Abstract
Centrifugal inoculation, or spinoculation, is widely used in virology research to enhance viral infection. However, the mechanism remained obscure. Using HIV-1 infection of human T cells as a model, we demonstrate that spinoculation triggers dynamic actin and cofilin activity, probably resulting from cellular responses to centrifugal stress. This actin activity also leads to the upregulation of the HIV-1 receptor and coreceptor, CD4 and CXCR4, enhancing viral binding and entry. We also demonstrate that an actin inhibitor, jasplakinolide, diminishes spin-mediated enhancement. In addition, small interfering RNA (siRNA) knockdown of LIMK1, a cofilin kinase, decreases the enhancement. These results suggest that spin-mediated enhancement cannot be explained simply by a virus-concentrating effect; rather, it is coupled with spin-induced cytoskeletal dynamics that promote receptor mobilization, viral entry, and postentry processes. Our results highlight the importance of cofilin and a dynamic cytoskeleton for the initiation of viral infection. Our results also indicate that caution needs to be taken in data interpretation when cells are spinoculated; some of the spin-induced cellular permissiveness may be beyond the natural capacity of an infecting virus.
INTRODUCTION
Centrifugation of cells to study stress response in cell differentiation has a long history dating back to the early 20th century (9, 33). The method was later applied to research in virology, presumably for directly depositing viral particles on cells to enhance infection (13). The mechanical stress introduced during spinning was also believed to be somewhat beneficial to infection, increasing cell permissiveness (17, 18). The technique, now termed spinoculation (12), has been widely used with a variety of viruses, such as cytomegalovirus (23), herpes simplex virus (HSV) (31), bluetongue virus (29), hepatitis C virus (43), retroviruses (5, 12), and the related HIV-1 (14, 22). In general, during spinoculation, cells are centrifuged at a low speed (approximately 1,000 to 2,000 × g) for several hours. This leads to a dramatic enhancement of viral replication of up to several-hundred-fold. Although used for over a half-century, the mechanism of how low-speed spin enhances viral infection remained largely obscure.
Several attempts have been made to elucidate the mechanisms. Early studies suggested that spinoculation may result in a concentrating effect that helps the virus penetrate cells, although the gravity force normally required for viral sedimentation is approximately 50,000 × g (31, 32). Others have suggested that centrifugation may increase the binding of virus to cells, removing a major rate-limiting step (3, 15, 22, 26). Intriguingly, different viruses responded differently to the same centrifugal conditions and these differences were not based on the weight or size of the viral particles (16, 17, 23). In addition, even with the same virus and the same centrifugal conditions, the enhancement was not always the same when measured on different cell lines (23, 26). In one report, spinoculation even replaced the requirement for the heparan sulfate coreceptor of HSV-1 in glycosylaminoglycan-deficient cells (26). These results suggest that spinoculation may not only concentrate viruses but also exert some physiological or biochemical effects on cells, rendering them more receptive for infection (17); presumably, this increase in cellular permissiveness may involve a membrane-mediated change in cells' response to viruses or a disruption of a cellular component that normally restricts viruses (17). Nevertheless, attempts to identify responsible host factors through multiple approaches have failed to reach any conclusion (16, 17).
The cortical actin is a major cytoskeletal support of the cell. It is expected that the gravity force generated from spinoculation would have a major impact on the cortical actin. Recently, it has been demonstrated that the cortical actin is a major barrier for HIV infection of resting CD4 T cells (44) and is involved in regulating CXCR4 cycling, viral DNA synthesis, and nuclear migration (8, 35, 36, 42, 44). Therefore, we examined the natural response of the cortical actin to centrifugal pressure and the ensuing effects on viral infection.
MATERIALS AND METHODS
Isolation of resting CD4 T cells from peripheral blood.
Resting CD4 T cells were purified from peripheral blood by two rounds of negative selection as previously described (41). Briefly, for the first-round depletion, we used monoclonal antibodies against human CD14, CD56, and HLA-DR, -DP, and -DQ (BD Biosciences). For the second-round depletion, we used monoclonal antibodies against human CD8, CD11b, and CD19 (BD Biosciences). Antibody-bound cells were depleted by using Dynabeads pan mouse IgG (Invitrogen). Purified cells were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen), penicillin (50 U/ml) (Invitrogen), and streptomycin (50 μg/ml) (Invitrogen). Cells were rested overnight before infection or treatment.
Virus preparation and infection of T cells.
Virus stocks of HIV-1NL4-3 (1) and HIV-1AD8 were prepared by transfection of HeLa cells with cloned proviral DNA as described previously (41). Supernatant was harvested at 48 h and filtered through a 0.45-μm nitrocellulose membrane. The virus titer (50% tissue culture infectious dose [TCID50]) was measured by infection of a Rev-dependent green fluorescent protein (GFP) indicator cell line, Rev-CEM (38, 39).
For infection of resting CD4 T cells, 103.5 to 104.5 TCID50 units of HIV-1 were used to infect 106 cells. For infection, CD4 T cells were incubated with the virus for 2 h at room temperature or centrifuged at 300 × g or 1,200 × g for 2 h at room temperature and then washed twice with medium to remove unbound virus. Infected cells were resuspended into fresh RPMI-1640 medium supplemented with 10% heat-inactivated fetal bovine serum, penicillin (50 U/ml), and streptomycin (50 μg/ml) at a density of 106 per ml and incubated for 5 days without stimulation. Cells were activated at day 5 with anti-CD3/CD28 magnetic beads at 4 beads per cell. For the viral replication assay, 10% of the infected cells was taken at days 1, 3, 5, 6, 7, 8, and 9 postinfection. Cells were pelleted, and the supernatant was saved for p24 enzyme-linked immunosorbent assay (ELISA). The levels of p24 in the supernatant were measured using a Coulter HIV-1 p24 assay kit (Beckman Coulter) or a Perkin Elmer Alliance p24 antigen ELISA kit (Perkin Elmer). The plates were read kinetically using an ELx808 automatic microplate reader (Bio-Tek Instruments) at 630 nm.
For infection of Rev-CEM cells (38), 200 to 500 ng of HIV-1NL4-3 was used to infect 2 × 105 cells. Cells were incubated with the virus for 2 h at room temperature or centrifuged at 300 × g, 600 × g, or 1,200 × g for 2 h at room temperature and then washed twice with medium to remove unbound virus. Infected cells were resuspended into 1 ml of fresh RPMI-1640 medium supplemented with 10% heat-inactivated fetal bovine serum, penicillin (50 U/ml), and streptomycin (50 μg/ml). The HIV-1 replication efficiency was measured by flow cytometry with propidium iodide (PI) added to exclude dead cells.
The LIMK1 knockdown cell line shLIMK 007 and the control cell line shNTC were generated as described previously (35). All cells were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen), penicillin (50 U/ml), streptomycin (50 μg/ml), and puromycin (1 μg/ml). Cells were infected or spinoculated with HIV-1NL4-3 for 2 h at room temperature, washed twice, and then resuspended in fresh medium at 2 × 105 per ml. Supernatant was taken daily to monitor viral replication by p24 ELISA.
Conjugation of antibodies to magnetic beads and stimulation of resting CD4 T cells.
Monoclonal antibodies against human CD3 (clone UCHT1) and CD28 (clone CD28.2) were from BD Pharmingen (BD Biosciences). These antibodies were conjugated to magnetic beads and used to stimulate resting CD4 T cells as previously described (44).
Pretreatment of Rev-CEM cells with the inhibitor jasplakinolide.
Cells were pretreated with jasplakinolide (Jas) (Invitrogen) (4) for 1 h at 37°C and then infected with HIV-1 in the continuous presence of Jas for 2 h. Following infection, cells were washed twice with medium, resuspended into fresh medium, and then cultured in the absence of Jas.
Surface staining of CD4 and CXCR4 receptors.
Phycoerythrin (PE)-labeled monoclonal antibody against human CD4 (clone RPA-T4) or CXCR4 (clone 12G5) was purchased from BD Biosciences. One-half million cells were incubated with 1 μg mouse IgG (Jackson ImmunoResearch Laboratories) and then stained with the labeled antibodies on ice in phosphate-buffered saline (PBS)−0.1% bovine serum albumin (BSA) for 30 min. Cells were washed with cold PBS−0.1% BSA and then analyzed on a FACSCalibur (BD Biosciences).
FITC-phalloidin staining of F-actin and flow cytometry.
One million cells were centrifuged at 300 × g for various time periods. F-actin staining was carried out as previously described (44). Briefly, staining was carried out using 1 × 106 cells. Cells were pelleted, fixed, and permeabilized with CytoPerm/Cytofix buffer (BD Biosciences) for 20 min at room temperature and washed with cold Perm/Wash buffer (BD Biosciences) twice, followed by staining with 5 μl of 0.3 mM fluorescein isothiocyanate (FITC)-labeled phalloidin for 30 min on ice in the dark. After being washed twice with cold Perm/Wash buffer, cells were resuspended in 1% paraformaldehyde and analyzed on a FACSCalibur (BD Biosciences). For the time course experiment, multiple centrifuges were used to ensure equal fixation and washing times between samples. Our preliminary study found that varied fixation times can affect staining intensity, whereas the time in cold Perm/Wash buffer had a minimal impact on staining.
Western blotting to measure cofilin activation.
One million cells were lysed in NuPAGE LDS (lithium dodecyl sulfate) sample buffer (Invitrogen), followed by sonication. Samples were heated at 70°C for 10 min, separated by SDS-PAGE, and then transferred onto nitrocellulose membranes (Invitrogen). The membranes were washed in TBST (50 mM Tris·HCl [pH 7.4], 150 mM NaCl, 0.1% Tween 20) for 3 min and then blocked for 30 min at room temperature with StartingBlock blocking buffer (Pierce). The blots were incubated with a mouse anti-cofilin antibody (1:1,000 dilution) (BD Biosciences) and a rabbit anti-phospho-cofilin (ser3) antibody (1:500 dilution) (Cell Signaling) diluted in 2.5% milk-TBST and rocked overnight at 4°C. The blots were washed three times for 15 min and then incubated with goat anti-mouse 680-labeled and goat anti-rabbit 800cw-labeled antibodies (Li-Cor Biosciences) (1:5,000 diluted in blocking buffer) for 1 h at 4°C. The blots were washed three times for 15 min and scanned with an Odyssey infrared imager (Li-Cor Biosciences).
Real-time PCR amplification of viral DNA.
Real-time PCR quantification of viral late DNA and 2-long terminal repeat (2-LTR) circles was performed as described previously (40, 44). For measuring 2-LTR circles, a nested PCR approach was used. The DNA was first amplified with the primers LTR-nef2 and LTR-gag (40), followed by real-time PCR with primers MH536, MH535, and MH603 (6).
Alu-PCR amplification of viral integration.
Alu-PCR quantification of viral integration was performed as described previously (44). Briefly, DNA from HeLa-NLneo cells was extracted and diluted into uninfected HeLa cellular DNA to serve as a standard. For measurement of integration, total genomic DNA was purified and PCR amplified using Alu-forward primer (5′-GCCTCCCAAAGTGCTGGGATTACAG-3′) and Alu-gag-reverse primer (5′-GCTCTCGCACCCATCTCTCTCC-3′) as previously described (44). Background amplification from unintegrated viral DNA was also measured as a control, using the Alu-gag-reverse primer alone (gag-only primer). Following the first-round PCR amplification, an aliquot equivalent to one-fifth the volume of the Alu-PCR products was used for real-time PCR analysis of viral DNA as described above. The copy number of integrated viral DNA was calculated based on the integrated viral DNA standard prepared from HeLa-NLneo cells.
RESULTS
Spinoculation enhances HIV infection of transformed and resting CD4 T cells.
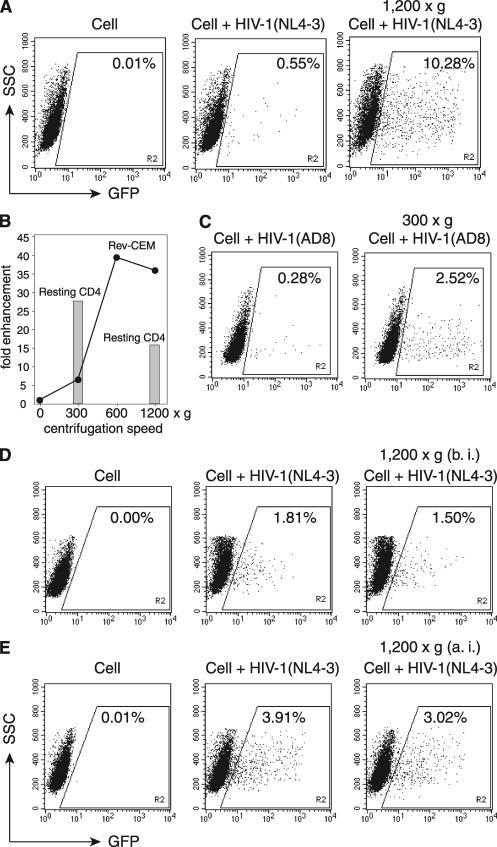
To demonstrate spin-mediated enhancement of HIV-1 infection of human CD4 T cells, we used an HIV-1 Rev-dependent GFP indicator cell line, Rev-CEM, to measure HIV-1 infection (38). Cells were infected either directly or under conditions of 1,200 × g for 2 h. As shown in Fig. 1A, we observed an approximately 18-fold increase in the percentage of GFP-positive cells. We also performed spinoculation under different gravity forces. In general, the enhancement was speed dependent, but it could be seen at a speed as low as 300 × g (Fig. 1B). The experiments were repeated multiple times, and the enhancement at 300 × g was about 5- to 10-fold for Rev-CEM cells. This enhancement was not HIV-1 tropism dependent, as we observed spin-mediated enhancement using HIV-1AD8 (Fig. 1C). Similar enhancement was also observed in HIV-1 latent infection of resting CD4 T cells purified from peripheral blood (Fig. 1B). Surprisingly, the enhancement at 300 × g was 4-fold greater than that of Rev-CEM cells, and further spinning of resting T cells at 1,200 × g reduced rather than increased the enhancement. These results suggested that concentrating viruses may not solely explain the enhancement. Possible impacts of persistent low-speed spin on cellular conditions may also contribute to the process. Nevertheless, spinning Rev-CEM cells before (Fig. 1D) or after infection (Fig. 1E) had minimal effects on HIV infection, suggesting that if there is any cellular effect, it would probably be transient and concurrent with the early processes of viral infection.
Fig. 1.
Spinoculation enhances HIV-1 infection of transformed and resting CD4 T cells. (A) To demonstrate spin-mediated enhancement of HIV-1 infection, Rev-CEM cells (2 × 105 cells) were not infected (Cell) or were infected with HIV-1NL4-3 for 2 h in the absence or presence of spin at 1,200 × g. SSC, side scatter. (B) Rev-CEM cells were infected similarly at different centrifugal speeds. Shown are the levels of enhancement (fold) based on the percentages of GFP-positive cells spun as shown. Cells were washed twice and cultured for 2 days, and then GFP-positive cells were measured by flow cytometry. Resting CD4 T cells (1 × 106) were also spinoculated at 300 × g or 1,200 × g for 2 h, washed, cultured for 5 days, and then activated with CD3/CD28 stimulation. Shown are the levels of enhancement (fold) of viral replication at day 9 postinfection for cells spun as shown. (C) Rev-CEM cells were infected similarly with HIV-1AD8 at 300 × g. (D) Effects of spinning cells prior to HIV infection. Rev-CEM cells were directly infected with HIV-1NL4-3 or centrifuged at 1,200 × g for 2 h before infection (b. i.) and then infected with HIV-1NL4-3 for 2 h in the absence of spin. (E) Effects of spinning cells after HIV infection. In a separate experiment, Rev-CEM cells were directly infected with HIV-1 for 2 h or infected with HIV-1 for 2 h, washed twice with medium, and then centrifuged at 1,200 × g for 2 h. Cells were cultured for 2 days, and then GFP-positive cells were measured by flow cytometry.
The concentrating effect of spinoculation.
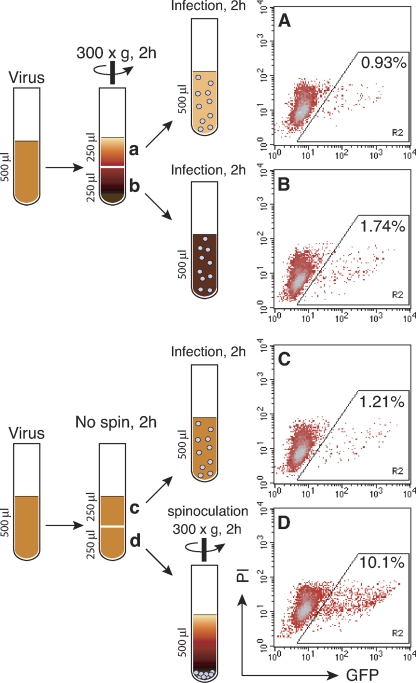
Given the complications of concurrent events in spinoculation, we attempted to separate the virus-concentrating effect from possible spin-imposed effects on cells. We decided to use a minimal speed of 300 × g. This would permit us to study cellular responses in a low background of virus sedimentation. To determine whether 300 × g was sufficient to pellet viruses, we first centrifuged the virus at this speed for 2 h (Fig. 2), and then, equal volumes of the top and bottom portions were used to infect Rev-CEM cells. From the top layer, we observed a minimal decrease of the GFP-positive cells, to 0.93% (Fig. 2A) in comparison with the not-spun control (1.21%) (Fig. 2C). This was roughly a 20% depletion of viruses from the top layer, which is consistent with an increase in the GFP-positive cells infected from the bottom layer to 1.74% (Fig. 2B). These results suggested that there was a low-level concentrating effect induced by the spin. Nevertheless, most of the viruses remained in the supernatant after 2 h at 300 × g. We further measured direct sedimentation of viruses on cells. As shown in Fig. 3A, cells were first pelleted by a brief centrifugation at 300 × g for 5 min, and then viruses were laid on top of the supernatant and centrifuged at 300 × g for an additional 2 h. As a control, identical amounts of cells and viruses were mixed and then infected or spinoculated (300 × g) for 2 h (Fig. 3B and C). We did not observe significant enhancement of infection (from 0.72% to 0.73%) when the virus and cells were separated before centrifugation (Fig. 3A); again, the 2-h spin at 300 × g did not bring down excessive amounts of viruses. On the other hand, when the cells and virus were premixed and then spinoculated (Fig. 3C), we observed an approximately 10-fold enhancement (from 0.72% to 7.12%). These results suggested that the initial binding of virus to cell is important for the subsequent enhancement by spin. It is possible that the centrifugal force introduced to the virus-cell complex may trigger cellular processes to facilitate viral infection.
Fig. 2.
The concentrating effect of spinoculation. (A and B) Viral supernatant (500 μl, 1,000 ng p24) was centrifuged at 300 × g for 2 h at 4°C, and then equal volumes of the top (A) and bottom (B) portions were used to infect 2 × 105 cells for 2 h at room temperature. Cells were washed twice and cultured for 2 days, and then GFP-positive cells were measured by flow cytometry. (C and D) As controls, viral supernatants were incubated for 2 h at 4°C, and then equal volumes were used to infect 2 × 105 cells for 2 h at room temperature (C) or spinoculated at 300 × g (D) PI, propidium iodide.
Fig. 3.
Viral sedimentation in spinoculation. (A) Rev-CEM cells (250 μl, 2 × 105 cells) were centrifuged at 300 × g for 5 min, and then viral supernatant (250 μl, 500 ng p24) was gently loaded on top of the cell supernatant and centrifuged at 300 × g for 2 h. Cells were washed twice and cultured for 2 days, and then GFP-positive cells were measured by flow cytometry. (B and C) As controls, cells and viruses were mixed and infected for 2 h (B) or spinoculated at 300 × g (C).
Spinoculation triggers actin and cofilin activity.
The actin cytoskeleton provides major structural support for cells and is directly involved in the early processes of viral infection, specifically, viral entry, DNA synthesis, and postentry migration (8, 35, 36, 42, 44). We speculated that the gravity force generated from spin would have a direct impact on the actin cytoskeleton. Thus, we performed a time course analysis to monitor actin dynamics in Rev-CEM cells centrifuged at 300 × g. We observed robust actin activity, which is characterized by cycles of actin polymerization and depolymerization (Fig. 4A), probably resulting from cellular responses to persistent centrifugal pressure on cells. Because cofilin is a major regulator of actin dynamics (19), we also examined the activity of cofilin and found that cofilin is activated through phosphorylation and dephosphorylation (Fig. 4B). This result suggested that cofilin is directly involved in the spin-mediated actin dynamics.
Fig. 4.
Spinoculation triggers actin and cofilin activity. (A and B) Rev-CEM cells were centrifuged at 300 × g in 2-ml round-bottom tubes using fixed-angle microcentrifuges for various times, fixed, and then stained with FITC-phalloidin for analysis of F-actin by flow cytometry (A). Cells were also directly lysed following spin and measured by Western blotting using anti-phospho-cofilin (p-cofilin) or anti-cofilin antibodies. The relative intensity of F-actin staining was plotted (B). (C to F) The effects of spin (at 300 × g or 1,200 × g) on CD4 and CXCR4 expression were also measured by surface staining and flow cytometry.
The cortical actin also directly regulates CXCR4 cycling in T cells (35). Thus, we further examined the effect of spinoculation on HIV receptors, both CD4 and CXCR4, and found that the surface density of CXCR4 was upregulated (Fig. 4C). The increase in surface CXCR4 can be detected as early as 15 min (Fig. 4D). On the other hand, the CD4 receptor was not upregulated at 300 × g (Fig. 4E) but was upregulated at 1,200 × g after spinning for 1 h (Fig. 4F), suggesting that higher centrifugal stress could nonspecifically upregulate multiple receptors on the cell surface.
The actin inhibitor jasplakinolide diminishes spin-mediated enhancement.
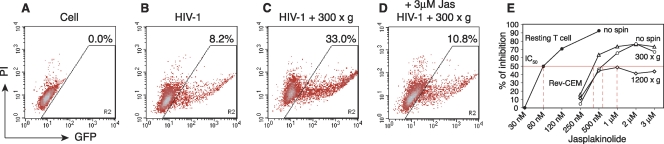
We sought to confirm a definitive role of actin in spin-mediated enhancement. To this end, we used a filamentous-actin (F-actin)-stabilizing agent, jasplakinolide (Jas). Jas binds to F-actin irreversibly and prevents F-actin depolymerization (4). We speculated that Jas could diminish spin-mediated enhancement through direct inhibition of actin activity. As shown in Fig. 5, we observed that 3 μM Jas largely diminished the spin-mediated enhancement of HIV-1 infection, demonstrating the direct involvement of F-actin in this process.
Fig. 5.
Effects of jasplakinolide on spinoculation. (A to D) Rev-CEM cells (2 × 105 cells) were not infected (A) or were infected with HIV-1NL4-3 for 2 h (B), spinoculated at 300 × g for 2 h (C), or spinoculated in the presence of 3 μM Jas for 2 h (D). Following infection or spinoculation, cells were washed twice and cultured in fresh medium in the absence of Jas. GFP-positive cells were measured at 48 h. To exclude cytotoxicity, propidium iodide (PI) was added before flow cytometry, and GFP-positive cells were counted only in the PI-negative, viable-cell population. (E) Rev-CEM cells were also pretreated with different dosages of Jas for 1 h and then infected (no spin) or spinoculated. The 50% inhibition dosage was calculated based on the percentage of inhibition of GFP-positive cells in comparison with the inhibition of cells not treated with Jas. Resting CD4 T cells were also treated with Jas for 1 h and infected in the presence of Jas for 2 h. Cells were washed and incubated in the absence of Jas for 5 days and then activated with CD3/CD28 stimulation. Viral replication was measured by p24 release and compared with that in infected CD4 T cells not treated with Jas. Shown are the average results for blood samples from two donors.
Previously, we had observed that HIV-1 infection of resting CD4 T cells in blood was more sensitive to Jas inhibition than that of transformed T cells (44). We further postulated that the cortical actin in resting T cells may be relatively static in the absence of cell cycle or chemotactic stimulation and, thus, can be effectively inhibited by low dosages of Jas. On the other hand, in transformed T cells, the cell cycle regulates actin dynamics, and to some extent, the relative resistance to Jas inhibition could be used to reflect the relative actin dynamics in cells. Thus, we performed a Jas dosage-dependent inhibition of HIV-1 infection. As shown in Fig. 5E, the Jas dosage for 50% inhibition of HIV infection (50% inhibitory concentration [IC50]) was 60 nM for resting T cells. As expected, the Jas IC50 for HIV-1 infection of transformed Rev-CEM cells was increased to 250 to 500 nM. In addition, spinning of Rev-CEM cells at 300 × g further increased the IC50 to above 500 nM. Moreover, when Rev-CEM cells were spun at 1,200 × g, the IC50 approached approximately 1 μM, and even higher Jas dosages, such as 3 μM, did not inhibit HIV-1 by more than 50%. These results confirmed that spinoculation triggers actin dynamics and that this actin activity can partially overcome the inhibitory effect of the actin inhibitor.
LIMK1 knockdown decreases spin-mediated enhancement.
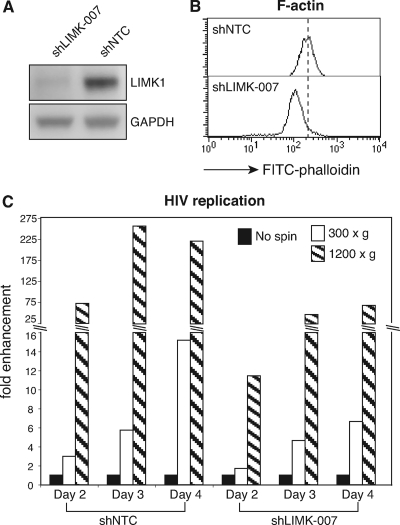
As a complementary approach to address the role of actin in spinoculation, we took advantage of a recent siRNA knockdown study on the cofilin kinase LIMK1 (35). It has been shown that knockdown of LIMK1 through small interfering RNA (siRNA) decreases filamentous actin and reduces actin dynamics and the ability of cells to migrate toward SDF-1 (21, 35). We speculated that with a decreased cortical actin density and actin dynamics in the LIMK1 knockdown cells (Fig. 6A and B), spin-mediated enhancement would also be diminished. As shown in Fig. 6C, we observed a substantial reduction in spin-mediated enhancement of HIV replication in shLIMK-007 cells in comparison with the control shNTC cells (short hairpin RNA [shRNA] nontargeting control) (a 257-fold enhancement in shNTC versus a 46-fold enhancement in shLIMK-007 at 1,200 × g on day 3). These results suggested again that the cortical actin plays a critical role in the enhancement of viral infection by spinoculation. These data also indicated that the spin-mediated enhancement may not be attributed simply to a virus-concentrating effect; otherwise, similar enhancement would be seen for both shLIMK-007 and shNTC cells.
Fig. 6.
LIMK1 knockdown decreases spin-mediated enhancement. (A) The LIMK1 knockdown cell line, shLIMK-007, and the control cell line, shNTC, were examined by Western blotting using an anti-LIMK1/2 antibody or an anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody for the loading control. (B) These cells were also examined by F-actin staining with FITC-phalloidin. (C) shLIMK-007 or shNTC cells (2 × 105) were infected with HIV-1NL4-3 (200 ng p24) or spinoculated (at 300 × g or 1,200 × g) for 2 h, washed, and then incubated for 4 days. Viral replication was monitored by p24 release. The fold enhancement of viral replication by spinoculation was calculated using not-spun infected cells as “1.”
Spinoculation induces HIV-1 integration in resting CD4 T cells.
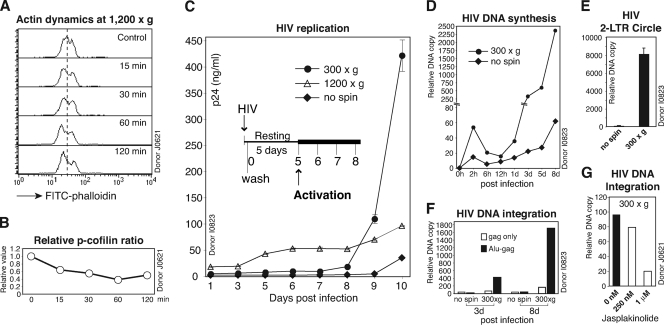
Several recent studies have demonstrated an important role of the actin cytoskeleton in the establishment of HIV-1 latent infection of resting CD4 T cells (8, 35, 44, 45). It has been suggested that the cortical actin in resting T cells is relatively static and represents a restriction and that HIV-1 utilizes gp120-CXCR4 interaction to activate cofilin to overcome this restriction (44). Consistently, chemokines such as CCL19 activate cofilin and actin activity, promoting efficient HIV-1 nuclear localization and DNA integration in resting CD4 T cells (8). Given that spinoculation triggers dynamic actin and cofilin activity (Fig. 7A and B), we performed spinoculation of resting T cells and observed a great enhancement of HIV replication (Fig. 7C). Spinoculation at 1,200 × g did not enhance viral replication as much as that at 300 × g (day 9 and day 10), although there was a higher early release of viral p24 (from day 5). The reason for this lower enhancement at 1,200 × g remains unknown, but it could result from possible cytopathic effects of persistent centrifugal stress on resting T cells.
Fig. 7.
Spinoculation induces HIV-1 integration in resting CD4 T cells. (A and B) Spinoculation triggers actin and cofilin activity in resting CD4 T cells. Cells were centrifuged at 1,200 × g in 2-ml round-bottom tubes in fixed-angle microcentrifuges for various times, fixed, and then stained with FITC-phalloidin for analysis of F-actin by flow cytometry (A). Cells were also directly lysed following spin and measured by Western blotting using anti-phospho-cofilin (p-cofilin) and an anti-GAPDH antibody for the loading control. The relative p-cofilin ratio was plotted (B). (C) Resting CD4 T cells (1 × 106) were infected (no spin) or spinoculated with HIV-1NL4-3 (200 ng p24) at 300 × g or 1,200 × g for 2 h, washed, cultured for 5 days, and then activated with CD3/CD28 stimulation. Viral replication was monitored by p24 release. (D) Viral DNA synthesis was also measured by real-time PCR (using 100 ng total cellular DNA). (E) The synthesis of viral 2-LTR circles at day 3 postinfection was measured by a nested real-time PCR method. (F) Viral DNA integration at day 3 and day 8 postinfection was measured by real-time Alu-PCR in the presence (Alu-gag) or absence (gag only) of the Alu primer (using 200 ng total cellular DNA). (G) Inhibition of spin-mediated HIV-1 integration by jasplakinolide. Resting CD4 T cells were treated with Jas for 1 h, spinoculated with HIV-1NL4-3 (300 × g) for 2 h, washed, and cultured in the absence of Jas for 3 days. Viral DNA integration at day 3 postinfection was measured by real-time Alu-PCR.
We also measured viral DNA synthesized following spinoculation at 300 × g and observed higher levels of viral DNA at an early time (2 h) (Fig. 7D). Nevertheless, within 12 h, most of the DNA was degraded to a level comparable to that of the not-spun control cells. The faster DNA synthesis and decay rate at the early time suggested that spinoculation probably enhanced viral uptake via both the productive and nonproductive pathways, such as endocytotic entry (45). Viral DNA synthesis following day 1 was also considerably higher in spinoculated cells. This was accompanied by higher amounts of 2-LTR circles in these cells (Fig. 7E). These results demonstrated that spinoculation mediated higher levels of viral DNA synthesis and nuclear migration. Consistently, measurement of viral DNA integration by Alu-PCR revealed that a detectable amount of viral DNA integrated at day 3 postinfection, even before T cell activation at day 5 (Fig. 7F). The result is in agreement with a previous demonstration that HIV can integrate into resting CD4 T cells when spinoculated (30). Nevertheless, in the absence of spinoculation or T cell activation, HIV integration in resting T cells has been difficult to detect with the standard Alu-PCR (41, 44) but could be detectable by repeated sampling and ultrasensitive PCR (2). Collectively, our data demonstrated that spinoculation shares certain similarities with chemokines such as CCL19 in triggering actin and cofilin activity, which greatly promoted viral nuclear migration and DNA integration in resting CD4 T cells (8).
DISCUSSION
In this article, we demonstrate that spin-mediated enhancement of HIV-1 infection cannot be explained by a single concentrating effect (30); rather, it is coupled with spin-induced actin and cofilin activity that enhances viral binding, entry, postentry DNA synthesis, and nuclear migration. Our study highlights the importance of a dynamic actin cytoskeleton for the initiation of viral infection.
The spin-triggered cofilin activation seems to be different from ligand-mediated activation. The early cofilin activation via dephosphorylation (at 15 min and 30 min) (Fig. 4B) did not concur with the actin depolymerization; rather, it followed the actin activity (at 10 min and 25 min) (Fig. 4B). It is possible that activation of cofilin may result from a cellular response to balance spin-induced actin activities. This “compensatory signaling” may begin with pressure-induced breakage of actin filaments, followed by actin polymerization for pressure resistance. This actin polymerization, in turn, may trigger a default feedback pathway for cofilin activation to depolymerize actin. After several cycles, cells may eventually reach a balance between actin polymerization and depolymerization, with an increased treadmilling speed (Fig. 4B). For HIV-1 infection, such spin-induced actin and cofilin dynamics are aligned with the evolving capacity of the virus (37) to modulate actin and cofilin activities through viral proteins, such as gp120 (42, 44) and Nef (28). In vivo, it is likely that such viral capacity is greatly aided by cytokines and chemokines, such as CCL19 and CCL21, to facilitate infection (8, 10, 11, 25, 36).
The limited ability of HIV-1 to infect resting CD4 T cells in the absence of chemotactic stimulation or T cell activation has been clearly demonstrated by in vitro studies (27, 41). Although HIV-1 can trigger actin and cofilin activity in resting T cells through gp120 binding to the chemokine coreceptors (44), this transit activity facilitates viral nuclear migration but permits minimal (2) or no DNA integration (41). However, both spinoculation and chemokines can mediate efficient viral DNA integration in resting T cells (8, 30, 34). A remaining question is whether spinoculation or chemokines can also mediate a cellular condition (in addition to triggering actin and cofilin activity) to favor integration. It is tempting to speculate that both treatments may also modulate chromatin to permit efficient integration. Certainly, actin and actin-related proteins, such as Arp7, Arp9, and Baf53, have been found to be a part of the chromatin-remodeling complexes RSC and SWI/SNF (7, 46). These proteins may serve to connect the cytoskeletal processes to chromatin remodeling. Nevertheless, it is also possible that there is no real restriction for HIV to integrate in resting CD4 T cells; the only limitation would be the inability of HIV to translocate a sufficient amount of DNA into the nucleus to find an open chromatin location. Once this limitation is overcome by chemokines or spinoculation, integration occurs naturally.
In this study, we only examined spin-mediated actin and cofilin activity. Likewise, spinoculation may also affect other components of the cytoskeleton, such as microtubules. In addition, we did not examine surface receptors other than CD4/CXCR4. We also expect that multiple receptors may be affected by spinoculation. It has been suggested that cortical actin is a barrier to cellular uptake and secretory processes (24). Thus, it is possible that the spin-mediated actin dynamics would have a large impact on receptor cycling. Given that the spin-mediated receptor dynamics concur with a low-level virus-concentrating effect, the combined effect would have a dramatic impact on viral entry.
We did not examine intracellular signaling molecules other than cofilin and LIMK. It has been known that mechanical shear stress can activate multiple surface receptors and intracellular kinases, such as protein kinase C (PKC), focal adhesion kinases (FAKs), Rho family small GTPases, phosphatidylinositol 3-kinase (PI3K), mitogen-activated protein kinases (MAPKs), and NF-κB (20). Although the role of these signaling molecules in spin-mediated enhancement of HIV-1 infection was not investigated in our study, we do not exclude their possible involvement in this process. Nevertheless, spinning cells before or after HIV-1 infection did not significantly enhance HIV-1 replication (Fig. 1D and E), suggesting that the enhancement is mainly associated with promoting the early steps of viral infection and may not be caused by a general upregulation of the cellular signaling and transcriptional environment. Finally, given the broad effects of spinoculation on the cytoskeleton, cell signaling, and receptor dynamics, caution needs to be taken in data interpretation when cells are spinoculated; certain viral requirements for signal pathways can be forfeited. In addition, some of the spin-induced cellular permissiveness may be beyond the natural capacity of an infecting virus.
ACKNOWLEDGMENTS
We thank the George Mason University (GMU) Student Health Center for blood donations, U. O'Doherty for Alu/real-time PCR protocols, C. Stell for editorial assistance, and the NIH AIDS Research and Reference Reagent Program, NIAID, NIH, for reagents.
This work was supported by NIH Public Health Service Grant 1R01AI081568 from NIAID to Y.W. W.W. was supported in part by the 2010 NYCDC AIDS Ride organized by M. Rosen.
Footnotes
Published ahead of print on 27 July 2011.
REFERENCES
- 1. Adachi A., et al. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Agosto L. M., et al. 2007. HIV-1 integrates into resting CD4(+) T cells even at low inoculums as demonstrated with an improved assay for HIV-1 integration. Virology 368:60–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bahnson A. B., et al. 1995. Centrifugal enhancement of retroviral mediated gene transfer. J. Virol. Methods 54:131–143 [DOI] [PubMed] [Google Scholar]
- 4. Bubb M. R., Spector I., Beyer B. B., Fosen K. M. 2000. Effects of jasplakinolide on the kinetics of actin polymerization. An explanation for certain in vivo observations. J. Biol. Chem. 275:5163–5170 [DOI] [PubMed] [Google Scholar]
- 5. Bunnell B. A., Muul L. M., Donahue R. E., Blaese R. M., Morgan R. A. 1995. High-efficiency retroviral-mediated gene transfer into human and nonhuman primate peripheral blood lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 92:7739–7743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Butler S. L., Hansen M. S., Bushman F. D. 2001. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 7:631–634 [DOI] [PubMed] [Google Scholar]
- 7. Cairns B. R., Erdjument-Bromage H., Tempst P., Winston F., Kornberg R. D. 1998. Two actin-related proteins are shared functional components of the chromatin-remodeling complexes RSC and SWI/SNF. Mol. Cell 2:639–651 [DOI] [PubMed] [Google Scholar]
- 8. Cameron P. U., et al. 2010. Establishment of HIV-1 latency in resting CD4+ T cells depends on chemokine-induced changes in the actin cytoskeleton. Proc. Natl. Acad. Sci. U. S. A. 107:16934–16939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Conklin E. G. 1910. The effects of centrifugal force upon the organization and development of the eggs of fresh water pulmonates. J. Exp. Zool. 9:417–454 [Google Scholar]
- 10. Damas J. K., et al. 2009. Enhanced levels of the CCR7 ligands CCL19 and CCL21 in HIV infection: correlation with viral load, disease progression and response to highly active antiretroviral therapy. AIDS 23:135–138 [DOI] [PubMed] [Google Scholar]
- 11. Damas J. K., et al. 2009. Homeostatic chemokines CCL19 and CCL21 promote inflammation in human immunodeficiency virus-infected patients with ongoing viral replication. Clin. Exp. Immunol. 157:400–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Forestell S. P., Dando J. S., Bohnlein E., Rigg R. J. 1996. Improved detection of replication-competent retrovirus. J. Virol. Methods 60:171–178 [DOI] [PubMed] [Google Scholar]
- 13. Gey G. O., Bang F. B., Gey M. K. 1954. Responses of a variety of normal and malignant cells to continuous cultivation, and some practical applications of these responses to problems in the biology of disease. Ann. N. Y. Acad. Sci. 58:976–999 [DOI] [PubMed] [Google Scholar]
- 14. Ho W. Z., et al. 1993. Centrifugal enhancement of human immunodeficiency virus type 1 infection and human cytomegalovirus gene expression in human primary monocyte/macrophages in vitro. J. Leukoc. Biol. 53:208–212 [DOI] [PubMed] [Google Scholar]
- 15. Hodgkin P. D., Scalzo A. A., Swaminathan N., Price P., Shellam G. R. 1988. Murine cytomegalovirus binds reversibly to mouse embryo fibroblasts: implications for quantitation and explanation of centrifugal enhancement. J. Virol. Methods 22:215–230 [DOI] [PubMed] [Google Scholar]
- 16. Hudson J. B. 1988. Further studies on the mechanism of centrifugal enhancement of cytomegalovirus infectivity. J. Virol. Methods 19:97–108 [DOI] [PubMed] [Google Scholar]
- 17. Hudson J. B., Misra V., Mosmann T. R. 1976. Cytomegalovirus infectivity: analysis of the phenomenon of centrifugal enhancement of infectivity. Virology 72:235–243 [DOI] [PubMed] [Google Scholar]
- 18. Hughes J. H. 1993. Physical and chemical methods for enhancing rapid detection of viruses and other agents. Clin. Microbiol. Rev. 6:150–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lappalainen P., Drubin D. G. 1997. Cofilin promotes rapid actin filament turnover in vivo. Nature 388:78–82 [DOI] [PubMed] [Google Scholar]
- 20. Li Y. S., Haga J. H., Chien S. 2005. Molecular basis of the effects of shear stress on vascular endothelial cells. J. Biomech. 38:1949–1971 [DOI] [PubMed] [Google Scholar]
- 21. Nishita M., Aizawa H., Mizuno K. 2002. Stromal cell-derived factor 1alpha activates LIM kinase 1 and induces cofilin phosphorylation for T-cell chemotaxis. Mol. Cell. Biol. 22:774–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. O'Doherty U., Swiggard W. J., Malim M. H. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74:10074–10080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Osborn J. E., Walker D. L. 1968. Enhancement of infectivity of murine cytomegalovirus in vitro by centrifugal inoculation. J. Virol. 2:853–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pfeiffer J. R., Seagrave J. C., Davis B. H., Deanin G. G., Oliver J. M. 1985. Membrane and cytoskeletal changes associated with IgE-mediated serotonin release from rat basophilic leukemia cells. J. Cell Biol. 101:2145–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Saleh S., et al. 2007. CCR7 ligands CCL19 and CCL21 increase permissiveness of resting memory CD4+ T cells to HIV-1 infection: a novel model of HIV-1 latency. Blood 110:4161–4164 [DOI] [PubMed] [Google Scholar]
- 26. Scanlan P. M., Tiwari V., Bommireddy S., Shukla D. 2005. Spinoculation of heparan sulfate deficient cells enhances HSV-1 entry, but does not abolish the need for essential glycoproteins in viral fusion. J. Virol. Methods 128:104–112 [DOI] [PubMed] [Google Scholar]
- 27. Spina C. A., Guatelli J. C., Richman D. D. 1995. Establishment of a stable, inducible form of human immunodeficiency virus type 1 DNA in quiescent CD4 lymphocytes in vitro. J. Virol. 69:2977–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stolp B., et al. 2009. HIV-1 Nef interferes with host cell motility by deregulation of cofilin. Cell Host Microbe 6:174–186 [DOI] [PubMed] [Google Scholar]
- 29. Sundin D. R., Mecham J. O. 1989. Enhanced infectivity of bluetongue virus in cell culture by centrifugation. J. Clin. Microbiol. 27:1659–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Swiggard W. J., et al. 2005. Human immunodeficiency virus type 1 can establish latent infection in resting CD4+ T cells in the absence of activating stimuli. J. Virol. 79:14179–14188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tenser R. B. 1978. Ultracentrifugal inoculation of herpes simplex virus. Infect. Immun. 21:281–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tenser R. B., Dunstan M. E. 1980. Mechanisms of herpes simplex virus infectivity enhanced by ultracentrifugal inoculation. Infect. Immun. 30:193–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tschopp A., Cogoli A. 1983. Hypergravity promotes cell proliferation. Experientia 39:1323–1329 [DOI] [PubMed] [Google Scholar]
- 34. Unutmaz D., KewalRamani V. N., Marmon S., Littman D. R. 1999. Cytokine signals are sufficient for HIV-1 infection of resting human T lymphocytes. J. Exp. Med. 189:1735–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vorster P. J., et al. 2011. LIM kinase 1 modulates cortical actin and CXCR4 cycling and is activated by HIV-1 to initiate viral infection. J. Biol. Chem. 286:12554–12564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu Y. 2010. Chemokine control of HIV-1 infection: beyond a binding competition. Retrovirology 7:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu Y. 2009. The co-receptor signaling model of HIV-1 pathogenesis in peripheral CD4 T cells. Retrovirology 6:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu Y., Beddall M. H., Marsh J. W. 2007. Rev-dependent indicator T cell line. Curr. HIV Res. 5:395–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu Y., Beddall M. H., Marsh J. W. 2007. Rev-dependent lentiviral expression vector. Retrovirology 4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu Y., Marsh J. W. 2003. Early transcription from nonintegrated DNA in human immunodeficiency virus infection. J. Virol. 77:10376–10382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wu Y., Marsh J. W. 2001. Selective transcription and modulation of resting T cell activity by preintegrated HIV DNA. Science 293:1503–1506 [DOI] [PubMed] [Google Scholar]
- 42. Wu Y., et al. 2008. Cofilin activation in peripheral CD4 T cells of HIV-1 infected patients: a pilot study. Retrovirology 5:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ye L., et al. 2008. Centrifugal enhancement of hepatitis C virus infection of human hepatocytes. J. Virol. Methods 148:161–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yoder A., et al. 2008. HIV envelope-CXCR4 signaling activates cofilin to overcome cortical actin restriction in resting CD4 T cells. Cell 134:782–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yu D., Wang W., Yoder A., Spear M., Wu Y. 2009. The HIV envelope but not VSV glycoprotein is capable of mediating HIV latent infection of resting CD4 T cells. PLoS Pathog. 5:e1000633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhao K., et al. 1998. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 95:625–636 [DOI] [PubMed] [Google Scholar]