Abstract
Defense mechanisms against intracellular bacterial pathogens are incompletely understood. Our study characterizes a type I IFN-dependent cell-autonomous defense pathway directed against Legionella pneumophila, an intracellular model organism and frequent cause of pneumonia. We show that macrophages infected with L. pneumophila produced IFNβ in a STING- and IRF3-dependent manner. Paracrine type I IFNs stimulated up-regulation of IFN-stimulated genes and a cell-autonomous defense pathway acting on replicating and non-replicating Legionella within their specialized vacuole. Our infection experiments in mice lacking receptors for type I and/or II IFNs show that type I IFNs contribute to expression of IFN-stimulated genes and to bacterial clearance as well as resistance in L. pneumophila pneumonia in addition to type II IFN. Overall, our study shows that paracrine type I IFNs mediate defense against L. pneumophila, and demonstrates a protective role of type I IFNs in in vivo infections with intracellular bacteria.
Introduction
The innate immune system utilizes pattern recognition receptors (PRRs) for sensing conserved microbial molecules such as bacterial cell wall components or nucleic acids. PRR activation directly or indirectly stimulates antimicrobial responses that function either cell-autonomously within infected cells, depend on secretion of antimicrobial molecules, or are mediated by recruited leukocytes (Ishii et al., 2008;Opitz et al., 2010;Radtke and O'Riordan, 2006).
PRRs include the membrane-bound Toll-like receptors (TLR) as well as the cytosolic Nod-like receptors (NLR) and retinoic acid-induced gene I (RIG-I)-like receptors (RLR) (Ishii et al., 2008;Vance et al., 2009). In addition, the existence of a heterogeneous group of cytosolic DNA sensors is emerging (Ishii et al., 2006;Stetson and Medzhitov, 2006) that consist of absent in melanoma (AIM)2 (Burckstummer et al., 2009;Fernandes-Alnemri et al., 2009;Hornung et al., 2009;Roberts et al., 2009), DNA-dependent activator of IFN-regulatory factors (DAI)/Z-DNA-binding protein (ZBP)1 (Takaoka et al., 2007), RNA polymerase III/RIG-I (Ablasser et al., 2009;Chiu et al., 2009) and most likely other yet-to-be-identified molecules.
Some PRRs are capable of activating IRF transcription factors that regulate expression of type I interferons (IFNα/β). This response is induced by TLR3, 4, 7, and 9, as well as TLR2 in certain cell types, that engage the adapters TRIF and/or MyD88 (Barbalat et al., 2009;Takeuchi and Akira, 2010), by RIG-I signaling via MAVS (also known as IPS-1, Cardif and VISA) and STING (also known as MITA), and by MDA5 signaling through MAVS (Kawai et al., 2005;Meylan et al., 2005;Seth et al., 2005;Xu et al., 2005). Moreover, most cytosolic DNA sensors activate IFNα/β responses via STING (Ishikawa et al., 2009). IFNα/β bind to a common IFNα/β receptor (IFNAR) and induce the expression of hundreds of IFN-stimulated genes (ISGs), several of which have antiviral functions. Moreover, an increasing number of studies indicate that type I IFNs also regulate host responses to bacterial infections. Studies showed that type I IFNs are detrimental in infections with Listeria monocytogenes and Staphylococcus aureus (Auerbuch et al., 2004;Carrero et al., 2004;Martin et al., 2009;O'Connell et al., 2004), but beneficial in infections with different other extracellular bacteria (Mancuso et al., 2007).
The Gram-negative bacterium Legionella pneumophila is a frequent cause of the severe pneumonia Legionnaire`s disease, and a useful model for investigating intracellular innate immune mechanisms. After phagocytosis by macrophages, intracellular growth of L. pneumophila requires the Dot/Icm-encoded type IV secretion system (T4SS), which is used to inject bacterial effector molecules into the host cytosol that orchestrate the creation of a specialized replication vacuole (Isberg et al., 2009). In permissive cells, this Legionella-containing vacuole (LCV) escapes fusion with lysosomes and instead recruits secretory vesicles from the endoplasmic reticulum (ER), mitochondria and ribosomes. Innate immune defense to L. pneumophila infection depends on the TLRs 2, 5 and 9 (Archer et al., 2009;Bhan et al., 2008;Hawn et al., 2003;Hawn et al., 2007), the NLRs NAIP5 and NLRC4 (Molofsky et al., 2006;Zamboni et al., 2006), and on type II IFN (IFNγ) (Heath et al., 1996;Sporri et al., 2006). NAIP5 and NLRC4, for example, detect Legionella flagellin, leading to caspase-1-dependent pyroptosis as well as IL-1β and IL-18 secretion (Lightfield et al., 2008;Molofsky et al., 2006;Zamboni et al., 2006), and to enforced trafficking of the LCV towards the endocytic pathway (Amer et al., 2006;Fortier et al., 2007). IFNγ, which is mainly produced by natural killer (NK) cells, activates macrophages through the IFNγ receptor (IFNGR) leading to restriction of intracellular Legionella growth (Heath et al., 1996;Sporri et al., 2006).
Besides, we and others previously showed that host cells infected with L. pneumophila produced type I IFNs (Chiu et al., 2009;Lippmann et al., 2008;Monroe et al., 2009;Opitz et al., 2006;Stetson and Medzhitov, 2006). In the present study, we further examined this signalling pathway and demonstrate that the type I IFN production activates an intracellular defense pathway that contributes to bacterial restriction in vitro and in vivo.
Results
L. pneumophila-infected cells produced IFNβ depending on the bacterial T4SS, bacterial uptake, recognition of bacterial DNA, and on the host cell molecules STING and IRF3
First we examined the expression of IFNβ in Legionella-infected macrophages. L. pneumophila infection led to robust expression of IFNβ mRNA (Fig. 1A). A bacterial mutant defective in the dotA gene, which encodes an essential component of the T4SS, and thus does not translocate any Dot/Icm substrates, was unable to activate IFNβ expression. An icmS mutant, which has a functional T4SS but fails to translocate a subset of bacterial effector proteins, was capable of inducing type I IFN expression. Flagellin-negative and replication-deficient thymidine auxotroph mutants induced strong IFNβ responses.
Figure 1. Role of bacterial and host cell factors in stimulation of IFNβ expression in L. pneumophila-infected cells.
(A) B6 BMMs were infected with L. pneumophila wt, mutants deficient for flaA, dotA and icmS or the thy mutant at a MOI of 25 for indicated time intervals. (B) B6 BMMs were treated with either bafilomycin A (Baf), chymostatin (Chy) or cytochalasin D (Cyt) or were left untreated and stimulated with viableor heat-killed (HK) L.p. wt for 4 hrs. (C) BMMs were transfected for 4 hrs with bacterial extracts that were either left untreated or were incubated with DNase I, RNase A, RNase H, DNase and RNases (D+R) or Proteinase K. (D, E) Wt BMMs and respective knockout BMMs were infected with L. pneumophila wt and/or ΔflaA (flaA) at a MOI of 25 for 4 hrs. (F-H) Wt or MAVS-/- BMMs were transfected with control siRNA (C) or siRNA against STING or DAI. After 48 hrs cells were infected with L. pneumophila for 4 hrs. (I) BMMs from wt and MAVS-/- mice were transfected with control siRNA (C) or siRNA against STING. After 48 hrs, cells were additionally transfected with Legionella DNA. Relative expression of IFNβ mRNA (A-E, H, I), STING (F) and DAI (G) was determined by quantitative RT-PCR. IFNβ mRNA values of untreated cells were set as 1. Data shown are representatives of at least three independent experiments carried out in triplicates. Unless indicated, statistics refer to the wt at the respective time point (A), to the untreated control (B) or to the untreated, transfected control (C) (*p<0.05; **p<0.01; ***p<0.001; relative to # no bacteria detected).
IFNβ expression was dependent on uptake of viable bacteria, as treatment of cells prior to infection with cytochalasin D or heat-inactivation of bacteria blocked IFNβ production (Fig. 1B). Bafilomycin A and chymostatin, substances that interfere with endosomal acidification or lysosomal proteinases, respectively, led to reduced but not blunted IFNβ expression. Recent studies have suggested that sensing of bacterial DNA and/or RNA by cytosolic PRRs activate the pathway leading to type I IFN production in L. pneumophila-infected cells (Chiu et al., 2009;Monroe et al., 2009;Stetson and Medzhitov, 2006), although experimental proofs for both hypotheses are incomplete, and many of the molecular mechanisms underlying this pathway remain unclear. We found that treatment of bacterial extracts with DNase I, but not with RNase A, RNase H or Proteinase K, reduced their capability to induce IFNβ expression when delivered into BMMs (Fig. 1C and Fig. S1). Moreover, transfection of Legionella DNA into BMMs led to IFNβ production (see below, Fig. 1I, Fig. S2I).
We next investigated which host cell proteins were involved in IFNβ production in L. pneumophila-infected cells. In accordance with earlier studies (Opitz et al., 2006;Stetson and Medzhitov, 2006), we found that IFNβ expression in L. pneumophila-infected BMMs depended on transcription factor IRF3 (Fig. 1D). The type I IFN response was not inhibited by a lack of TLRs 2, 3, 4, 7, and 9 or NOD2 expression (Fig. S2A, Fig. S2B). We did not observe a contribution of MAVS to the IFNβ response in BMMs infected with different strains of L. pneumophila wt, L. pneumophila thy or ΔflaA at different MOIs, time intervals and infection conditions, such as bacterial growth phases (Fig. 1E, Fig. S2C-G), whereas intracellular delivery of the MDA5 ligand polyI:C induced a MAVS-dependent IFNβ response (Fig. S2H). Moreover, BMMs deficient for RIG-I or MDA5 were capable of producing IFNβ after Legionella infection (data not shown). In contrast, knock-down of STING partially reduced the type I IFN response in Legionella-infected cells (Fig. 1F, H). siRNA-mediated knock-down of DAI did not decrease IFNβ response upon confrontation with the bacteria (Fig. 1G, H). Moreover, concomitant knock-out/knock-down of STING and MAVS or of DAI and MAVS did not lead to additional effects on the Legionella-activated IFNβ response as compared to the single knock-outs/knock-downs alone (Fig. 2H). Moreover, STING, but not MAVS, was involved in the IFNβ response induced by intracellular transfection of L. pneumophila DNA or poly(dA-dT) into BMMs (Fig. 1I, Fig. S2I-K). Collectively, our data suggest that the IFNβ response to L. pneumophila depends on the uptake of viable bacteria, phagosomal function, bacterial T4SS, detection of microbial DNA, and the host cell molecules STING as well as IRF3.
Figure 2. Type I IFNs activate a cell-autonomous defense independently of NAIP5/NLRC4-mediated recognition of flagellin and of IFNγ.
(A-C) Intracellular growth of L.p. wt or ΔflaA in BMMs from wt (B6) and IRF3-/- (A), and IFNAR-/- (B, C) mice was monitored over 72 hrs by determining CFU per well at the indicated time points. BMMs were infected with L. pneumophila at a MOI of 0.1. Where indicated, cells were treated with 5 or 50 U/ml IFNβ 18 hrs prior and during the course of infection or were left untreated. Data shown are representatives of at least three independent experiments carried out in triplicates. (*p<0.05, **p<0.01, ***p<0.001, no indication if not significant)
Type I IFNs activate a cell-autonomous resistance pathway against L. pneumophila that is independent of flagellin recognition by NAIP5/NLRC4 and of IFNγ
Next, we examined the effect of type I IFNs on L. pneumophila growth. We found that growth of wt L. pneumophila was rapidly terminated in C57BL/6 (B6) BMMs, whereas these bacteria showed substantial replication in B6 IRF3-/- BMMs (Fig. 2A). Flagellin-deficient L. pneumophila, which evades NAIP5/NLRC4-mediated cell-autonomous resistance, replicated intracellularly to high numbers in wt BMMs, and their replication was further enhanced in IRF3-/- BMMs. In contrast, intracellular replication of wt and ΔflaA were identical in B6 wt and MAVS-deficient cells (Fig. S3A). BMMs lacking IFNAR were permissive for wt L. pneumophila, and replication of the flagellin-deficient mutant was enhanced as compared to their replication in B6 wt BMMs (Fig. 2B). Replication of ΔflaA was dose-dependently inhibited by exogenous IFNβ in B6 wt, but not IFNAR-/- cells (Fig. 2C). We did not observe enhanced bacterial growth in BMMs lacking IFNGR (Fig. S3B), and IFNβ treatment inhibited L. pneumophila replication in IFNGR-/- BMMs as effectively as in B6 wt BMMs (Fig. S3C). These data indicate that IRF3-regulated production of type I IFNs as well as exogenous IFNβ treatment activate an intracellular resistance pathway against L. pneumophila that is dependent on IFNAR but independent of flagellin recognition by NAIP5/NLRC4 and of IFNγ.
The type I IFN-mediated resistance pathway inhibits L. pneumophila in their replication vacuoles after establishment of the specialized vacuole
For further elucidation of the mechanism underlying the type I IFN-mediated cell-autonomous resistance pathway against L. pneumophila, we quantified bacteria by counting them in single vacuoles. We found higher numbers of L. pneumophila Δfla per replication vacuole at 10 hrs p.i. as compared to 4 hrs p.i., whereas wt bacteria did not replicate in their vacuoles in B6 wt macrophages (Fig. 3A, B). Importantly, L. pneumophila wt and ΔflaA vacuole loads were increased in IFNAR-/- as compared to B6 wt BMMs. IFNβ treatment of wt BMMs significantly affected numbers of L. pneumophila ΔflaA per replication vacuole. Next, we asked whether the effect of type I IFNs on bacterial numbers in replication vacuoles was due to a failure to remodel the LCV into an ER-derived organelle or enforced fusion of the LCV with lysosomes. We thus isolated LCVs from B6 wt and IFNAR-/- BMMs treated or not treated with IFNβ and assessed recruitment of the ER marker Calnexin or the endolysosomal marker Lamp-1. As expected, most Legionella ΔdotA vacuoles were Calnexin-negative, but stained positive for Lamp-1, whereas vacuoles containing wt or ΔflaA bacteria were mostly positive for Calnexin and negative for Lamp-1 (Fig. 3C, D). We did found, however, no effect of type I IFNs on either ER recruitment or lysosomal fusion to the LCV. Moreover, we did not find any evidence for involvement of caspase-1-dependent cell death (pyroptosis), apoptosis, autophagy, reactive nitrogen or oxygen species, iron deprivation and tryptophan depletion (e.g. by indolamine 2,3-dioxygenase (IDO)) in type I IFN-mediated bacterial restriction (Fig. S4 and data not shown). Overall, the type I IFN-mediated intracellular defense appears to influence bacterial numbers in their vacuoles without influencing vacuole formation itself or involvement of several antimicrobial mechanisms known to affect intracellular bacteria.
Figure 3. Type I IFN affects bacterial numbers after establishment of the specialized Legionella-containing vacuole.
(A,B) BMMs from B6 wt or IFNAR-/- mice were infected with L.p. wt or ΔflaA for 4 and 10 hrs and treated with IFNβ where indicated. (A) The percentage of vacuoles containing 1, 2-5 or more than 6 bacteria was determined from three individually performed experiments carried out in duplicates (*p<0.05, **p<0.01, no indication if not significant). (B) Representative images of Z-stack projections of macrophages infected with L.p. ΔflaA for 10 hrs are depicted. Zoom demonstrates individual vacuoles containing 1, 2-5 or more than 6 bacteria that were counted and classified. Scale bars correspond to 10 μm. (C, D) LCVs were isolated from untreated or IFNβ-stimulated wt (B6) or IFNAR-/- BMMs that were infected with GFP-expressing L.p. wt, ΔflaA or ΔdotA. LCVs were stained with antibodies against Calnexin or Lamp-1, respectively, and visualized by fluorescence microscopy. Results shown in (C) comprise data from three (Calnexin) and two (Lamp-1) individually performed experiments. Scale bars correspond to 5 μm (D).
The type I IFN-dependent cell-autonomous resistance against L. pneumophila acts on replicating and non-replicating bacteria, and is partly dependent on Irgm1
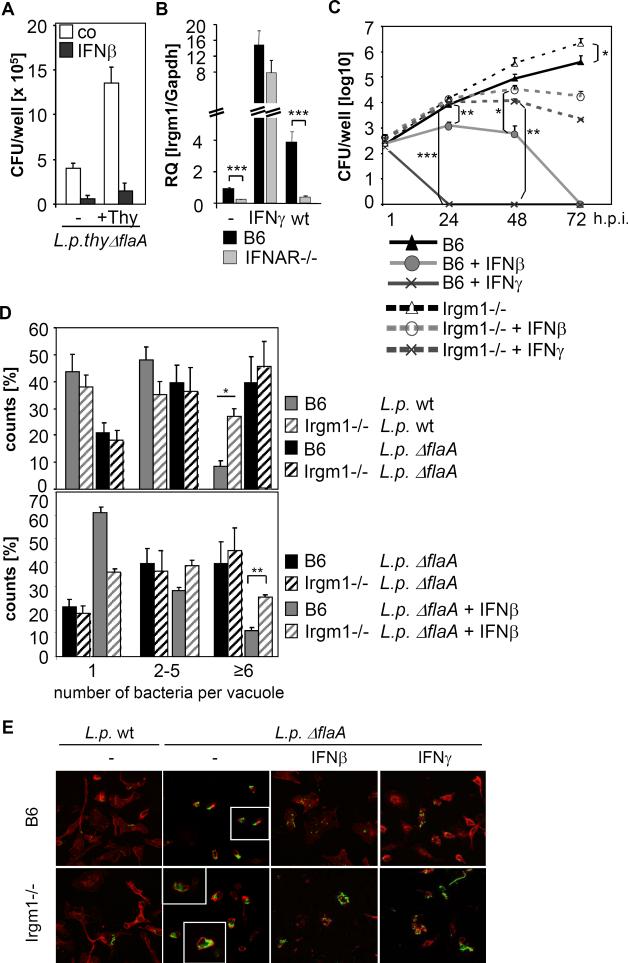
Given that intracellular L. pneumophila replication was inhibited by type I IFNs, we next investigated whether type I IFNs affected bacterial viability by examining recovery of non-replicating bacteria. Reduced numbers of the non-replicating thy mutant were recovered from IFNβ-treated BMMs compared to untreated cells (Fig. 4A), suggesting that type I IFN-mediated resistance inhibits non-replicating as well as replicating L. pneumophila. Consequently, we examined expression of various potentially antimicrobial ISGs in B6 wt and IFNAR-/- BMMs infected with L. pneumophila. mRNA of the immunity-related GTPases (IRGs) Irgm1, Irgm3, and Irgd, but not of Irga6 were IFNAR-dependently up-regulated upon Legionella infection (Fig. 4B and data not shown). Moreover, we found IFNAR-dependent up-regulation of the ubiquitin-like protein ISG15, which is known to mediate antiviral defence (data not shown). Since Irgm1 is known to mediate IFNγ-induced resistance to a broad range intracellular pathogens by targeting and disrupting pathogen-containing vacuoles (Howard, 2008), we performed replication assays in BMMs lacking Irgm1. As depicted in Fig. 4C, inhibition of bacterial replication by IFNβ (and IFNγ, which was used as a control) was strongly impaired in Irgm1-/- BMMs. Moreover, replication of L. pneumophila ΔflaA was slightly enhanced (approximately four-fold) in untreated Irgm1-deficient cells 72 hrs p.i. compared to wt BMMs. We did not, however, observe a clear-cut effect of Irgm1 on the replication of wt bacteria over a time period of 72 hrs (data not shown). When examining L. pneumophila infection after 10 hrs, we found more wt and ΔflaA L. pneumophila residing in each vacuole (Fig. 4D) and cell (Fig. 4E) in Irgm1-/- compared to wt BMMs. IFNβ treatment was less effective in reducing bacterial numbers in Irgm1-/- BMMs (Fig. 4E). Collectively, these data suggest that the type I IFN-mediated cell-autonomous defense against L. pneumophila is partly dependent on the antimicrobial effects of Irgm1 and most likely other ISGs.
Figure 4. The type I IFN-dependent resistance pathway inhibits replicating and non-replicating L. pneumophila, and is partly dependent on Irgm1.
(A) BMMs were treated with 100 U/ml IFNβ as indicated, infected with L.p. thy ΔflaA in the presence or absence of supplemented thymidin, and intracellular bacterial numbers were determined 10 hrs p.i.. Data shown are representative of three independent experiments carried out in triplicates. (B) B6 and IFNAR-/- BMMs were stimulated with IFNγ or infected with L.p. wt for 24 hrs. Relative Irgm1 mRNA levels were quantified by Q-PCR. (C) BMMs from wt and Irgm1-/- mice were treated with IFNβ or IFNγ prior and during infection as indicated, and infected with L.p. ΔflaA. Bacterial replication was monitored over 72 hrs by determining CFU per well at the indicated time points. Data shown are representatives of four independent experiments carried out in triplicates. (D) BMMs from B6 and Irgm1-/- mice were treated with IFNβ as indicated prior and during infection, infected with L.p. wt or ΔflaA for 10 hrs, and the percentage of vacuoles containing 1, 2-5 or more than 6 bacteria was determined. Results shown comprise data from at least three individually performed experiments carried out in duplicates. (E) BMMs from B6 and Irgm1-/- mice were treated with IFNβ or IFNγ as indicated, and infected with L.p. wt or ΔflaA for 10 hrs. Bacteria were stained with an anti-Legionella-LPS antibody (green) and actin with phalloidin (red), visualization was done by fluorescence microscopy with an 63x objective. Inlets depict zoom of individual vacuoles. (*p<0.05, **p<0.01, ***p<0.001, no indication if not significant)
Expression of ISGs in L. pneumophila infection in vivo
Next, we investigated the expression of IFNβ as well as of the ISGs Irgm1, Irga6 and ISG15 in L. pneumophila lung infection. Wt, IFNAR-/-, IFNGR-/- and IFNAR/IFNGR-/- mice were infected with bacteria lacking flagellin in order to circumvent NAIP5/NLRC4-mediated resistance. Expressions of IFNβ and Irgm1, Irga6 and ISG15 were up-regulated in L. pneumophila Δfla-infected lungs (Fig. 5-D). Expression of Irgm1 in lungs of L. pneumophila-infected mice was dependent on both IFNGR and IFNAR (Fig. 5B). In contrast, Irga6 expression was solely dependent the type II IFN, and ISG15 mRNA levels were dependent on type I IFNs only (Fig. 5C, D). Thus, both type I and type II IFNs cooperatively affect expression of molecules that may fulfil antibacterial tasks.
Figure 5. Expression of IFNβ and ISGs in L. pneumophila lung infection.
Wt, IFNAR-/-, IFNGR-/- and IFNAR/IFNGR-/- mice were intranasally infected with L.p. ΔflaA (1×106 bacteria/mouse) or received PBS instillation respectively. Mice were sacrificed on day 2, 4 or 6 post infection or 2 days after PBS instillation, and IFNβ (A), Irgm1 (B), Irga6 (C) and ISG15 (D) mRNA expression levels in lungs of infected mice were determined by quantitative RT-PCR relative to PBS-treated mice that were set as 1 (not shown) (n=4). (*p<0.05, **p<0.01, ***p<0.001, no indication if not significant)
Role of type I IFNs in L. pneumophila infection in vivo
Finally, we investigated the role of type I IFN alone and in cooperation with type II IFN in L. pneumophila lung infection. In wt and IFNAR-/- mice, pulmonary bacterial burden decreased from day 2 till 6 p.i. (Fig. 6A), and no systemic bacterial dissemination into the spleen was observed (Fig. 6B). IFNGR-/- mice displayed a higher pulmonary bacterial burden compared to wt mice and in one out of four mice bacteria were recovered from the spleen. Notably, bacterial burden in lung and spleen of mice deficient in both IFNAR and IFNGR were further enhanced compared to IFNGR-/- mice at day 6 p.i. IFNAR/IFNGR-/- mice were unable to suppress replication and systemic dissemination of L. pneumophila Δfla. A significant weight loss on day 6, displaying a still active course of the disease, was measured in IFNAR/IFNGR-/- mice exclusively (Fig. 6C). In the lungs of wt, IFNAR-/- and IFNGR-/- mice, infection with L. pneumophila led to the recruitment of neutrophils until day 2 p.i. In the following neutrophils declined from day 2 p.i. until day 6 p.i. almost reaching basal levels. In contrast the percentage of neutrophil in lungs of IFNAR/IFNGR-/- mice further increased over the time period examined (Fig. 6D). Pulmonary neutrophil recruitment was paralleled by an elevated fraction of neutrophils in the circulation of mice (Fig. 6E). In summary, both type I and II IFNs have protective roles in L. pneumophila lung infections.
Figure 6. Protective role of type I IFNs in L. pneumophila lung infection.
Wt, IFNAR-/-, IFNGR-/- and IFNAR/IFNGR-/- mice were intranasally infected with L.p. ΔflaA (1×106 bacteria/mouse) or received PBS instillation, respectively. Mice were sacrificed on day 2, 4 or 6 post infection or 2 days after PBS instillation, and bacterial loads in the lung (A) and spleen (B) were quantified. Gain/Loss of bodyweight post infection was determined at day 6 (C). Neutrophils in the lung (D) and in the blood (E) were determined at day 2, 4 and 6 post infection and 2 days after PBS instillation. (n=4; *p<0,05; # no bacteria detected).
Discussion
Here we characterized a defense mechanism against L. pneumophila that involves cytosolic recognition of the bacteria, autocrine production of type I IFNs, up-regulation of antimicrobial ISGs, and inhibition of the bacteria within their specialized vacuoles (Fig. 7). The type I IFN-mediated defense contributes to antibacterial clearance in L. pneumophila pneumonia in addition to type II IFN.
Figure 7.
Overview of the type I IFN-mediated cell-autonomous resistance pathway that controls L. pneumophila infection together with NAIP5/NLRC4 in vitro and together with NAIP5/NLRC4 and the type II IFN IFNγ in vivo.
Our results showing IFNβ production after transfection of Legionella DNA, and reduced IFNβ-stimulatory activities of bacterial extracts after DNase treatment are in agreement with recent studies that suggested that DNA is the major bacterial ligand that triggers the type I IFN response to different bacterial infections (Charrel-Dennis et al., 2008;Chiu et al., 2009;Leber et al., 2008;Stetson and Medzhitov, 2006). During infection, Legionella DNA may be translocated via the Dot/Icm-encoded T4SS into the host cell cytosol, as has been previously proposed (Segal et al., 1998;Vogel et al., 1998). This leakage of bacterial DNA appears to be intact in the icmS mutant, which has a functional T4SS but fails to translocate a subset of bacterial effector proteins. In fact, macrophages infected with Δicms bacteria showed an slightly enhanced I IFN production, which might be related to a defect in translocating antagonistic effector proteins or to the rapid fusion of Δicms bacteria-containing vacuoles with lysosomes (Coers et al., 2000) possibly leading to enhanced release of bacterial nucleic acids. Similar to one study examining L. pneumophila infection (Monroe et al., 2009), we also observed IFNβ production after transfection of Legionella RNA (unpublished data). Our experiments using different nucleases, however, suggested that bacterial DNA rather than RNA may be the major bacterial component involved in stimulating IFNβ production in response to L. pneumophila, although sensing of additional Legionella molecules such as cyclic diGMP might also play a role (Sauer et al., 2011).
We observe that L. pneumophila-infected cells produced IFNβ in a partially STING- and IRF3-dependent manner, but independent of MAVS. Moreover, concomitant knockout/knock-down of MAVS and DAI did not impair L. pneumophila-induced IFNβ production. These results, along with recent studies (Chiu et al., 2009;Wang et al., 2008), indicate the existence of at least three redundant cytosolic DNA-sensing pathways comprising of (i) DAI, (ii) RNA-polymerase III, RIG-I and MAVS, and (iii) one or more pathway(s) yet-to-be-identified. Postulation of the latter explains IFNβ induction in L. pneumophila-infected BMMs, which appears to involve the adapter molecule STING. STING, however, might also be involved in sensing of additional bacterial molecules such as cyclic dinucleotides, which are also known to trigger type I IFN responses (Sauer et al., 2011). Our data oppose own and other previous studies performed on human lung epithelial cells and murine macrophages that indicated a contribution of the MAVS-dependent pathway in IFNβ responses to L. pneumophila infection (Chiu et al., 2009;Monroe et al., 2009;Opitz et al., 2006). Respective differences of human and murine cells might underlie differential expression/function of DNA-sensing proteins in diverse cells as has been indicated before (Chiu et al., 2009;Wang et al., 2008). It appears more difficult to explain the discrepancies between our and previous studies in respect to MAVS involvement in mouse BMMs. As we have tested all different infection conditions (MOI, time interval, bacterial growth phase) as well as bacterial strains used in the previous papers (Chiu et al., 2009;Monroe et al., 2009), differences in these parameters do unlikely account for the different results. We think that varying conditions of BMM culture or different sources of the knockout mice (mice from Chen group vs. mice from Tschopp group) might lead to partly opposing results.
Here we show that resistance against L. pneumophila infection in macrophages in vitro relies on flagellin recognition by NAIP5/NLRC4 and on the type I IFNs which is in line with own and others previous findings (Coers et al., 2007;Opitz et al., 2006;Plumlee et al., 2009;Schiavoni et al., 2004). Whereas activity of both pathways efficiently suppresses bacterial replication, functional defects in either pathway allow for substantial growth of L. pneumophila. Uncontrolled bacterial multiplication was observed in IRF3- or IFNAR-deficient cells infected with a L. pneumophila ΔflaA mutant that evades NAIP5/NLRC4. Our results further indicate that the type I IFN-mediated resistance pathway affects bacterial numbers in replication vacuoles by activating bacterial killing rather than affecting the replication of L. pneumophila. This pathway most likely involves Irgm1 and other genes that are type I IFN-dependently up-regulated in L. pneumophila-infected cells. We speculate that Irgm1, together with other IRG proteins, might inhibit Legionella by deforming and/or disrupting their vacuole, as it has been indicated for other intracellular pathogens (Howard, 2008). Considering that type I IFNs are produced after infection with basically all intracellular bacteria tested so far, and that some of the up-regulated ISGs possess antimicrobial activity against a broad range of bacteria, it is reasonable to assume that a similar type I IFN-mediated intracellular defense pathway might also be functional in some other bacterial infections (Buss et al., 2010).
Intracellular host defense to L. pneumophila infection in vivo has been known to rely on type II IFN and NAIP5/NLRC4 (Heath et al., 1996;Molofsky et al., 2006;Sporri et al., 2006;Zamboni et al., 2006). Our experiments indicate that host innate immunity additionally involves type I IFNs for bacterial clearance and control of disease. Type I IFNs have been associated with antiviral immunity for several decades. Only recently have they been additionally attributed to bacterial infections. In infections with bacteria, they appear to positively or negatively regulate different host defense pathways including cell death (Carrero et al., 2004;O'Connell et al., 2004;Stockinger et al., 2002), inflammasome activation (Henry et al., 2007), chemokine production and leukocyte recruitment (Shahangian et al., 2009;Watanabe et al., 2010), IL-17A secretion by γδ T cells (Henry et al., 2010), and type II IFN signalling (Rayamajhi et al., 2010). The individual role of each of the type I IFN-regulated cell-autonomous and non-cell-autonomous immune mechanisms might determine whether the overall function of the type I IFNs is detrimental or beneficial for the host in the bacterial infection examined (Auerbuch et al., 2004;Mancuso et al., 2007;Martin et al., 2009;O'Connell et al., 2004).
In conclusion, this study demonstrates a protective role of type I IFNs in infections with intracellular bacteria.
Experimental procedures
Ethics Statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Committee of animal welfare commissioners in Berlin, Germany. The protocol was approved by the Committee on the Ethics of Animal Experiments of the Charité-Universitätsmedizin Berlin and the governmental institutions (Permit Number: G0446/08). All efforts were made to minimize suffering.
Bacterial strains and reagents
L. pneumophila serogroup 1 strains (Lp01), the isogenic mutant strains (Lp01 ΔflaA, ΔdotA and ΔicmS), or Lp02, a thymidine auxotroph Lp01 derivative (thy), with or without 10 μg/ml thymidine, were used for in vitro infections. Mice were infected with JR32 ΔflaA. Bacteria were cultured on buffered charcoal-yeast extract (BCYE) agar for 2 days at 37°C before use. GFP-expressing bacteria were selected with 6.25 μg/ml chloramphenicol and GFP expression was induced by 0.1-0.2 mM IPTG. Bacterial extracts were prepared as described previously (Charrel-Dennis et al., 2008;Stetson and Medzhitov, 2006) and treated either with DNase I (100 U/ml, Sigma), RNase A (100 μg/ml, Sigma), RNaseH (100 μg/ml, Ambion) for 45 min at 37°C or Proteinase K (108 μAU/ml, Calbiochem) for 45 min at 54°C, or left untreated. DNA and RNA from L. pneumophila were purified using the DNA preparation kit (Qiagen) or RNA isolation kit (Macherey&Nagel), respectively.
Bone marrow-derived macrophages
Bone marrow-derived macrophages (BMMs) were prepared from femurs and tibiae of wild-type, Cardif-/- (Michallet et al., 2008) (MAVS-/-), IRF3-/- (Sato et al., 2000), NOD2-/-(Kobayashi et al., 2005), TLR2,3,4,7,9-/- (Conrad et al., 2009), IFNAR-/- (Stetson and Medzhitov, 2006), IFNGR-/- (Muller et al., 1994), Irgm1-/- (Collazo et al., 2001) that were all on a C57BL/6 background (B6).
Macrophage transfection, infection and stimulation
BMMs were transfected with siRNA (Ambion) targeting STING (pool of sequence a and b; a: sense GGAUCCGAAUGUUCAAUCAtt, antisense UGAUUGAACAUUCGGAUCCgg and b: sense GGUCCUCUAUAAGUCCCUAtt, antisense UAGGGACUUAUAGAGGACCag) or DAI (sense ggagcucaguacaucuacatt, antisense uguagauguacugagcuccgt) using HiPerfect (Qiagen). BMMs were infected with L. pneumophila wt or ΔflaA at indicated MOIs, spun at 200 × g for 5 min and incubated for indicated time intervals. Where indicated, cells were incubated either with bafilomycin A (200 nM), chymostatin (100 μM) or cytochalasin D (2 μM) 30 min prior and during infection. Bacterial or synthetic nucleic acids and bacterial lysates were transfected into cells using Lipofectamine 2000 (Invitrogen) and incubated for 4 hrs at 37°C. For intracellular replication assays, BMMs were infected with L. pneumophila at a MOI of 0.1 or 10 as stated, centrifuged at 200 × g for 5 min and incubated at 37°C for 30 min. Cells were washed with PBS and were further incubated with RPMI containing 50 μg/ml gentamicin for 1 h in order to kill extracellular bacteria. Subsequently, cells were washed with PBS and incubated in BMM culture medium for different time intervals and/or were lysed with sterile water. Serial dilutions were plated on BCYE agar for 3 d at 37°C to determine bacterial CFU.
In vivo infection
Wildtype B6 mice were purchased from Jackson Laboratories. WT 129Sv, IFNAR-/- 129Sv (Muller et al., 1994), and IFNGR-/- B6 mice (Muller et al., 1994) were provided by Uwe Klemm (MPI-IB, Berlin, Germany). IFNAR/IFNGR-/- double-knockout mice were generated by breeding IFNAR-/- with IFNGR-/- and the genotypes were determined by PCR from DNA prepared from tail biopsies. All procedures were approved by institutional and governmental authorities. Although the mice strains used were on different backgrounds, we found that bacterial growth, course of bodyweight and pulmonary neutrophil recruitment were not different between B6 and 129Sv infected with ΔflaA (Fig. S5). Anaesthetized mice were intranasally infected with 1×106 ΔflaA in 40μl PBS (Archer et al., 2009). Control mice received 40μl PBS intranasally and were sacrificed two days after instillation. On day 2, 4 or 6 p.i., mice were anaesthetized, tracheotomized, ventilated, and blood was drawn from the vena cava inferior. After exsanguination the lung was flushed with sterile 0.9% NaCl via the pulmonary artery before lung and spleen were removed. Organs were homogenized using a cell-strainer (100μM, BD Bioscience). For determination of bacterial counts, the homogenates were lysed with 0.2% Triton-X100 and serial dilutions were plated on BCYE agar plates. mRNA from lung homogenates was prepared as described below. Pulmonary leukocytes were differentiated manually by light microscopy of May-Grünwald-Giemsa-stained cytospin preparations from lung homogenate prior to lysis. Leukocytes in the blood were analysed by flow cytometry (FACScalibur, BD Bioscience) according to their side-scatter/forward-scatter properties and CD45 and Gr-1 expression (BD Bioscience).
Immunofluorescence assay
To count replicating bacteria inside replication vacuoles (RV), BMMs were seeded onto glass coverslips and infected at a MOI of 20. At 4 and 10 hrs p.i., cells were washed with PBS and fixed with 3% PFA in PBS. Cells were permeabilized with 1% Triton X-100 in PBS/BSA and blocked with 5% goat serum followed by exposure to a Legionella-specific antibody and subsequent incubation with an Alexa Fluor 488-conjugated goat anti-mouse IgG. F-actin was stained with phalloidin Alexa Fluor 546. Coverslips were mounted on slides using PermaFluor and examined under a LSM5 Pascal fluorescence microscope or an inverted spinning disc confocal microscope (Zeiss A1m) with an Andor EMCCD iXon+DV885 camera. Z-stacks were taken with a 63x objective in 200 nm plane distance. Images were processed by Z-stack projection using ImageJ.
LCV isolation assay
For isolation of LCV, BMMs were seeded and infected with GFP-encoding L. pneumophila at a MOI of 20. At indicated time points cells were scraped in cold PBS, spun for 5 min at 200 × g and cell pellets were resuspended in protease inhibitor cocktail-containing buffer (20 mM HEPES/KOH, pH 7.2; 250 mM sucrose, 0.5 mM EGTA, pH 8) and lysed in a ball bearing homogenizer with 15-20 passages. After centrifugation at 1500 rpm for 3 min, post-nuclear supernatants (PNS) were diluted in homogenization buffer and subsequently spun onto 0.01% poly-L-lysine coated coverslips at 150 × g for 5 min at 4°C. PNS were fixed with PLP-sucrose fixative reagent (75 mM lysine, 30 mM NaPO4 pH 7.4, 3.4% sucrose, 2.5 mg/ml Naperiodate, 4% PFA) and subsequently washed with PBS and stained for Lamp-1 or Calnexin in 0.1% saponine/2% BSA.
RNA preparation and quantitative RT-PCR
Total RNA was isolated from BMMs or lung homogenates using RNeasy Mini kit (Qiagen) and reverse-transcribed using high capacity reverse transcription kit (Applied Biosystems). Quantitative PCR (Q-PCR) was performed on an ABI 7300 instrument using TaqMan gene expression assays (Applied Biosystems). The input was normalized to the average expression of GAPDH and relative expression of the respective gene in untreated cells or PBS-treated mice was set as 1.
Immunoblotting
For immunoblotting cells were lysed in SDS- and 1% NP40-containing lysis buffer, cleared extracts were separated by SDS-PAGE and SDS-gels were blotted onto Hybond nitrocellulose membranes. Antibodies against LC3 (nanoTools) and actin were used.
Statistics
Results were statistically evaluated using Student's t-test or Whitney U-test and data are expressed as means +/- SD. p values < 0.001 are indicated by three asterisks (***), p values < 0.01 by two asterisks (**), p values < 0.05 by one asterisk (*).
Supplementary Material
Acknowledgements
We are grateful to J. Tschopp, University of Lausanne, for providing Cardif-/- mice, to R. Flavell for permission to use Nod2-/- mice, and A. Dorhoi for providing Nod2-/- mice. For invaluable support with image acquisition as well as helpful discussions we would like to thank Jost Enninga, Institute Pasteur. We would like to thank Kristy Archer, Eric D. Cambronne, Natalia Tschowri, Julia Polansky, Ivo Gomperts Boneca and Jean-Marc Ghigo for helpful discussions and invaluable advice. We thank B. Gutbier, C. Nogueira, E. Campodonico, C. Case, C. Thöne-Reineke, U. Klemm and R. Vance for technical help and stimulating discussions. We thank D. Stoll and J. Hellwig for excellent technical assistance. Parts of this work will be included in the PhD thesis of J.L. This work was supported in part by the International Max Planck Research School for Infectious Diseases and Immunology to J.L., by a NIH grant AI57831 to G.T., by a VA Merit Review grant to G.T., by a Jürgen Manchot Stiftung grant to B.O. and by Deutsche Forschungsgemeinschaft grants OP 86/5-1 to B.O., SFB/TR84 project A1 to S.B. and B.O..
References
- Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amer A, Franchi L, Kanneganti TD, Body-Malapel M, Ozoren N, Brady G, Meshinchi S, Jagirdar R, Gewirtz A, Akira S, Nunez G. Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J Biol Chem. 2006 17 11;281:35217–35223. doi: 10.1074/jbc.M604933200. [DOI] [PubMed] [Google Scholar]
- Archer KA, Alexopoulou L, Flavell RA, Roy CR. Multiple MyD88-dependent responses contribute to pulmonary clearance of Legionella pneumophila. Cell Microbiol. 2009;11:21–36. doi: 10.1111/j.1462-5822.2008.01234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbuch V, Brockstedt DG, Meyer-Morse N, O'Riordan M, Portnoy DA. Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. J Exp Med. 2004 16 8;200:527–533. doi: 10.1084/jem.20040976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbalat R, Lau L, Locksley RM, Barton GM. Toll-like receptor 2 on inflammatory monocytes induces type I interferon in response to viral but not bacterial ligands. Nat Immunol. 2009;10:1200–1207. doi: 10.1038/ni.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhan U, Trujillo G, Lyn-Kew K, Newstead MW, Zeng X, Hogaboam CM, Krieg AM, Standiford TJ. Toll-like receptor 9 regulates the lung macrophage phenotype and host immunity in murine pneumonia caused by Legionella pneumophila. Infect Immun. 2008;76:2895–2904. doi: 10.1128/IAI.01489-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, Superti-Furga G. An orthogonal proteomic genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- Buss C, Opitz B, Hocke AC, Lippmann J, van, Laak V, Hippenstiel S, Krull M, Suttorp N, Eitel J. Essential role of mitochondrial antiviral signaling, IFN regulatory factor (IRF)3, and IRF7 in Chlamydophila pneumoniae-mediated IFN-beta response and control of bacterial replication in human endothelial cells. J Immunol. 2010 15 3;184:3072–3078. doi: 10.4049/jimmunol.0902947. [DOI] [PubMed] [Google Scholar]
- Carrero JA, Calderon B, Unanue ER. Type I interferon sensitizes lymphocytes to apoptosis and reduces resistance to Listeria infection. J Exp Med. 2004 16 8;200:535–540. doi: 10.1084/jem.20040769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrel-Dennis M, Latz E, Halmen KA, Trieu-Cuot P, Fitzgerald KA, Kasper DL, Golenbock DT. TLR-independent type I interferon induction in response to an extracellular bacterial pathogen via intracellular recognition of its DNA. Cell Host Microbe. 2008 Nov 12;4:543–554. doi: 10.1016/j.chom.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009 Jul 8;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coers J, Kagan JC, Matthews M, Nagai H, Zuckman DM, Roy CR. Identification of Icm protein complexes that play distinct roles in the biogenesis of an organelle permissive for Legionella pneumophila intracellular growth. Mol Microbiol. 2000;38:719–736. doi: 10.1046/j.1365-2958.2000.02176.x. [DOI] [PubMed] [Google Scholar]
- Coers J, Vance RE, Fontana MF, Dietrich WF. Restriction of Legionella pneumophila growth in macrophages requires the concerted action of cytokine and Naip5/Ipaf signalling pathways. Cell Microbiol. 2007;9:2344–2357. doi: 10.1111/j.1462-5822.2007.00963.x. [DOI] [PubMed] [Google Scholar]
- Collazo CM, Yap GS, Sempowski GD, Lusby KC, Tessarollo L, Woude GF, Sher A, Taylor GA. Inactivation of LRG-47 and IRG-47 reveals a family of interferon gamma-inducible genes with essential, pathogen-specific roles in resistance to infection. J Exp Med. 2001 16 7;194:181–188. doi: 10.1084/jem.194.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad ML, Ferstl R, Teich R, Brand S, Blumer N, Yildirim AO, Patrascan CC, Hanuszkiewicz A, Akira S, Wagner H, Holst O, von Mutius E, Pfefferle PI, Kirschning CJ, Garn H, Renz H. Maternal TLR signaling is required for prenatal asthma protection by the nonpathogenic microbe Acinetobacter lwoffii F78. J Exp Med. 2009 21 12;206:2869–2877. doi: 10.1084/jem.20090845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009 26 3;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortier A, de Chastellier C, Balor S, Gros P. Birc1e/Naip5 rapidly antagonizes modulation of phagosome maturation by Legionella pneumophila. Cell Microbiol. 2007;9:910–923. doi: 10.1111/j.1462-5822.2006.00839.x. [DOI] [PubMed] [Google Scholar]
- Hawn TR, Berrington WR, Smith IA, Uematsu S, Akira S, Aderem A, Smith KD, Skerrett SJ. Altered inflammatory responses in TLR5-deficient mice infected with Legionella pneumophila. J Immunol. 2007 15 11;179:6981–6987. doi: 10.4049/jimmunol.179.10.6981. [DOI] [PubMed] [Google Scholar]
- Hawn TR, Verbon A, Lettinga KD, Zhao LP, Li SS, Laws RJ, Skerrett SJ, Beutler B, Schroeder L, Nachman A, Ozinsky A, Smith KD, Aderem A. A common dominant TLR5 stop codon polymorphism abolishes flagellin signaling and is associated with susceptibility to legionnaires' disease. J Exp Med. 2003 17 11;198:1563–1572. doi: 10.1084/jem.20031220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath L, Chrisp C, Huffnagle G, LeGendre M, Osawa Y, Hurley M, Engleberg C, Fantone J, Brieland J. Effector mechanisms responsible for gamma interferon-mediated host resistance to Legionella pneumophila lung infection: the role of endogenous nitric oxide differs in susceptible and resistant murine hosts. Infect Immun. 1996;64:5151–5160. doi: 10.1128/iai.64.12.5151-5160.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry T, Brotcke A, Weiss DS, Thompson LJ, Monack DM. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. J Exp Med. 2007 14 5;204:987–994. doi: 10.1084/jem.20062665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry T, Kirimanjeswara GS, Ruby T, Jones JW, Peng K, Perret M, Ho L, Sauer JD, Iwakura Y, Metzger DW, Monack DM. Type I IFN signaling constrains IL-17 A/F secretion by gammadelta T cells during bacterial infections. J Immunol. 2010 Jan 4;184:3755–3767. doi: 10.4049/jimmunol.0902065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009 26 3;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J. The IRG proteins: a function in search of a mechanism. Immunobiology. 2008;213:367–375. doi: 10.1016/j.imbio.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Isberg RR, O'Connor TJ, Heidtman M. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol. 2009;7:13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, Ludwig H, Sutter G, Suzuki K, Hemmi H, Sato S, Yamamoto M, Uematsu S, Kawai T, Takeuchi O, Akira S. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- Ishii KJ, Koyama S, Nakagawa A, Coban C, Akira S. Host innate immune receptors and beyond: making sense of microbial infections. Cell Host Microbe. 2008 Dec 6;3:352–363. doi: 10.1016/j.chom.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009 Aug 10;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nunez G, Flavell RA. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005 Apr 2;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- Leber JH, Crimmins GT, Raghavan S, Meyer-Morse NP, Cox JS, Portnoy DA. Distinct TLR- and NLR-mediated transcriptional responses to an intracellular pathogen. PLoS Pathog. 2008;4:e6. doi: 10.1371/journal.ppat.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfield KL, Persson J, Brubaker SW, Witte CE, von Moltke J, Dunipace EA, Henry T, Sun YH, Cado D, Dietrich WF, Monack DM, Tsolis RM, Vance RE. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat Immunol. 2008;9:1171–1178. doi: 10.1038/ni.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippmann J, Rothenburg S, Deigendesch N, Eitel J, Meixenberger K, van, Laak V, Slevogt H, N'Guessan PD, Hippenstiel S, Chakraborty T, Flieger A, Suttorp N, Opitz B. IFNbeta responses induced by intracellular bacteria or cytosolic DNA in different human cells do not require ZBP1 (DLM-1/DAI). Cell Microbiol. 2008;10:2579–2588. doi: 10.1111/j.1462-5822.2008.01232.x. [DOI] [PubMed] [Google Scholar]
- Mancuso G, Midiri A, Biondo C, Beninati C, Zummo S, Galbo R, Tomasello F, Gambuzza M, Macri G, Ruggeri A, Leanderson T, Teti G. Type I IFN signaling is crucial for host resistance against different species of pathogenic bacteria. J Immunol. 2007 Jan 3;178:3126–3133. doi: 10.4049/jimmunol.178.5.3126. [DOI] [PubMed] [Google Scholar]
- Martin FJ, Gomez MI, Wetzel DM, Memmi G, O'Seaghdha M, Soong G, Schindler C, Prince A. Staphylococcus aureus activates type I IFN signaling in mice and humans through the Xr repeated sequences of protein A. J Clin Invest. 2009;119:1931–1939. doi: 10.1172/JCI35879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005 20 10;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Michallet MC, Meylan E, Ermolaeva MA, Vazquez J, Rebsamen M, Curran J, Poeck H, Bscheider M, Hartmann G, Konig M, Kalinke U, Pasparakis M, Tschopp J. TRADD protein is an essential component of the RIG-like helicase antiviral pathway. Immunity. 2008;28:651–661. doi: 10.1016/j.immuni.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Molofsky AB, Byrne BG, Whitfield NN, Madigan CA, Fuse ET, Tateda K, Swanson MS. Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J Exp Med. 2006 17 4;203:1093–1104. doi: 10.1084/jem.20051659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe KM, McWhirter SM, Vance RE. Identification of Host Cytosolic Sensors and Bacterial Factors Regulating the Type I Interferon Response to Legionella pneumophila. PLoS Pathog. 2009;5:e1000665. doi: 10.1371/journal.ppat.1000665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994 24 6;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- O'Connell RM, Saha SK, Vaidya SA, Bruhn KW, Miranda GA, Zarnegar B, Perry AK, Nguyen BO, Lane TF, Taniguchi T, Miller JF, Cheng G. Type I interferon production enhances susceptibility to Listeria monocytogenes infection. J Exp Med. 2004 16 8;200:437–445. doi: 10.1084/jem.20040712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opitz B, van, Laak V, Eitel J, Suttorp N. Innate immune recognition in infectious and noninfectious diseases of the lung. Am J Respir Crit Care Med. 2010 15 6;181:1294–1309. doi: 10.1164/rccm.200909-1427SO. [DOI] [PubMed] [Google Scholar]
- Opitz B, Vinzing M, van, Laak V, Schmeck B, Heine G, Gunther S, Preissner R, Slevogt H, N'Guessan PD, Eitel J, Goldmann T, Flieger A, Suttorp N, Hippenstiel S. Legionella pneumophila induces IFNbeta in lung epithelial cells via IPS-1 and IRF3, which also control bacterial replication. J Biol Chem. 2006 24 11;281:36173–36179. doi: 10.1074/jbc.M604638200. [DOI] [PubMed] [Google Scholar]
- Plumlee CR, Lee C, Beg AA, Decker T, Shuman HA, Schindler C. Interferons direct an effective innate response to Legionella pneumophila infection. J Biol Chem. 2009 30 10;284:30058–30066. doi: 10.1074/jbc.M109.018283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke AL, O'Riordan MX. Intracellular innate resistance to bacterial pathogens. Cell Microbiol. 2006;8:1720–1729. doi: 10.1111/j.1462-5822.2006.00795.x. [DOI] [PubMed] [Google Scholar]
- Rayamajhi M, Humann J, Penheiter K, Andreasen K, Lenz LL. Induction of IFN-alphabeta enables Listeria monocytogenes to suppress macrophage activation by IFN-gamma. J Exp Med. 2010 15 2;207:327–337. doi: 10.1084/jem.20091746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, Hume DA, Stacey KJ. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009 20 2;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N, Taniguchi T. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- Sauer JD, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, Vance RE. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect Immun. 2011;79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavoni G, Mauri C, Carlei D, Belardelli F, Pastoris MC, Proietti E. Type I IFN protects permissive macrophages from Legionella pneumophila infection through an IFN-gamma-independent pathway. J Immunol. 2004 15 7;173:1266–1275. doi: 10.4049/jimmunol.173.2.1266. [DOI] [PubMed] [Google Scholar]
- Segal G, Purcell M, Shuman HA. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc Natl Acad Sci U S A. 1998 17 2;95:1669–1674. doi: 10.1073/pnas.95.4.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005 Sep 9;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Shahangian A, Chow EK, Tian X, Kang JR, Ghaffari A, Liu SY, Belperio JA, Cheng G, Deng JC. Type I IFNs mediate development of postinfluenza bacterial pneumonia in mice. J Clin Invest. 2009 Jan 6; doi: 10.1172/JCI35412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporri R, Joller N, Albers U, Hilbi H, Oxenius A. MyD88-dependent IFN-gamma production by NK cells is key for control of Legionella pneumophila infection. J Immunol. 2006 15 5;176:6162–6171. doi: 10.4049/jimmunol.176.10.6162. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Stockinger S, Materna T, Stoiber D, Bayr L, Steinborn R, Kolbe T, Unger H, Chakraborty T, Levy DE, Muller M, Decker T. Production of type I IFN sensitizes macrophages to cell death induced by Listeria monocytogenes. J Immunol. 2002 Jan 12;169:6522–6529. doi: 10.4049/jimmunol.169.11.6522. [DOI] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, Ohba Y, Taniguchi T. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007 26 7;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010 19 3;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009 23 7;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Andrews HL, Wong SK, Isberg RR. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998 Jun 2;279:873–876. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- Wang Z, Choi MK, Ban T, Yanai H, Negishi H, Lu Y, Tamura T, Takaoka A, Nishikura K, Taniguchi T. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc Natl Acad Sci U S A. 2008 Aug 4;105:5477–5482. doi: 10.1073/pnas.0801295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Asano N, Fichtner-Feigl S, Gorelick PL, Tsuji Y, Matsumoto Y, Chiba T, Fuss IJ, Kitani A, Strober W. NOD1 contributes to mouse host defense against Helicobacter pylori via induction of type I IFN and activation of the ISGF3 signaling pathway. J Clin Invest. 2010 Mar 5;120:1645–1662. doi: 10.1172/JCI39481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005 16 9;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Zamboni DS, Kobayashi KS, Kohlsdorf T, Ogura Y, Long EM, Vance RE, Kuida K, Mariathasan S, Dixit VM, Flavell RA, Dietrich WF, Roy CR. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat Immunol. 2006;7:318–325. doi: 10.1038/ni1305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.