Abstract
The RNA genome of the hepatitis E virus (HEV) contains a hypervariable region (HVR) in ORF1 that tolerates small deletions with respect to infectivity. To further investigate the role of the HVR in HEV replication, we constructed a panel of mutants with overlapping deletions in the N-terminal, central, and C-terminal regions of the HVR by using a genotype 1 human HEV luciferase replicon and analyzed the effects of deletions on viral RNA replication in Huh7 cells. We found that the replication levels of the HVR deletion mutants were markedly reduced in Huh7 cells, suggesting a role of the HVR in viral replication efficiency. To further verify the results, we constructed HVR deletion mutants by using a genetically divergent, nonmammalian avian HEV, and similar effects on viral replication efficiency were observed when the avian HEV mutants were tested in LMH cells. Furthermore, the impact of complete HVR deletion on virus infectivity was tested in chickens, using an avian HEV mutant with a complete HVR deletion. Although the deletion mutant was still replication competent in LMH cells, the complete HVR deletion resulted in a loss of avian HEV infectivity in chickens. Since the HVR exhibits extensive variations in sequence and length among different HEV genotypes, we further examined the interchangeability of HVRs and demonstrated that HVR sequences are functionally exchangeable between HEV genotypes with regard to viral replication and infectivity in vitro, although genotype-specific HVR differences in replication efficiency were observed. The results showed that although the HVR tolerates small deletions with regard to infectivity, it may interact with viral and host factors to modulate the efficiency of HEV replication.
INTRODUCTION
Hepatitis E virus (HEV), the causative agent of hepatitis E, is classified in the genus Hepevirus of the family Hepeviridae (11, 17). It is now known that hepatitis E is a zoonotic disease and that animal reservoirs exist for HEV (13, 26–29, 38, 40). The inefficient replication of HEV in cell cultures has hindered progress in understanding the biology of HEV. The problem has been overcome partially by either characterizing individually expressed proteins from expression vectors or transfecting cells and intrahepatically inoculating animals with capped RNA transcripts generated in vitro from infectious clones (16, 17, 32, 33). More recently, efficient in vitro HEV replication systems have been reported (3, 31, 35, 37), which may aid in future understanding of HEV replication.
The 7.2-kb RNA genome of HEV contains three open reading frames (ORFs), namely, ORF1, ORF2, and ORF3, flanked by 5′- and 3′-nontranslated regions (2). The putative functional domains in the ORF1 protein include methyltransferase, protease, helicase, and RNA-dependent RNA polymerase (RdRp) domains (2). ORF2 encodes the major capsid protein, whereas ORF3 encodes a small multifunctional protein (2, 6, 18, 30, 39). The methyltransferase and guanylyltransferase activities in capping of the viral RNA (25), the role of RdRp in viral RNA synthesis (1), the helicase-associated 5′-triphosphatase activity (20), and NTPase and RNA duplex-unwinding activities have all been demonstrated for HEV (19).
HEV encodes replication factors with the conserved ORF1 protein domains that have similarities with those of rubella virus and alphaviruses (21). The highly conserved X domain in the ORF1 protein of HEV, which flanks the papain-like protease domain, is preceded by a proline-rich hinge region that may constitute a flexible hinge between the X domain and the upstream domains (21). The hypervariable region (HVR) of the ORF1 protein in HEV overlaps the proline-rich sequence located between the N terminus of the X domain and the C-terminal portion of the putative papain-like protease domain (22). The HVR varies both in length and in sequence among different HEV strains. We recently demonstrated that HEV infectivity can tolerate small deletions in the HVR and that amino acid residues in this region are dispensable for virus infectivity (32). We previously predicted that amino acids (aa) 557 to 641 of avian HEV are hypervariable (32). However, since that initial publication, two additional genotypes of avian HEV have been identified from chickens worldwide (3), which allowed us to more precisely predict the HVR of avian HEV, which spans amino acids 557 to 603 of the ORF1 protein. The objective of this study was to assess the roles of the HVR in the efficiency of HEV replication.
MATERIALS AND METHODS
Cell lines and HEV infectious cDNA clones.
A subclone of the Huh7 human liver cell line, S10-3 (9, 10, 14), and a genotype 1 human HEV (Sar55 strain) infectious clone (12) were gifts from Suzanne Emerson and Robert Purcell at NIH (Bethesda, MD). HepG2/C3A (ATCC CRL-10741) cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum (FBS) in a humidified atmosphere of 5% CO2 at 37°C. Leghorn male hepatoma (LMH) chicken liver cells (ATCC CRL-2117) were maintained at 37°C with 5% CO2 in Waymouth's MB752/1 medium containing 10% FBS. The infectious cDNA clones of the genotype 3 swine HEV and avian HEV strains used in this study were reported previously (15, 16).
Generation of a genotype 1 human HEV luciferase replicon.
A genotype 1 human HEV replicon expressing firefly luciferase (pSK-REP) was constructed by replacing the amino terminus-encoding portion of the ORF2 gene (nucleotides [nt] 5148 to 5816) of the genotype 1 human HEV (strain Sar55) pSK-HEV-2 infectious clone (12) with a PCR product of the firefly luciferase gene amplified from the pGL4.10[luc2] vector (Promega) (from nt 100 to 1752) by using primer set HuLP4/HuLP2 (Table 1). As a control, a null replication mutant of the genotype 1 HEV replicon, pSK-GAA, was constructed by mutating the conserved GDD motif of RdRp to GAA, using a QuikChange mutagenesis kit (Stratagene) with primers HuGAAF and HuGAAR (Table 1).
Table 1.
Oligonucleotide primers used for construction of genotype 1 human HEV luciferase replicon, avian HEV luciferase replicon, HVR deletion mutants, and chimeric HEV replicons
| Primer | Sequencea (5′ → 3′) | Polarity |
|---|---|---|
| Primers for genotype 1 human HEV constructs | ||
| HuLP1 | GATTGGCATGCTACAGGCTGTTGCTGATGG | + |
| HuLP2 | GCGAATTCCTATTATTCTTGCCGCCCTTCTTGG | − |
| HuLP3 | TTTGGCGTCTTCCATGGTCGCGAACCCATG | − |
| HuLP4 | CATGGGTTCGCGACCATGGAAGATGCCAAAAAC | + |
| HuGAAF | GCTGCCTTTAAAGGTGCTGCTTCGATAGTGCT | + |
| HuGAAR | AGCACTATCGAAGCAGCACCTTTAAAGGCAGC | − |
| HuF | GGGAGCATGCTCAGAAGTTTATAACACGCC | + |
| HuR | GTACCTCTGGTAAAATGCATGACAGAGCCC | − |
| Hurr | AGCATCAACCTCCGACCAAGTGCGGGTGTA | − |
| Huf1 | TCGGAGGTTGATGCTTCTATACCTAGTAGG | + |
| Huf2 | TCGGAGGTTGATGCTCCCCCCCCTGCACCG | + |
| Huf3 | TCGGAGGTTGATGCTCTCTCTGCTCCGGCG | + |
| Huf4 | GAGCCTTCTATACCTGCTCCTGGCGCTACC | + |
| Huf5 | CAGCCCGACTTAGGTGCCCCAGCCATAACC | + |
| Huf6 | GTTCCTAGTCCAGCCCACCAGACGGCCCGG | + |
| Hur1 | AGGTATAGAAGGCTCAGATGTAAAACCTAA | − |
| Hur2 | ACCTAAGTCGGGCTGGGCTGGACTAGGAAC | − |
| Hur3 | GGCTGGACTAGGAACAGCATCAACCTCCGA | − |
| Huf7 | GAGCCGGCTCCTGGCCGGCATCGCCGCCTG | + |
| Huf8 | CCTGCACCGGATCCTCGGCATCGCCGCCTG | + |
| Huf9 | ACACCTACCCCGGCGCGGCATCGCCGCCTG | + |
| Hur4 | GCCAGGAGCCGGCTCACCACGCGCCGGAGC | − |
| Hur5 | AGGATCCGGTGCAGGGGGGGGTAGAGGGGC | − |
| Hur6 | CGCCGGGGTAGGTGTGGCGGCCCTACTAGG | − |
| HuHVRf | CGTCTCAGTCGCGGCATCGCCGCCTGCTCTTT | + |
| HuHVRr | CGTCTCACGACCAAGTGCGGGTGTAAAGTG | − |
| Primers for avian HEV constructs | ||
| Av-p1A | CCCGTTAACAATATGCCCTTGCCG | + |
| Av-p1F | CCAGGCGCCCCGTTACCAATATGCCCTTGCC | + |
| Av-p1R | GGTCCGCGGGGCAATGGTTATACGGGAACGG | − |
| Av-p2A | AGCGGAGACGGCCGGTGGTGCGCAGTT | + |
| Av-p2B | CCGGCCGTCTCCGCTGGCTTGACTCCGACGCGC | − |
| Av-p3A | CGGGGGTGCCGGTACAGGGCCAGCGGA | + |
| Av-p3B | GTACCGGCACCCCCGCCTGAGGTCAGTGAGTCC | − |
| AvFF | GGTACGAAGTCTGCAGTTAGCAAGTGG | + |
| AvR | CGGGTTAACAAGCCAGTCGGCGGCA | − |
| AvEcoRVF | GGGGATATCTGCGTGGCCGAAAACTTTG | + |
| AvSalIR | GCGGTCGACTTATTACACGGCGATCTTG | − |
| AvBsmBIf | CGTCTCAATGGAAGATGCCAAAAACA | + |
| AvBsmBIr | CGTCTCACCATCCCACCCCACTTTCCT | − |
| AvGAAF | GTTGTGTTTCAAAGGTGCTGCTAGTGTCGTTGTCTGTG | + |
| AvGAAR | CACAGACAACGACACTAGCAGCACCTTTGAAACACAAC | − |
| Primers for chimeric HEV constructs | ||
| Huf | CGTCTCATTCCCGGCATCGCCGCCTGCTCTT | + |
| Hur | CGTCTCACGACCAAGTGCGGGTGTAAAGTG | − |
| Swf | CGTCTCAGTCGACATCTGGCTTTTCTAGCG | + |
| Swr | CGTCTCAGGAAGGCGGGGGTGTTGGTGGC | − |
| Avf | CGTCTCAGTCGACTTGGTCAAACTGCGCACCACCGGCC | + |
| Avr | CGTCTCAGGAAGCGAACCTCGCGCGTCGGAGTCAA | − |
| SwSMF | AGGGGACTTTGTATACGCGTACTTGGTCAACATCTG | + |
| SwSMR | CCCGCCTTCCCGCACGCGTCGTCTCCTCTAC | − |
| HuHVRf | GGGACGCGTACTTGGTCAGAGGTTGATGCTGTTCCT | + |
| HuHVRr | CCGACGCGTGCGGGCCGTCTGGTGGGTTATGGC | − |
Underlined nucleotides were required for cloning or mutagenesis purposes.
Construction of HVR deletion mutants of the genotype 1 human HEV replicon.
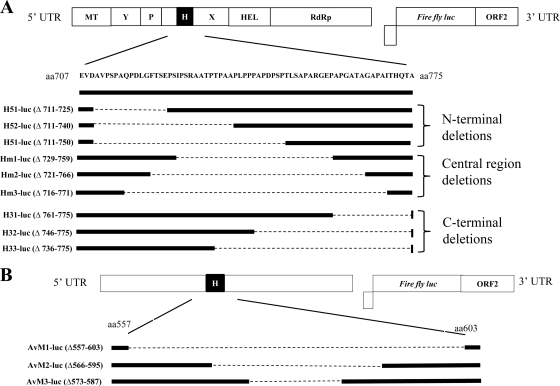
A total of nine overlapping HVR deletion mutants of the genotype 1 human HEV luciferase replicon were constructed using fusion PCR (Fig. 1 A). Amino acid residues 711 to 725, 711 to 740, and 711 to 750 in ORF1 were deleted from the N-terminal region of the HVR to generate deletion mutants H51-luc, H52-luc, and H53-luc, respectively; amino acid residues 729 to 759, 721 to 766, and 716 to 771 were deleted from the central region of the HVR to construct the deletion mutants Hm1-luc, Hm2-luc, and Hm3-luc, respectively; and amino acid residues 761 to 775, 746 to 775, and 736 to 775 were deleted from the C-terminal region of the HVR to produce the deletion mutants H31-luc, H32-luc, and H33-luc, respectively. In addition to the 9 mutants with various lengths of HVR deletion, we also generated a complete HVR deletion mutant of genotype 1 human HEV, pSKHEV2-ΔHVR-luc, by engineering an in-frame deletion (Δ707-775) into the backbone of the genotype 1 HEV replicon (pSK-REP). The primers used to generate the deletion mutants are listed in Table 1.
Fig. 1.
Schematic diagrams of the organization of HEV luciferase replicons and their derived HVR deletion mutants. (A) Subgenomic luciferase replicon of genotype 1 human HEV. The putative functional domains are indicated at the top. MT, methyltransferase; Y, Y domain; P, papain-like cysteine protease; H, hypervariable region; X, X domain; HEL, helicase; RdRp, RNA-dependent RNA polymerase. The various HEV HVR deletion mutants and the relative positions of the deletions are indicated schematically. (B) Subgenomic luciferase replicon of avian HEV, with the HVR (aa 557 to 603) indicated as a solid box. In-frame deletions of the HVR were generated by fusion PCR. The deleted amino acids are shown as a dashed line, and the relative deleted amino acid positions are indicated for each mutant.
Generation of an avian HEV luciferase replicon.
To verify the role of the HVR in HEV replication from the results obtained with the genotype 1 human HEV replicon, we constructed an avian HEV replicon (Av-HEV-luc) expressing firefly luciferase. The firefly luciferase gene was inserted in frame with the start codon of ORF2 to replace nt 3752 to 6090 of the avian HEV infectious clone pT7-aHEV (15). A BsmBI site engineered into primers (Table 1) was used to ligate the amplified fragments. The final fusion PCR product was substituted for the EcoRV-to-SalI region in the avian HEV infectious clone. As a control, a null replication mutant of the avian HEV replicon, AvGAA-luc, was also constructed by mutating the conserved GDD motif of the avian HEV RdRp to GAA by using a QuikChange mutagenesis kit (Stratagene) with primers AvGAAF and AvGAAR (Table 1).
Construction of HVR deletion mutants of avian HEV luciferase replicon.
In-frame amino acid deletions Δ557-603, Δ566-595, and Δ573-587 in ORF1 were engineered into the backbone of the avian HEV luciferase replicon to generate HVR deletion mutants Avm1-luc, Avm2-luc, and Avm3-luc, respectively (Fig. 1B). The final PCR products were digested with HpaI after purification and were ligated into the backbone of the avian HEV luciferase replicon. Site-directed mutagenesis was performed to complete the construction of the mutant Avm1-luc, using the primer Av-p1F and its complement, Av-p1R (Table 1), to remove a nonsilent mutation introduced during the amplification step. For the in vivo chicken infectivity study, amino acid deletion (Δ557-603) of the HVR was engineered into the infectious clone of avian HEV to construct a complete HVR deletion mutant of the avian HEV clone, namely, cAvm1.
Construction of chimeric hepatitis E viruses with swapped HVRs between different HEV genotypes.
Infectious clones of genotype 1 human HEV (pSK-HEV2), genotype 3 swine HEV (pSHEV3), and their respective replicon derivatives, pSK-REP and pSHEV3-luc, were used to construct HEV chimeras. Briefly, aa 707 to 775 in ORF1 of genotype 1 human HEV and its replicon derivative were replaced with aa 707 to 790 of genotype 3 swine HEV to construct the chimeric clone pSKHEV2-Sw and the chimeric replicon pSKHEV2-Sw-luc. Anchor primers HuF and HuR (Table 1), containing SphI and NsiI restriction enzyme sites, respectively, were used to amplify the final ligation product, which was subsequently ligated into the backbone of genotype 1 human HEV and its replicon derivative. To construct the chimeric HEV HVR clone pSKHEV2-Av and the chimeric HEV HVR replicon pSKHEV2-Av-luc, aa 707 to 775 in ORF1 of genotype 1 human HEV and its replicon derivative were replaced with aa 557 to 603 of avian HEV by using a similar strategy. The chimeric HVR clone pSHEV3-Hu and the chimeric HVR replicon pSHEV3-Hu-luc were constructed by replacing aa 707 to 790 in ORF1 of genotype 3 swine HEV and its replicon derivative with aa 707 to 775 of genotype 1 human HEV. An MluI restriction site that was engineered to flank the HVR of genotype 3 swine HEV was used to construct the chimera. Primers used to construct the chimeras are shown in Table 1.
In vitro transcription.
The plasmid constructs with the backbone of genotype 1 human HEV were linearized using BglII. The plasmid constructs with the backbone of genotype 3 swine HEV were linearized using XbaI, and the plasmid constructs with the avian HEV luciferase replicon and full-length avian HEV infectious clone were linearized using XhoI. In vitro RNA transcripts were subsequently produced using an mMessage mMachine T7 kit (Ambion) as previously described (15–17).
In vitro transfection of Huh7 and LMH cells.
The in vitro-generated capped RNA transcripts were used for transfection of Huh7-S10-3 cells and LMH cells as previously described (8, 15, 32). For transfection of Huh7 cells with RNA transcripts generated from plasmid constructs with the backbone of the genotype 1 human HEV replicon (firefly luciferase replicon), the RNA transcripts synthesized from the T7 promoter of a Renilla luciferase vector (pRL-TK; Promega) were used for normalization and as an internal control. The RNA transcripts generated from the T7 promoter of a firefly luciferase gene were used as an internal control for the normalization of luciferase values obtained from cells transfected with RNA transcripts generated from plasmids with the backbone of the genotype 3 swine HEV replicon (Renilla luciferase replicon). Transfections were performed in quadruplicate for each sample, and the plates were incubated at 34.5°C. LMH chicken liver cells, which support the replication of avian HEV (15), were transfected at approximately 85% confluence with RNA transcripts generated from avian HEV replicon constructs in a 12-well plate by using a Lipofectamine LTX kit (Invitrogen) essentially as previously described (15, 32). The replication competency of HEV chimeras with swapped HVRs was determined by transfecting 10 μg of the transcribed RNA from each of the chimeras as well as wild-type HEV into Huh7 cells, using the DMRIE-C transfection reagent as previously described (8).
IFA and confocal microscopy.
At 5 days posttransfection, LMH cells were trypsinized and replated on 24-well plates. On day 6, the LMH cells were rinsed with phosphate-buffered saline (PBS), fixed with a solution containing 70% acetone and 30% ethanol, and stained by immunofluorescence assay (IFA) with a 1:500-diluted anti-avian HEV convalescent-phase serum as previously described (8, 15, 32). For Huh7 cells, at 3 days posttransfection, the cells were trypsinized and replated onto wells of LabTek chamber slides. On day 6, the Huh7 cells were rinsed with PBS, fixed with acetone, and stained by IFA with a 1:200-diluted chimpanzee anti-HEV convalescent-phase serum (chimp 1313 serum) as previously described (8, 9). Vectashield (Vector Laboratories) mounting medium was added to the washed wells and viewed under a Zeiss LSM 510 laser scanning confocal microscope.
Intrahepatic inoculation of SPF chickens with capped RNA transcripts from avian HEV clones.
To assess the effect of complete HVR deletion on the infectivity of avian HEV, a chicken bioassay involving direct intrahepatic inoculation of in vitro-transcribed RNAs was utilized (4, 15, 23). Briefly, 30 4-week-old specific-pathogen-free (SPF) chickens that were negative for avian HEV were divided into three groups, with 10 chickens in each group. With birds under full anesthesia (isoflurane), a 2-cm parasternal incision was made to visualize the right lobe of the liver. The RNA transcripts were injected into two different sites of the liver, with approximately 200 μl (approximately 75 μg) per injection site. The 10 chickens in group A were each injected with a total of 400 μl of RNA transcripts from the complete HVR deletion mutant clone cAvm1. Chickens in group B were each injected intrahepatically with RNA transcripts from the wild-type avian HEV infectious clone as positive controls. The 10 chickens in group C were each injected similarly with PBS buffer as negative controls. Fecal swabs and sera were collected from each chicken prior to inoculation and weekly thereafter and were tested by reverse transcription-PCR (RT-PCR) for avian HEV RNA. Weekly serum samples were also tested by enzyme-linked immunosorbent assay (ELISA) for seroconversion to avian HEV antibodies as previously described (23, 32). All inoculated chickens were necropsied at 5 weeks postinoculation.
In vitro infectivity assay.
An in vitro HEV infectivity assay was used to assess the viability of HEV mutants essentially as reported previously (8–10). Briefly, confluent monolayers of Huh7 cells in a T25 flask were trypsinized at 9 days posttransfection with various constructs and pelleted by centrifugation, and the cell pellets were lysed by 3 freeze-thaw cycles. The cell lysates were then used in the infectivity assay using HepG2/C3A cells as previously described (8–10).
Luciferase assay.
Huh7 cells at 4 days posttransfection and LMH cells at 5 days posttransfection were washed with PBS and lysed. The cell lysates were centrifuged briefly, and 20 μl of supernatant was used for luciferase assays with a dual-luciferase reporter assay system (Promega) according to the manufacturer's instructions. Luciferase activities were measured using a Tecan Safire2 microplate reader.
Statistical analysis.
One-way analysis of variance (ANOVA) followed by Tukey's post hoc test was used for multiple comparisons of the experimental groups based on the level of their replication in Huh7 or LMH cells. Significant differences were defined as those having a P value of ≤0.05.
RESULTS
N-terminal and central region deletions of the HVR significantly reduce the replication efficiency of genotype 1 human HEV.
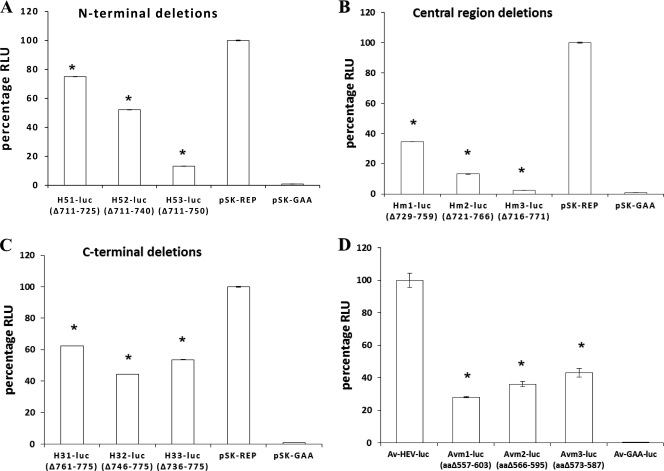
To examine the possible contribution of the HVR to HEV replication efficiency, a series of in-frame HVR deletion mutations were introduced into the genotype 1 human HEV pSK-HEV2 luciferase replicon. The replication levels of N-terminal HVR deletion mutants H51-luc (Δ711-725), H52-luc (Δ711-740), and H53-luc (Δ711-750) were compared with that of the wild-type HEV replicon pSK-REP in Huh7 cells. The percentages of viral replication compared to the wild-type HEV level were approximately 85%, 50%, and 13% for the H51-luc (Δ15 aa), H52-luc (Δ40 aa), and H53-luc (Δ50 aa) mutants, respectively (Fig. 2 A). The results showed that deletions in the N-terminal region of the HVR did not abolish viral replication but substantially reduced the replication efficiencies of HEV RNA, as evidenced by significantly lower levels of luciferase activity in all three deletion mutants than that of wild-type pSK-REP (Fig. 2A). Furthermore, mutants with larger deletions in the HVR replicated at significant lower levels than those with smaller deletions (Fig. 2A).
Fig. 2.
Luciferase activities in cells transfected with RNA transcripts from parental wild-type HEV replicons and their HVR deletion mutant derivatives. (A) Relative replication levels in Huh7 cells of mutants with deletions in the N terminus of the HVR (H51-luc, H52-luc, and H53-luc), the parental genotype 1 HEV replicon (pSK-REP), and the null mutant (pSK-GAA). (B) Relative replication levels in Huh7 cells of mutants with deletions in the central region of the HVR (Hm1-luc, Hm2-luc, and Hm3-luc), the positive control pSK-REP, and the negative control pSK-GAA. (C) Relative replication levels in Huh7 cells of mutants with deletions within the C terminus of the HVR (H31-luc, H32-luc, and H33-luc), along with positive and negative controls. The luciferase activity in Huh7 cells, which reflects the replication levels of viral RNAs, was measured by determining the firefly luciferase activity at 4 days posttransfection, and normalization for transfection efficiency was performed by using the Renilla luciferase activity measured at the same time. The value determined with the parental wild-type genotype 1 HEV replicon (pSK-REP) was set as 100% and used as a reference to normalize the replication of the other mutant replicons. A replicon carrying an inactivating mutation (GAA) in RdRp served as a negative control. Values are means and standard deviations for at least two independent experiments, each performed in quadruplicate. In most cases, the variations are very small and therefore the error bars are not visible. (D) Relative replication levels in LMH chicken liver cells of the parental avian HEV replicon (Av-HEV-Luc) and its derived HVR deletion mutants (Avm1-luc, Avm2-luc, and Avm3-luc) at 5 days posttransfection. The luciferase activity in LMH cells was measured similarly to that described above for the genotype 1 human HEV replicon and its mutants in Huh7 cells. The differences in luciferase activities produced by the wild-type replicons and their mutant derivatives were compared by one-way ANOVA and Tukey's multiple comparison test. Asterisks (*) indicate statistical differences compared to the parental wild-type replicon. RLU, relative light units.
Analogous to the results for N-terminal HVR deletion mutants, the three mutants with various deletions in the central region (Hm1-luc, Hm2-luc, and Hm3-luc) also showed a gradient of reduced viral replication activity, where mutants with larger deletions replicated at significant lower levels than those with smaller deletions (Fig. 2B). The replication level of each deletion mutant was drastically reduced, as evidenced by the significantly lower levels of luciferase activity: compared to wild-type HEV, the replication levels for the Hm1-luc, Hm2-luc, and Hm3-luc mutants were only 34.5%, 13%, and 2.5%, respectively (Fig. 2B), indicating more deleterious effects of deletions in the central region of the HVR on viral replication.
Deletions in the C-terminal region of the HVR have relatively smaller effects on the efficiency of genotype 1 HEV RNA replication.
When the replication levels of the C-terminal HVR deletion mutants H31-luc (62%), H32-luc (44%), and H33-luc (53%) were compared to that of wild-type HEV, the replication levels of the C-terminal deletion mutants were reduced by about 2-fold, and the differences between wild-type pSK-REP and each of the three mutants were statistically significant (Fig. 2C). However, there was no significant difference in luciferase activity between the H31-luc and H33-luc mutants, although significant differences were evident between the H31-luc and H32-luc mutants and between the H32-luc and H33-luc mutants.
Deletions of the avian HEV HVR of various lengths reduce the efficiency of avian HEV replication in LMH chicken liver cells.
To further verify the role of the HVR in modulating the efficiency of HEV replication observed with genotype 1 human HEV, we subsequently constructed an HEV replicon from avian HEV and evaluated the replication efficiencies of three HVR deletion mutants of avian HEV in LMH cells. Similar to the results observed for genotype 1 human HEV, all three avian HEV mutants (Avm1-luc, Avm2-luc, and Avm3-luc) replicated at significant lower levels in LMH cells than wild-type Av-HEV-luc did, and gradient reductions in viral replication efficiency were also observed for the three avian HEV mutants in LMH cells (Fig. 2D). Mutant Avm1-luc with a larger HVR deletion replicated at a significantly lower level (28%) than the Avm2-luc (36%) and Avm3-luc (43%) mutants with smaller HVR deletions.
Deletion of the complete HVR of avian HEV abolishes virus infectivity in vivo.
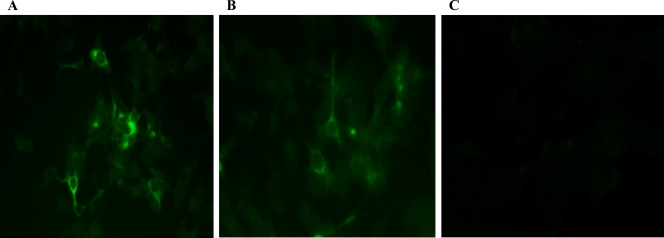
To determine whether deletion of the complete HVR would affect virus infectivity in vivo, we tested the deletion mutant cAvm1, with a complete deletion of the HVR region (aa 557 to 603) in the backbone of an avian HEV infectious clone, for its replication competency in vitro and for infectivity in chickens. Capped RNA transcripts from the cAvm1 mutant and the wild-type avian HEV infectious clone were transfected into LMH cells. On day 6 posttransfection, avian HEV-specific viral antigen was detected in transfected LMH cells (Fig. 3). Intracytoplasmic fluorescent signals were detected in cells transfected with RNA transcripts from both wild-type avian HEV and the complete HVR deletion mutant (cAvm1), indicating that the mutant was still replication competent in LMH cells.
Fig. 3.
IFA of LMH chicken liver cells following transfection with capped RNA transcripts from an avian HEV infectious clone and its derived complete HVR deletion mutant. (A) LMH cells transfected with RNA transcripts from wild-type avian HEV at 6 days posttransfection. (B) LMH cells transfected with RNA transcripts from the complete HVR deletion mutant of avian HEV (cAvm1). (C) Mock-transfected LMH cells.
The ability of this complete HVR deletion mutant of avian HEV to infect chickens was tested by direct intrahepatic inoculation of SPF chickens (4, 15, 23) with capped RNA transcripts from the cAvm1 mutant. Avian HEV-specific RNA was detected variably in fecal and serum samples from chickens inoculated with wild-type avian HEV. Six of the 10 chickens in the wild-type avian HEV-inoculated group seroconverted to IgG avian HEV antibodies (data not shown). However, none of the 10 chickens in the complete HVR deletion mutant-inoculated group had detectable avian HEV RNA in feces or serum, and the chickens remained seronegative throughout the study (data not shown). Negative-control chickens remained negative for avian HEV infection. The results from the chicken study showed that the complete HVR deletion mutant of avian HEV is noninfectious in vivo.
Intergenotypic chimeric hepatitis E viruses with swapped HVRs are replication competent in vitro.
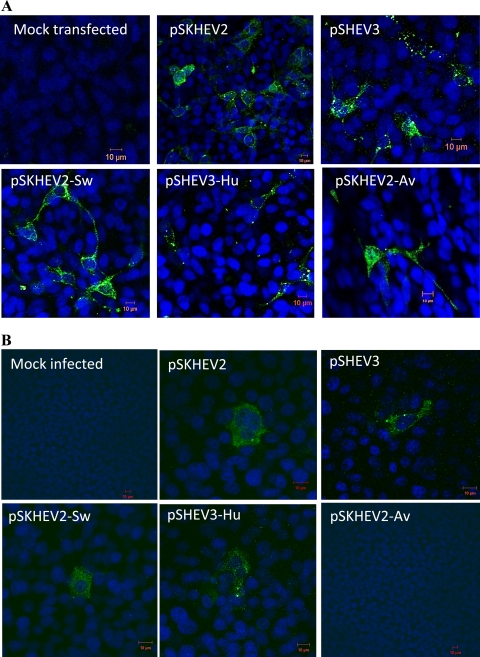
Since the HVRs among different HEV genotypes varied extensively in sequence and length, we sought to determine if the HVRs from different HEV genotypes possess similar biological functions by studying the interchangeability of HVR sequences between HEV genotypes. For this purpose, we first analyzed if the HVRs could be exchanged functionally with respect to the ability to replicate in cells. Three chimeric hepatitis E viruses with swapped HVRs (pSKHEV2-Sw, pSHEV3-Hu, and pSKHEV2-Av) were constructed, and their replication competency was assessed in Huh7-S10-3 cells, along with the wild-type genotype 1 human HEV (pSKHEV2) and genotype 3 swine HEV (pSHEV3) infectious clones. ORF2-specific viral antigens were detected in transfected Huh7 cells by use of an ORF2-specific HEV antibody (chimp 1313 serum). Cytoplasmic fluorescent signals were detected in Huh7 cells transfected with RNA transcripts from both wild-type HEVs and the chimeras (Fig. 4 A). Mock-transfected cells were negative for viral antigen. Detection of viral antigens in cells transfected with the capped RNAs from the chimeric viruses indicated that substitution of the HVRs between HEV genotypes did not affect the replication competency of HEV.
Fig. 4.
Replication competency of chimeric hepatitis E viruses with swapped HVRs in Huh7 cells and infectivity in HepG2 cells. (A) Immunofluorescence staining of Huh7 cells transfected with capped RNA transcripts from infectious cDNA clones of wild-type HEV (genotype 1 HEV [pSKHEV2] and genotype 3 swine HEV [pSHEV3]) and their derivative chimeric viruses (pSKHEV2-Sw, pSKHEV2-Av, and pSHEV3-Hu). At 6 days posttransfection, Huh7 cells were stained by IFA for HEV ORF2 antigen (green) and were viewed by confocal microscopy. (B) Immunofluorescence staining of HepG2 cells infected with lysates of Huh7 cells transfected with RNA transcripts from the wild-type and chimeric virus clones in panel A. Lysates were collected from Huh7 cells transfected with wild-type HEVs and their derivative chimeras on day 9 posttransfection and were used to infect HepG2 cells. The HepG2 cells were stained by IFA for HEV ORF2 antigen (green) and then viewed by confocal microscopy.
Intergenotypic chimeric hepatitis E viruses with swapped HVRs are capable of producing infectious virus in vitro.
Although viral RNA replication was not affected by the HVR exchange, the effects of HVR swap on the steps downstream of viral RNA replication could not be ruled out. Thus, we performed an in vitro infectivity assay to determine if the HVR exchange had any effect on virus infectivity. HepG2/C3A cells were infected with lysates of transfected Huh7 cells collected at 9 days posttransfection (8, 9). The inoculated cells were examined by IFA for the presence of viral antigen. Cytoplasmic fluorescent signals were detected in cells infected with the lysates of Huh7 cells transfected with the pSKHEV2-Sw and pSHEV3-Hu chimeras, as well as with wild-type genotype 1 human HEV (pSKHEV2) and genotype 3 swine HEV (pSHEV3), indicating the production of infectious chimeric virus particles in the Huh7 cell lysates. However, the HepG2/C3A cells infected with lysates of Huh7 cells that were transfected with the RNA transcripts from the pSKHEV2-Av chimera containing the avian HEV HVR sequence did not show any detectable evidence of viral infection (Fig. 4B).
The HVR contains genotype-specific sequences important for the efficiency of HEV RNA replication.
To further analyze whether the HVR contains genotype-specific elements that may affect the efficiency of HEV replication, we constructed four chimeric HEV HVR replicons and analyzed the impact of heterologous HVRs on HEV replication efficiency. Huh7 cells were transfected with the capped RNA transcripts from the mutant replicons along with the RNAs from the wild-type HEV replicons. A wild-type HEV replicon carrying amino acid substitutions which altered the conserved GDD motif of the RdRp active site to GAA was transfected into Huh7 cells in parallel as a negative control. Cells were harvested at daily intervals until 4 days posttransfection, and the luciferase activities were measured to assess the replication levels of the chimeric HEV HVR replicon RNAs. The firefly luciferase activity was monitored to compare the replication levels of mutant replicons pSHEV3-Hu-luc, pSKHEV2-Av-luc, and pSKHEV2-ΔHVR with that of the wild-type HEV replicon pSK-REP. Similarly, Renilla luciferase activity was monitored to compare the replication levels of the chimeric HEV replicon pSHEV3-Hu-luc with that of the wild-type HEV replicon pSHEV3-luc.
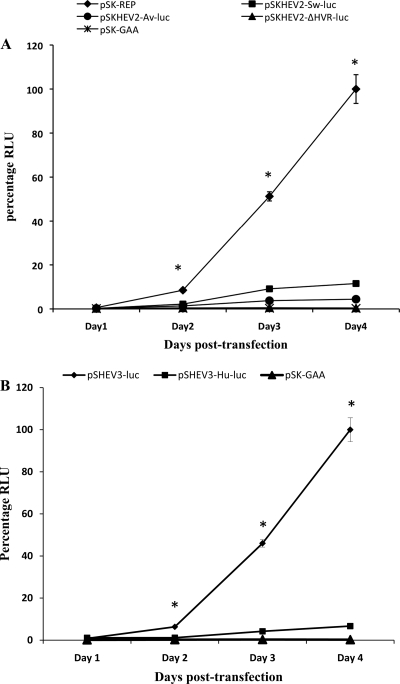
Within the first 24 h after transfection, no significant difference in replication levels was observed between the wild-type HEV replicons (pSK-REP and pSHEV3-luc) and the negative-control mutant pSK-GAA (Fig. 5). For the wild-type genotype 1 replicon pSK-REP, the luciferase activity was increased 14-fold, 88-fold, and 172-fold over the input values on days 2, 3, and 4 posttransfection, respectively. Similarly, the luciferase activity was increased 8-fold, 58-fold, and 125-fold in the case of the wild-type genotype 3 replicon pSHEV3-luc during the same periods. The replication kinetics observed with the chimeric HEV HVR mutant replicons were different from those of the wild-type replicons during the 4-day period (Fig. 5), and the results showed that heterologous HVRs had an impact on the efficiency of HEV replication. At each time point (except for day 1 posttransfection), the luciferase activities obtained with the chimeric mutant replicons were significantly lower than those of wild-type replicons. The chimeric replicon pSKHEV2-Sw-luc showed approximately 4-fold, 5-fold, and 9-fold reductions in virus RNA replication levels compared to the wild-type replicon pSK-REP on days 2, 3, and 4, respectively. The luciferase expression level of the pSKHEV2-Av-luc replicon was reduced 6-fold, 13-fold, and 22-fold during the same period compared to the wild-type HEV replicon. Similarly, the replication levels of the pSHEV3-Hu-luc replicon had 6-fold, 11-fold, and 15-fold reductions compared to the wild-type replicon pSHEV3-luc on days 2, 3, and 4 posttransfection, respectively. The replication of HEV RNA was essentially abolished when the complete HVR was deleted in the mutant replicon pSKHEV2-ΔHVR, and its replication level was similar to that of the negative control pSK-GAA. The results suggest that the HVR, although exchangeable between genotypes, contains genotype-specific sequences that can affect the efficiency of HEV RNA replication.
Fig. 5.
Replication kinetics of chimeric HEV luciferase replicons with swapped HVRs. (A) Replication kinetics of the wild-type genotype 1 HEV replicon pSK-REP, the chimeric replicon pSKHEV2-Sw-luc, and the mutant replicon pSKHEV2-ΔHVR-luc. The firefly luciferase activity in cell lysates of Huh7 cells was determined at 1, 2, 3, and 4 days posttransfection. Data were normalized for transfection efficiency among the chimeric replicons as determined by measurement of the Renilla luciferase activity. The value determined with the parental HEV replicon (pSK-REP) was set as 100% and used as a reference to normalize the replication of other mutant replicons. A replicon (pSK-GAA) carrying an inactivating mutation (GAA) in RdRp served as a negative control. (B) Replication of the genotype 3 wild-type HEV replicon pSHEV3-luc and the chimeric replicon pSHEV3-Hu-luc in Huh7 cells. The Renilla luciferase activity was determined at daily intervals for 4 days posttransfection. The luciferase value determined for the parental wild-type HEV replicon pSHEV3-luc was set as 100% and used to normalize the replication of pSHEV3-Hu-luc, while the pSK-GAA replicon served as a negative control. Values are means and standard deviations for at least two independent experiments, each performed in quadruplicate. Asterisks (*) indicate statistical differences compared to the parental wild-type HEV replicons. RLU, relative light units.
DISCUSSION
Recently, we demonstrated that small deletions in the HVR are tolerated by HEV in terms of its infectivity (32), suggesting structural flexibility of the HVR. However, the importance of the HVR for the efficiency of HEV replication remains largely unknown. Thus, in the present study, we first aimed to determine the effects of HVR deletions of various lengths and locations on the efficiency of HEV replication. We also sought to determine if the sequence variability observed in the HVR is genotype specific with respect to virus replication and infectivity.
The importance of HVR for efficient HEV RNA replication was studied by introducing overlapping deletions of various lengths into the HEV luciferase replicon pSK-REP of genotype 1 human HEV. The results showed that small deletions in the N-terminal region and central region of the HVR were associated with marked reductions in HEV replication efficiency, as evidenced by significantly reduced luciferase activity. Larger deletions in these regions resulted in more drastic reductions in viral RNA replication efficiency. In contrast to the relatively gradual decline in replication levels exhibited by mutants with N-terminal and central region deletions of the HVR, the size of the deletions in the C-terminal region of HVR apparently had a relatively smaller effect on viral RNA replication efficiency than that for deletions in the N-terminal and central regions. The results suggested that all of the HVR deletion mutants were replication competent in vitro, although deletions in the HVR reduced the efficiency of viral RNA synthesis in Huh7 cells.
Avian HEV, a nonmammalian member of the Hepeviridae family, was used for further confirmation of the results obtained with the genotype 1 human HEV. Compared to wild-type avian HEV, the viral RNA replication levels of the three avian HEV HVR deletion mutants were reduced significantly, and progressively larger deletions resulted in corresponding decreases in replication levels of avian HEV. Thus, the results for the avian HEV HVR deletion mutants are similar to those for genotype 1 human HEV. Although the ORF1 HVR (aa 707 to 775) contains residues that are dispensable for viral infectivity (32), the association of various length deletions of the HVR with different reduced levels of viral RNA replication observed from this study suggests a role for the HVR in modulating the efficiency of HEV replication.
It is known that the HVR of HEV overlaps with the proline-rich hinge region (22, 32). Proline is the most common terminal linker residue located between functionally relevant regions of protein structures. We speculate that deletions introduced into linkers such as the HVR of HEV may alter specific interactions with viral and/or host factors and thus modulate the efficiency of viral replication. This may also explain the observed replication gradient among HVR deletion mutants of various lengths, since a larger deletion could interfere with the interaction of domains with their substrates, exerting a size limit to maintain correct folding of the functional domain. Deletions in the C-terminal region of the HVR did not show typical replication gradients that were seen for the N-terminal and central region deletions, suggesting that the deletions in the C-terminal region of the HVR might not interfere with folding of the HVR domain and/or polyprotein. Sequence analyses identified the presence of SH3 motifs in the HVRs of all four genotypes of mammalian HEVs and avian HEV. Although these SH3 motifs are not absolutely required for virus replication or infectivity, they have been shown to play an important role in viral pathogenesis by enhancing their replication efficiency and infectivity in other virus systems (5, 24, 34). Therefore, HEV may similarly exploit these SH3-mediated interactions to enhance its replication and/or infectivity.
To assess the effect of complete HVR deletion on avian HEV infectivity, we generated a complete HVR deletion mutant of avian HEV. The replication competency of the complete HVR deletion mutant was verified in LMH cells. To determine the impact of the complete HVR deletion on the infectivity of avian HEV in vivo, 4-week-old SPF chickens were inoculated intrahepatically with capped RNA transcripts from the complete HVR deletion mutant. The results showed that the complete HVR deletion mutant of avian HEV was noninfectious during the 5 weeks of the study, as evidenced by the lack of fecal virus shedding, viremia, or seroconversion in inoculated chickens, although chickens inoculated with the capped RNA from wild-type avian HEV became infected. The results from this chicken study confirmed our previous finding from a small animal study in which we showed that 3 chickens inoculated with the RNA transcripts from a larger HVR deletion mutant of avian HEV (aa 557 to 641) had no detectable viremia or fecal virus shedding and that 2 of the 3 chickens were seronegative, whereas the third chicken had a borderline optical density (32). The observation that a complete HVR deletion mutant is replication competent in vitro but noninfectious in vivo suggests a less critical role for the HVR in cells with innate immunity defects. Nonstructural viral genes that are dispensable for virus replication are known to play a role in modulating host immune responses such as deubiquitinating and interferon antagonism activities (7, 36). It will be interesting to determine if the HVR of HEV also contains critical residues that may play a role in antagonizing or evading host immune responses in vivo.
The impact of genetic variability of the HVR on virus replication and infectivity was also analyzed in this study. We found that all three chimeric hepatitis E viruses with swapped HVRs were replication competent in Huh7 cells and that the HEV HVR chimeras pSKHEV2-Sw and pSHEV3-Hu yielded infectious viruses that were able to infect HepG2 cells as well as the wild-type virus. However, infection was not detected in lysates from cells transfected with the pSKHEV2-Av chimera, containing the avian HEV HVR sequence. The lack of evidence of infection in HepG2 cells infected with the pSKHEV2-Av chimera could be explained by the presence of very few infectious particles in the cell lysates, or more likely, the HVR from avian HEV is not compatible with the mammalian HEV backbone. This observation further supports the proposed classification of avian HEV as a separate genus within the Hepeviridae family.
Knowing that the chimeric viruses with swapped HVRs were replication competent and infectious, we further analyzed the impact of the exchanged HVRs on the efficiency of HEV replication. Three chimeric HEV luciferase replicons with swapped HVRs were constructed. The intergenotypic chimeric mammalian HEV HVR replicons, pSKHEV2-Sw-luc and pSHEV3-Hu-luc, replicated at significantly lower levels than those of wild-type HEV replicons, suggesting a genotype specificity of the HVR. The pSKHEV2-Av-luc replicon, harboring the HVR of avian HEV in the backbone of genotype 1 human HEV, was barely replication competent in Huh7 cells. In addition, we also compared the replication efficiency of the complete HVR deletion mutant of the genotype 1 human HEV replicon (pSKHEV2-ΔHVR) with that of the wild-type replicon (pSK-REP) and showed that the replication efficiency of pSKHEV2-ΔHVR was essentially abolished to the levels of the negative control. The observed lower levels of replication as a result of the HVR exchange suggested that the composition of the HVR sequence may influence the replication efficiency of HEV RNA. We speculate that the chimeras with swapped HVRs did not affect the secondary structure and thus preserved the overall genomic structure and folding of the ORF1 functional domains, but the swaps may have disrupted genotype-specific protein-protein interactions.
In summary, the results from this study demonstrate structural and functional flexibility of the HVR of HEV. The results also reveal the importance of the HVR for efficient replication of HEV RNA. Although the HVR can be exchanged functionally, there exists a degree of genotype specificity with respect to the efficiency of virus replication and infectivity. Further in-depth structural and functional studies of the HVR are required to definitively identify potential physiologically relevant interaction sites of the HVR and their viral and/or cellular counterparts, which is beyond the scope of the present study.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (AI074667 and AI050611).
We thank Suzanne U. Emerson and Robert H. Purcell at the NIAID, NIH, Bethesda, MD, for generously providing us with the genotype 1 human HEV infectious clone, a subclone of the Huh7 cell line, and the anti-HEV chimp 1313 antiserum. We thank Pete Jobst and the animal care staff at Virginia Tech for their assistance in the animal study.
Footnotes
Published ahead of print on 20 July 2011.
REFERENCES
- 1. Agrawal S., Gupta D., Panda S. K. 2001. The 3′ end of hepatitis E virus (HEV) genome binds specifically to the viral RNA-dependent RNA polymerase (RdRp). Virology 282:87–101 [DOI] [PubMed] [Google Scholar]
- 2. Ahmad I., Holla R. P., Jameel S. 21 February 2011. Molecular virology of hepatitis E virus. Virus Res. doi: 10.1016/j.virusres.2011.02.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bilic I., Jaskulska B., Basic A., Morrow C. J., Hess M. 2009. Sequence analysis and comparison of avian hepatitis E viruses from Australia and Europe indicate the existence of different genotypes. J. Gen. Virol. 90:863–873 [DOI] [PubMed] [Google Scholar]
- 4. Billam P., et al. 2005. Systematic pathogenesis and replication of avian hepatitis E virus in specific-pathogen-free adult chickens. J. Virol. 79:3429–3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bliska J. 1996. How pathogens exploit interactions mediated by SH3 domains. Chem. Biol. 3:7–11 [DOI] [PubMed] [Google Scholar]
- 6. Chandra V., Kalia M., Hajela K., Jameel S. 2010. The ORF3 protein of hepatitis E virus delays degradation of activated growth factor receptors by interacting with CIN85 and blocking formation of the Cbl-CIN85 complex. J. Virol. 84:3857–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen Z., et al. 2010. Immunodominant epitopes in nsp2 of porcine reproductive and respiratory syndrome virus are dispensable for replication, but play an important role in modulation of the host immune response. J. Gen. Virol. 91:1047–1057 [DOI] [PubMed] [Google Scholar]
- 8. Córdoba L., et al. 2011. Three amino acid mutations (F51L, T59A, and S390L) in the capsid protein of the hepatitis E virus collectively contribute to virus attenuation. J. Virol. 85:5338–5349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Emerson S. U., et al. 2006. Putative neutralization epitopes and broad cross-genotype neutralization of hepatitis E virus confirmed by a quantitative cell-culture assay. J. Gen. Virol. 87:697–704 [DOI] [PubMed] [Google Scholar]
- 10. Emerson S. U., Nguyen H., Torian U., Purcell R. H. 2006. ORF3 protein of hepatitis E virus is not required for replication, virion assembly, or infection of hepatoma cells in vitro. J. Virol. 80:10457–10464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Emerson S. U., Purcell R. H. 2003. Hepatitis E virus. Rev. Med. Virol. 13:145–154 [DOI] [PubMed] [Google Scholar]
- 12. Emerson S. U., et al. 2001. Recombinant hepatitis E virus genomes infectious for primates: importance of capping and discovery of a cis-reactive element. Proc. Natl. Acad. Sci. U. S. A. 98:15270–15275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Feagins A. R., Opriessnig T., Huang Y. W., Halbur P. G., Meng X. J. 2008. Cross-species infection of specific-pathogen-free pigs by a genotype 4 strain of human hepatitis E virus. J. Med. Virol. 80:1379–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Graff J., Torian U., Nguyen H., Emerson S. U. 2006. A bicistronic subgenomic mRNA encodes both the ORF2 and ORF3 proteins of hepatitis E virus. J. Virol. 80:5919–5926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang F. F., Pierson F. W., Toth T. E., Meng X. J. 2005. Construction and characterization of infectious cDNA clones of a chicken strain of hepatitis E virus (HEV), avian HEV. J. Gen. Virol. 86:2585–2593 [DOI] [PubMed] [Google Scholar]
- 16. Huang Y. W., et al. 2005. Capped RNA transcripts of full-length cDNA clones of swine hepatitis E virus are replication competent when transfected into Huh7 cells and infectious when intrahepatically inoculated into pigs. J. Virol. 79:1552–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang Y. W., Opriessnig T., Halbur P. G., Meng X. J. 2007. Initiation at the third in-frame AUG codon of open reading frame 3 of the hepatitis E virus is essential for viral infectivity in vivo. J. Virol. 81:3018–3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kalia M., Chandra V., Rahman S. A., Sehgal D., Jameel S. 2009. Heparan sulfate proteoglycans are required for cellular binding of the hepatitis E virus ORF2 capsid protein and for viral infection. J. Virol. 83:12714–12724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Karpe Y. A., Lole K. S. 2010. NTPase and 5′ to 3′ RNA duplex-unwinding activities of the hepatitis E virus helicase domain. J. Virol. 84:3595–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Karpe Y. A., Lole K. S. 2010. RNA 5′-triphosphatase activity of the hepatitis E virus helicase domain. J. Virol. 84:9637–9641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Koonin E. V. 1991. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 72:2197–2206 [DOI] [PubMed] [Google Scholar]
- 22. Koonin E. V., et al. 1992. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-strand RNA plant and animal viruses. Proc. Natl. Acad. Sci. U. S. A. 89:8259–8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kwon H. M., et al. 2011. Construction of an infectious cDNA clone of avian hepatitis E virus (avian HEV) recovered from a clinically healthy chicken in the United States and characterization of its pathogenicity in specific-pathogen-free chickens. Vet. Microbiol. 147:310–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Macdonald A., et al. 2005. Further studies on hepatitis C virus NS5A-SH3 domain interactions: identification of residues critical for binding and implications for viral RNA replication and modulation of cell signalling. J. Gen. Virol. 86:1035–1044 [DOI] [PubMed] [Google Scholar]
- 25. Magden J., et al. 2001. Virus-specific mRNA capping enzyme encoded by hepatitis E virus. J. Virol. 75:6249–6255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Meng X. J. 2010. Hepatitis E virus: animal reservoirs and zoonotic risk. Vet. Microbiol. 140:256–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Meng X. J. 2010. Recent advances in hepatitis E virus. J. Viral Hepat. 17:153–161 [DOI] [PubMed] [Google Scholar]
- 28. Meng X. J., et al. 1998. Genetic and experimental evidence for cross-species infection by swine hepatitis E virus. J. Virol. 72:9714–9721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meng X. J., et al. 2002. Prevalence of antibodies to hepatitis E virus in veterinarians working with swine and in normal blood donors in the United States and other countries. J. Clin. Microbiol. 40:117–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moin S. M., Chandra V., Arya R., Jameel S. 2009. The hepatitis E virus ORF3 protein stabilizes HIF-1alpha and enhances HIF-1-mediated transcriptional activity through p300/CBP. Cell. Microbiol. 11:1409–1421 [DOI] [PubMed] [Google Scholar]
- 31. Okamoto H. 2011. Efficient cell culture systems for hepatitis E virus strains in feces and circulating blood. Rev. Med. Virol. 21:18–31 [DOI] [PubMed] [Google Scholar]
- 32. Pudupakam R. S., et al. 2009. Deletions of the hypervariable region (HVR) in open reading frame 1 of hepatitis E virus do not abolish virus infectivity: evidence for attenuation of HVR deletion mutants in vivo. J. Virol. 83:384–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ropp S. L., Tam A. W., Beames B., Purdy M., Frey T. K. 2000. Expression of the hepatitis E virus ORF1. Arch. Virol. 145:1321–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Saksela K., Cheng G., Baltimore D. 1995. Proline-rich (PxxP) motifs in HIV-1 Nef bind to SH3 domains of a subset of Src kinases and are required for the enhanced growth of Nef+ viruses but not for down-regulation of CD4. EMBO J. 14:484–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shukla P., et al. 2011. Cross-species infections of cultured cells by hepatitis E virus and discovery of an infectious virus-host recombinant. Proc. Natl. Acad. Sci. U. S. A. 108:2438–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sun Z., Chen Z., Lawson S. R., Fang Y. 2010. The cysteine protease domain of porcine reproductive and respiratory syndrome virus nonstructural protein 2 possesses deubiquitinating and interferon antagonism functions. J. Virol. 84:7832–7846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tanaka T., Takahashi M., Kusano E., Okamoto H. 2007. Development and evaluation of an efficient cell-culture system for hepatitis E virus. J. Gen. Virol. 88:903–911 [DOI] [PubMed] [Google Scholar]
- 38. Tei S., Kitajima N., Takahashi K., Mishiro S. 2003. Zoonotic transmission of hepatitis E virus from deer to human beings. Lancet 362:371–373 [DOI] [PubMed] [Google Scholar]
- 39. Yamada K., et al. 2009. ORF3 protein of hepatitis E virus is essential for virion release from infected cells. J. Gen. Virol. 90:1880–1891 [DOI] [PubMed] [Google Scholar]
- 40. Yazaki Y., et al. 2003. Sporadic acute or fulminant hepatitis E in Hokkaido, Japan, may be food-borne, as suggested by the presence of hepatitis E virus in pig liver as food. J. Gen. Virol. 84:2351–2357 [DOI] [PubMed] [Google Scholar]