Abstract
The Acheta domesticus densovirus (AdDNV), isolated from crickets, has been endemic in Europe for at least 35 years. Severe epizootics have also been observed in American commercial rearings since 2009 and 2010. The AdDNV genome was cloned and sequenced for this study. The transcription map showed that splicing occurred in both the nonstructural (NS) and capsid protein (VP) multicistronic RNAs. The splicing pattern of NS mRNA predicted 3 nonstructural proteins (NS1 [576 codons], NS2 [286 codons], and NS3 [213 codons]). The VP gene cassette contained two VP open reading frames (ORFs), of 597 (ORF-A) and 268 (ORF-B) codons. The VP2 sequence was shown by N-terminal Edman degradation and mass spectrometry to correspond with ORF-A. Mass spectrometry, sequencing, and Western blotting of baculovirus-expressed VPs versus native structural proteins demonstrated that the VP1 structural protein was generated by joining ORF-A and -B via splicing (splice II), eliminating the N terminus of VP2. This splice resulted in a nested set of VP1 (816 codons), VP3 (467 codons), and VP4 (429 codons) structural proteins. In contrast, the two splices within ORF-B (Ia and Ib) removed the donor site of intron II and resulted in VP2, VP3, and VP4 expression. ORF-B may also code for several nonstructural proteins, of 268, 233, and 158 codons. The small ORF-B contains the coding sequence for a phospholipase A2 motif found in VP1, which was shown previously to be critical for cellular uptake of the virus. These splicing features are unique among parvoviruses and define a new genus of ambisense densoviruses.
INTRODUCTION
Insect parvoviruses (densoviruses) belong to the Densovirinae subfamily of the Parvoviridae and are small, isometric, nonenveloped viruses (diameter, ∼25 nm) that contain a linear single-stranded DNA of 4 to 6 kb (2, 3, 27). These viruses can be subdivided into two large groups, those with ambisense genomes and those with monosense genomes. Like vertebrate parvoviruses, all densoviruses have a genomic DNA with hairpins at both ends, often (but not necessarily for all genera) as inverted terminal repeats (ITRs). All densoviruses with ambisense genomes package both complementary strands in equimolecular ratios as single strands in separate capsids (27). The nonstructural (NS) gene cassette is found in the 5′ half of one genome strand, and the structural protein (VP) gene cassette is found in the 5′ half of the complementary strand. By convention, the genome is oriented so that the NS cassette is found in the left half. Expression strategies of densoviruses often involve (alternative) splicing and leaky scanning translation mechanisms (28). So far, the near-atomic structures of three densoviruses, Penaeus stylirostris densovirus (PstDNV), Bombyx mori densovirus 1 (BmDNV-1), and Galleria mellonella densovirus (GmDNV), have been solved (10, 11, 21). The capsid of densoviruses consists of 60 subunits (T=1) of identical proteins that may contain N-terminal extensions not involved in capsid formation but that confer additional functions to the capsid. One of these functions is a phospholipase A2 (PLA2) activity that is required for genome delivery during infection (34). Densoviruses are usually highly pathogenic for their natural hosts (5).
The monosense densoviruses have been classified into 3 uniform genera, i.e., Iteravirus, with a 5.0-kb genome, 0.25-kb ITRs, and a PLA2 motif in VP; Brevidensovirus, with a 4.0-kb genome, no ITRs but terminal hairpins, and no PLA2 motif; and Hepanvirus, with a single member, hepatopancreatic parvovirus, with a 6.3-kb genome also lacking a PLA2 motif and ITRs but with 0.2-kb terminal hairpins (23, 27). In contrast, the ambisense densoviruses have been classified into one uniform genus, Densovirus, with a 6-kb genome and 0.55-kb ITRs, and a second genus, Pefudensovirus, with only Periplaneta fuliginosa densovirus (PfDNV) as a member, with a 5.5-kb genome, 0.2-kb ITRs, and a split VP gene cassette (2, 26). Ribosome frameshifts have been proposed to connect its VP open reading frames (ORFs) (33). So far, all ambisense densoviruses have an N-terminal PLA2 motif in their largest VP. Some sequenced ambisense densoviruses, e.g., Myzus persicae densovirus (MpDNV) (32), Blattella germanica densovirus (BgDNV) (18), and Planococcus citri densovirus (PcDNV) (25), are as yet unclassified. The ambisense virus Culex pipiens densovirus (CpDNV) has a different genome organization for both the NS and VP proteins and will have to be classified in a different genus (1).
Acheta domesticus densovirus (AdDNV) was isolated from diseased Acheta domesticus L. house crickets from a Swiss commercial mass rearing facility (16). The virus spread rapidly and was responsible for high mortality rates, such that the rearing could not be saved. This was the first observation of a densovirus in an orthopteran species. Infected tissues included adipose tissue, the midgut, the hypodermis, and particularly the Malpighi tubules, but the most obvious pathological change was the completely empty digestive cecae (24). The cecae, which flank the proventriculus, are the sites where most enzymes are released and most absorption of nutrients occurs. Feulgen-positive masses were observed in the nuclei of infected cells (16). Commercial production facilities for the pet industry or for research mass rearings of house crickets in Europe are frequently affected by this pathogen. This virus was previously not known to circulate in North America, except for a small epidemic in the Southern United States in the 1980s (22). Beginning in 2009, sudden, severe outbreaks were observed in commercial facilities in Canada and the United States, leading to losses of hundreds of millions of dollars and to an acute crisis in the pet food industry (24). In this study on AdDNV, we observed that over the last 34 years the annual replacement rate was about 2.45 × 10−4 substitution/nucleotide (nt) and that the VP gene cassette consists of two ORFs, a characteristic of the Pefudensovirus genus (24).
In the present study, the complete genome and the expression strategy of AdDNV were investigated and showed features not yet described for other densoviruses or vertebrate parvoviruses. The most striking observation was the intricate splicing pattern of its VP ORFs, resulting, in contrast to the case for all parvoviruses studied so far, in unrelated N-terminal extensions of its two largest structural proteins and in the probable production of several supplementary NS proteins from the VP cassette.
MATERIALS AND METHODS
Rearing of crickets.
A. domesticus L. house crickets were obtained from a commercial supplier and were reared under conditions of about 60% relative humidity, 28°C, and a 16-h-8-h light-dark cycle. Diet conditions and drinking water supply, as well as conditions for perching, hiding, and oviposition, were as described previously (24).
Infection techniques.
The visceral cavity of nymphs of about 1.5 to 2 cm was injected with an inoculum consisting of a viral suspension obtained by grinding an infected cricket in 1× phosphate-buffered saline (PBS) plus 2% ascorbic acid, clarifying the mixture by centrifugation for 10 min at 8,000 × g, and filtering it through 450-nm membranes. Mortality was usually 100% within 2 weeks. Alternatively, infection was achieved by feeding with a virus-contaminated diet as previously described (24).
Virus and DNA preparation.
Virus was purified as previously described (29). Lysis buffer [300 μl of 6 M guanidine-HCl, 10 mM urea, 10 mM Tris-HCl, 20% Triton X-100, pH 4.4, containing 80 μg/ml poly(A) carrier RNA] and 200 μl sample were mixed and incubated for 10 min at 70°C. The sample was vortexed after adding 125 μl isopropanol, and the DNA was then purified on High-Pure plasmid spin columns (Roche Molecular Biochemicals) according to the supplier's instructions.
Cloning, mutation analysis, and sequencing of viral DNA.
The 1977 isolate of AdDNV was cloned into the pCR-XL-TOPO vector (Invitrogen Life Sciences), using supercompetent Sure 2 Escherichia coli cells (Stratagene) at 30°C. Point mutations in the AdDNV genome were generated with a QuikChange site-directed mutagenesis kit (Stratagene), whereas deletion mutants were obtained via the type IIb restriction endonuclease strategy (7). Independent clones were sequenced in both directions by primer walking. The terminal hairpins yielded compressions that were difficult to sequence; however, inclusion of 1 M betaine (Sigma) and 3% dimethyl sulfoxide (DMSO) or restriction in the hairpin by DraI yielded clean sequence reads. DNAs from subsequent isolates were amplified by PCR and sequenced between the ITRs.
Isolation and characterization of viral RNA.
Total RNAs were isolated from 30 mg adipose tissue from infected cricket larvae (2 to 5 days postinfection [p.i.]) and from recombinant baculovirus-infected cells at 48 h p.i. by use of an RNeasy minikit from Qiagen. The DNase I treatment was extended from 15 to 30 min or repeated twice. A PCR test was included to verify the absence of DNA. Total extracted RNA was subjected to mRNA purification using an mRNA isolation kit (Roche).
Northern blots.
About 20 to 30 μg total RNA in a 6-μl volume was added to 18 μl buffer (1× MOPS [20 mM morpholinepropanesulfonic acid, 5 mM sodium acetate, 0.5 mM EDTA adjusted to pH 8 with NaOH], 18.5% formaldehyde, 50% formamide), 5 μl loading buffer was added, and the mixture was incubated for 5 to 10 min at 65 to 70°C and separated by electrophoresis on a 1% formaldehyde-agarose gel. Parallel lanes contained RNA size markers (Promega). After migration and washing, RNAs were transferred to positively charged nylon membranes (Roche) by capillary blotting overnight. The blotted membranes were prehybridized with 10 mg/ml herring sperm DNA in 50% formamide before hybridization with 32P-labeled probes. The probes corresponded to a 1.5-kb BglII-SalI restriction fragment specific for the VP coding sequence and a 0.87-kb Eco47III-DraI restriction fragment specific for NS. Hybridized probes were visualized with a Storm 840 phosphorimager.
Mapping of 5′ ends, 3′ ends, and introns of viral transcripts.
The most probable locations of the transcripts were predicted from the ORFs obtained by sequence analysis. A 3′ rapid amplification of cDNA ends (3′-RACE) system was used to characterize the 3′ ends of the polyadenylated transcripts, using the RNAtag and ADAP primers (Table 1) and PCR (28), whereas the 5′ ends were determined with a FirstChoice RLM RACE kit (Ambion) according to the instructions of the supplier. The locations of introns were determined after reverse transcription of the transcripts by use of avian myeloblastosis virus (AMV) reverse transcriptase (Promega) in a final volume of 20 μl for 1 h at 42°C, PCR using internal DNV-specific primers for overlapping regions, and dideoxy sequencing of the amplicons according to standard methods (28).
Table 1.
PCR primers used in this study
| Primer | Sequencea | Position (nt) in AdDNV | Target or use |
|---|---|---|---|
| AdVPR | TTTGTTGCAATCCCATAATAGTAC | 2610–2633 | Near 3′ end of VP mRNA |
| NAdR | gctctagatCATCTTGAACGTTTACCACCACT | 3892–3915 | Just upstream of VP4 |
| Adsp | tcggaattcCACGTTCTTGTGGATGAGG | 4362–4380 | 19-37 nt into VP2 |
| Adsvp | gccTACCAGAAATCCGTGTAATGACA | 4546-4534/4403-4393 | Small intron splice |
| Ad3s | CGTGAGTACTGATACTTTTTATTT | 4435–4412 | End of ORF-B (TGA) |
| Ads | gccCCTCAACAACCTAAAAAACGTGAGTACTGA | 4453–4424 | End of ORF-B (TGA) |
| Ad6s | CCTAAAAAACGTGAGTACTGA | 4444–4424 | End of ORF-B (TGA) |
| Adl | gccGACGTAATTGGTGGACCTGTATATCCT | 4477–4451 | End of ORF-B (−8 aa) |
| Adm | gccCCTGTATATCCTCAACAACCTAAA | 4462–4439 | End of ORF-B (−4 aa) |
| AdlgF | tcggaattcATGTCTGGCGTCTTTACA | 5230–5213 | Start of VP1 (ORF-B) |
| RNAtag | gggtctagagctcgagT17 | Poly(A) sequence | 3′/5′-RACE (first round) |
| ADAP | gggtctagagctcgagT | Subsequent rounds of RACE | |
| ProNSF | acggtaccGATATAAAGAGCAAGCACCC | 109–128 | NS1 promoter (KpnI) |
| ProNSR | gaagatCTGTGCTGGAGGCGCTTCTACTGCAGCGAACAACGTACCTGAGTTCCAGAACAC | 240–207 | NS1 promoter (reverse) |
| ProVP1R | gaagatctAAGACGCCAGAGATTTAATACT | 5217–5238 | VP1 (BglII site; reverse) |
| ProVP1F | gtaggtaccGATATAAAGAGCAAGCACCCA | 5323–5300 | VP1(KpnI site; forward) |
| PrNSMf | ACGGTACCGACTATAAATAGAGCTGAGC | MlDNV (forward) | |
| PrNSMr | GAAGATCTATCTTGCAATAGATATACCTA | MlDNV (reverse) | |
| pFECRIF | CGCAAATAAATAAGAATTCTACTGTTTTCGTAAC | Mutation in pFastBac1 | |
| pFECRIR | GTTACGAAAACAGTAGAATTCTTATTTATTTGCG | Mutation in pFastBac1 | |
| Ad1HAR | GCTCTAGATCAAGCGTAATCTGGAACATCGTATGGGTATTTTTGTTGCAATCCCATAATA | VP products (reverse primer [HA]) | |
| AdATG1B | ggaattcATGTCCGTCTTTACAGATCTCAC | ||
| AdmATG1B | ggaattcACCTCCGTCTTTACAGATCTCTCAC |
The AdDNV sequence is shown in capital letters. Underlined nucleotides indicate stop codons (e.g., TGA) or restriction sites (e.g., ggtacc [KpnI]).
Promoter activity in AdDNV genome.
Promoter regions were amplified by PCR and cloned upstream of the luciferase gene in the pGL3-basic system. The ProNSF and ProNSR primers were used for the NS promoter, the ProVP1F and ProVP1R primers were used for the VP1 promoter, and the PrNSMf and PrNSMr primers were used for the Mythimna loreyi densovirus (MlDNV) NS promoter (control) (Table 1). Sequencing was performed to confirm the promoter direction. For the assay, Ld652 cells were seeded into wells of 24-well cell culture plates. Each well contained about 0.5 ml of cells at 5 × 105 cells/ml. The cells were cultured overnight. Transfection was performed with 2.5 μl DOTAP reagent and 0.6 μg DNA in 15 μl HEPES, and the mixture was added to 245 μl medium (without antibiotic or fetal bovine serum [FBS]) per well. Cells were harvested at 48 or 60 h posttransfection, washed twice with PBS, and resuspended in 100 μl of Bright-Glo lysis buffer (Promega). Cell lysates were quickly centrifuged to remove cell debris, and 25-μl aliquots of the cell extract were used to determine luciferase activity according to the instructions for a luciferase assay system (Promega).
Expression of structural proteins and analysis of VP ORFs by use of a baculovirus system.
The potential VP coding sequences (see below) were cloned into the Autographica californica nuclear polyhedrosis virus (AcNPV) downstream of the polyhedrin promoter by use of the Bac-To-Bac baculovirus expression system (14) (Invitrogen) via the pFastBac1 and pFastBacHT vectors according to the supplier's instructions. In constructs involving expression of VP1, the initiation codon had to be moved closer to the start of the transcript. For this purpose, an EcoRI site was introduced 100 bp upstream of the multiple cloning site (MCS), using the pFECRF and pFECRIR mutation primers (Table 1), followed by removal of the small EcoRI fragment between the new and MCS EcoRI sites. Inserts were generated by PCR (28) with the primers given in Table 1, using the wild-type (wt) template or a template in which intron II (see below) splicing sites had been mutated. The forward primer with an EcoRI site was either AdATG1B, which coincided with the initiation codon of VP1, or an equivalent in which the initiation codon ATG was mutated to ACC (AdmATG1B), and the reverse primer Ad1HAR, containing an XbaI site, was used (Table 1). All pFastBac recombinant constructs were verified by sequencing.
Protein analysis by SDS-PAGE, Western blotting, and N-terminal amino acid sequencing.
Capsid proteins were analyzed by SDS-PAGE (13), using the structural proteins of Junonia coenia densovirus (JcDNV) or broad-range standards (Bio-Rad) as size markers. Expressed proteins were analyzed by Western blotting (28, 30), using polyvinylidene difluoride (PVDF) membranes and Roche blocking reagent. For amino acid sequencing, structural proteins from AdDNV were separated by SDS-PAGE on 10% polyacrylamide gels and were electroblotted onto nitrocellulose membranes (Westran, Schleicher & Schuell, Keene, NH) and sequenced according to the method of Matsudaira (15).
MS.
Expressed proteins from baculovirus constructs and native proteins from the virus were analyzed by mass spectrometry (MS) after separation by SDS-PAGE. The proteins, dissociated with 2% SDS at 95°C for 5 min, were run in a 10% acrylamide gel (13). The protein bands were cut from the gel and destained with water-sodium bicarbonate buffer and acetonitrile. Each protein was reduced with dithiothreitol (DTT) and alkylated with iodoacetamide prior to in-gel digestion with trypsin (8). The tryptic peptides were eluted from the gel with acetonitrile containing 0.1% trifluoroacetic acid. The tryptic peptides were then separated on an Agilent Nanopump instrument using a C18 Zorbax trap and an SB-C18 Zorbax 300 reversed-phase column (150 mm × 75 μm; 3.5-μm particle size) (Agilent Technologies, Inc.). All mass spectra were recorded on a hybrid linear ion trap-triple-quadrupole mass spectrometer (Q-Trap; Applied Biosystems/MDS Sciex Instruments, CA) equipped with a nano-electrospray ionization source. The analysis of MS-MS data was performed with Analyst software, version 1.4 (Applied Biosystems/MDS Sciex Instruments, CA). MASCOT (Matrix Science, London, United Kingdom) was used to create peak lists from MS and MS/MS raw data.
Nucleotide sequence accession number.
The AdDNV sequence is available in the GenBank database under accession number HQ827781.
RESULTS
AdDNV infection of A. domesticus.
AdDNV is a frequent cause of epizootics in commercial or research mass rearing facilities for house crickets in Europe. The highest mortality is observed in the last larval stage and in young adults. These crickets die slowly over a period of several days; although they appear healthy, they lie on their backs and do not move. The guts of infected A. domesticus crickets that are still alive and no longer move are almost always completely empty. Beginning in September 2009, mass epizootics have also occurred in rearing facilities throughout North America.
DNA sequence and organization of AdDNV isolates.
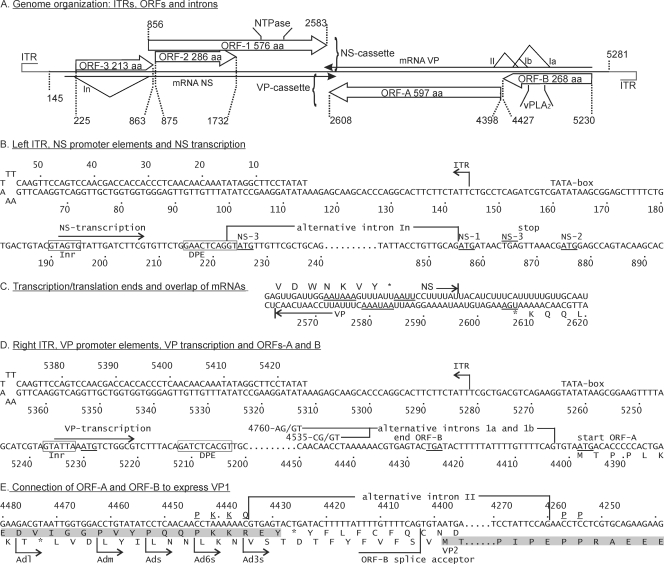
Three full-length genomic clones in the pCR-XL-TOPO vector, namely, pAd22, pAd25, and pAd35, were obtained from the 1977 AdDNV isolate. Both strands of the viral genomes were sequenced (for the full annotated sequence, see Fig. S1 in the supplemental material). Nucleotide substitutions in more recent isolates have been reported elsewhere (24). The total length of the genome was 5,425 nt and contained ITRs of 144 nt, of which the distal 114 nt could fold into a perfect I-type palindromic hairpin (Fig. 1A, B, and D). The side arms in the typical Y-shaped terminal palindromes of many parvoviruses were missing in the case of AdDNV.
Fig. 1.
Genome organization of AdDNV. (A) Overview of genome organization, positions, and sizes of ITRs, ORFs, and introns. The In intron in the NS mRNA, between nt 223 and nt 855, occurs in about half of the NS transcripts. Ia (nt 4403 to 4758), Ib (nt 4403 to 4533), and II (nt 4260 to 4434) are introns that occur in alternative VP transcripts. vPLA2 indicates the position of the viral phospholipase A2 motif. (B) Left ITR and regulation of production of NS transcripts. NS transcripts start at 192-A and yield NS3 from 225-A. However, a fraction of these transcripts are spliced just upstream of this start codon (intron In), leading to translation of NS1 from 856-A (AUG with a poor initiation environment) and, through leaky scanning, of NS2 from 875-A. Inr and DPE are promoter elements. (C) Like the case for all members of the Densovirus genus, the 3′ ends of AdDNV NS and VP transcripts overlap in the middle of the genome. The stop codons and AATAAA motifs are underlined. (D) Right ITR, VP transcription sites, and splicing in ORF-B on the complementary strand. Transcription starts at nt 5235, and VP1 initiation is at nt 5230. The short 5′-UTR predicts an inefficient initiation (leaky scanning) and could be responsible for the production of a nested set of N-terminally extended viral proteins. However, removal of either of the two alternative introns in ORF-B (Ia or Ib) did not connect the exons in ORF-B and ORF-A in frame, so only nonstructural proteins could be produced from nt 5230 and VP2 could be produced directly from the first AUG in ORF-A when this splicing occurred. (E) An alternative intron II, which is mutually exclusive with introns Ia and Ib because the ORF-B splice acceptor is removed, connects ORF-B and ORF-A (both shaded) in frame so that VP1 can be produced from nt 5230. The VP1 sequence around the splicing site is underlined and shown above the nt sequence.
Both complementary strands contained large ORFs in their 5′ halves; one strand had 3 large ORFs (ORFs 1 to 3), 2 of which were overlapping, and its complementary strand had 2 large ORFs (ORF-A and -B) (Fig. 1A). ORFs 1 to 3 potentially code for proteins consisting of 576, 286, and 213 amino acids (aa), respectively, whereas ORFs A and B potentially code for proteins of 597 and 268 aa, respectively. nBLAST analysis (http://www.ncbi.nih.gov) of the 5 ORFs revealed that the ORF-1 product is a homologue of densovirus NS1 proteins, the ORF-2 product is a homologue of densovirus NS2 proteins, the ORF-A product is a homologue of densovirus VP proteins, the ORF-B product is a homologue of parvoviral phospholipase A2 proteins (N-terminal sequence of VP1), and the ORF-3 product does not have homologous proteins. NS1 ORF-1 can also be recognized by the presence of rolling circle replication and Walker A and B motifs, and the VP ORF can be recognized by the presence of a PLA2 motif (Fig. 1A; see Fig. S1 in the supplemental material). Since the convention for all parvoviruses is to have the genes coding for the nonstructural proteins in the left half of the genome, it was decided to define the strand of the ambisense AdDNV genome containing the ORFs for the NS genes as the plus strand so the genes would be located similarly.
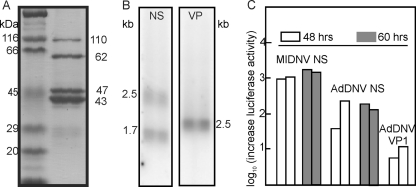
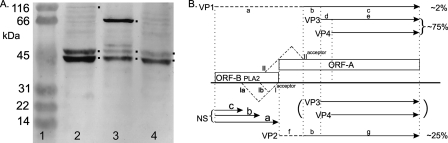
SDS-PAGE revealed that the capsid is composed of 4 structural proteins with estimated molecular masses ranging from 43 to 110 kDa (Fig. 2A), although a fifth protein may arise during purification, probably due to proteolysis (not shown). Attempts to obtain N-terminal sequences failed for VP1, VP3, and VP4, but the sequence TPPLKPHP(I)(E) was obtained for VP2, which indicated that its translation started at the AUG start codon of ORF-A, at nt 4398 to 4396 (Fig. 1D; see Fig. S1 in the supplemental material), and predicted a molecular mass of 65.3 kDa for VP2. ORF-B encoded the PLA2 motif recently identified in the structural proteins of most parvoviruses (4, 6, 28, 34) but was too small to code for a VP1 of 110 kDa as estimated by SDS-PAGE (Fig. 2A). Therefore, splicing of ORF-A and ORF-B seemed necessary to code for the largest AdDNV structural protein. Since the N-terminal coding region of ORF-A before its first ATG overlapped with the C-terminal coding region of ORF-B (Fig. 1E; see Fig. S1 in the supplemental material), an unspliced transcript could also code for VP1 by a ribosome frameshift in the ORF-A-ORF-B overlapping region, as suggested for PfDNV (33). These hypotheses were investigated further by transcript mapping and expression studies.
Fig. 2.
(A) SDS-PAGE analysis of structural proteins of AdDNV. Lane 1, Bio-Rad broad-range standard proteins as markers; lane 2, four proteins of AdDNV (VP1 to -4, in decreasing size). The estimated masses corresponded reasonably with the sequence-predicted masses (88, 65, 51, and 47 kDa, respectively). (B) Northern blotting of NS and VP transcripts. The two NS transcripts corresponded to a spliced and an unspliced form (see the text for further details). The single VP band consisted of at least three forms of almost identical size. (C) Promoter activities of the predicted NS and VP promoters in two independent experiments using the pGL3 vector, lepidopteran LD cells, and the Promega luciferase assay. The NS promoter was assayed at both 48 and 60 h. The NS promoter of MlDNV, a lepidopteran virus, was used as a control in the lepidopteran cells.
Northern blotting and mapping of viral transcripts.
Northern blotting of RNAs obtained from infected Acheta larvae revealed two bands of NS transcripts (about 2.5 and 1.8 kb) and one band of VP transcripts (about 2.5 kb) (Fig. 2B). The transcript maps for RNAs isolated from both diseased crickets and recombinant baculovirus-infected cells were established by 5′- and 3′-RACE and are shown in Fig. 1B to D. The 3′ termini of the NS and VP transcripts had a 34-nt overlap (Fig. 1C), similar to the situation observed with members of the Densovirus genus (27). NS transcription and splicing followed the same strategy as that previously described for GmDNV (28). A large unspliced transcript (nt 192 to 2596) was found to code for NS3 (first AUG in ORF-3), starting at nt 225. The NS3 coding sequence was removed in the spliced form in roughly half of these transcripts, with an intron from 221-AG/GT to 853-CAG, resulting in a 1,770-nt transcript that was able to code for NS1, starting at the new first codon (856-AUG), and NS2, starting at 875-AUG, by a leaky scanning mechanism due to the poor environment of the NS1 AUG codon (cagAUGa) and the strong environment of the downstream NS2 initiation codon (AcgAUGG). These two maps confirmed the sizes of the mRNAs observed by Northern blotting and indicated that this virus expressed NS1-3 in a fashion identical to that for other Densovirus members.
The single band of VP transcripts observed by Northern blotting could actually represent different forms of mRNA with similar intron sizes. The unspliced form, starting at nt 5235, would be 2,672 nt [plus the poly(A) tail] long. First, we determined whether ORF-A and ORF-B could be connected in frame by splicing, using RT-PCR with primers Adsp and AdlgF (Table 1). For all parvoviruses studied in this respect, VP1 is an N-terminally extended form of VP2, and the position of the Adsp primer was therefore chosen about 25 nt downstream of the VP2 start codon in ORF-A, with AdlgF located at the start of ORF-B. Two alternative introns, with sizes of 131 and 356 nt, were found with the same splice acceptor at codon 4405-CAG (Fig. 1D and 3A and B). Both introns failed to yield an in-frame coding sequence with ORF-A. The stop codon in the spliced ORF-B overlapped the start codon for VP2 (ugUAAUGa) (Fig. 1D). In some systems, e.g., influenza B virus (19, 20) and some non-long-terminal-repeat (non-LTR) retrotransposons (12), reinitiation occurs at such stop-start sequences. Expression of the intronless sequence from nt 4546 (before the small intron) to nt 3892 in the baculovirus system via pFastbacHTb, using primers Adsvp and NAdR (Table 1) and cloning using EheI and EcoRI sites, did not yield products larger than the 29 aa expected from the baculovirus/ORF-B construct, arguing against reinitiation. These results gave credence to the previously suggested ribosome frameshift for PfDNV to generate a nested set of N-terminally extended structural proteins, as observed for all parvoviruses studied thus far (33). To test this hypothesis, we made several recombinant baculovirus constructs such that their expression products would be amenable to N-terminal sequencing in the potential frameshift region and they would be of reasonable size for mass spectrometry (Fig. 1E).
Fig. 3.
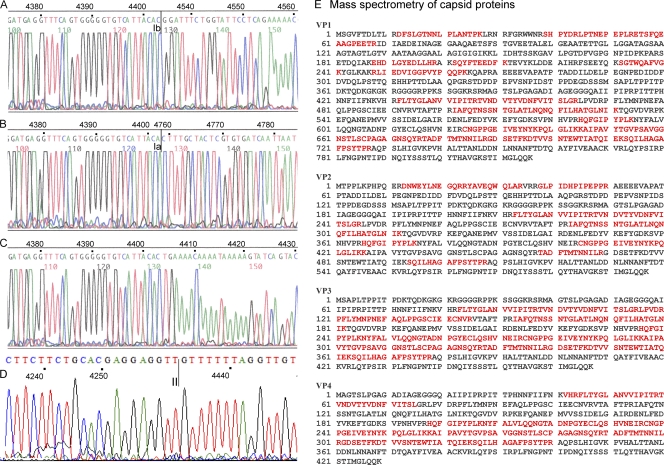
(A to D) Sequences of mRNAs in the VP ORFs, i.e., removal of the Ib intron in ORF-B (A), removal of the Ia intron in ORF-B (B), unspliced mRNA (C), and the connection of ORF-A with ORF-B by splice II (D). (E) Mascot search results after mass spectrometry for capsid proteins from viruses in natural infections. The proteins were treated with trypsin, which cuts at the C-terminal side of K or R unless the next residue is P. Matched peptides are shown in bold red.
Expression of VP1.
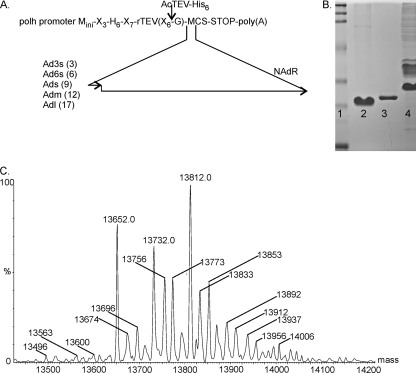
Figure 1E shows the 43-nt overlap of ORF-B with the N-terminal extension of ORF-A and the positions of primers (after the ORF-B and before the ORF-A stop codon) used to study the potential translational frameshift. The pFastbacHTb vector was used to yield products that could be purified via their N-terminal His tails and cleaved with the tobacco etch virus (TEV) protease, leaving only one codon (Gly) upstream of the insertion at the blunt-end EheI restriction site (Fig. 4A). The PCR products obtained using the forward primers Ad3s, Ad6s, Ads, Adm, and Adl (Table 1) (the distance from the ORF-B stop codon is indicated, in codons, in Fig. 4A) and the reverse primer NAdR, chosen at the beginning of the extension of VP4 to ensure stable products, were cloned into the EheI site, and the clones were selected for orientation by XbaI analysis (the XbaI sites in the MCS and the NAdR primer should be close to each other) and then verified by sequencing.
Fig. 4.
(A) Construction of various ORF-A and ORF-B constructs with different Ad primers (Ad3s, Ad6s, etc.) linking a short part of ORF-B to a larger part of ORF-A, using the NAdR primer and the pFastbacHTb baculovirus system. Upstream of the insert is a TEV proteolytic site and a histidine tail (H6); N-terminal sequencing of the digested purified protein would reveal the connecting sequences. The numbers in parentheses with the different primers (Ad3s, Ad6s, etc.) denote the distances in codons from the ORF-B stop codon (small arrow). (B) SDS-PAGE analysis of recombinant Ads3 protein generated with Ad3s and NAdR primers, within ORF-B and ORF-A, respectively, using the pFastbacHTb baculovirus system. Lane 1, protein markers; lane 2, Ads3 protein after AcTEV protease digestion and purification on a Ni column; lane 3, AcTEV protease (contains a His tag for easy removal after the reaction); lane 4, Ads3 protein prior to cleavage with AcTEV. (C) Mass spectrometry of TEV-treated and purified Ads3 protein, which was predicted from its sequence to have a mass of 13,650.0 Da. A major peak at 13,652 Da verified the predicted sequence. Two other major peaks, at 13,732 Da and 13,812 Da, had an additional 80 and 160 Da, indicating the addition of one and two phosphate groups, respectively. Minor bands had one or more protons replaced by sodium, each adding an additional 22 Da.
All constructs (His6-TEV recognition site-EheI site-insert-XbaI site) yielded proteins that could be purified via their His tails and that had molecular masses indicating that the two frames were connected. The Ad3s and Ads products were treated with TEV protease and were subjected to N-terminal sequencing after the N-terminal His tail/TEV site fragments and the protease were removed by affinity chromatography on Ni-nitrilotriacetic acid (Ni-NTA) columns and analyzed by SDS-PAGE (Fig. 4B). The sequences obtained (Ad3s sequence, GQP?RAEE; and Ads sequence, GAPQQP??QPPaAE), containing an N-terminal G and GA that were introduced via the primers and remained after TEV cleavage, indicated that nt 4435 in ORF-B was connected in frame with nt 4259 in ORF-A. Mass spectrometry analysis of the purified Ads product yielded a mass of 13,652 Da, with masses of 13,732 and 13,812 Da for phosphorylated species, for a predicted mass of 13,649.64 Da for the sequence GAPQQP…GGKRSR (Fig. 4C). These results confirmed the occurrence of a splice between nt 4435 and nt 4259. The predicted mass of VP1 is thus 88 kDa less than that estimated by SDS-PAGE (Fig. 2A), which may be explained by the phosphorylation observed by mass spectrometry.
Splicing was further investigated by RT-PCR of mRNAs extracted from infected crickets, using 2 sets of primers, namely, AdlgF/NAdR and Ads/AdVPR (Table 1), covering the whole coding sequence of VP1 except for the common VP4 sequence, with estimated products of 1,357 and 1,847 bp without splicing and 1,182 and 1,672 bp after intron II splicing, respectively. The intron II splice was also confirmed by sequencing of the VP1 cDNA (Fig. 3D). As illustrated in Fig. 1D and E, the Ia or Ib intron and the intron II splice were mutually exclusive, i.e., the intron II splice removed the acceptor site for the two intron I splices, whereas the intron I splices removed the donor site for intron II splicing. This expression strategy was further confirmed using recombinant baculovirus constructs. The VP1 sequence from which intron II was removed, and which was thus rendered resistant to ORF-B splicing (Ia and Ib), did not yield VP2 (Fig. 5A). In contrast, constructs with a mutated VP1 initiation codon and a normal VP template yielded a strong VP2 band but also some VP3 and VP4 (Fig. 5A), because type II splicing could remove the VP2 initiation codon and thus allow downstream initiation. When the template without intron II was used in combination with the mutated VP1 sequence, it yielded, as expected, VP3 and VP4 only. The leaky scanning of the VP3 initiation codon can probably be explained by its weak environment (uccAUGa), in contrast to the strong environment of the VP4 initiation codon (agaAUGg). Therefore, the VP multicistronic cassette yielded 2 sets of structural proteins (Fig. 5B).
Fig. 5.
(A) Western blot of pFastbac VP constructs. Lane 1, prestained SDS-PAGE protein standards (Bio-Rad). Lane 2, expression products from the ORF-A-ORF-B construct from which intron II was deleted. Strong bands were observed for VP3 and VP4, with a weak band for VP1 (all indicated with dots), in addition to some spurious bands. Lane 3, expression products from the ORF-A-ORF-B construct with a mutated VP1 initiation codon. A strong band was observed for VP2, but relatively strong bands were also seen for VP3 and VP4, since some type II splicing occurred that removed the VP2 initiation codon. A weak band was also observed just above VP3. Lane 4, ORF-A-ORF-B construct with a mutated VP1 initiation codon and with intron II deleted, yielding only VP3 (weak initiation codon) and VP4. This lane also contained some of the spurious, nonspecific bands seen in lane 2. (B) Schematic representation of expression products of the VP cassette. Depending on the splice (Ia and Ib versus II), different sets of structural proteins (VP1, VP3, and VP4 versus VP2) were generated. VP3 and VP4 only were expressed in specific constructs (see panel A, lane 4). In addition, expression products solely from ORF-B (NSa, NSb, and NSc) could be expected if initiation at the VP1 codon was combined with Ia or Ib or if no splicing occurred. The predicted pIs of the common protein and N-terminal extensions differed considerably (a, 4.90; b, 3.55; c, 8.58; d, 11.60; e, 6.21; f, 6.72) and may have been a factor in the difference in observed and predicted masses of the capsid proteins (see Fig. 2A). The dashed N-terminal extensions of VP1 and VP2 denote their unique sequences.
Analysis of promoter activity.
The promoter elements as well as the poly(A) signals were predicted by the mapping of transcription starts and polyadenylation sites of both NS and VP transcripts (Fig. 1B to D). To assess and compare their functionality, promoter regions (including the start of transcription) were amplified by PCR and cloned upstream of the luciferase gene in the pGL3-basic system. Their relative activities were determined by luciferase assays in independent duplicates at either 40 or 60 h posttransfection. The activity of the NS promoter of AdDNV was very significant in Ld652 cells from gypsy moths but was lower than that of the NS promoter of MlDNV, a lepidopteran densovirus (Fig. 2C). However, the VP1 promoter activity was significantly lower, suggesting the need for trans-activation, the absence of a critical factor reacting with the non-ITR region of the VP1 promoter, or differences in transcription factors between the cricket and gypsy moth systems.
Mass spectrometry of AdDNV capsid proteins.
AdDNV was purified and the proteins separated by SDS-PAGE and subjected to mass spectrometry analysis in order to confirm the results obtained by analyzing the baculovirus constructs of the viral proteins. The proteins purified from the gel were digested with trypsin, and sequences of the peptides were determined. Analysis of VP1 and VP2 confirmed the results obtained with the baculovirus expression experiments with respect to the in-frame linking of ORF-B and ORF-A. The peptide sequences obtained covered 33% of VP1, 26% of VP2, 50% of VP3, and 42% of VP4 (Fig. 3E). One outlier peptide identified for VP3 was found in VP2, but with an ion score of 4, this was considered background.
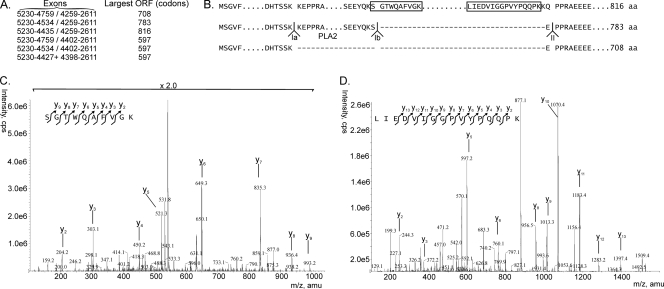
Splicing of the Ia and Ib donor sites with the intron II acceptor site would theoretically also be possible and would yield additional products of 783 and 708 amino acids, close to the 816 amino acids observed for VP1. However, only 4 structural proteins were observed, and mass spectrometry demonstrated the presence of two VP1-specific peptides located in the introns of these potential supplementary products, of 708 and 783 amino acids (Fig. 6), confirming that the 816-aa species corresponded to VP1.
Fig. 6.
Potential VP1 proteins encoded by the VP gene cassette. (A) Different potential splice combinations of the ORFs in the VP gene cassette and the largest protein that could then be coded. N-terminal amino acid sequencing showed that the 597-codon ORF corresponded to VP2 and, theoretically, that the three larger ORFs could code for the larger VP1 protein. (B) Schematic representation of potential sequences of large structural proteins, using the three splice combinations Ia donor-II acceptor (708 aa), Ib donor-II acceptor (783 aa), and II donor-II acceptor (816 aa). Mass spectrometry of VP1 proteins from viruses obtained after natural infections revealed the boxed sequences, which were not present in the potential 783-aa and 708-aa proteins. Therefore, the 816-aa protein corresponded to VP1. (C) Mass spectrometry identifying the unique peptides in the boxed sequences of the 816-aa protein in panel B as part of VP1. Peptides are shown with the charged C-terminal lysine, e.g., y2 is GK (m/z = 204.2), y3 is VGK (m/z = 303.1), etc., for the SGTWQAFVGK peptide. The ion score of this peptide is 61, indicating an excellent matching of the MS/MS spectrum to the identified peptide. (D) Mass spectrometry of the other peptide, similar to the peptide in panel C. The ion score is very high for this peptide, namely, 87. A detailed analysis of the peptides in panels C and D is given in Fig. S2 in the supplemental material.
DISCUSSION
The 1977 isolate of AdDNV was cloned and its expression strategy analyzed. Additional AdDNV isolates from Europe, isolated in 2004, 2006, 2007, and 2009, and from North America, isolated in 1988 (Tennessee) and 2009 (Quebec, Alberta, British Columbia, and Washington State), were amplified by PCR targeting the region between the ITRs and then sequenced (reported elsewhere [24]). All 2009 North American isolates had identical sequences, suggesting a common source, and differed from the 1977, 2004, and 2006 isolates by 49, 18, and 16 nt substitutions, respectively. The genome organizations of these isolates were identical.
The sequences of the AdDNV ORFs were compared, using nBLAST, with those of other densoviruses, in particular with those of viruses such as PfDNV, PcDNV, BgDNV, MpDNV, and Dysaphis plantaginea densovirus (DplDNV), which also have split VP ORFs (see Fig. S3 in the supplemental material). Surprisingly, the highest identities by far for the AdDNV NS1 and ORF-A proteins were found for proteins of PcDNV from Planococcus citri (a citrus mealybug belonging to the Pseudococcidae family of the Hemiptera insect order, whereas Acheta domesticus belongs to the Gryllidae family of the insect order Orthoptera).
The VP transcript was found to start 23 nt upstream of the 3′-ITR, at nt 5467, and the starts of both NS transcripts were at nt 573, 23 nt downstream of the 5′-ITR. This suggested that many promoter elements would be located within the ITRs and would be identical for the VP and NS promoters. The sequence context of both starts corresponded reasonably well with the consensus sequence for Inr boxes (TCAGTG); however, the promoter activity in lepidopteran cells differed considerably (Fig. 2C). The region from the 5′-untranslated region (5′-UTR) in the VP mRNA to the putative VP1 AUG was only 5 nt long, whereas for the two NS transcripts, the 5′-UTRs were 32 (1.8-kb transcript) and 30 (2.5-kb transcript) nt long.
The expression strategy of the NS cassette is identical to that for the other members of the Densovirus genus. In the unspliced transcript (Fig. 1A), NS3 is translated, whereas in the spliced form the ORF for NS3 is removed and translation starts at the weak initiation codon of NS1 or, due to leaky scanning, at the coding sequence for NS2, 19 nt downstream.
In contrast to this conserved strategy, VP expression has unique features that so far have not been observed for vertebrate parvoviruses and densoviruses, which all have a perfect nested set of N-terminally extended structural proteins. AdDNV displays split VP ORFs, and its two largest structural proteins have different extensions to which no roles have yet been assigned. PfDNV (33), PcDNV (25), BgDNV (9, 17), and MpDNV (31, 32), which all have a split VP ORF, with the smaller ORF encoding the PLA2 motif, probably have similar expression strategies (27). For PfDNV, a large number of donor and acceptor splicing sites have also been identified in the VP ORFs. Splicing of the sd3 and sa3 splicing sites in cDNA11 of PfDNV could also yield a large VP1 protein. Like the case for AdDNV, many of the splicing combinations could be inconsequential for generation of the structural proteins. For BgDNV (Dmitry V. Mukha, personal communication), the larger of the two VP ORFs also codes for VP2, and splicing that is slightly different from the strategy in AdDNV also brings the two ORFs in frame and codes for VP1.
Furthermore, it was expected that initiation at the VP1 start codon, which was leaky due to the short 5′-UTR, did not depend on the presence of a downstream splice and that the various spliced and unspliced forms of mRNA were equally probable to be translated. This would lead to premature termination of translation in the case of unspliced mRNA (Fig. 3C), yielding a 268-aa ORF-B product (NSa) that is not involved in capsid formation (Fig. 1A). The 2 ORF-B introns would be responsible for 2 additional, minor nonstructural proteins, of 233 (NSb) and 158 (NSc) aa. Splicing of the intron II acceptor site with the Ia and Ib donor sites was not observed (Fig. 6).
The genome organization and expression strategy of AdDNV place it in a separate genus from Densovirus, and probably from Pefudensovirus. Unfortunately, a definitive description of the VP expression of PfDNV is still lacking to resolve whether these viruses should be coclassified. The NS cassette structure of AdDNV, PfDNV, BgDNV, and PcDNV is identical to that of members of the Densovirus genus but different from that of the CpDNV ambisense densovirus (1), which has been proposed to be classified in a new genus, Cupidensovirus (26). However, these viruses with split VP ORFs are unique among all ambisense densoviruses studied so far in that their ITRs form perfect hairpins of about 150 to 200 nt and they have genomes of about 5,450 nt (27). The low sequence identity among the AdDNV-like viruses may not be evocative of a need to classify them into diverse genera.
Supplementary Material
ACKNOWLEDGMENTS
P.T. acknowledges support from the Natural Sciences and Engineering Research Council of Canada.
We thank the professionals in the cricket-rearing industry for their work.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 20 July 2011.
REFERENCES
- 1. Baquerizo-Audiot E., et al. 2009. Structure and expression strategy of the genome of Culex pipiens densovirus, a mosquito densovirus with an ambisense organization. J. Virol. 83:6863–6873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bergoin M., Tijssen P. 2010. Densoviruses: a highly diverse group of arthropod parvoviruses, p. 57–90 In Asgari N., Johnson K. N. (ed.), Insect virology. Horizon Scientific Press, Norwich, United Kingdom [Google Scholar]
- 3. Bergoin M., Tijssen P. 2008. Parvoviruses of arthropods, p. 76–85 In Mahy B., van Regenmortel M. (ed.), Encyclopedia of virology, 2nd ed., vol. 4 Elsevier, Oxford, United Kingdom [Google Scholar]
- 4. Farr G. A., Zhang L. G., Tattersall P. 2005. Parvoviral virions deploy a capsid-tethered lipolytic enzyme to breach the endosomal membrane during cell entry. Proc. Natl. Acad. Sci. U. S. A. 102:17148–17153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fédière G. 2000. Epidemiology and pathology of Densovirinae, p. 1–11 In Faisst S., Rommelaere J. (ed.), Parvoviruses. From molecular biology to pathology and therapeutic uses. Karger, Basel, Switzerland [Google Scholar]
- 6. Fediere G., Li Y., Zadori Z., Szelei J., Tijssen P. 2002. Genome organization of Casphalia extranea densovirus, a new iteravirus. Virology 292:299–308 [DOI] [PubMed] [Google Scholar]
- 7. Fernandes S., Tijssen P. 2009. Seamless cloning and domain swapping of synthetic and complex DNA. Anal. Biochem. 385:171–173 [DOI] [PubMed] [Google Scholar]
- 8. Hellman U., Wernstedt C., Gonez J., Heldin C. H. 1995. Improvement of an “in-gel” digestion procedure for the micropreparation of internal protein fragments for amino acid sequencing. Anal. Biochem. 224:451–455 [DOI] [PubMed] [Google Scholar]
- 9. Kapelinskaya T. V., Martynova E. U., Korolev A. L., Schal C., Mukha D. V. 2008. Transcription of the German cockroach densovirus BgDNV genome: alternative processing of viral RNAs. Dokl. Biochem. Biophys. 421:176–180 [DOI] [PubMed] [Google Scholar]
- 10. Kaufmann B., et al. 2010. Structure of Penaeus stylirostris densovirus, a shrimp pathogen. J. Virol. 84:11289–11296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kaufmann B., et al. 2011. Structure of Bombyx mori densovirus 1, a silkworm pathogen. J. Virol. 85:4691–4697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kojima K. K., Matsumoto T., Fujiwara H. 2005. Eukaryotic translational coupling in UAAUG stop-start codons for the bicistronic RNA translation of the non-long terminal repeat retrotransposon SART1. Mol. Cell. Biol. 25:7675–7686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 14. Luckow V. A., Lee S. C., Barry G. F., Olins P. O. 1993. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J. Virol. 67:4566–4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Matsudaira P. 1987. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J. Biol. Chem. 262:10035–10038 [PubMed] [Google Scholar]
- 16. Meynardier G., et al. 1977. Virus de type densonucléose chez les orthoptères. Ann. Soc. Entomol. Fr. 13:487–493 [Google Scholar]
- 17. Mukha D. V., Chumachenko A. G., Dykstra M. J., Kurtti T. J., Schal C. 2006. Characterization of a new densovirus infecting the German cockroach, Blattella germanica. J. Gen. Virol. 87:1567–1575 [DOI] [PubMed] [Google Scholar]
- 18. Mukha D. V., Schal K. 2003. A densovirus of German cockroach Blatella germanica: detection, nucleotide sequence and genome organization. Mol. Biol. (Moscow) 37:607–618 [PubMed] [Google Scholar]
- 19. Powell M. L., Brown T. D., Brierley I. 2008. Translational termination-re-initiation in viral systems. Biochem. Soc. Trans. 36:717–722 [DOI] [PubMed] [Google Scholar]
- 20. Powell M. L., Napthine S., Jackson R. J., Brierley I., Brown T. D. 2008. Characterization of the termination-reinitiation strategy employed in the expression of influenza B virus BM2 protein. RNA 14:2394–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Simpson A. A., Chipman P. R., Baker T. S., Tijssen P., Rossmann M. G. 1998. The structure of an insect parvovirus (Galleria mellonella densovirus) at 3.7 A resolution. Structure 6:1355–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Styer E. L., Hamm J. J. 1991. Report of a densovirus in a commercial cricket operation in the Southeastern United-States. J. Invertebr. Pathol. 58:283–285 [Google Scholar]
- 23. Sukhumsirichart W., Attasart P., Boonsaeng V., Panyim S. 2006. Complete nucleotide sequence and genomic organization of hepatopancreatic parvovirus (HPV) of Penaeus monodon. Virology 346:266–277 [DOI] [PubMed] [Google Scholar]
- 24. Szelei J., et al. 2011. Susceptibility of North-American and European crickets to Acheta domesticus densovirus (AdDNV) and associated epizootics. J. Invertebr. Pathol. 106:394–399 [DOI] [PubMed] [Google Scholar]
- 25. Thao M. L., Wineriter S., Buckingham G., Baumann P. 2001. Genetic characterization of a putative densovirus from the mealybug Planococcus citri. Curr. Microbiol. 43:457–458 [DOI] [PubMed] [Google Scholar]
- 26. Tijssen P. Parvoviridae. In King A. M. Q., Adams M. J., Carstens E., Lefkowitz E. J. (ed.), Virus taxonomy: classification and nomenclature of viruses: ninth report of the International Committee on Taxonomy of Viruses, in press. Elsevier, San Diego, CA [Google Scholar]
- 27. Tijssen P., et al. 2006. Evolution of densoviruses, p. 55–68 In Kerr J. R., et al. (ed.), Parvoviruses. Hodder Arnold, London, United Kingdom [Google Scholar]
- 28. Tijssen P., et al. 2003. Organization and expression strategy of the ambisense genome of densonucleosis virus of Galleria mellonella. J. Virol. 77:10357–10365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tijssen P., Tijssen-van der Slikke T., Kurstak E. 1977. Biochemical, biophysical, and biological properties of densonucleosis virus (paravovirus). II. Two types of infectious virions. J. Virol. 21:225–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Towbin H., Staehelin T., Gordon J. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. U. S. A. 76:4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. van Munster M., et al. 2003. Characterization of a new densovirus infecting the green peach aphid Myzus persicae. J. Invertebr. Pathol. 84:6–14 [DOI] [PubMed] [Google Scholar]
- 32. van Munster M., et al. 2003. A new virus infecting Myzus persicae has a genome organization similar to the species of the genus Densovirus. J. Gen. Virol. 84:165–172 [DOI] [PubMed] [Google Scholar]
- 33. Yamagishi J., Hu Y., Zheng J., Bando H. 1999. Genome organization and mRNA structure of Periplaneta fuliginosa densovirus imply alternative splicing involvement in viral gene expression. Arch. Virol. 144:2111–2124 [DOI] [PubMed] [Google Scholar]
- 34. Zadori Z., et al. 2001. A viral phospholipase A2 is required for parvovirus infectivity. Dev. Cell 1:291–302 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.