxed. While most kinases favor certain residues flanking the phosphorylation site [3, 39], this ideal context may not be present or tolerated in all relevant substrates. Indeed, proteome-wide analysis [37] suggests that roughly two thirds of Cdk sites in yeast are not “consensus” sites (S/T-P-x-K/R). In Ste5, none of the 8 N-terminal Cdk sites matches this consensus. The presence of a Cln2 docking site converts the Ste5 N-terminus from a weak substrate into a better substrate, but only for Cln1/2-Cdc28. This enhancement may compensate for poor sequence context, which in turn could minimize use by other cyclin-Cdks, thus providing a regulatory benefit. Other possible benefits of cyclin docking interactions include: (i) they may enhance Cdk phosphorylation even for substrates that are not cyclin specific; (ii) they may help drive multi-site phosphorylation of Cdk substrates [40]; and (iii) they may impart useful regulatory behavior by fostering interplay with competitors or other bound factors. Indeed, the Cln2 docking site in Ste5 overlaps a binding site for the phosphatase Ptc1 [28], and hence these two factors may compete for access to Ste5 in vivo. Overlapping cyclin and phosphatase binding sites have also been found in the mammalian protein Rb [41], suggesting a common theme.
Differences in docking strength may impact the extent and/or timing of substrate phosphorylation [40]. For example, the docking sites in Ste5 and Ste20 bind stronger to Cln2 than Cln1, and such differences could contribute to disparities in efficacy seen for these two cyclins [16, 42, 43]. We also observed varying degrees of potency for different Cln2 docking sites, though the responsible sequence features remain uncertain. The Ste5 and Ste20 sites share an LxxPΦxΦ motif and an overall enrichment in Leu, Pro, and/or hydrophobic residues, which was used as a criterion to identify additional sites. An LLPP motif is shared by several of the strong sites, but the docking site in Ste20 does not match this motif and yet is very potent. Thus, deducing the key sequence features of Cln2 docking sites will require subsequent study, as will determination of whether they bind directly to the cyclin versus the cyclin-Cdk complex.
Subcellular localization can also contribute to functional specialization of cyclins [44–46]. Hence, in addition to driving phosphorylation in cis, some cyclin docking sites may help localize cyclins and/or promote phosphorylation in trans of other proteins in the same complex or subcellular locale [47]. In fact, prior to our discovery that it binds Cln2, we originally found that the Cln2 docking site in Ste20 could trigger hyperpolarized growth when over-expressed and membrane-localized [33]; in retrospect, this phenotype could result from the generation of excess cortical binding sites for Cln2, which promotes apical polarized growth [48]. Several Cln1/2-Cdc28 substrates are involved in cell polarity [4] (see Figure S4 legend), so it will be of interest to determine whether they each contain Cln1/2 docking sites or if docking sites in some can serve a scaffolding role that promotes Cdk phosphorylation of co-bound or co-localized substrates. Synthetic approaches, such as those described here, can be used to discover these docking sites as well as to characterize their functional properties and activities in a standardized setting, which ultimately can illuminate how the regulatory behavior of native proteins and pathways arises from the combined properties of individual motifs.
EXPERIMENTAL PROCEDURES
Detailed methods are described in the Supplemental Material, which also lists all yeast strains and plasmids plus describes which were used for each experiment.
Phosphorylation Assays
Cells harboring epitope-tagged proteins and PGAL1-GST-cyclin constructs were induced with galactose for 1–3 hr. Cell extracts were prepared by glass-bead lysis in trichloroacetic acid solution [49], and proteins were analyzed by SDS-PAGE and immunoblotting. To follow phosphorylation in synchronous cultures, a cdc15-2 strain harboring tagged proteins was arrested at 37°C for 3 hr, then released at 25°C. Aliquots were taken at 20 min intervals to prepare protein samples and score bud emergence.
Signaling Assays
To measure effects of PGAL1-CLN2 on pheromone response, cells were grown in raffinose media, induced with galactose, and then treated with α factor. MAPK phosphorylation was measured by anti-phospho-p44/42 immunoblots [50]. FUS1-lacZ expression was measured by β-galactosidase assay [12, 51].
GST Binding Assays
Cultures harboring PGAL1-GST fusion proteins and cyclin-myc13 constructs were induced with 2% galactose for 3 hr. Extracts were prepared by glass bead lysis in a non-ionic detergent buffer [51]. Aliquots were reserved, then GST fusions and co-bound proteins were collected with glutathione-sepharose beads and analyzed by SDS-PAGE and immunoblotting.
Supplementary Material
HIGHLIGHTS.
Cdk phosphorylation of Ste5 and Ste20 depends on binding sites for the cyclin Cln2
Functionally similar Cln2 docking sites exist in several other Cdk substrates
Specific docking sites can dictate the cyclin preference of phosphorylation in vivo
Exchange of portable motifs allows re-wiring of a Cdk regulatory circuit
Acknowledgments
We are grateful to Matthew Winters and Rachel Lamson for technical assistance, and to Mart Loog for discussions and communication of unpublished results. We also thank Jenny Benanti for the Cln310A allele, Patty Pope for cyclin-myc13 constructs, Wendell Lim for the Ste5 ND mutant, and Dan McCollum and Jenny Benanti for comments on the manuscript. This work was supported by a grant from the NIH (GM57769) to P.M.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Morgan DO. The Cell Cycle: Principles of Control. London: New Science Press; 2007. [Google Scholar]
- 2.Bloom J, Cross FR. Multiple levels of cyclin specificity in cell-cycle control. Nat Rev Mol Cell Biol. 2007;8:149–160. doi: 10.1038/nrm2105. [DOI] [PubMed] [Google Scholar]
- 3.Ubersax JA, Ferrell JE., Jr Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol. 2007;8:530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- 4.Enserink JM, Kolodner RD. An overview of Cdk1-controlled targets and processes. Cell Div. 2010;5:11. doi: 10.1186/1747-1028-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cross FR. DAF1, a mutant gene affecting size control, pheromone arrest, and cell cycle kinetics of Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4675–4684. doi: 10.1128/mcb.8.11.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nash R, Tokiwa G, Anand S, Erickson K, Futcher AB. The WHI1+ gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J. 1988;7:4335–4346. doi: 10.1002/j.1460-2075.1988.tb03332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moffat J, Andrews B. Late-G1 cyclin-CDK activity is essential for control of cell morphogenesis in budding yeast. Nat Cell Biol. 2004;6:59–66. doi: 10.1038/ncb1078. [DOI] [PubMed] [Google Scholar]
- 8.Skotheim JM, Di Talia S, Siggia ED, Cross FR. Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature. 2008;454:291–296. doi: 10.1038/nature07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCusker D, Denison C, Anderson S, Egelhofer TA, Yates JR, 3rd, Gygi SP, Kellogg DR. Cdk1 coordinates cell-surface growth with the cell cycle. Nat Cell Biol. 2007;9:506–515. doi: 10.1038/ncb1568. [DOI] [PubMed] [Google Scholar]
- 10.Oehlen LJ, Cross FR. Potential regulation of Ste20 function by the Cln1-Cdc28 and Cln2-Cdc28 cyclin-dependent protein kinases. J Biol Chem. 1998;273:25089–25097. doi: 10.1074/jbc.273.39.25089. [DOI] [PubMed] [Google Scholar]
- 11.Wu C, Leeuw T, Leberer E, Thomas DY, Whiteway M. Cell cycle- and Cln2p-Cdc28p-dependent phosphorylation of the yeast Ste20p protein kinase. J Biol Chem. 1998;273:28107–28115. doi: 10.1074/jbc.273.43.28107. [DOI] [PubMed] [Google Scholar]
- 12.Strickfaden SC, Winters MJ, Ben-Ari G, Lamson RE, Tyers M, Pryciak PM. A mechanism for cell-cycle regulation of MAP kinase signaling in a yeast differentiation pathway. Cell. 2007;128:519–531. doi: 10.1016/j.cell.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loog M, Morgan DO. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–108. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
- 14.Dohlman HG, Thorner JW. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu Rev Biochem. 2001;70:703–754. doi: 10.1146/annurev.biochem.70.1.703. [DOI] [PubMed] [Google Scholar]
- 15.Bardwell L. A walk-through of the yeast mating pheromone response pathway. Peptides. 2005;26:339–350. doi: 10.1016/j.peptides.2004.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oehlen LJ, Cross FR. G1 cyclins CLN1 and CLN2 repress the mating factor response pathway at Start in the yeast cell cycle. Genes Dev. 1994;8:1058–1070. doi: 10.1101/gad.8.9.1058. [DOI] [PubMed] [Google Scholar]
- 17.Wassmann K, Ammerer G. Overexpression of the G1-cyclin gene CLN2 represses the mating pathway in Saccharomyces cerevisiae at the level of the MEKK Ste11. J Biol Chem. 1997;272:13180–13188. doi: 10.1074/jbc.272.20.13180. [DOI] [PubMed] [Google Scholar]
- 18.McKinney JD, Chang F, Heintz N, Cross FR. Negative regulation of FAR1 at the Start of the yeast cell cycle. Genes Dev. 1993;7:833–843. doi: 10.1101/gad.7.5.833. [DOI] [PubMed] [Google Scholar]
- 19.Oda Y, Huang K, Cross FR, Cowburn D, Chait BT. Accurate quantitation of protein expression and site-specific phosphorylation. Proc Natl Acad Sci U S A. 1999;96:6591–6596. doi: 10.1073/pnas.96.12.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henchoz S, Chi Y, Catarin B, Herskowitz I, Deshaies RJ, Peter M. Phosphorylation- and ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor Far1p in budding yeast. Genes Dev. 1997;11:3046–3060. doi: 10.1101/gad.11.22.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng KY, Noble ME, Skamnaki V, Brown NR, Lowe ED, Kontogiannis L, Shen K, Cole PA, Siligardi G, Johnson LN. The role of the phospho-CDK2/cyclin A recruitment site in substrate recognition. J Biol Chem. 2006;281:23167–23179. doi: 10.1074/jbc.M600480200. [DOI] [PubMed] [Google Scholar]
- 22.Schulman BA, Lindstrom DL, Harlow E. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc Natl Acad Sci U S A. 1998;95:10453–10458. doi: 10.1073/pnas.95.18.10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilmes GM, Archambault V, Austin RJ, Jacobson MD, Bell SP, Cross FR. Interaction of the S-phase cyclin Clb5 with an “RXL” docking sequence in the initiator protein Orc6 provides an origin-localized replication control switch. Genes Dev. 2004;18:981–991. doi: 10.1101/gad.1202304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dowdy SF, Hinds PW, Louie K, Reed SI, Arnold A, Weinberg RA. Physical interaction of the retinoblastoma protein with human D cyclins. Cell. 1993;73:499–511. doi: 10.1016/0092-8674(93)90137-f. [DOI] [PubMed] [Google Scholar]
- 25.Kelly BL, Wolfe KG, Roberts JM. Identification of a substrate-targeting domain in cyclin E necessary for phosphorylation of the retinoblastoma protein. Proc Natl Acad Sci U S A. 1998;95:2535–2540. doi: 10.1073/pnas.95.5.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harvey SL, Charlet A, Haas W, Gygi SP, Kellogg DR. Cdk1-dependent regulation of the mitotic inhibitor Wee1. Cell. 2005;122:407–420. doi: 10.1016/j.cell.2005.05.029. [DOI] [PubMed] [Google Scholar]
- 27.Bhattacharyya RP, Remenyi A, Good MC, Bashor CJ, Falick AM, Lim WA. The Ste5 scaffold allosterically modulates signaling output of the yeast mating pathway. Science. 2006;311:822–826. doi: 10.1126/science.1120941. [DOI] [PubMed] [Google Scholar]
- 28.Malleshaiah MK, Shahrezaei V, Swain PS, Michnick SW. The scaffold protein Ste5 directly controls a switch-like mating decision in yeast. Nature. 2010;465:101–105. doi: 10.1038/nature08946. [DOI] [PubMed] [Google Scholar]
- 29.Archambault V, Chang EJ, Drapkin BJ, Cross FR, Chait BT, Rout MP. Targeted proteomic study of the cyclin-Cdk module. Mol Cell. 2004;14:699–711. doi: 10.1016/j.molcel.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 30.Pryciak PM. Designing new cellular signaling pathways. Chem Biol. 2009;16:249–254. doi: 10.1016/j.chembiol.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim WA. Designing customized cell signalling circuits. Nat Rev Mol Cell Biol. 2010;11:393–403. doi: 10.1038/nrm2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winters MJ, Lamson RE, Nakanishi H, Neiman AM, Pryciak PM. A membrane binding domain in the ste5 scaffold synergizes with gbetagamma binding to control localization and signaling in pheromone response. Mol Cell. 2005;20:21–32. doi: 10.1016/j.molcel.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 33.Takahashi S, Pryciak PM. Identification of novel membrane-binding domains in multiple yeast Cdc42 effectors. Mol Biol Cell. 2007;18:4945–4956. doi: 10.1091/mbc.E07-07-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peter M, Neiman AM, Park HO, van Lohuizen M, Herskowitz I. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 1996;15:7046–7059. [PMC free article] [PubMed] [Google Scholar]
- 35.Leberer E, Wu C, Leeuw T, Fourest-Lieuvin A, Segall JE, Thomas DY. Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J. 1997;16:83–97. doi: 10.1093/emboj/16.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- 37.Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koivomagi M, Valk E, Venta R, Iofik A, Lepiku M, Morgan DO, Loog M. Dynamics of Cdk1 Substrate Specificity during the Cell Cycle. Mol Cell. 2011;42:610–623. doi: 10.1016/j.molcel.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mok J, Kim PM, Lam HY, Piccirillo S, Zhou X, Jeschke GR, Sheridan DL, Parker SA, Desai V, Jwa M, et al. Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Sci Signal. 2010;3:ra12. doi: 10.1126/scisignal.2000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salazar C, Brummer A, Alberghina L, Hofer T. Timing control in regulatory networks by multisite protein modifications. Trends Cell Biol. 2010;20:634–641. doi: 10.1016/j.tcb.2010.08.012. [DOI] [PubMed] [Google Scholar]
- 41.Hirschi A, Cecchini M, Steinhardt RC, Schamber MR, Dick FA, Rubin SM. An overlapping kinase and phosphatase docking site regulates activity of the retinoblastoma protein. Nat Struct Mol Biol. 2010;17:1051–1057. doi: 10.1038/nsmb.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang F, Herskowitz I. Phosphorylation of FAR1 in response to alpha-factor: a possible requirement for cell-cycle arrest. Mol Biol Cell. 1992;3:445–450. doi: 10.1091/mbc.3.4.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Queralt E, Igual JC. Functional distinction between Cln1p and Cln2p cyclins in the control of the Saccharomyces cerevisiae mitotic cycle. Genetics. 2004;168:129–140. doi: 10.1534/genetics.104.029587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller ME, Cross FR. Distinct subcellular localization patterns contribute to functional specificity of the Cln2 and Cln3 cyclins of Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:542–555. doi: 10.1128/mcb.20.2.542-555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edgington NP, Futcher B. Relationship between the function and the location of G1 cyclins in S. cerevisiae. J Cell Sci. 2001;114:4599–4611. doi: 10.1242/jcs.114.24.4599. [DOI] [PubMed] [Google Scholar]
- 46.Draviam VM, Orrechia S, Lowe M, Pardi R, Pines J. The localization of human cyclins B1 and B2 determines CDK1 substrate specificity and neither enzyme requires MEK to disassemble the Golgi apparatus. J Cell Biol. 2001;152:945–958. doi: 10.1083/jcb.152.5.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pascreau G, Churchill ME, Maller JL. Centrosomal localization of cyclins E and A: structural similarities and functional differences. Cell Cycle. 2011;10:199–205. doi: 10.4161/cc.10.2.14444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lew DJ, Reed SI. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee MJ, Dohlman HG. Coactivation of G protein signaling by cell-surface receptors and an intracellular exchange factor. Curr Biol. 2008;18:211–215. doi: 10.1016/j.cub.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takahashi S, Pryciak PM. Membrane localization of scaffold proteins promotes graded signaling in the yeast MAP kinase cascade. Curr Biol. 2008;18:1184–1191. doi: 10.1016/j.cub.2008.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lamson RE, Winters MJ, Pryciak PM. Cdc42 regulation of kinase activity and signaling by the yeast p21-activated kinase Ste20. Mol Cell Biol. 2002;22:2939–2951. doi: 10.1128/MCB.22.9.2939-2951.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.