Abstract
Disentangling cellular heterogeneity is a challenge in many fields, particularly in the stem cell and cancer biology fields. Here, we demonstrate how to combine viral genetic barcoding with high-throughput sequencing to track single cells in a heterogeneous population. We use this technique to track the in vivo differentiation of unitary hematopoietic stem cells (HSCs). The results are consistent with single cell transplantation studies, but require two orders of magnitude fewer mice. In addition to its high throughput, the high sensitivity of the technique allows for a direct examination of the clonality of sparse cell populations such as HSCs. We show how these capabilities offer a clonal perspective of the HSC differentiation process. In particular, our data suggests that HSCs do not equally contribute to blood cells after irradiation-mediated transplantation, and that two distinct HSC differentiation patterns co-exist in the same recipient mouse post irradiation. This technique can be applied to any viral accessible cell type for both in vitro and in vivo processes.
While the mammalian organism consists of more than a hundred cell types, many tissues are sustained by relatively few varieties of multi-potent stem cells1–3. For instance, hematopoietic stem cells (HSCs) are responsible for replenishing many types of functional blood and immune system cells4,5. Given their importance, a comprehensive understanding of stem cells is crucial for advancing the development of regenerative medicine. However, stem cells are usually sparsely dispersed within heterogeneous tissue matrices. Measurements may be diluted by the presence of other cells. Similarly, signals from other cells may be misinterpreted as emanating from stem cells. In addition, recent studies suggest that the stem cell population itself may be heterogeneous6–12 and that their heterogeneity may play important roles in aging, myelodysplastic syndrome, and leukemia6,11,13–15. Cellular heterogeneity has also been proposed to exist in many classes of cancers13–18. Current failures in cancer therapy may arise from the inability to target every self-renewing cell of the cancerous mass15,16,19–21.
Heterogeneous cell populations are frequently separated using monoclonal antibodies conjugated with fluorescent dyes4,5,22–24. Tagged cells can be analyzed and sorted based on their fluorescent color(s). Using this strategy, our lab and others have managed to isolate mouse HSCs using a cocktail composed of more than 12 antibodies4,5,22–24. The discovery of new antibodies further increases the purity of the isolation. However, the discovery process is not deterministic and antibodies that target the intended or putative cell population may not exist. Thus, it is difficult to determine whether or not the isolated cell population remains heterogeneous. Ultimately, this problem can only be resolved by analyzing the cell population with single cell precision.
Conventional methods for studying HSCs at the single cell level rely on single cell transplantation8,10. It is very costly, time consuming and technically challenging to collect sufficient data so as to be representative of the entire cell population. The inefficiencies prevent us from carrying out single cell studies for many crucial biological and clinical questions1–3,15–17,21. To improve experimental productivity, a few groups have developed a strategy to trace cells using distinct viral insertion sites9,25–30. The genomic locations of the insertion sites are assayed using Southern blots. This technique relies on restriction enzymes to cleave the genomic DNA into different lengths and inherently suffers from low resolution, poor quantification and sensitivity. Moreover, the millions of cells required for Southern blotting are unobtainable for sparse cell populations such as HSCs. To increase the sensitivity and resolution, several adjunct approaches have been applied using PCR based strategies31–33, Sanger sequencing detection34 and microarray detection35,36. Despite improvements, these methods still suffer from limited resolution, poor quantification and are unable to directly address the clonality of pure stem cells37.
In this study, we combine three previously separate technologies viral cellular labeling, high throughput sequencing, and DNA barcoding to overcome the aforementioned limitations. Viral cellular labeling has been applied to trace the in vivo development of single cells9,25–30,33. High throughput sequencing has been used for many quantitative genetic and epigenetic studies32,38–40. DNA barcoding has been used to mark reactions, genes and cells34,41,42. Here, we show how a novel combination of these three technologies can offer high throughput, single cell sensitivity, and precise quantitative results. We demonstrate the applicability of this technique by tracking the in vivo differentiation of mouse HSCs, and show how it offers a clonal perspective of the HSC differentiation process.
RESULTS
Experimental workflow
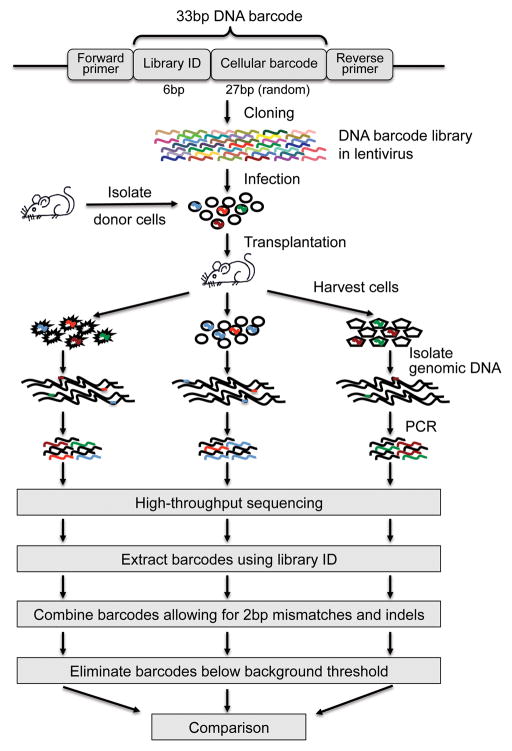
The experimental system utilizes synthesized barcodes drawn from a large semi-random 33mer DNA barcode library to uniquely label and track individual cells (Fig. 1). The barcode library is cloned into a lentiviral construct and is delivered to cells at a titer low enough such that most cells receive a single barcode. The DNA barcodes are integrated into the cellular genomes by the lentivirus43–45, which allows them to be replicated alongside the host cells’ genomes during cell division. Progeny descending from a labeled cell can thus be readily identified by the same DNA barcode. In our in vivo demonstration experiments, barcoded HSCs were transplanted into mice. 22 weeks post transplantation, the recipient mice were sacrificed and several hematopoietic populations including HSCs were isolated from the blood and bone marrow using conventional cell surface markers4,5,24,46,47. The genomic DNA of each cell population was extracted. DNA barcodes were recovered using PCR and analyzed using an Illumina GA II high-throughput sequencer.
Figure 1.
Experimental workflow. A DNA barcode consists of a common 6bp library ID at the 5′ end followed by a random 27bp cellular barcode. In the figure, different colors represent different barcode sequences. A lentiviral vector delivers a large library of barcodes into a small number of cells such that each cell receives a unique barcode. Barcodes replicate with the cells in the recipient mice after transplantation. Afterwards, the progeny of the donor cells are harvested. Barcodes are recovered from the genomic DNA using PCR and analyzed using high throughput sequencing (Illumina GA II). The 6bp library ID helps to identify barcodes in the sequencing result. Identical 33bp barcodes are combined allowing for mismatches and indels up to 2bp in total. The barcodes are then compared across different cell populations that originate from the same starting cell population.
The high sensitivity and throughput of next generation sequencing allow for the accurate identification and quantification of every barcode recovered from a cell population. Barcode sequences can be separated from background noise sequences by their library ID, a 6bp signature sequence that identifies the barcode library of origin. Sequences belonging to the same barcode are combined to provide a copy number for each barcode. Because of inherent sequencing error, we allow for mismatches and indels up to 2bp in total at this step. No mismatch or indel is allowed for the 6bp library ID. We exclude barcodes whose copy numbers are below a background noise threshold that is automatically defined using a computer algorithm. Finally, barcode compositions from different cell populations are compared to address specific questions. In the HSC demonstration experiment, the presence of DNA barcodes in different hematopoietic populations provides information on HSC proliferation and differentiation.
The key steps of this clonal tracking technology are ensuring single cell representations of the barcodes and handling sequencing errors. In the following sections, we will discuss how we overcome these two technical barriers, starting with the construction of the lentivirus barcode library.
Lentivirus barcode library construction and delivery
The 33bp DNA barcode consists of a common 6bp sequence at the 5′ end followed by a random 27bp sequence. The latter uniquely labels each cell while the former acts as a library ID that identifies the barcode library of origin. The library ID also allows several experiments using different barcode libraries to be combined in a single sequencing run. The DNA barcodes are cloned into the non-expressing region of the lentivirus. Linkers complementary to the primers required for the high-throughput sequencing are appended to both ends of the barcode sequence in the lentiviral construct. Thus, a simple PCR step is sufficient to prepare the barcodes from the genomic DNA for loading onto the high throughput sequencer. This single-step design eliminates handling errors. Rare barcodes could thus be easily amplified and detected in the system. The lentivirus also carries a reporter GFP to allow for the easy monitoring of infection efficiency. So far, eighteen barcode libraries with eighteen different library IDs have been constructed (Supplementary Table 1).
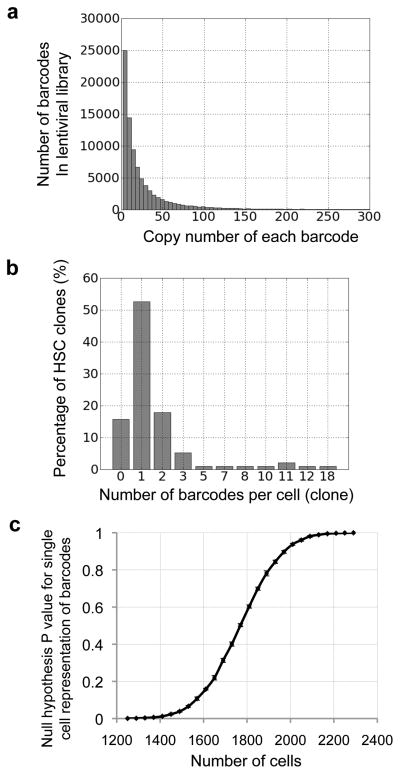
The barcode library must be efficiently cloned into the lentiviral construct. Each batch of barcode virus was evaluated against a control BB88 cell line (ATCC number: TIB-55) before use. 100,000 BB88 cells were infected at MOI=1 and cultured for one week before harvesting. More than 80,000 different barcodes were recovered with the vast majority possessing low copy numbers (Fig. 2a and Supplementary Fig. 1). This indicates that most barcodes are equally represented within the lentiviral library, and that the sequencing result does not cover the entire virus library. Thus, this experiment establishes a lower bound on the true diversity of the library.
Figure 2.
DNA barcode library and delivery. (a) Histogram displaying barcode copy numbers from a lentiviral library. Additional lentiviral libraries are shown in Supplementary Fig. 1, together with the negative controls to demonstrate the level of background noise for this experiment. (b) Histogram showing the number of barcode(s) that each HSC clone receives after infection. 95 HSC clones were examined in total. This distribution fits a normal distribution shown in Supplementary Fig. 3. (c) Monte Carlo simulation of the null hypothesis that more than 95% of the barcodes represent single cells. The P value is plotted against the size of the cell population whose barcodes are recovered in the result.
To assess how many barcodes were delivered to each cell, mouse HSCs (lineage (CD3, CD4, CD8, B220, Gr1, Mac1, Ter119)-/ckit+/Sca1+/CD34−/CD150+/Flk2−) were infected with barcode-bearing virus (Supplementary Fig. 2)22–24. Double FACS sorted HSCs were co-cultured with the virus for 10 hours under conditions that maintained their stem cell characteristics. The infection titer was chosen such that half of the HSCs would express GFP post infection. After three rounds of washing post infection, single HSCs were sorted into individual culture wells on a 96-well plate. The HSC clones were then cultured in differentiation-inducing media in order to increase the cell population. Ten days later, genomic DNA was extracted from each colony. Limiting dilution QPCR (digital PCR) were performed to compare the barcode copy number with the genomic DNA copy number for each HSC clone. The results suggest that most HSCs received a single copy of the barcode (Fig. 2b). The number of barcodes per cell appears to be a normal distribution centered around one barcode per cell (Supplementary Fig. 3). Multiple labeling of the same cell occurs randomly at a low frequency. A lower virus titer could reduce the number of cells labeled by multiple barcodes, but it will also reduce the total number of cells labeled.
Conditions for single cell representation
The ideal barcode lentivirus library should be large enough to ensure that the chance for any particular barcode sequence to be delivered to multiple cells is very low. In real experiments, the library has a finite size and viruses carrying the same barcode may infect multiple target cells if the ratio of barcode library size to target cell population size is small. We calculated the probability of single cell representation for a given cell population size using a Monte Carlo simulation (Fig. 2c).
The calculations are based on experimentally measured distributions of barcode frequencies (Fig. 2a) and of the number of barcodes per cell (Fig. 2b). This takes into account both the real size of the barcode library and any inherent viral copy and infection variability (Fig. 2a). In the mean time, the simulation also takes into consideration that some cells may receive zero or multiple barcodes (Fig. 2b). Given these conditions, we calculated the null hypothesis P values for different target cell population sizes such that more than 95% of the barcodes represent single cells (Fig. 2c). For the eighteen virus libraries that we generated, the maximum number of cells that can be used to ensure that with greater than 95% probability more than 95% of the barcodes represent single cells is on average 500–1500 cells (Supplementary Table 2). As mentioned previously, the experimentally observed barcode library size is a lower bound of the actual size. Therefore, the calculated cell population size is also a lower bound on the actual number of cells that can be used to ensure single cell representation.
Eliminating sequencing background
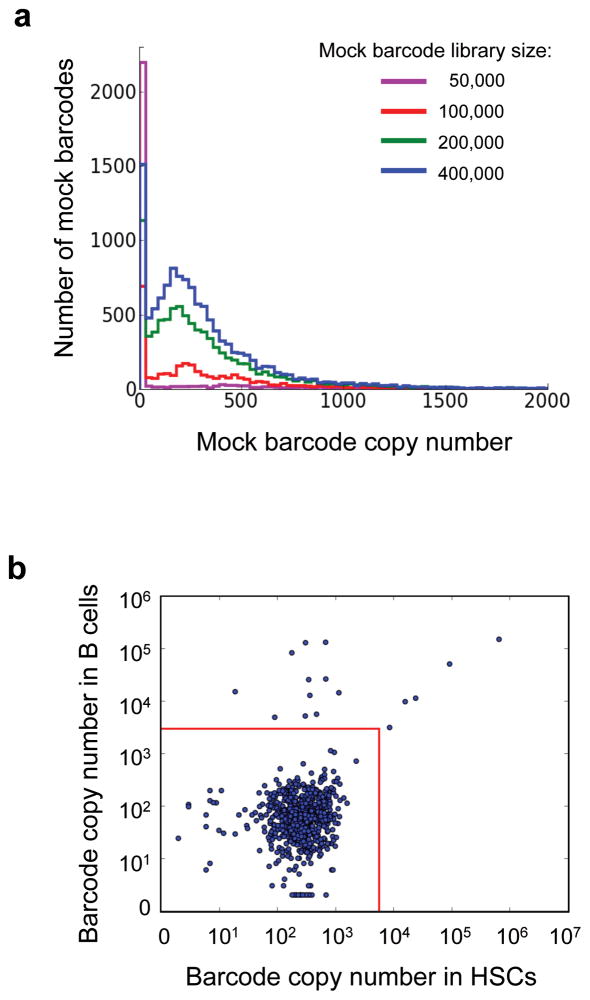
Approximately 20% of the sequences recovered by the sequencer in our study lacked a valid 6bp library ID. They arose from sequencing errors and from other background noise of the experimental system (Supplementary Fig. 4). These sequences can be identified because their first 6bp do not correspond to any legitimate library ID (Fig. 3a). The background noise sequences are also observed among sequences with an intact 6bp library ID (Fig. 3b). Their abundance is much greater than expected based on the viral infection rate (Fig. 2b). The background noise sequences usually number fewer than 1,000 copies (Fig. 3a); In contrast, real barcodes typically number more than 10,000 copies (Fig. 3b). Background noise sequences appear randomly in different samples without exhibiting any distinct patterns, contrary to real barcodes with high copy numbers (Fig. 3b). Using the background noise sequences missing the expected library IDs (Fig. 3a), we have developed an algorithm to determine the background threshold for each sequencing result.
Figure 3.
Background noise sequences. (a) Background noise sequences without the expected 6bp library IDs. Sequences with identical 6bp at the 5′ end are clustered. Mock barcode libraries are constructed from clustered sequences whose initial 6bp are not among the expected library IDs. Mock barcodes from 20 mock barcode libraries are plotted as one line to demonstrate their copy number distribution. Different lines display the mock barcode libraries with different sizes. (b) Log scale plot of barcode copy numbers in HSCs and in B cells from one irradiated mouse transplanted with 1000 donor HSCs. Each dot represents a distinct barcode. Barcodes with copy numbers below 1000 appear randomly in the two cell populations whereas barcodes with copy numbers higher than 10,000 form a distinct pattern. Red lines illustrate background thresholds as calculated by our algorithm.
The algorithm first bins the background noise sequences using random 6bp mock library IDs, and calculates the distribution of their copy numbers per barcode (Fig. 3a). These copy number distributions are used to estimate the false positive rate for barcodes with expected 6bp library IDs within the same sequencing sample. This assumes that the copy number distribution for background noise sequences without the 6bp library ID represents the distribution of the background noise sequences with the 6bp library ID in the same sequencing sample. We define the background cutoff such that the threshold copy number has less than 1% possibility of being part of the background (Fig. 3b).
The copy number threshold may eliminate some real barcodes with low copy numbers. This problem can be reduced when multiple cell populations derived from the same infected cell population are analyzed. All of the barcodes that are above the background copy number threshold in the different cell populations can be combined to form a list of ‘original barcodes’ that represent the barcodes from the original common infected cell population. The comprehensive ‘original barcode’ list can be used to identify low copy number barcodes from some of the harvested cell populations. The barcode tracking system may miss barcodes/cells that are consistently under-represented in all cell populations. However, the use of the comprehensive ‘original barcode’ list can help to preserve barcodes that are significantly present in any of the cell populations relevant to the experimental question.
Tracking mouse HSCs in vivo
To demonstrate the applicability of the barcode tracking system, we used it to track the in vivo differentiation of mouse HSCs in lethally irradiated recipient mice. 9000 HSCs were transplanted into each mouse immediately after 10 hours of lentiviral infection for barcode labeling (Fig. 2b). DNA barcodes from ten different types of the hematopoietic cell populations were analyzed 22 weeks post transplantation4,5,24,46,47 (Supplementary Fig. 5–6 and Supplementary Table 3). Typically 30–50 barcodes were recovered from each mouse. This suggests that around 50–80 HSCs out of the initial pool had been successfully engrafted and had actively proliferated and/or differentiated (Fig. 2b). The engrafted cell numbers are well within the range that ensures single cell representation of the barcodes (Fig. 2c and Supplementary Table 2). Barcodes from cells that fail to engraft are not recovered, and therefore do not affect the single cell representation of the recovered barcodes.
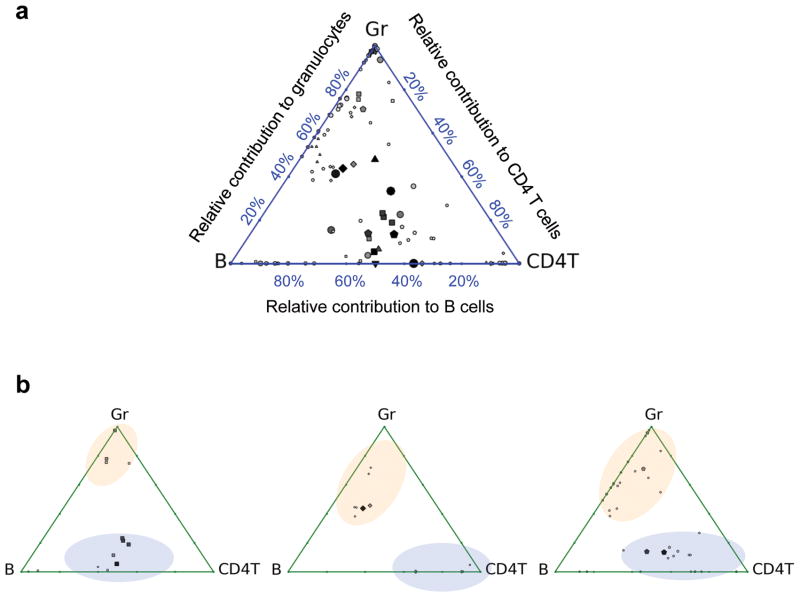
We first compare the results from our barcode tracking system with the results from single cell transplantation studies (Fig. 4)8. In our result, barcodes representing HSC clones demonstrate lineage biases as reported by previous single cell transplantation experiments (Fig. 4a)8. This validates the single cell precision of our barcode tracking system. While the single cell transplantation studies used 352 mice8, we were able to track many more HSC clones using only 7 mice. The barcode tracking system also provides information previously unattainable with conventional single cell transplantation. For instance, our data reveals two clearly separated HSC populations with distinct lineage biases in each irradiated recipient mouse (Fig. 4b and Supplementary Fig. 7). This suggests that there exist two subpopulations of HSCs6–12 or differentiation regulatory mechanisms in the same organism after irradiation-mediated transplantation. Some HSCs’ differentiation is biased towards B cells and T cells whereas others’ is biased towards B cells and granulocytes. We were able to identify these two HSC differentiation regulatory modes in the same mouse without having to discover the markers for the cells that are regulated under each mode. This demonstrates the power of this technique for identifying new regulatory mechanisms in a heterogeneous cell population.
Figure 4.
Lineage bias of HSC differentiation after irradiation. Triangle plots8 show the relative proportion of barcodes in granulocytes (Gr), B cells (B) and CD4+ T cells (CD4T) 22 weeks after lethal irradiation mediated transplantation. Each dot within the triangle represents a distinct barcode. Bigger and darker dots represent more abundant barcodes. The distance of a dot to the three vertices of the triangle is inversely correlated with the relative abundance of the barcode within the particular cell populations. For example, if a barcode is only found in one cell population, the dot is plotted at the corresponding vertex; if a barcode appears equally in all three populations, the dot is plotted in the middle of the triangle. (a) Barcodes from seven mice are plotted in one triangle. The barcodes from each mouse are represented by a particular shape: circle, square, triangle pointing up, triangle pointing down, diamond, pentagon, and octagon. (b) Each triangle plot depicts a single mouse. Distinct barcode groups are highlighted with blue and orange ellipses. Plots for all the seven mice are shown in Supplementary Fig. 7.
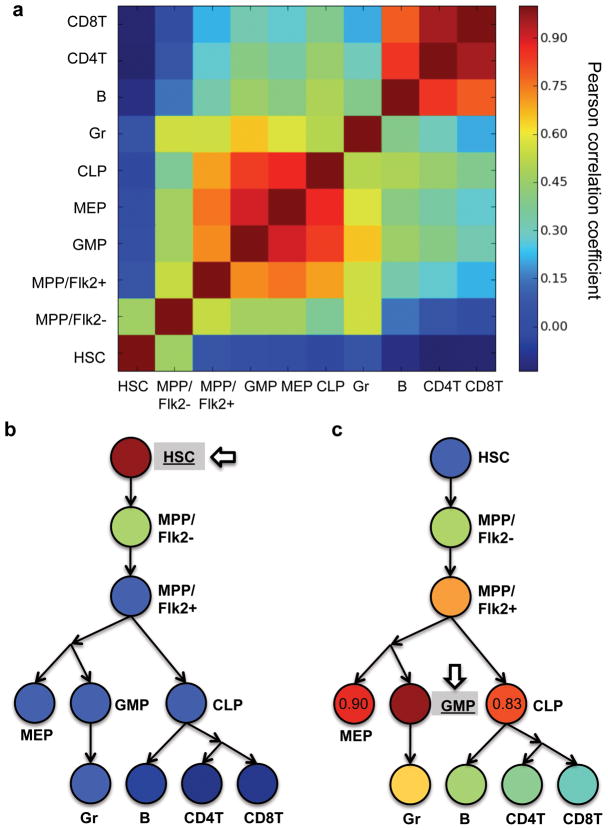
The barcode tracking system is sensitive enough to directly measure the clonality of HSCs, and affords a clonal perspective of the HSC differentiation process (Fig. 5). We calculated the Pearson correlation coefficients of the DNA barcode compositions to measure the clonal correlations between the major hematopoietic stages4,5,24,46,47 (Fig. 5 and Supplementary Table 4). This parameter quantifies the similarity of the barcode distribution in different cell populations. There are two groups of closely correlated hematopoietic populations in the Pearson correlation comparison matrix (Fig. 5a). One group consists of progenitor cell populations including granulocytic/monocytic progenitors (GMP), megakaryotic/erythroid progenitors (MEP), and common lymphocyte progenitors (CLP); the other group consists of mature lymphoid blood cells including B cells, CD4 T cells and CD8 T cells. When we compared barcodes from HSCs with barcodes from other cell populations (Fig. 5b), we found that HSC barcodes are poorly correlated with barcodes from mature blood cells at the bottom of the hematopoietic hierarchy. This suggests that HSCs do not equally contribute to blood cells in irradiated recipient mice. Nonetheless, HSC barcodes correlate better with those from its immediate downstream multipotent progenitor (MPP/Flk2−).
Figure 5.
Clonal correlations of hematopoietic populations. Pearson correlation coefficients of barcode representations (copy numbers) are calculated to quantify the clonal correlations. The colors are assigned based on the mean correlations from seven mice. Raw data for individual mouse are shown in Supplementary Table 4. (a) Clonal correlations of extracted hematopoietic populations. (b) Clonal correlations of the hematopoietic populations compared with HSCs. (c) Clonal correlations of the hematopoietic populations compared with granulocyte/monocytic progenitor (GMP). Pearson correlations coefficient values are labeled for MEP and CLP to highlight the difference. (b–c) The circles and arrows were arranged based on the general model of hematopoiesis to depict the developmental relationships of the hematopoietic populations4,5,24,46,47. Abbreviations: hematopoietic stem cell (HSC), Flk2− multipotent progenitor (MPP/Flk2−), Flk2+ multipotent progenitor (MPP/Flk2+), granulocyte/monocytic progenitor (GMP), megakaryotic/erythroid progenitor (MEP), common lymphocyte progenitor (CLP), granulocyte (Gr), B cell (B), CD4 T cells (CD4T) and CD8 T cells (CD8T).
Interesting clonal correlations were observed between granulocytic/monocytic progenitor (GMP) and other hematopoietic populations (Fig. 5c). When compared to its upstream progenitors, GMP correlates the best with its immediate upstream progenitor, Flk2+ multipotent progenitor (MPP/Flk2+) (Fig. 5c)24. When compared to intermediate progenitor cells from similar differentiation stages in the hematopoietic hierarchy, GMP is correlated more closely with the megakaryotic/erythroid progenitor (MEP) than with the common lymphocyte progenitor (CLP) (Fig. 5c)46,47. This is consistent with previous studies that suggest GMP and MEP both belong to myeloid lineages and share a common myeloid progenitor (CMP)47. When compared to blood cells, GMP correlates most closely with its progeny granulocyte (Gr) (Fig. 5c). In general, the clonal correlations of the hematopoietic populations reflect the progression and divergence of the hematopoietic hierarchy as characterized by previous studies4,5,24,46,47. This clonal correlation parameter can be used as a novel indicator to determine the developmental relationships of different cell populations. For instance, it can be applied to studies of cancer metastasis to identify the primary and secondary metastatic sites. As we have shown, the relationship between various cell populations can be directly inferred from comparisons between their barcode distributions.
DISCUSSION
In this barcode tracking system, the mapping between DNA barcodes and target cells are randomly established and the exact linkages cannot be a priori determined. Conclusions can only be drawn after the experimental condition has been applied. For example, we recovered barcodes after the infected HSCs had undergone proliferation and differentiation in vivo, and we drew conclusions based on comparisons between different hematopoietic populations originating from a common starting HSC population engrafted in each mouse (Fig. 4–5).
Comparisons are most informative when identical barcodes are compared between cell populations harvested from a common mouse. It may not be appropriate to directly compare different barcodes from the same cell population as the PCR and sequencing steps can introduce uncontrolled biases for different barcode sequences48. To confirm quantitative behaviors of particular cell clones, it is necessary to perform GC content correction48 and QPCR verification.
The barcode labeling process requires several hours of culture and lentiviral infection prior to transplantation. It has been shown that this process does not alter HSC function9,44,45. In addition, our result using the barcode method is consistent with conventional studies absent of any culturing or viral infection8 (Fig. 4a). Nonetheless it is possible that the cell culture and lentiviral infection may alter the infected HSCs in manners undetected, as the lentiviral vector inserts the DNA barcode at dispersed sites in the host cell’s genome. It is always recommended to carry out several repeat experiments to exclude rare events arising from the viral insertion (Supplementary Fig. 7), which can be easily accomplished using multiple barcode virus libraries with different library IDs. Future versions of this system may avoid this problem by integrating the tracking barcode in a specific genomic location. This will also strictly enforce a one-to-one correspondence between genetic barcodes and individual cells.
In summary, we have demonstrated a novel experimental system that offers a simple way to study unitary cells mixed within a heterogeneous population. Using the HSC transplantation system as an example, we have shown how this system can be used to simultaneously track the proliferation and development of hundreds of cells in vivo with single cell precision. High throughput is critical for studying rare or stochastic cellular events. It also reveals novel features that are not apparent at low cell numbers. For instance, our data reveals the existence of two clearly separated hematopoietic stem cell populations that possess distinct lineage biases in each irradiated mouse (Fig. 4b). This key feature was missed in earlier studies that were limited by the number of cell clones that could be tracked in a single mouse.
In addition, the high sensitivity of the barcode tracking system allows for the first time a direct examination of the entire hematopoietic process starting with the hematopoietic stem cells themselves. We are now able to ask and answer many new questions that were previously impractical to address. For instance, comparisons between the clonal composition of HSCs with their down stream hematopoietic progenitors suggest that HSCs are not equally involved in differentiation after irradiation-mediated transplantation (Fig. 5b). This key feature of HSC differentiation was again missed in earlier studies that lacked the sensitivity to directly examine the clonality of HSCs.
The methods used to deliver and eventually recover the barcodes in our system can be easily extended to the clonal tracking of both in vitro and in vivo processes for virtually any cell types that can be infected by a lentivirus. It provides a convenient way to study cell populations with potential heterogeneity. For instance, this barcode tracking system can be applied to cell and gene therapy to track and quantify the fate and distribution of transplanted cells49. The high sensitivity of this technique allows for the analysis of clinical samples with very low cell numbers and for the identification of early stage malignance before the subsequent expansion or metastasis. The integrated barcodes may provide the location and target of malignant cells. Early oncogenic events can also be easily identified in both in vitro and in vivo therapeutic safety assessments using this barcode tracking system. In addition, the high throughput of the barcode tracking system allows for the extension of gene target and drug screens to the clonal level. Different drug candidates can be applied to cells barcoded with different library IDs. All of the cells can then be combined and sequenced together. Clonal level information can be easily recovered using this barcode tracking system for many different applications.
METHODS
Construction of DNA barcode library
Semi random 33-base-pair DNA sequences were generated by the protein and nucleic acid facility (PAN) at the Stanford University School of Medicine and were cloned into the non-expressing region of the lentivirus pCDH from System Biosciences (catalog number: CD523A-1). Oligos are synthesized on an ABI 3900 DNA/RNA instrument using beta-cyanoethyl phosphoramidite chemistry. Mixed nucleotide monomers were added at each position of the 27bp cellular barcode during the oligo synthesis (Fig. 1).
Mice
The donor mice used in the experiments were 8–12 weeks old C57BL6/Ka (CD45.1+). The recipient mice were 8–12 weeks old C57B L6/Ka (CD45.2+). Mice were bred and maintained at Stanford University’s Research Animal Facility. Animal procedures were approved by the International Animal Care and Use Committee.
HSC transplantation
Bone marrow cells were obtained from the crushed bones of donor mice using PBS with 2% fetal bovine serum. Bone debris was removed by density gradient centrifugation using histopaque 1119 (Sigma, St. Louis, MO; product number: 11191). The cells were then c-kit enriched with CD117 microbeads (AutoMACS, Miltenyi Biotec, Auburn, CA; order number: 130-091-224) before staining with monoclonal antibodies (Supplementary Table 3). HSCs were isolated using double FACS sortings with the FACS-Aria II (BD Biosciences, San Jose, CA). Cells were transplanted via retro-orbital injection after recipient mice were lethally irradiated at 950 cGy. 250,000 whole bone marrow cells were injected as helper cells together with the donor HSCs.
HSC culture and lentivirus infection
HSCs (ckit+/lineage(CD3, CD4, CD8, B220, Gr1, Mac1, Ter119)-/Sca1+/CD34−/CD150+/Flk2−) were cultured in the presence of 20 ng/ml SCF (R&D Systems) and 20 ng/ml TPO (R&D Systems) for 10 hours during lentivirus infection50. 8ng/ul polybrene were added into the culture to facilitate virus infection. HSCs were washed three times before transplantation. The virus infection titer was set such that half of the donor HSCs will express GFP after transplantation. This helps to ensure that most HSCs receive a single copy of the barcode (Fig. 2b).
Cell harvest
Recipient mice were euthanized 22 weeks after transplantation. Whole blood was obtained by collecting the perfusate from the heart. The blood was separated using 2% dextran at 37°C for 20 min, and subsequently lysed using ACK lysis buffer (150mM NH4Cl, 1mM KHCO3, and 0.1mM EDTA) for 5 minutes on ice. Cells were stained with antibodies (Supplementary Table 3) and sorted on the BD FACS-Aria II.
Progenitor cells from the bone marrow were harvested using the same method as the preparation for donor HSCs described above. However, no histopaque was applied in order to maximize the retrieval of progenitor cells from the same mouse. To reduce the number of cells during sorting, the cells were enriched with CD117/ckit microbeads (AutoMACS, Miltenyi Biotec, Auburn, CA; order number: 130-091-224) and IL7Rα antibody (ebioscience, San Diego, CA; catalog number: 13-0161-82) followed by anti-Rat IgG microbeads (AutoMACS, Miltenyi Biotec, Auburn, CA; order number: 130-048-502). Donor cells were sorted based on the CD45 marker.
Cell surface markers for the harvested hematopoietic populations are summarized as following24,46,47:
Granulocytes: CD4−/CD8−/B220−/CD19−/Mac1+/Gr1+/side scatterhigh;
B cells: CD4−/CD8−/Gr1−/Mac1−/B220+/CD19+;
CD4T cells: B220−/CD19−/Mac1−/Gr1−/TCRab+/CD4+/CD8−;
CD8 T cells: B220−/CD19−/Mac1−/Gr1−/TCRab+/CD4−/CD8+;
HSC (hematopoietic stem cells): lineage (CD3, CD4, CD8, B220, Gr1, Mac1, Ter119)-/IL7Ra−/ckit+/Sca1+/Flk2−/CD34−/CD150+;
MPP/Flk2− (multipotent progenitor Flk2−): lineage (CD3, CD4, CD8, B220, Gr1, Mac1, Ter119)-/IL7Ra−/ckit+/Sca1+/Flk2−/CD34+;
MPP/Flk2+ (multipotent progenitor FLk2+): lineage (CD3, CD4, CD8, B220, Gr1, Mac1, Ter119)-/IL7Ra−/ckit+/Sca1+/Flk2+;
CLP (common lymphocyte progenitor): lineage (CD3, CD4, CD8, B220, Gr1, Mac1, Ter119)-/IL7Ra+/Flk2+;
MEP (megakaryotic/erythroid progenitor): lineage (CD3, CD4, CD8, B220, Gr1, Mac1, Ter119)-/IL7Ra−/ckit+/Sca1−/CD34−/FcgR−;
GMP (granulocyte/monocytic progenitor): lineage (CD3, CD4, CD8, B220, Gr1, Mac1, Ter119)-/IL7Ra−/ckit+/Sca1−/CD34+/FcgR+.
DNA barcode extraction and sequencing
Genomic DNA was extracted from the cells using the DNeasy Blood & Tissue kit (Qiagen, catalog number: 69504). PCR was used to amplify the DNA barcode and in the same step add linkers necessary for the high throughput sequencing (Finnzymes, catalog number: F-530L). The PCR product at the correct size was cut from 3% agar gel (Lonza, catalog number: 50080). Sequencing was performed using the Illumina GA II sequencer by the core sequencing facility at the Institute for Stem Cell Biology and Regenerative Medicine at Stanford University School of Medicine.
DNA barcodes analysis
Sequencing data was processed using custom python code (available upon request). We first combined raw sequencing data with library IDs allowing for mismatches and indels up to 2bp in total, a standard way to handle Illumina GAII sequences. Barcodes with copy numbers lower than the background noise threshold were eliminated, before we combined the barcodes of different cell populations from the same mouse to create a comprehensive list of ‘original barcodes’ for each mouse. Finally, we went through the raw sequencing data again to pull out the sequences that represent these “original barcodes” allowing for mismatches and indels up to 2bp in total.
Quantifying virus barcode copy number per HSC clone (Fig. 2b)
Genomic DNA was extracted from each HSC clones using the DNeasy Blood & Tissue kit (Qiagen, catalog number: 69504). QPCR were performed in triplicate using the SYBR green PCR master mix (ABI, catalog number: 4367659) on the ABI PRISM 7900HT Sequence Detection System. Primers for genomic DNA were designed at the Sox2 promoter (Forward: taggaaaaggctgggaacaa; Reverse: cactcaccccctcttctcac). Primers for DNA barcode recovery PCR came from designs by Illumina Inc. A standard curve using a control sample was used to measure all of the HSC clones. The control sample was quantified using limiting dilution QPCR (digital PCR) to obtain the ratio of DNA barcode to genomic DNA.
Monte Carlo simulations to determine limits of single cell representation (Fig. 2c)
Monte Carlo simulations were performed using custom python code. Raw data from the barcode libraries (Fig. 2a) and from the HSC infection rates (Fig. 2b) were directly used for this simulation. For each target cell population size (1240 to 2300 cells with step size 40 cells), 1000 experiments were conducted for each round during which a DNA barcode was randomly assigned for each cell. 10 rounds of simulations were performed in total to generate the standard deviations.
Supplementary Material
Acknowledgments
We thank G. Mantalas, T. Snyder and B. Passarelli for helping with the high-throughput sequencing; K. Schepers, I. Dimov, M. Drukker, and D. Sahoo for helpful discussions; P. Lovelace for FACS core management. We also thank L. Jerabek and T. Storm for laboratory management; C. Muscat and T. Naik for antibody conjugation; A. Mosley for animal supervision. This work is supported by NIH-R01-CA86065 and NIH-U01-HL099999. R.L. is supported by CIRM-TG2-01159.
Footnotes
AUTHOR CONTRIBUTIONS R.L. and I.L.W. designed the experiments. R.L. performed the experiments. N.F.N. and S.R.Q. set up and carried out the high-throughput sequencing. R.L. analyzed the data and wrote the manuscript. All authors edited the manuscript.
References
- 1.Weissman IL. Stem cells: units of development, units of regeneration, and units in evolution. Cell. 2000;100:157–168. doi: 10.1016/s0092-8674(00)81692-x. S0092-8674(00)81692-X [pii] [DOI] [PubMed] [Google Scholar]
- 2.Blanpain C, Horsley V, Fuchs E. Epithelial stem cells: turning over new leaves. Cell. 2007;128:445–458. doi: 10.1016/j.cell.2007.01.014. S0092-8674(07)00070-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Snippert HJ, Clevers H. Tracking adult stem cells. EMBO Rep. 2011;12:113–122. doi: 10.1038/embor.2010.216. embor2010216 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seita J, Weissman IL. Hematopoietic stem cell: self-renewal versus differentiation. Wiley Interdiscip Rev Syst Biol Med. 2010;2:640–653. doi: 10.1002/wsbm.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryder D, Rossi DJ, Weissman IL. Hematopoietic stem cells: the paradigmatic tissue-specific stem cell. Am J Pathol. 2006;169:338–346. doi: 10.2353/ajpath.2006.060312. S0002-9440(10)62717-4 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beerman I, et al. Functionally distinct hematopoietic stem cells modulate hematopoietic lineage potential during aging by a mechanism of clonal expansion. Proc Natl Acad Sci U S A. 2010;107:5465–5470. doi: 10.1073/pnas.1000834107. 1000834107 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morita Y, Ema H, Nakauchi H. Heterogeneity and hierarchy within the most primitive hematopoietic stem cell compartment. J Exp Med. 2010;207:1173–1182. doi: 10.1084/jem.20091318. jem.20091318 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dykstra B, et al. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell. 2007;1:218–229. doi: 10.1016/j.stem.2007.05.015. S1934-5909(07)00021-5 [pii] [DOI] [PubMed] [Google Scholar]
- 9.McKenzie JL, Gan OI, Doedens M, Wang JC, Dick JE. Individual stem cells with highly variable proliferation and self-renewal properties comprise the human hematopoietic stem cell compartment. Nat Immunol. 2006;7:1225–1233. doi: 10.1038/ni1393. ni1393 [pii] [DOI] [PubMed] [Google Scholar]
- 10.Sieburg HB, et al. The hematopoietic stem compartment consists of a limited number of discrete stem cell subsets. Blood. 2006;107:2311–2316. doi: 10.1182/blood-2005-07-2970. 2005-07-2970 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho RH, Sieburg HB, Muller-Sieburg CE. A new mechanism for the aging of hematopoietic stem cells: aging changes the clonal composition of the stem cell compartment but not individual stem cells. Blood. 2008;111:5553–5561. doi: 10.1182/blood-2007-11-123547. blood-2007-11-123547 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weksberg DC, Chambers SM, Boles NC, Goodell MA. CD150- side population cells represent a functionally distinct population of long-term hematopoietic stem cells. Blood. 2008;111:2444–2451. doi: 10.1182/blood-2007-09-115006. blood-2007-09-115006 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rossi DJ, Jamieson CH, Weissman IL. Stems cells and the pathways to aging and cancer. Cell. 2008;132:681–696. doi: 10.1016/j.cell.2008.01.036. S0092-8674(08)00137-2 [pii] [DOI] [PubMed] [Google Scholar]
- 14.Lapidot T, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 15.Jordan CT, Guzman ML, Noble M. Cancer stem cells. N Engl J Med. 2006;355:1253–1261. doi: 10.1056/NEJMra061808. 355/12/1253 [pii] [DOI] [PubMed] [Google Scholar]
- 16.Dick JE. Looking ahead in cancer stem cell research. Nat Biotechnol. 2009;27:44–46. doi: 10.1038/nbt0109-44. nbt0109-44 [pii] [DOI] [PubMed] [Google Scholar]
- 17.Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science. 2009;324:1670–1673. doi: 10.1126/science.1171837. 324/5935/1670 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. 0530291100 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bao S, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. nature05236 [pii] [DOI] [PubMed] [Google Scholar]
- 20.Diehn M, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–783. doi: 10.1038/nature07733. nature07733 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dick JE. Stem cell concepts renew cancer research. Blood. 2008;112:4793–4807. doi: 10.1182/blood-2008-08-077941. 112/13/4793 [pii] [DOI] [PubMed] [Google Scholar]
- 22.Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 23.Kiel MJ, Yilmaz OH, Iwashita T, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. S0092-8674(05)00540-4 [pii] [DOI] [PubMed] [Google Scholar]
- 24.Christensen JL, Weissman IL. Flk-2 is a marker in hematopoietic stem cell differentiation: a simple method to isolate long-term stem cells. Proc Natl Acad Sci U S A. 2001;98:14541–14546. doi: 10.1073/pnas.261562798. 261562798 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dick JE, Magli MC, Huszar D, Phillips RA, Bernstein A. Introduction of a selectable gene into primitive stem cells capable of long-term reconstitution of the hemopoietic system of W/Wv mice. Cell. 1985;42:71–79. doi: 10.1016/s0092-8674(85)80102-1. S0092-8674(85)80102-1 [pii] [DOI] [PubMed] [Google Scholar]
- 26.Keller G, Paige C, Gilboa E, Wagner EF. Expression of a foreign gene in myeloid and lymphoid cells derived from multipotent haematopoietic precursors. Nature. 1985;318:149–154. doi: 10.1038/318149a0. [DOI] [PubMed] [Google Scholar]
- 27.Lemischka IR, Raulet DH, Mulligan RC. Developmental potential and dynamic behavior of hematopoietic stem cells. Cell. 1986;45:917–927. doi: 10.1016/0092-8674(86)90566-0. 0092-8674(86)90566-0 [pii] [DOI] [PubMed] [Google Scholar]
- 28.Jordan CT, Lemischka IR. Clonal and systemic analysis of long-term hematopoiesis in the mouse. Genes Dev. 1990;4:220–232. doi: 10.1101/gad.4.2.220. [DOI] [PubMed] [Google Scholar]
- 29.Mazurier F, Gan OI, McKenzie JL, Doedens M, Dick JE. Lentivector-mediated clonal tracking reveals intrinsic heterogeneity in the human hematopoietic stem cell compartment and culture-induced stem cell impairment. Blood. 2004;103:545–552. doi: 10.1182/blood-2003-05-1558. 2003-05-1558 [pii] [DOI] [PubMed] [Google Scholar]
- 30.Drize NJ, Keller JR, Chertkov JL. Local clonal analysis of the hematopoietic system shows that multiple small short-living clones maintain life-long hematopoiesis in reconstituted mice. Blood. 1996;88:2927–2938. [PubMed] [Google Scholar]
- 31.Schmidt M, et al. High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR) Nat Methods. 2007;4:1051–1057. doi: 10.1038/nmeth1103. nmeth1103 [pii] [DOI] [PubMed] [Google Scholar]
- 32.Maetzig T, et al. Polyclonal fluctuation of lentiviral vector-transduced and expanded murine hematopoietic stem cells. Blood. 2011;117:3053–3064. doi: 10.1182/blood-2010-08-303222. [DOI] [PubMed] [Google Scholar]
- 33.Laukkanen MO, et al. Low-dose total body irradiation causes clonal fluctuation of primate hematopoietic stem and progenitor cells. Blood. 2005;105:1010–1015. doi: 10.1182/blood-2004-04-1498. [DOI] [PubMed] [Google Scholar]
- 34.Gerrits A, et al. Cellular barcoding tool for clonal analysis in the hematopoietic system. Blood. 2010;115:2610–2618. doi: 10.1182/blood-2009-06-229757. blood-2009-06-229757 [pii] [DOI] [PubMed] [Google Scholar]
- 35.Schepers K, et al. Dissecting T cell lineage relationships by cellular barcoding. J Exp Med. 2008;205:2309–2318. doi: 10.1084/jem.20072462. jem.20072462 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Heijst JW, et al. Recruitment of antigen-specific CD8+ T cells in response to infection is markedly efficient. Science. 2009;325:1265–1269. doi: 10.1126/science.1175455. 325/5945/1265 [pii] [DOI] [PubMed] [Google Scholar]
- 37.Harkey MA, et al. Multiarm high-throughput integration site detection: limitations of LAM-PCR technology and optimization for clonal analysis. Stem Cells Dev. 2007;16:381–392. doi: 10.1089/scd.2007.0015. [DOI] [PubMed] [Google Scholar]
- 38.Roulet E, et al. High-throughput SELEX SAGE method for quantitative modeling of transcription-factor binding sites. Nat Biotechnol. 2002;20:831–835. doi: 10.1038/nbt718. nbt718 [pii] [DOI] [PubMed] [Google Scholar]
- 39.Sebat J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. 1138659 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim S, et al. High-throughput, sensitive quantification of repopulating hematopoietic stem cell clones. J Virol. 2010;84:11771–11780. doi: 10.1128/JVI.01355-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Craig DW, et al. Identification of genetic variants using bar-coded multiplexed sequencing. Nat Methods. 2008;5:887–893. doi: 10.1038/nmeth.1251. nmeth.1251 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berns K, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. nature02371 [pii] [DOI] [PubMed] [Google Scholar]
- 43.Kustikova O, et al. Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking. Science. 2005;308:1171–1174. doi: 10.1126/science.1105063. 308/5725/1171 [pii] [DOI] [PubMed] [Google Scholar]
- 44.Gonzalez-Murillo A, Lozano ML, Montini E, Bueren JA, Guenechea G. Unaltered repopulation properties of mouse hematopoietic stem cells transduced with lentiviral vectors. Blood. 2008;112:3138–3147. doi: 10.1182/blood-2008-03-142661. blood-2008-03-142661 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montini E, et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nature biotechnology. 2006;24:687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- 46.Karsunky H, Inlay MA, Serwold T, Bhattacharya D, Weissman IL. Flk2+ common lymphoid progenitors possess equivalent differentiation potential for the B and T lineages. Blood. 2008;111:5562–5570. doi: 10.1182/blood-2007-11-126219. blood-2007-11-126219 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 48.Fan HC, Quake SR. Sensitivity of noninvasive prenatal detection of fetal aneuploidy from maternal plasma using shotgun sequencing is limited only by counting statistics. PLoS One. 2010;5:e10439. doi: 10.1371/journal.pone.0010439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barese CN, Dunbar CE. Contributions of gene marking to cell and gene therapies. Hum Gene Ther. 2011;22:659–668. doi: 10.1089/hum.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seita J, et al. Lnk negatively regulates self-renewal of hematopoietic stem cells by modifying thrombopoietin-mediated signal transduction. Proc Natl Acad Sci U S A. 2007;104:2349–2354. doi: 10.1073/pnas.0606238104. 0606238104 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.