Abstract
A new drug target-- the "switch region"--has been identified within bacterial RNA polymerase (RNAP), the enzyme that mediates bacterial RNA synthesis. The new target serves as the binding site for compounds that inhibit bacterial RNA synthesis and kill bacteria. Since the new target is present in most bacterial species, compounds that bind to the new target are active against a broad spectrum of bacterial species. Since the new target is different from targets of other antibacterial agents, compounds that bind to the new target are not cross-resistant with other antibacterial agents. Four antibiotocs that function through the new target have been identified: myxopyronin, corallopyronin, ripostatin, and lipiarmycin. This review summarizes the switch region, switch-region inhibitors, and implications for antibacterial drug discovery.
Introduction
Bacterial RNAP as a target for antibacterial therapy
Bacterial RNAP is a proven target for broad-spectrum antibacterial therapy [reviewed in 1–4] The suitability of bacterial RNAP as a target for broad-spectrum antibacterial therapy follows from the fact that bacterial RNAP is an essential enzyme (permitting efficacy), the fact that bacterial RNAP subunit sequences are highly conserved (providing a basis for broad-spectrum activity), and the fact that bacterial RNAP-subunit sequences are not highly conserved in eukaryotic RNAP I, RNAP II, and RNAP III (providing a basis for therapeutic selectivity).
The rifamycin antibacterial agents--notably rifampin, rifapentine, rifabutin, and rifamixin--function by binding to and inhibiting bacterial RNAP [1–6]. The rifamycins bind to a site on bacterial RNAP adjacent to the RNAP active center and prevent extension of RNA chains beyond a length of 2–3 nt. The rifamycins are in current clinical use in treatment of both Gram-positive and Gram-negative bacterial infections [1–6]. The rifamycins are of particular importance in treatment of tuberculosis; the rifamycins are first-line anti-tuberculosis agents and are among the few antituberculosis agents able to kill non-replicating tuberculosis bacteria [7]. The rifamycins also are of importance in treatment of bacterial infections relevant to biowarfare or bioterrorism; combination therapy with ciprofloxacin, clindamycin, and rifampicin was successful in treatment of inhalational anthrax following the 2001 anthrax attacks [8], and combination therapy with ciprofloxacin and rifampicin, or doxycycline and rifampicin, is recommended for treatment of future cases of inhalational anthrax [9].
The clinical utility of the rifamycin antibacterial agents is threatened by the existence of bacterial strains resistant to rifamycins [1–6]. Resistance to rifamycins typically involves substitution of residues in or immediately adjacent to the rifamycin binding site on bacterial RNAP--i.e., substitutions that directly decrease binding of rifamycins [1–6].
In view of the public-health threat posed by rifamycin-resistant and multidrug-resistant bacterial infections, there is an urgent need for new classes of antibacterial agents that (i) inhibit bacterial RNAP (and thus have the same biochemical effects as rifamycins), but that (ii) inhibit bacterial RNAP through binding sites that do not overlap the rifamycin binding site (and thus do not share cross-resistance with rifamycins.
Bacterial RNAP "switch-region" as a target for antibacterial therapy
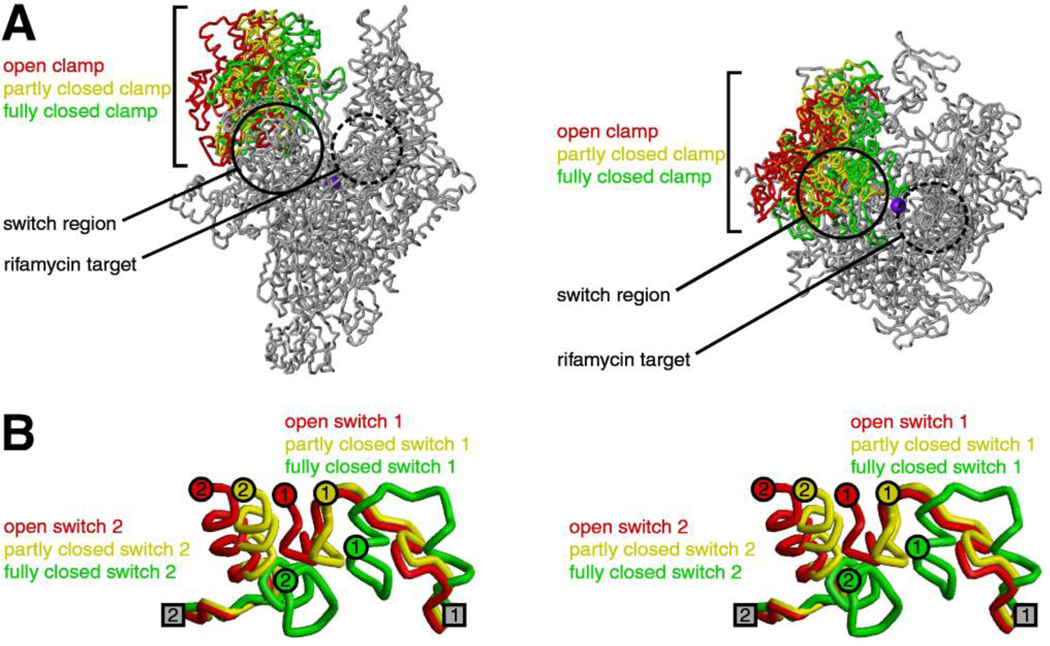
Recent work has identified a new drug target--the "switch region"--within bacterial RNAP [10–14; reviewed in 15–17]. The switch region is a structural element that mediates conformational changes and contacts required for RNAP to load DNA into the RNAP active-center cleft during transcription initiation (Fig. 1; [11–20]). The switch region is located at the base of the RNAP "clamp" and serves as the "hinge" that mediates opening of the RNAP clamp to load DNA into the RNAP active-center cleft and mediates closing of the RNAP clamp to retain DNA in the RNAP active-center cleft (Fig. 1A; [11–20; A.C. and R.H.E., unpublished]). Five segments of the switch region, termed "switch 1" through "switch 5," undergo changes in local conformation upon clamp opening and closing (Fig. 1B; [11,12,18–20]); switch 1 and switch 2 undergo particularly large changes in local conformation (Fig. 1B). Residues of switch 1, switch 2, and switch 3 make direct contacts with the loaded, unwound DNA template strand inside the RNAP active-center cleft [20–22], raising the possibility that direct contacts between the switch region and the loaded, unwound DNA template strand may coordinate, and mechanically couple, DNA loading, DNA unwinding, and clamp closure [18–20,23]. Residues of switch 2 and switch 3 also make up one wall of the RNAP RNA exit channel [20–22] and make direct contacts with the nascent RNA product in transcription elongation complexes [21,22].
Figure 1. RNAP clamp and RNAP switch region.
(A) Conformational states of the RNAP clamp (two orthogonal views) [11,12]. Structure of RNAP showing open (red), partly closed (yellow), and fully closed (green) clamp conformations, as observed in crystal structures (PDB 1I3Q, PDB 1HQM, PDB 1I6H). Circle, switch region; dashed circle, binding site for rifamycins; violet sphere, active-center Mg2+.
(B) Conformational states of the RNAP switch region (stereoview) [11,12]. Structure of RNAP switch 1 and RNAP switch 2 (β’ residues 1304–1329 and β’ residues 330–349; residues numbered as in E. coli RNAP) showing conformational states associated with open (red), partly closed (yellow), and fully closed (green) clamp conformations, as observed in crystal structures (PDB 1I3Q, PDB 1HQM, PDB 1I6H). Gray squares, points of connection of switch 1 and switch 2 to the RNAP main mass. Colored circles, points of connection of switch 1 and switch 2 to the RNAP clamp.
Compounds that bind to the switch region and interfere with an essential switch-region-dependent conformational change, DNA contact, or RNA contact will inhibit bacterial RNAP [10–17]. Since the switch region is highly conserved across both Gram-positive and Gram-negative bacterial species, inhibitors that function through the switch region typically will inhibit RNAP from both Gram-positive and Gram-negative bacterial species [10–12]. Since the switch region does not overlap the rifamycin binding site, inhibitors that function through the switch region typically will not share cross-resistantance with rifamycins (Fig. 1A; [10–12]).
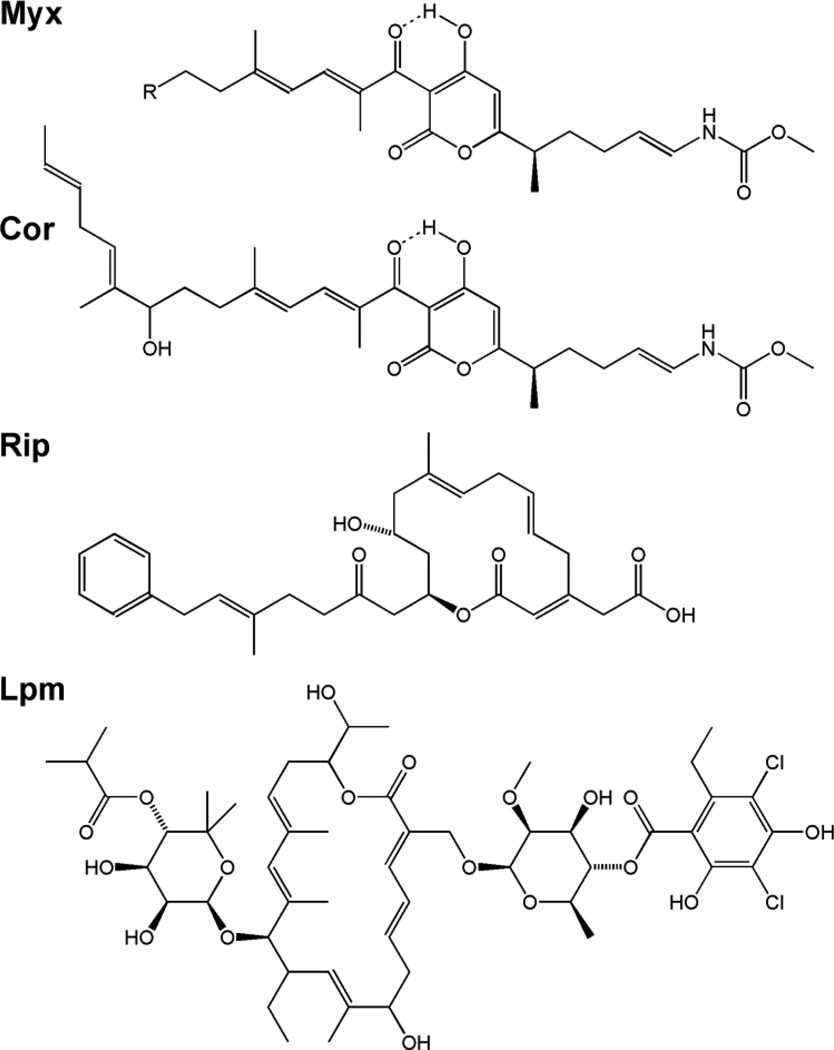
Four natural products that bind to the switch region, inhibit bacterial RNAP, and exhibit broad-spectrum antibacterial activity have been identified: myxopyronin (Myx), corallopyronin (Cor), ripostatin (Rip), and lipiarmycin (Lpm) (Figure 2; [10–14]).
Figure 2. Structures of inhibitors that function through the RNAP switch-region.
(A) Myx A (R = n-propyl) and Myx B (R = n-butyl). (B) Cor A. (C) Rip A. (C) Lpm A3.
Myxopyronin (Myx)
RNAP-inhibitory and antibacterial activities
Myx is an α-pyrone antibiotic (MW = 417 Da for Myx A and 431 Da for Myx B) produced by the Myxobacterium Myxococcus fulvus Mf50 (Figure 4A; [24–26]). Myx potently inhibits both Gram-positive bacterial RNAP and Gram-negative bacterial RNAP (IC50s in the low nanomolar range; Table 1; [11,12,24,27]) but does not inhibit eukaryotic RNAP [24]. Myx exhibits potent antibacterial activity against drug-sensitive and drug-resistant strains of a broad spectrum of Gram-positive bacteria, including Staphylococcus aureus, Enterococcus faecalis, Enterococcus faecium, Streptococcus pneumoniae, Clostridium difficile, Mycobacterium tuberculosis, and Bacillus anthracis (MICs = 1–13 µg/ml; Table 2; [12,24,27,28]). Myx also exhibits potent antibacterial activity against some Gram-negative bacteria, including Haemophilus influenzae, Moraxella catarrhalis, and Francisella tularensis (MICs = 6.1–16 µg/ml; Table 2). The absence of antibacterial activity against other Gram-negative species is attributable to cellular-uptake barriers and efflux, as illustrated by the absence of activity against wild-type E. coli but high activity against an rfa tolC mutant of E. coli, which has defects in cellular-uptake barriers and efflux (Table 2; [11,12,27]). Effects of Myx are bacteriocidal at concentrations ~2 to ~4 times MIC [A.S., J.M., and R.H.E., unpublished]. Myx exhibits no cross-resistance with rifampin (Table 2; [11,12,27,28]), and Myx exhibits no cross-resistance with other small-molecule inhibitors of RNAP that function through sites other than the switch region (CBR703, MccJ25, GE23077, streptolydigin, sorangicin, and salinamide; [D.D., J.M., and R.H.E., unpublished]) When co-administered with rifampin, Myx exhibits superadditive ("synergistic") antibacterial activity [A.S. and R.H.E., unpublished].
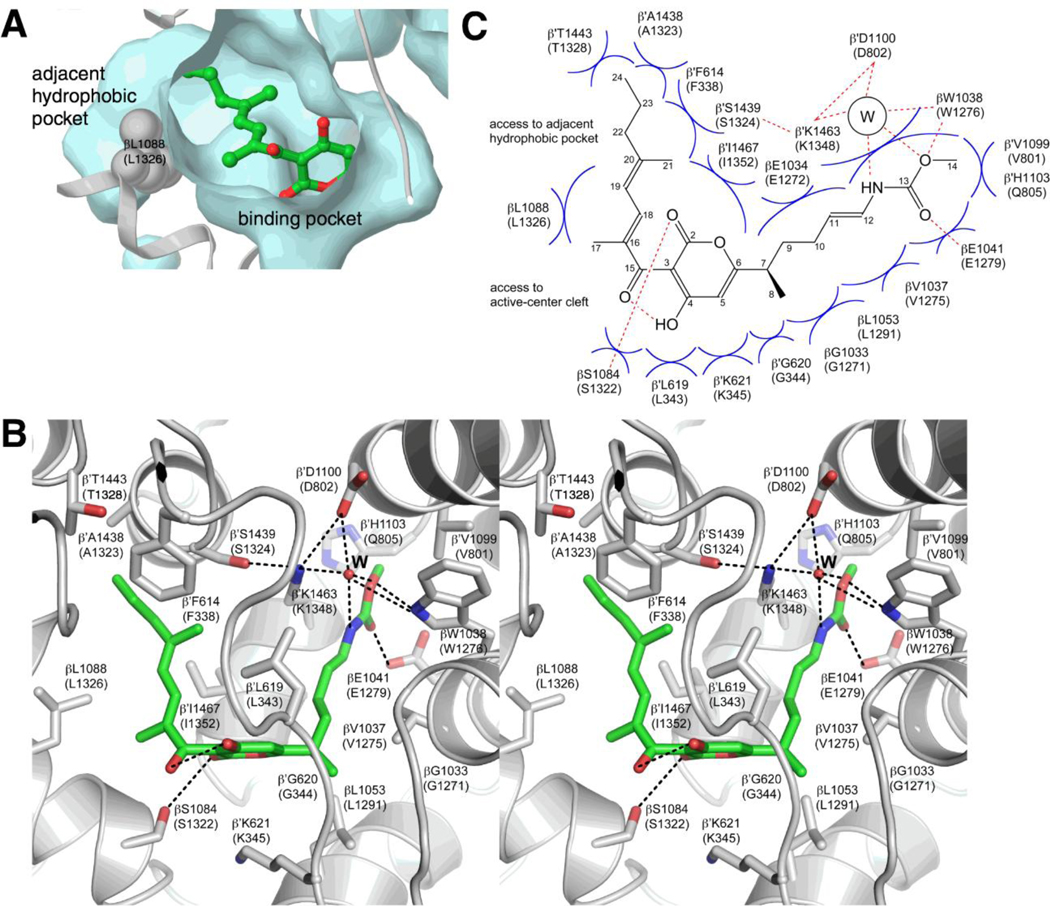
Figure 4. Crystal structure of the RNAP-Myx complex (PDB 3DXJ).
(A) Binding pocket for Myx [12]. Cyan, surface representation of the binding pocket and the adjacent hydrophobic pocket. Gray, ribbon representation of RNAP backbone. Green, Myx carbon atoms; red, Myx oxygen atoms. RNAP residues are numbered as in T thermophilus RNAP and, in parentheses, as in E. coli RNAP.
(B) Contacts between RNAP and Myx (stereoview) [12]. Gray, RNAP backbone (ribbon representation) and RNAP sidechain carbon atoms (stick representation); green, Myx carbon atoms; red, oxygen atoms; blue, nitrogen atoms. “W,” ordered bound water molecule. Dashed lines, H-bonds.
(C) Schematic summary of contacts between RNAP and Myx [12]. “W”, ordered bound water molecule. Red dashed lines, H-bonds. Blue arcs, van der Waals interactions.
Table 1.
RNAP-inhibition efficacies of inhibitors that function through the RNAP switch region
| RNAP | IC50 (µM)a,b |
|||
|---|---|---|---|---|
| Myx | Cor | Rip | Lpm | |
|
Gram-positive bacterial RNAP Staphylococcus aureus RNAP |
0.01 | 0.2 | 4 | 0.4 |
|
Gram-negative bacterial RNAP Escherichia coli RNAP |
0.01 | 0.2 | 0.6 | 6 |
Compounds were Myx B (synthesized as in [27]; provided by Y.W.E.), Cor A (isolated as in [37]; provided by R.J. and H.I.), Rip A (isolated as in [41]; provided by R.J. and H.I.), and Lpm A3 (isolated as in [48]; provided by S.D.).
Data are from fluorescence-detected transcription assays [73], using a DNA fragment containing the bacteriophage T4 N25 promoter, recombinant S. aureus RNAP σA holoenzyme (produced by co-expressing the S. aureus rpoA, rpoB, rpoC, and rpoZ genes in E. coli, expressing the S. aureus sigA gene in E. coli; and combining purified products in vitro), and E. coli RNAP σ70 holoenzyme (Epicentre, Inc.) [S.L. and R.H.E., unpublished].
Table 2.
Antibacterial efficacies of inhibitors that function through the RNAP switch region
| Organism | Minimum Inhibitory Concentration (µg/ml)a-f |
|||
|---|---|---|---|---|
| Myx | Cor | Rip | Lpm | |
| Gram-positive bacteria | ||||
| Staphylococcus aureus ATCC12600 MSSA | 1.0 | 4.9 | 13 | 3.5 |
| Staphylococcus aureus BAA-1707 MRSA (MW2) | 1.3 | 4.9 | 13 | 4.0 |
| Staphylococcus aureus BAA-1717 MRSA (USA300) | 1.3 | 4.9 | 16g | 3.2 |
| Staphylococcus aureus ATCC12600-Rif RRSA (βH516V) | 1.0 | 8.0 | 13 | 4.5 |
| Staphylococcus aureus ATCC12600-Rif RRSA (βH526N) | 1.0 | 10 | 13 | 4.5 |
| Staphylococcus aureus ATCC12600-Rif RRSA (βH526Y) | 1.0 | 8.0 | 13 | 2.8 |
| Staphylococcus aureus ATCC12600-Rif RRSA (βS531L) | 1.0 | 8.0 | 13 | 2.3 |
| Enterococcus faecalis ATCC19433 | 7.8 | >40 | >40 | 7.4 |
| Enterococcus faecium ATCC19434 | 1.1 | 7.1 | 13 | 2.1 |
| Streptococcus pneumoniae ATCC49619 | 2.9 | 5.5 | >40 | 4.1 |
| Clostridium difficile ATCC9689 | 3.1 | 0.78 | 3.1 | 0.012 |
| Mycobacterium tuberculosis H37Rv | 1.6 | 3.1 | >40 | 3.1 |
| Bacillus anthracis Vollum-1B | 13 | 13 | >40 | 0.78 |
| Gram-negative bacteria | ||||
| Escherichia coli DH5α | >40 | >40 | >40 | >40 |
| Escherichia coli D21f2tolC (rfa tolC) | 0.4 | 0.8 | 1.1 | 2 |
| Haemophilus influenzae ATCC33391 | 16 | >40 | >40 | >40 |
| Moraxella catarrhalis ATCC25238 | 6.1 | 12 | 13 | 11 |
| Francisella tularensis SCHU4 | 13 | >40 | >40 | 25 |
Compounds were Myx B (synthesized as in [27]; provided by Y.W.E.), Cor A (isolated as in [37]; provided by R.J. and H.I.), Rip A (isolated as in [41]; provided by R.J. and H.I.), and Lpm A3 (isolated as in [48]; provided by S.D.).
Data for S. aureus. E. faecalis., E. faecium., and E. coli are from spiral gradient endpoint assays performed essentially as in [74–76] (A.S., D.D., and R.H.E., unpublished data). Assays employed exponential-gradient plates containing 150 mm × 4 mm Mueller-Hinton II cation-adjusted agar (BD, Inc.) and 0.1–40 µg/ml of test compound. Plates were prepared using an Autoplate 4000 spiral plater (Spiral Biotech, Inc.). Saturated overnight cultures were swabbed radially onto plates, and plates were incubated for 16 h at 37°C. For each culture, the streak length was measured using a clear plastic template (Spiral Biotech, Inc.), the test-compound concentration at the streak endpoint was calculated using the program SGE (Spiral Biotech, Inc.), and the MIC was defined as the calculated test-compound concentration at the streak endpoint.
Data for S. pneumoniae, H. influenzae, and M. catarrhalis are from spiral gradient endpoint assays, performed as above, except that plates contained GC II agar (Teknova, Inc.)and were incubated in a 5% CO2/95% air atmosphere in an Anoxomat Mark II (Mart Microbiology, Inc.) (A.S. and R.H.E., unpublished data).
Data for C. difficile are from anaerobic broth microdilution assays [77] (A.S. and R.H.E., unpublished data).
Data for M. tuberculosis are from microplate Alamar Blue assays [78] (M.T., N.C., and R.H.E., unpublished data).
Data for B. anthracis and F. tularensis are from aerobic broth microdilution assays [79] (M.T., N.C., and R.H.E., unpublished data).
MIC50.
The frequency of spontaneous resistance to Myx is comparable to the frequency of spontaneous resistance to rifampin (4 × 10−8 vs. 5 × 10−8; measured in S. aureus [29; A.S. and R.H.E., unpublished]. However, the biological fitness of Myx-resistant mutants is lower than the biological fitness of rifampin-resistant mutants (measured in S. aureus; [A.S., D.D., and R.H.E, unpublished]. All Myx-resistant mutation confer non-zero fitness costs, and a majority of Myx-resistant mutations confer >10%/generation fitness costs [A.S. and R.H.E., unpublished]; in contrast, two rifampin-resistant mutations confer zero fitness costs, and only a minority of rifampin-resistant mutations confer >10%/generation fitness costs [30; A.S. and R.H.E., unpublished]. In view of the strong correlation between biological fitness of resistant mutations and clinical prevalence of resistance mutations [30–33], the finding that Myx-resistant mutants have lower biological fitness than rifampin-resistant mutants suggests that Myx-resistant mutants will have lower clinical prevalence than rifampin-resistant mutants.
A preliminary rat pharmacokinetics study indicates that Myx has good plasma half-life upon iv administration (t0.5 = 5.2 h) and good systemic bioavailability upon oral administration (F = 75%) [T.K. and R.H.E., unpublished]. Myx exhibits no acute toxicity in mice at doses of up to at least 100 mg/kg sc [24].
The above properties make Myx an attractive lead compound for antibacterial drug discovery, and, accordingly, efforts to optimize and validate Myx as an antibacterial therapeutic agent are in progress. Syntheses of Myx have been developed [27,34,35], and more than 100 novel Myx analogs have been synthesized [34,35; Y.W.E., A.Z., J.S., and R.H.E., unpublished]. The availability of high-resolution structural information for the RNAP-Myx complex (see below; [12,13]) has enabled structure-based optimization of Myx and has resulted in the design and synthesis of novel Myx analogs more potent than Myx B [Y.W.E., A.Z., J.S., and R.H.E., unpublished]. (One Myx analog synthesized prior to the availability of high-resolution structural information had been reported to be more potent than Myx B: 7-desmethyl-Myx B [34,35]. However, this compound in fact is less potent than Myx B [Y.W.E., A.Z., J.S., and R.H.E., unpublished].) The Myx biosynthetic locus of M. fulvus Mf50 has been cloned and sequenced, and efforts to accomplish efficient surrogate-host production of Myx and Myx analogs are in progress [S.S., R.R.N., and R.H.E., unpublished]. Antibacterial efficacy studies in animal models of infection are in progress [Y.W.E. and R.H.E., unpublished].
Target and mechanism
The Myx target within RNAP was identified by isolation and sequencing of Myx-resistant mutants following high-level mutagenesis of E. coli RNAP subunit genes in vitro [11,12]. Substitutions conferring Myx-resistance were obtained in RNAP β subunit (RpoB) residues 1255, 1275, 1278, 1279, 1283, 1285, 1291, 1298, 1315, 1320, 1322, and 1326, and in RNAP β' subunit (RpoC) residues 345, 1351, 1352, and 1354 [11,12]. Sequenced Myx-resistant mutants of S. aureus map to equivalent residues of S. aureus RNAP, indicating that the Myx target is the same in E. coli and in S. aureus [29; A.S., D.D., and R.H.E., unpublished]. In the three-dimensional structure of RNAP, the sites of substitutions conferring Myx-resistance define a single determinant with dimensions of ~20 Å × ~20 Å × ~10 Å ("Myx target"; Fig. 3A; [11,12]). The Myx target is located in the RNAP switch region and encompasses a significant fraction of the RNAP switch region, including switch 2 and segments of β and β' adjacent to switch 2. The Myx target does not overlap the rifampin target, consistent with the observation that Myx does not exhibit cross-resistance with rifampin (Fig. 3A; [11,12]). All residues of the Myx target are conserved in bacterial RNAP, consistent with the observation that Myx exhibits broad-spectrum activity against bacterial RNAP [11,12]. Three residues of the Myx target are not conserved--and indeed are radically different--in eukaryotic RNAP (β residues 1275, 1279, and 1322), consistent with observation that Myx does not exhibit activity against eukaryotic RNAP [11,12]. Transcription initiation involves: (i) binding of RNAP to promoter DNA to yield an RNAP-promoter closed complex, RPc, in which DNA is outside the RNAP active-center cleft and fully double-stranded; (ii) isomerization of the RNAP-promoter closed complex to yield a catalytically competent RNAP-promoter open complex, RPo, in which DNA is inside the RNAP active-center cleft and 14 bp of DNA are unwound; (iii) synthesis of the first ~10–15 nt of the RNA product as an RNAP-promoter initial transcribing complex, RPitc; and (iv) breakage of RNAP-promoter interactions to yield an RNAP-DNA elongation complex, RDe [reviewed in 36]. Biochemical experiments indicate that Myx inhibits transcription initiation by inhibiting the isomerization of RPc to RPo [12,13]. (Rifamycins inhibit transcription initiation at a later stage: synthesis of RNA by RPitc [1–6].) Experiments with promoter subfragments indicate that Myx interferes with interactions between RNAP and the promoter DNA segment comprising positions −11 to +15, which is the DNA segment that must be loaded into the RNAP active-center cleft and unwound during the isomerization of RPc to RPo [12]. Single-molecule fluorescence resonance energy transfer (smFRET) experiments show that Myx interferes with opening of the RNAP clamp, show that the clamp is predominantly open in RPc in the absence of Myx, and show that the clamp is closed in RPc in the presence of Myx.[A.C., D. Wang, Y. Korlann, B.T. Nixon, S. Weiss, and R.H.E,, unpublished].
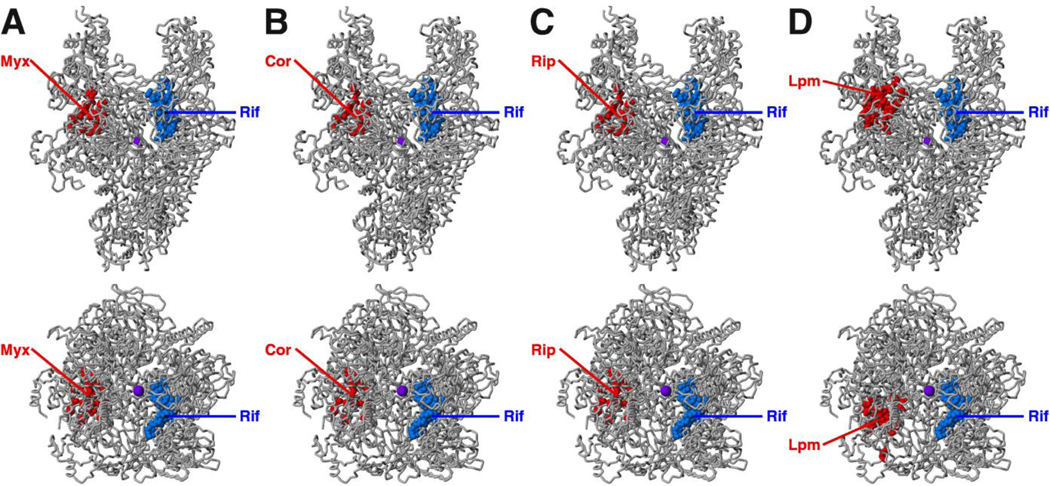
Figure 3. Targets of inhibitors that function through the RNAP switch-region.
(A) Target of Myx [11,12]. (B) Target of Cor [11,12]. (C) Target of Rip [11,12]. (D) Target of Lpm [10; D.D., R.Y.E., E.S., S.D., and R.H.E., unpublished]. Each panel shows two orthogonal views of RNAP. Red, sites of substitutions conferring moderate- to high-level resistance (≥4-fold resistance) to the specified inhibitor; blue, sites of substitutions conferring resistance to rifamycins; violet sphere, active-center Mg2+. The Myx, Cor, and Rip targets overlap essentially in toto. The Lpm target is adjacent to, but does not overlap, or only minimally overlaps, the Myx, Cor, and Rip targets (compare bottom subpanels of D to bottom subpanels of A–C).
Crystal structures have been determined of Thermus thermophilus RNAP holoenzyme in complex with Myx A (Fig. 4; [12]) and in complex with 7-desmethyl-Myx B [13]. The structures show definitively that Myx binds in the RNAP switch region, define contacts between Myx and RNAP, and define effects of Myx on RNAP conformation [12,13]. Myx binds within a nearly completely enclosed, predominantly hydrophobic, binding pocket formed by switch 1, switch 2, and adjacent segments of β and β' (Fig. 4A,B). The binding pocket is crescent-shaped, has dimensions of ~25 Å (measured along the curve of the crescent) × ~5 Å × ~4 Å, and has a volume of ~500 Å3 (Fig. 4A,B). The binding pocket connects to an adjacent hydrophobic pocket having a volume of ~120 Å3 (Fig. 4A). The adjacent hydrophobic pocket is located close to the terminus of the Myx dienone sidechain, and, as such, potentially is able to accommodate the sidechain extension present in Cor (see below). The Myx α-pyrone ring contacts β residue 1322 and β’ residues 343–345 and 1352 (Fig. 4B,C; residues numbered as in E. coli RNAP). The Myx dienone sidechain contacts β residue 1326 and β’ residues 334–338, 1323–1328, and 1352 (Fig. 4B,C; residues numbered as in E. coli RNAP). The Myx enecarbamate sidechain makes direct or water-mediated contacts with β residues 1271–1279 and 1291 and β’ residues 343–344, 801–805, and 1348–1351 (Fig. 4B,C; residues numbered as in E. coli RNAP). Myx alters the local conformation of a nine-residue segment of switch 2 (β' residues 336–344; residues numbered as in E. coli RNAP). This segment of switch 2 previously has been shown to adopt different conformations in open, partly closed, and fully closed clamp conformational states (Fig. 1B). This segment of switch 2 also previously has been shown to contact the DNA template strand in transcription elongation complexes [20–22]. Based on the available functional and data, two mutually non-exclusive models have been proposed for the mechanism by which Myx inhibits the isomerization of RPc to RPo. According to the first model, Myx interferes with switch-region conformational changes required for opening of the RNAP clamp to load DNA into the RNAP active-center cleft ("hinge jamming" model; [12]). According to the second model, Myx interferes with contacts between the switch region and the unwound DNA template strand required for DNA unwinding [13].
Corallopyronin (Cor)
RNAP-inhibitory and antibacterial activities
Cor is an α-pyrone antibiotic (MW = 542 Da) structurally related to Myx, differing by possession of a seven-carbon side-chain extension (Fig. 2B; [37]). Cor is produced by the Myxobacterium Corallococcus coralloides Cc c127 [37]. The RNAP-inhibitory and antibacterial activities of Cor parallel those of Myx, but generally are lower in potency (Table 1,Table 2; [11,12,37]). The rifampin-cross-resistance, spontaneous-resistance-frequency, and resistance-fitness-costs properties of Cor are essentially identical to those of Myx (Table 2; [11,12,28,38]).
Progress toward a synthesis of Cor has been reported [39].
The Cor biosynthetic locus of C. coralloides Cc c127 has been cloned and sequenced [40]. The Cor biosynthetic locus exhibits striking similarities in gene sequence and gene order to the Myx biosynthetic locus [40; S.S., R.R.N., and R.H.E., unpublished]. The proposed pathway of Cor biosynthesis entails synthesis of a first polyketide-derived chain by a hybrid methyloxycarbonyl-glycine-specific nonribosomal peptide synthase (NRPS) and Type-I polyketide synthase (PKS), synthesis of a second polyketide-derived chain by a Type-I PKS, and cleavage and joining of the two polyketide-derived chains [40].
Target and mechanism
The Cor target within RNAP was identified by analysis of Cor-cross-resistance properties of previously identified Myx-resistant mutants of E. coli and by direct isolation and sequencing of Cor-resistant mutants following high-level mutagenesis of E. coli RNAP subunit genes in vitro (Fig. 3B; [11,12]). The Cor target completely overlaps the Myx target and consists of all residues of the Myx target plus one additional residue: β residue 1326 [11,12]. The additional residue is inferred to interact with the seven-carbon sidechain extension present in Cor but not in Myx [11,12]. Consistent with this inference, in the structure of the RNAP-Myx complex, beta residue 1326 is located close to the ligand dienone sidechain terminus, the point of attachment of the seven-carbon sidechain extension present in Cor (Fig. 4A–C). Further consistent with this inference, in the structure of the RNAP-Myx complex, β residue 1326 forms one wall of an adjacent hydrophobic pocket that would have sufficient volume to accommodate the seven-carbon sidechain extension present in Cor if residue 1326 is Leu (as in wild-type RNAP) but not if residue 1326 is Trp (as in Cor-resistant RNAP substituted at position 1326) (Fig, 4A,C). Sequencing of spontaneous Cor-resistant mutants of S. aureus indicates that the Cor target is the same in S. aureus as in E. coli [38].
Cor, like Myx, inhibits the isomerization of RPc to RPo [12], interferes with interactions of RNAP with promoter positions −11 to +15 [12], and interferes with opening of the RNAP clamp [A.C., D. Wang, Y. Korlann, S. Weiss, and R.H.E,, unpublished]. The mechanism of action of Cor is likely to be identical to the mechanism of action of Myx [12].
Ripostatin (Rip)
RNAP-inhibitory and antibacterial activities
Rip is a 14-membered macrocylic-lactone antibiotic (MW = 494 Da) structurally unrelated to other switch-region-target inhibitors (Fig. 2C; [41,42]). Rip is produced by the Myxobacterium Sorangium cellulosum So ce377 [41,42]. The RNAP-inhibitory and antibacterial activities of Rip roughly parallel those of Myx, but are narrower in spectrum and generally lower in potency than those of Myx (Table 1,Table 2; [11,41]). The rifampin-cross-resistance, spontaneous-resistance-frequency, and resistance-fitness-costs properties of Rip are essentially identical to those of Myx (Table 2; [11,12,28,41]).
Several laboratories are at work on development of a synthesis for Rip [43–46].
Target and mechanism
The Rip target within RNAP was identified in the same manner as the Cor target (Fig. 3C; [11,12]). Surprisingly--in view of the lack of structural similarity between Rip, Myx, and Cor--the Rip target completely overlaps the Myx and Cor targets and is identical, residue for residue, to the Cor target (Fig. 3A–C; [11,12]).
Rip, like Myx and Cor, inhibits the isomerization of RPc to RPo [12], interferes with interactions of RNAP with promoter positions −11 to +15 [12], and interferes with opening of the RNAP clamp [A.C., D. Wang, Y. Korlann, S. Weiss, and R.H.E., unpublished]. The mechanism of action of Rip is likely to be similar or identical to the mechanisms of action of Myx and Cor [12].
Lipiarmycin (Lpm): RNAP-inhibitory and antibacterial activities
RNAP-inhibitory and antibacterial activities
Lpm (also known as clostomicin, tiacumicin, diffimicin, PAR-101, OPT-80, fidaxomicin, and Dificid; MW = 1,058 Da) is an 18-membered macrocyclic-lactone antibiotic produced by Actinomycete species, including Actinoplanes deccanensis, Micromonospora echinospora and Dactylosporangium aurantiacum hamdenensis (Fig. 2D; [47–54; reviewed in 55–60]).
(The proliferation of names is unfortunate. Lipiarmycin is the name applied to Lpm isolated from A deccanensis; clostomicin, is the name subsequently applied to Lpm isolated from M echinospora; tiacumicin is the name subsequently applied to Lpm isolated from D. aurantiacum; and diffimicin, PAR-101, OPT-80, fidaxomicin, and Dificid are successive trade names applied to Lpm isolated from D. aurantiacum. Lpm isolated from A. deccanensis and D. aurantiacum have the identical major component, Lpm A3 [52,54], have nearly identical minor components [52,54] and have indistinguishable RNAP-inhibitory and antibacterial activities [Y.W.E., A.S., and R.H.E., unpublished].) Lpm inhibits both Gram-positive bacterial RNAP and Gram-negative bacterial RNAP (Table 1; [49,50,61,62]). Lpm exhibits potent antibacterial activity against most Gram-positive bacteria and exhibits especially potent antibacterial activity against the Gram-positive bacterium Clostridium difficile (MIC = 0.012 µg/ml for C. difficile; Table 2; [28,48,51,53,63–66]). Lpm also exhibits antibacterial activity against some Gram-negative bacteria (Table 2). Lpm exhibits no cross-resistance with rifampin [10,28,59,65–67] or with other RNAP inhibitors that function through sites other than the switch region [D.D., and R.H.E., unpublished]. The frequency of spontaneous resistance to Lpm is comparable to, or lower than, the frequency of spontaneous resistance to rifampin (3 × 10−8 vs. 5 × 10−8; measured in S. aureus; A.S. and R.H.E., unpublished]).
Lpm exhibits an LD50 of ≥500 mg/kg upon ip or sc administration in mice [48,63] and an LD50 of >1000 mg/kg upon oral administration [55,56,60]. Systemically administered Lpm exhibits essentially zero systemic bioavailability (t0.5 ≪ 10 min; [55,56]). Orally administered Lpm also exhibits essentially zero systemic bioavailability but exhibits excellent gastrointestinal-tract bioavailability [55–60]. The combination of high antibacterial activity against C. difficile, high gastrointestinal-tract bioavailability upon oral administration, and low systemic bioavailability and systemic toxicity upon oral administration make Lpm extremely well suited for treatment of C. difficile-associated diarrhea [55–60] (Orally administered Lpm provides essentially a topical treatment of the gastrointestinal tract [55–60].) Results of a 2009 Phase III clinical trial indicated that orally administered Lpm is non-inferior to the current standard of care, vancomycin, for treatment of C. difficile-associated diarrhea [59,60]. Results of a 2011 Phase III clinical trial confirmed that orally administered Lpm is non-inferior to vancomycin and indicated that orally administered Lpm results in lower levels of recurrence [68]. On May 27, 2011, Lpm (fidaxomicin/Dificid) was approved by the US Food and Drug Administration (FDA) for treatment of C. difficile-associated diarrhea [69].
The near-zero systemic bioavailability of Lpm upon systemic or oral administration implies that Lpm is unlikely to be useful for treatment of systemic infections without structural optimization or formulation changes [69].
The Lpm biosynthetic loci of the D. aurantiacum and A. deccanensis producer strains have been characterized and exhibit strong similarities in gene sequence and gene order [70; S.S., R.R.N., and R.H.E., unpublished]. The proposed pathways of Lpm biosynthesis entail synthesis of a polyketide-derived macrocycle by a Type-I PKS, synthesis of a dichlorinated homoorsenellic acid moiety by an iterative Type-I PKS and a FADH2-dependent halogenase, and synthesis and incorporation of two carbohydrate moieties.
No synthesis of Lpm has been reported. However, more than ten Lpm analogs have been prepared by fermentation of the D. aurantiacum and A. deccanensis producer strains in the presence of unnatural precursors [71; Y.W.E., and R.H.E., unpublished], and more than twenty Lpm analogs, including analogs with higher potencies than Lpm against S. aureus and Enterococcus spp., have been prepared by fermentation of engineered gene-disruption mutants of the D. aurantiacum and A. deccanensis producer strains producer strains [70,72; S.S., and R.H.E., unpublished].
Target and mechanism
The Lpm target within RNAP was identified by the sequencing of spontaneous Lpm-resistant mutants of Bacillus subtilis [10,61], and by the isolation and sequencing of Lpm-resistant mutants following high-level mutagenesis of E. coli RNAP subunit genes in vitro [10]). Substitutions conferring Lpm-resistance were obtained in β residues 1251, 1256, 1302, 1319, 1321, and 1325, and in β' residues 249 and 337 (residues numbered as in E. coli RNAP [10]). The Lpm target was further defined by the isolation and sequencing of more than 100 additional Lpm-resistant mutants following high-level mutagenesis of E. coli RNAP subunit genes in vitro [D.D., R.Y.E., E.S., S.D., and R.H.E., unpublished]. The sequencing of Lpm-resistant mutants from a different B. subtilis strain, S. aureus, E. faecalis, and M. tuberculosis indicate that the Lpm target is the same in these organisms [65–67; A.S. and D.D., unpublished]. (It recently has been reported that deletion of residues 513–519 of E. coli transcription initiation factor σ70 confers resistance to Lpm [14], but this report is erroneous [S.C. and R.H.E. unpublished].)
In the three-dimensional structure of RNAP, the sites of substitutions conferring Lpm-resistance define a determinant with dimensions of ~35 Å × ~25 Å × ~20 Å ("Lpm target"; Fig. 3D; [10; D.D., R.Y.E., E.S., S.D., and R.H.E., unpublished]). The Lpm target encompasses the RNA exit channel and part of the switch region, including parts of switch 2 and switch 3.
The Lpm target is adjacent to the Myx/Cor/Rip target, but does not overlap, or only minimally overlaps, the Myx/Cor/Rip target (Fig. 3A–D). The Myx/Cor/Rip target is centered on switch 1 and the C-terminal part of switch 2. In contrast, the Lpm target is centered on the N-terminal part of switch 2 and switch 3.. No high-level Lpm-resistant mutant shows high-level cross-resistance to Myx, Cor, or Rip--and vice-versa [D.D. and R.H.E., unpublished].
The Lpm target does not overlap the rifampin target (Fig. 3D), consistent with the observation that Lpm does not exhibit cross-resistance with rifampin [10,65–67].
Based on the location of the Lpm target within the structure of RNAP, Lpm would be expected to interfere with RNAP switch-region function, RNAP RNA-exit-channel function, or both. Biochemical results indicate that Lpm--like Myx, Cor, and Rip--inhibits the isomerization of RPc to RPo [14; S.C., S.Y.D., J.M., and R.H.E., unpublished], interferes with interactions of RNAP with promoter positions −11 to +15 [S.C., J.M., and R.H.E., unpublished], and interferes with opening of the RNAP clamp [A.C., D. Wang, Y. Korlann, S. Weiss, and R.H.E., unpublished]. Therefore, the mechanism of action of Lpm may be similar, at least in part, to the mechanisms of action of Myx, Cor, and Rip.
Novel switch-region inhibitors
The finding that two structurally unrelated chemotypes--the α-pyrone chemotype represented by Myx and Cor and the 14-membered macrocyclic-lactone chemotype represented by Rip--inhibit RNAP through the Myx/Cor/Rip target suggests that the Myx/Cor/Rip target is highly "druggable" and provides a precedent for seeking additional chemotypes that inhibit RNAP through this target [12]. The availability of high-resolution structural information for the RNAP-Myx complex [12,13] enables structure-based in silico screening of virtual compound libraries in order to identify candidate compounds likely to inhibit RNAP through the Myx/Cor/Rip target [4]. In work to date--involving in silico screening for candidate ligands, followed by in vitro confirmation of RNAP inhibition, followed by in vitro confirmation of Myx-cross-resistance of RNAP inhibition--three novel, structurally unrelated, inhibitors that inhibit RNAP through the Myx/Cor/Rip target have been identified and confirmed [C.X., S.L., and R.H.E., unpublished]. Characterization of the antibacterial activities of the new inhibitors and determination of structures of the RNAP-inhibitor complexes are in progress.
Overview
With the 2011 FDA approval of Lpm (fidaxomicin/Dificid) for treatment of C. difficile-associated diarrhea [69], the RNAP switch region now is a proved target for antibacterial drug discovery and antibacterial therapy. With the recent determination of high-resolution structures of RNAP in complex with switch-region inhibitors [12,13], determination of targets of switch-region inhibitors [10–12], and determination of mechanisms of action of switch-region inhibitors [12–14], the tools now are in place to proceed rapidly with rational, structure- and mechanism-based, optimization of known switch-region inhibitors and discovery of new switch-region inhibitors.
Highlights.
Bacterial RNA polymerase is a proven target for antibacterial therapy.
The "switch region" is a new inhibitor binding site within bacterial RNA polymerase.
The "switch region" does not overlap the rifamycin binding site.
Inhibitors that function through the switch region are not cross-resistant with rifamycins.
Myxopyronin, corallopyronin, ripostatin, and lipiarmycin function through the switch region.
Acknowledgements
Preparation of this report was supported by NIH grants GM41376, AI072766, and AI90837, a Global Alliance for TB Drug Development contract, and a Howard Hughes Medical Institute Investigatorship to R.H.E.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended readings
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Chopra I. Bacterial RNA polymerase: a promising target for the discovery of new antimicrobial agents. Curr. Opin. Investig. Drugs. 2007;8:600–607. [PubMed] [Google Scholar]
- 2.Villain-Guillot P, Bastide L, Gualtieri M, Leonetti J. Progress in targeting bacterial transcription. Drug Discov. Today. 2007;12:200–208. doi: 10.1016/j.drudis.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 3.Mariani R, Maffioli S. Bacterial RNA polymerase inhibitors: an organized overview of their structure, derivatives, biological activity and current clinical development status. Curr. Med. Chem. 2009;16:430–454. doi: 10.2174/092986709787315559. [DOI] [PubMed] [Google Scholar]
- 4.Ho M, Hudson B, Das K, Arnold E, Ebright R. Structures of RNA polymerase-antibiotic complexes. Curr. Opin. Structl. Biol. 2009;19:715–723. doi: 10.1016/j.sbi.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Floss H, Yu T. Rifamycin: mode of action, resistance, and biosynthesis. Chem. Rev. 2005;105:621–632. doi: 10.1021/cr030112j. [DOI] [PubMed] [Google Scholar]
- 6.Campbell E, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, Darst S. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell. 2001;104:901–912. doi: 10.1016/s0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- 7.Mitchison D. Role of individual drugs in the chemotherapy of tuberculosis. Int. J. Tuberc. Lung Dis. 2000;4:796–806. [PubMed] [Google Scholar]
- 8.Mayer T, Bersoff-Matcha S, Murphy C, Earls J, Harper S, Pauze D, Nguyen M, Rosenthal J, Cerva D, Druckenbrod G, Hanfling D, Fatteh N, Napoli A, Nayyar A, Berman E. Clinical presentation of inhalational anthrax following bioterrorism exposure: report of two surviving patients. JAMA. 2001;286:2549–2553. doi: 10.1001/jama.286.20.2549. [DOI] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. Update: investigation of bioterrorism-related anthrax and interim guidelines for exposure management and antimicrobial therapy. JAMA. 2001 October;286:2226–2232. doi: 10.1001/jama.286.18.2226. [DOI] [PubMed] [Google Scholar]
- 10.Ebright R. RNA exit channel: target and method for inhibition of bacterial RNA polymerase. WO2005/001034. 2005 [Google Scholar]
- 11.Ebright R. Switch region: target and method for inhibition of bacterial RNA polymerase. WO2007/094799. 2007 [Google Scholar]
- 12.Mukhopadhyay J, Das K, Ismail S, Koppstein D, Jang M, Hudson B, Sarafianos S, Tuske S, Patel J, Jansen R, Irschik H, Arnold E, Ebright RH. The RNA polymerase "switch region" is a target for inhibitors. Cell. 2008;135:295–307. doi: 10.1016/j.cell.2008.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belogurov G, Vassylyeva M, Sevostyanova A, Appleman J, Xiang A, Lira R, Webber S, Klyuyev S, Nudler E, Artsimovitch I, Vassylyev D. Transcription inactivation through local refolding of the RNA polymerase structure. Nature. 2009;45:332–335. doi: 10.1038/nature07510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tupin A, Gualtieri M, Leonetti J, Brodolin K. The transcription inhibitor lipiarmycin blocks DNA fitting into the RNA polymerase catalytic site. EMBO J. 2010;29:2527–2537. doi: 10.1038/emboj.2010.135. This paper reports that Lpm inhibits the isomerization of RPc to RPo. (This paper should be read with caution. Parts of the paper addressing effects of Lpm on sub-reactions in the isomerization of RPc to RPo and addressing effects of deletion of residues 513–519 of transcription initiation factor σ are incorrect [S.Y.D., S.C., and R.H.E., unpublished].)
- 15.Sousa R. Inhibiting RNA polymerase. Cell. 2008;135:205–207. doi: 10.1016/j.cell.2008.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tupin A, Gualtieri M, Brodolin K, Leonetti JP. Myxopyronin: a punch in the jaws of bacterial RNA polymerase. Future Microbiol. 2009;4:145–149. doi: 10.2217/17460913.4.2.145. [DOI] [PubMed] [Google Scholar]
- 17.Haebich D, von Nussbaum F. Inhibition of RNA polymerase. Angew. Chem. Int. Ed. Engl. 2009;48:3397–3400. doi: 10.1002/anie.200900338. [DOI] [PubMed] [Google Scholar]
- 18.Cramer P, Bushnell D, Kornberg R. Structural basis of transcription: RNA polymerase II at 2.8 Å resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 19.Cramer P. Multisubunit RNA polymerases. Curr. Opin. Struct. Biol. 2002;12:89–97. doi: 10.1016/s0959-440x(02)00294-4. [DOI] [PubMed] [Google Scholar]
- 20.Gnatt A. Elongation by RNA polymerase II: structure-function relationship. Biochim. Biophys. Acta. 2002;1577:175–190. doi: 10.1016/s0167-4781(02)00451-7. [DOI] [PubMed] [Google Scholar]
- 21.Kettenberger H, Armache K, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 22.Vassylyev D, Vassylyeva M, Perederina A, Tahirov T, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 23. Pupov D, Miropolskaya N, Sevostyanova A, Bass I, Artsimovitch I, Kulbachinskiy A. Multiple roles of the RNA polymerase β’ SW2 region in transcription initiation, promoter escape, and RNA elongation. Nucl. Acids Res. 2010;38:5784–5796. doi: 10.1093/nar/gkq355. This paper shows that mutations within switch 2 affect multiple stages of transcription.
- 24.Irschik H, Gerth K, Höfle G, Kohl W, Reichenbach H. The myxopyronins, new inhibitors of bacterial RNA synthesis from Myxococcus fulvus (Myxobacterales) J. Antibiot. 1983;36:1651–1658. doi: 10.7164/antibiotics.36.1651. [DOI] [PubMed] [Google Scholar]
- 25.Kohl W, Irschik H, Reichenbach H, Höfle G. Antibiotics from gliding bacteria. XVII. Myxopyronin A and B - two novel antibiotics from Myxococcus fulvus strain Mx f50. Liebigs Ann. Chem. 1983;1983:1656–1667. [Google Scholar]
- 26.Kohl W, Irschik H, Reichenbach H, Höfle G. Antibiotics from gliding bacteria, XXII. Biosynthesis of myxopyronin A, an antibiotic from Myxococcus fulvus, strain Mx f50. Liebigs Ann. Chem. 1984;1984:1088–1093. [Google Scholar]
- 27.Hu T, Schaus J, Lam K, Palfreyman M, Wuonola M, Gustafson G, Panek J. Total synthesis and preliminary antibacterial evaluation of the RNA polymerase inhibitors (+/−)-Myxopyronin A and B. J. Org. Chem. 1998;63:2401–2406. [Google Scholar]
- 28.O'Neill A, Oliva B, Storey C, Hoyle A, Fishwick C, Chopra I. RNA polymerase inhibitors with activity against rifampin-resistant mutants of Staphylococcus aureus. Antimicrob. Agents Chemother. 2000;44:3163–3166. doi: 10.1128/aac.44.11.3163-3166.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moy T, Daniel A, Hardy C, Jackson A, Rehrauer O, Hwang Y, Zou D, Nguyen K, Silverman J, Li Q, Murphy C. Evaluating the activity of the RNA polymerase inhibitor myxopyronin B against Staphylococcus aureus. FEMS Microbiol. Lett. 2011;319:176–179. doi: 10.1111/j.1574-6968.2011.02282.x. [DOI] [PubMed] [Google Scholar]
- 30.Wichelhaus T, Böddinghaus B, Besier S, Schäfer V, Brade V, Ludwig A. Biological cost of rifampin resistance from the perspective of Staphylococcus aureus. Antimicrob. Agents. Chemother. 2002;46:3381–3385. doi: 10.1128/AAC.46.11.3381-3385.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Neill A, Huovinen T, Fishwick C, Chopra I. Molecular genetic and structural modeling studies of Staphylococcus aureus RNA polymerase and the fitness of rifampin resistance genotypes in relation to clinical prevalence. Antimicrob. Agents Chemother. 2006;50:298–309. doi: 10.1128/AAC.50.1.298-309.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Billington O, McHugh T, Gillespie S. Physiological cost of rifampin resistance induced in vitro in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 1999;43:1866–1869. doi: 10.1128/aac.43.8.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gagneux S, Long C, Small P, Van T, Schoolnik G, Bohannan B. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science. 2006;312:1944–1946. doi: 10.1126/science.1124410. [DOI] [PubMed] [Google Scholar]
- 34.Doundoulakis T, Xiang A, Lira R, Agrios K, Webber S, Sisson W, Aust R, Shah A, Showalter R, Appleman J, Simonsen K. Myxopyronin B analogs as inhibitors of RNA polymerase, synthesis and biological evaluation. Bioorg. Med. Chem. Lett. 2004;14:5667–5672. doi: 10.1016/j.bmcl.2004.08.045. [DOI] [PubMed] [Google Scholar]
- 35.Lira R, Xiang A, Doundoulakis T, Biller W, Agrios K, Simonsen K, Webber S, Sisson W, Aust R, Shah A, Showalter R, Banh V, Steffy K, Appleman J. Syntheses of novel myxopyronin B analogs as potential inhibitors of bacterial RNA polymerase. Bioorg. Med. Chem. Lett. 2007;17:6797–6800. doi: 10.1016/j.bmcl.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 36. Saecker R, Record MJ, Dehaseth P. Mechanism of bacterial transcription initiation: RNA polymerase-promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J. Mol. Biol. 2011 doi: 10.1016/j.jmb.2011.01.018. (in press) This paper provides a current and comprehensive review of the mechanism of transcription initiation
- 37.Irschik H, Jansen R, Höfle G, Gerth K, Reichenbach H. The corallopyronins, new inhibitors of bacterial RNA synthesis from Myxobacteria. J. Antibiot. 1985;38:145–152. doi: 10.7164/antibiotics.38.145. [DOI] [PubMed] [Google Scholar]
- 38.Mariner K, McPhillie M, Trowbridge R, Smith C, O'Neill A, Fishwick C, Chopra I. Activity and development of resistance to corallopyronin A, an inhibitor of RNA polymerase. Antimicrob. Agents Chemother. 2011;55:2413–2416. doi: 10.1128/AAC.01742-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wardenga G. Enwicklung eines synthetischen Zugangs zu potentiellen Antibiotika auf Basis der Naturstoffs Corallopyronin A. Hannover, Germany: Thesis, Gottfried Wilhelm Leibniz Universität; 2007. [Google Scholar]
- 40. Erol O, Schäberle T, Schmitz A, Rachid S, Gurgui C, El Omari M, Lohr F, Kehraus S, Piel J, Müller R, König G. Biosynthesis of the Myxobacterial antibiotic corallopyronin A. Chembiochem. 2010;11:1253–1265. doi: 10.1002/cbic.201000085. This paper reports cloning and sequencing of the putative biosynthetic locus for Cor.
- 41.Irschik H, Augustiniak H, Gerth K, Höfle G, Reichenbach H. The ripostatins, novel inhibitors of eubacterial RNA polymerase isolated from myxobacteria. J. Antibiot. 1995;48:787–792. doi: 10.7164/antibiotics.48.787. [DOI] [PubMed] [Google Scholar]
- 42.Augustiniak H, Irschik H, Reichenbach H, Höfle G. Antibiotics from gliding bacteria. Part LXXVIII. Ripostatin A, B, and C. Isolation and structure elucidation of novel metabolites from Sorangium cellulosum. Liebigs Ann. Chem. 1996;10:1657–1663. [Google Scholar]
- 43.Kujat C. Studien zur Totalsynthese von Ripostatin A & B. Hannover, Germany: Thesis, Gottfried Wilhelm Leibniz Universität; 2007. [Google Scholar]
- 44.Schleicher K. Nickel-catalyzed preparation of acrylamides from alpha olefins and isocyanates; Synthetic studies toward ripostatin A. Thesis, Massachusetts Institute of Technology; 2010. [Google Scholar]
- 45.http://www.scripps.edu/florida/chem/micalizio/research.html
- 46.http://www.haverford.edu/faculty/fblase
- 47.Parenti F, Pagani H, Beretta G. Lipiarmycin, a new antibiotic from Actinoplanes. I. Description of the producer strain and fermentation studies. J. Antibiot. 1975;28:247–252. doi: 10.7164/antibiotics.28.247. [DOI] [PubMed] [Google Scholar]
- 48.Coronelli C, White R, Lancini G, Parenti F. Lipiarmycin, a new antibiotic from Actinoplanes. II. Isolation, chemical, biological and biochemical characterization. J. Antibiot. 1975;28:253–259. doi: 10.7164/antibiotics.28.253. [DOI] [PubMed] [Google Scholar]
- 49.Sergio S, Pirali G, White R, Parenti F. Lipiarmycin, a new antibiotic from Actinoplanes III. Mechanism of action. J. Antibiot. 1975;1975:543–549. doi: 10.7164/antibiotics.28.543. [DOI] [PubMed] [Google Scholar]
- 50.Talpaert M, Campagnari F, Clerici L. Lipiarmycin: an antibiotic inhibiting nucleic acid polymerases. Biochem. Biophys. Res. Commun. 1975;63:328–334. doi: 10.1016/s0006-291x(75)80047-7. [DOI] [PubMed] [Google Scholar]
- 51.Omura S, Imamura N, Oiwa R, Kuga H, Iwata R, Masuma R, Iwai Y. Clostomicins, new antibiotics produced by Micromonospora echinospora subsp. armeniaca subsp. nov. I. Production, isolation, and physico-chemical and biological properties. J. Antibiot. 1986;39:1407–1412. doi: 10.7164/antibiotics.39.1407. [DOI] [PubMed] [Google Scholar]
- 52.Arnone A, Nasini G, Cavalleri B. Structure elucidation of the macrocyclic antibiotic lipiarmycin. J. Chem. Soc. Perkin Trans. I. 1987;6:1353–1359. [Google Scholar]
- 53.Theriault R, Karwowski J, Jackson M, Girolami R, Sunga G, Vojtko C, Coen L. Tiacumicins, a novel complex of 18-membered macrolide antibiotics. I. Taxonomy, fermentation and antibacterial activity. J. Antibiot. 1987;40:567–574. doi: 10.7164/antibiotics.40.567. [DOI] [PubMed] [Google Scholar]
- 54.Hochlowski J, Swanson S, Renfranz L, Whittern D, Buko A, McAlpine J. Tiacumicins, a novel complex of 18-membered macrolides. II. Isolation and structure determination. J. Antibiot. 1987;40:575–588. doi: 10.7164/antibiotics.40.575. [DOI] [PubMed] [Google Scholar]
- 55.Revill P, Serradell N. Tiacumicin B. Drugs of the Future. 2006;31:494–497. [Google Scholar]
- 56.Johnson A. Drug evaluation: OPT-80, a narrow-spectrum macrocyclic antibiotic. Curr. Opin. Investig. Drugs. 2007;8:168–173. [PubMed] [Google Scholar]
- 57.Gerber M, Ackermann G. OPT-80, a macrocyclic antimicrobial agent for the treatment of Clostridium difficile infections: a review. Expert Opin. Investig. Drugs. 2008;17:547–553. doi: 10.1517/13543784.17.4.547. [DOI] [PubMed] [Google Scholar]
- 58.Gerber M, Ackermann G. OPT-80, a macrocyclic antimicrobial agent for the treatment of Clostridium difficile infections. Expert Opin. Investig. Drugs. 2008;17:547–553. doi: 10.1517/13543784.17.4.547. [DOI] [PubMed] [Google Scholar]
- 59.Miller M. Fidaxomicin (OPT-80) for the treatment of Clostridium difficile infection. Expert Opin. Pharmacother. 2010;11:1569–1578. doi: 10.1517/14656566.2010.485614. [DOI] [PubMed] [Google Scholar]
- 60.Poxton I. Fidaxomicin: a new macrocyclic, RNA polymerase-inhibiting antibiotic for the treatment of Clostridium difficile infections. Future Microbiol. 2010;5:539–548. doi: 10.2217/fmb.10.20. [DOI] [PubMed] [Google Scholar]
- 61.Sonenshein A, Alexander H, Rothstein D, S, F Lipiarmycin-resistant ribonucleic acid polymerase mutants of Bacillus subtilis. J. Bacteriol. 1977;132:73–79. doi: 10.1128/jb.132.1.73-79.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sonenshein A, Alexander H. Initiation of transcription in vitro inhibited by lipiarmycin. J. Mol. Biol. 1979;127:55–72. doi: 10.1016/0022-2836(79)90459-5. [DOI] [PubMed] [Google Scholar]
- 63.Swanson R, Hardy D, Shipkowitz N, Hanson C, Ramer N, Fernades P, Clement J. In vitro and in vivo evaluation of tiacumincins B and C against Clostrdium difficile. Antimicrob. Agents Chemother. 1991;35:1108–1111. doi: 10.1128/aac.35.6.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ackermann G, Löffler B, Adler D, Rodloff A. In vitro activity of OPT-80 against Clostridium difficile. Antimicrob. Agents Chemother. 2004;48:2280–2282. doi: 10.1128/AAC.48.6.2280-2282.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kurabachew M, Lu S, Krastel P, Schmitt E, Suresh B, Goh A, Knox J, Ma N, Jiricek J, Beer D, Cynamon M, Petersen F, Dartois V, Keller T, Dick T, Sambandamurthy V. Lipiarmycin targets RNA polymerase and has good activity against multidrug-resistant strains of Mycobacterium tuberculosis. J. Antimicrob. Chemother. 2008;62:713–719. doi: 10.1093/jac/dkn269. [DOI] [PubMed] [Google Scholar]
- 66.Gualtieri M, Tupin A, Brodolin K, Leonetti J. Frequency and characterisation of spontaneous lipiarmycin-resistant Enterococcus faecalis mutants selected in vitro. Int. J. Antimicrob. Agents. 2009;34:605–606. doi: 10.1016/j.ijantimicag.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 67.Gualtieri M, Villain-Guillot P, Latouche J, Leonetti J, Bastide L. Mutation in the Bacillus subtilis RNA polymerase beta' subunit confers resistance to lipiarmycin. Antimicrob. Agents Chemother. 2006;50:401–402. doi: 10.1128/AAC.50.1.401-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Louie T, Miller M, Mullane K, Weiss K, Lentnek A, Golan Y, Gorbach S, Sears P, Shue Y. Fidaxomicin versus vancomycin for Clostridium difficile infection. N. Engl. J. Med. 2011;364:422–431. doi: 10.1056/NEJMoa0910812. This paper reports results of a Phase III clinical trial of Lpm (fidaxomicin/Dificid) for treatment of C. difficile-associated.diarrhea.
- 69. Traynor K. Fidaxomicin approved for C. difficile infections. Am. J. Health. Syst. Pharm. 2011;68:1276. doi: 10.2146/news110046. This short note reports FDA approval of Lpm (fidaxomicin/Dificid) for treatment of C. difficile-associated.diarrhea
- 70. Xiao Y, Li S, Niu S, Ma L, Zhang G, Zhang H, Zhang G, Ju J, Zhang C. Characterization of tiacumicin B biosynthetic gene cluster affording diversified tiacumicin analogues and revealing a tailoring dihalogenase. J. Am. Chem. Soc. 2011;133:1092–1105. doi: 10.1021/ja109445q. This paper reports cloning and sequencing of the Lpm biosynthetic locus of D. aurantiacum, construction of gene-disruption mutants of the locus, and isolation of novel Lpm analogs by fermentation using gene-disruption mutants of the locus.
- 71.Hochlowski J, Jackson M, Rasmussen R, Buko A, Clement J, Whittern D, McAlpine J. Production of brominated tiacumicin derivatives. J. Antibiot. 1997;50:201–205. [PubMed] [Google Scholar]
- 72. Niu S, Hu T, Li S, Xiao Y, Ma L, Zhang G, Zhang H, Yang X, Ju J, Zhang C. Characterization of a sugar-O-methyltransferase TiaS5 affords new tiacumicin analogues with improved antibacterial properties and reveals substrate promiscuity. ChemBioChem. 2011 doi: 10.1002/cbic.201100129. (in press) This paper reports construction of an additional gene-disruption mutant of the Lpm biosynthetic locus of D. aurantiacum, and isolation of additional novel Lpm analogs, including analogs exhibiting higher potencies against S aureus and Enterococcus spp., by fermentation using the gene-disruption mutant
- 73.Kuhlman P, Duff H, Galant A. A fluorescence-based assay for multisubunit DNA-dependent RNA polymerases. Anal. Biochem. 2004;324:183–190. doi: 10.1016/j.ab.2003.08.038. [DOI] [PubMed] [Google Scholar]
- 74.Wallace A, Corkill J. Application of the spiral plating method to study antimicrobial action. J. Microbiol. Meths. 1989;10:3030–3310. [Google Scholar]
- 75.Paton J, Holt A, Bywater M. Measurement of MICs of antibacterial agents by spiral gradient endpoint compared with conventional dilution methods. Int. J. Exp. Clin. Chemother. 1990;3:31–38. [Google Scholar]
- 76.Schalkowsky S. Measures of susceptibility from a spiral gradient of drug concentrations. Adv. Exp. Med. Biol. 1994;349:107–120. doi: 10.1007/978-1-4757-9206-5_10. [DOI] [PubMed] [Google Scholar]
- 77.Clinical and Laboratory Standards Institute (CLSI/NCCLS) Methods for Antimicrobial Susceptibility Testing of Anaerobic Bacteria; Approved Standard, Seventh Edition. CLIS Document M11-A7. Wayne, PA: CLSI; 2009. [Google Scholar]
- 78.Collins L, Franzblau S. Microplate Alamar Blue assay versus BACTEC 460 system for high-throughput screening of compounds against Mycobacterium tuberculosis and Mycobacterium avium. Antimicrob. Agents Chemother. 1997;41:1004–1009. doi: 10.1128/aac.41.5.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Clinical and Laboratory Standards Institute (CLSI/NCCLS) Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard, Eighth Edition. CLIS Document M07-A8. Wayne PA: CLIS; 2009. [Google Scholar]