Abstract
Simian immunodeficiency virus of chimpanzees (SIVcpz) has a significant negative impact on the health, reproduction, and life span of chimpanzees, yet the prevalence and distribution of this virus in wild-living populations are still only poorly understood. Here, we show that savanna chimpanzees, who live in ecologically marginal habitats at 10- to 50-fold lower population densities than forest chimpanzees, can be infected with SIVcpz at high prevalence rates. Fecal samples were collected from nonhabituated eastern chimpanzees (Pan troglodytes schweinfurthii) in the Issa Valley (n = 375) and Shangwa River (n = 6) areas of the Masito-Ugalla region in western Tanzania, genotyped to determine the number of sampled individuals, and tested for SIVcpz-specific antibodies and nucleic acids. None of 5 Shangwa River apes tested positive for SIVcpz; however, 21 of 67 Issa Valley chimpanzees were SIVcpz infected, indicating a prevalence rate of 31% (95% confidence interval, 21% to 44%). Two individuals became infected during the 14-month observation period, documenting continuing virus spread in this community. To characterize the newly identified SIVcpz strains, partial and full-length viral sequences were amplified from fecal RNA of 10 infected chimpanzees. Phylogenetic analyses showed that the Ugalla viruses formed a monophyletic lineage most closely related to viruses endemic in Gombe National Park, also located in Tanzania, indicating a connection between these now separated communities at some time in the past. These findings document that SIVcpz is more widespread in Tanzania than previously thought and that even very low-density chimpanzee populations can be infected with SIVcpz at high prevalence rates. Determining whether savanna chimpanzees, who face much more extreme environmental conditions than forest chimpanzees, are more susceptible to SIVcpz-associated morbidity and mortality will have important scientific and conservation implications.
INTRODUCTION
Among the many lentiviruses known to infect Old World primates in sub-Saharan Africa, simian immunodeficiency virus of chimpanzees (SIVcpz) is of particular interest because it is the immediate precursor of human immunodeficiency virus type 1 (HIV-1), the causative agent of AIDS (8, 16). However, SIVcpz is also an important pathogen in its own right, since it causes significant morbidity and mortality in its chimpanzee host (6, 15, 37). Recent natural history studies in Gombe National Park in western Tanzania showed that infected chimpanzees have a 10- to 16-fold increased risk of death compared to uninfected chimpanzees (15). SIVcpz-infected females were less likely to give birth and their infants had a much higher mortality rate than those of uninfected females (15). Most importantly, postmortem analyses showed that SIVcpz can cause CD4+ T lymphocyte depletion and histopathological findings consistent with end-stage AIDS (15). These results demonstrated that SIVcpz has a substantial negative impact on the health, reproduction, and life span of chimpanzees, a conclusion that was further substantiated by the identification of an SIVcpz-infected orphan chimpanzee in Cameroon, who suffered from progressive CD4+ T cell loss, severe thrombocytopenia, and clinical AIDS (6). Thus, current data indicate that SIVcpz infection may pose a significant threat to infected communities in the wild.
Chimpanzees (Pan troglodytes) are widely dispersed across sub-Saharan Africa and classified into four subspecies with nonoverlapping ranges (7, 9, 11). These include the western subspecies Pan troglodytes verus, the Nigeria-Cameroonian subspecies P. t. ellioti, the central subspecies P. t. troglodytes, and the eastern subspecies P. t. schweinfurthii. Thus far, SIVcpz infection has been documented only in members of the P. t. troglodytes and P. t. schweinfurthii subspecies (44). This is most likely because chimpanzees acquired SIV by cross-species transmission from monkeys on which they prey after the split of P. t. verus and P. t. ellioti from the other two subspecies. Indeed, phylogenetic analyses have shown that SIVcpz represents a complex mosaic, generated by recombination of two lineages of SIV infecting red-capped mangabeys and Cercopithecus monkey species (3). Since the current habitat of these species overlaps that of central chimpanzees, it is likely that SIVcpz first emerged in west central Africa and then spread eastward, either during or subsequent to the divergence of central and eastern chimpanzees.
Chimpanzees are highly endangered and reclusive, which precludes invasive testing of any kind (27). To screen wild-living populations for SIVcpz infection, our laboratory has developed diagnostic methods that detect virus-specific antibodies and nucleic acids in fecal and urine samples with high sensitivity and specificity (16, 41, 42). These noninvasive methods, combined with genotyping approaches for species confirmation and individual identification (5, 15, 37, 47, 50), have enabled analyses of wild-living chimpanzee populations throughout central Africa. Molecular epidemiological studies have shown that SIVcpz is endemic in both central and eastern chimpanzees, with virus-positive communities identified throughout their habitats (43, 44). Surprisingly, however, the distribution of SIVcpz in wild-living populations is uneven, with high prevalence rates (30% to 50%) in some communities and rare or absent infection in others (15, 16, 19, 37, 41, 49). While the reasons for this sporadic distribution remain to be determined, it is clear that the infection status of any one community cannot be predicted but has to be determined empirically.
The first wild chimpanzee with SIVcpz infection was identified in Gombe in 2000 (42). Since then, the resident Mitumba, Kasekela, and Kalande communities have been screened extensively, with most members tested biannually. These studies have revealed widespread SIVcpz infection, with prevalence rates of 13%, 12%, and 46% for the Mitumba, Kasekela, and Kalande communities, respectively (37). Behavioral and virological studies also showed that SIVcpz spreads primarily by sexual routes, with a probability of transmission per coital act of about 0.001, very similar to that for HIV-1-infected heterosexual humans (10, 37). The major route of virus spread between communities was found to occur via migration of infected females (37). Finally, demographic studies identified an association between SIVcpz prevalence and population size. The Kalande community, which consistently had the highest SIVcpz prevalence, suffered a catastrophic population decline, while the sizes of the Mitumba and Kasekela communities, which were infected at much lower levels, remained stable (37).
The finding of SIVcpz in Gombe prompted surveys of chimpanzee communities in other areas of East Africa (Fig. 1A). These included the Kanyawara and Ngogo communities in Kibale National Park in western Uganda, habituated chimpanzees in the Budongo Forest in northern Uganda, and members of the M group in Mahale Mountains National Park in western Tanzania (44). In addition, surveys were conducted in the Kyambura Gorge and the Bwindi Impenetrable Forest in southern Uganda and the Nyungwe Forest Reserve in western Rwanda (unpublished data). Although over 500 fecal and urine samples were screened, none of these was positive for SIVcpz antibodies and/or nucleic acids. In contrast, SIVcpz was detected at multiple field sites throughout the Democratic Republic of the Congo (19, 43). These data suggested that chimpanzees at the easternmost limits of the P. t. schweinfurthii range were largely free of SIVcpz infection, possibly because of severely fragmented habitats and/or other biogeographical barriers that impeded virus spread and/or had caused its extinction. The presence of SIVcpz in Gombe thus seemed to represent an isolated occurrence, perhaps a remnant from a past dispersal, with no demonstrable linkages to other communities.
Fig. 1.
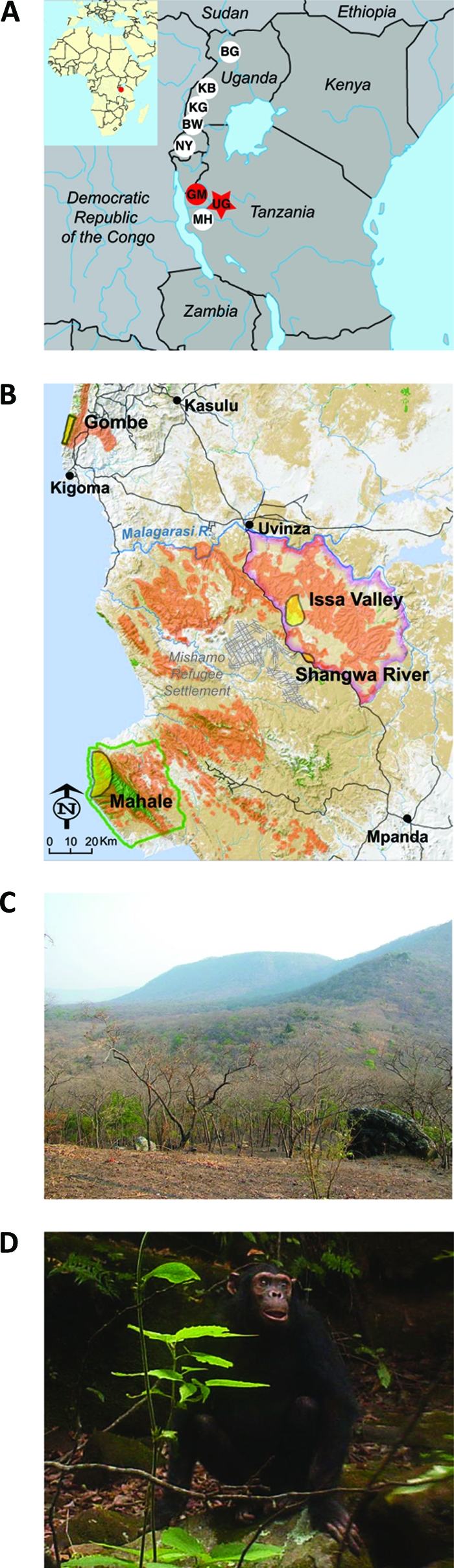
The Ugalla study site. (A) Location of Ugalla (UG) in western Tanzania (star) relative to other chimpanzee study sites (circles) in east Africa (BG, Budongo Forest; NY, Nyungwe Forest Reserve; KB, Kibale National Park; KG, Kyambura Gorge; BW, Bwindi Impenetrable Forest; GM, Gombe National Park; MH, Mahale Mountains National Park); sites where SIVcpz was found are highlighted in red. An inset depicts the location of this region within Africa. (B) Shaded reliefmap of northwestern Tanzania. The locations of the 85-km2 fecal collection area in the Issa Valley (yellow oval) and a single site near the Shangwa River site (yellow circle) within the greater Ugalla region (magenta) are shown in relation to the locations of Gombe and Mahale Mountains National Parks. The Mishamo Refugee Settlement is indicated by hatch marks. Green areas denote evergreen forested regions, while brown areas indicate open and dense Miombo woodlands derived from 2007 satellite imagery (unpublished data). Orange shaded areas indicate potential savanna chimpanzee nesting habitat predicted as described previously (26). (C) Typical Miombo woodlands in the Ugalla region during the dry season. (D) Picture of a savanna chimpanzee taken by a motion-triggered camera (photo credit: Ugalla Primate Project and the Max Plank Institute for Evolutionary Anthropology).
To examine whether the Gombe chimpanzees were indeed an exception, we surveyed apes in additional areas of Tanzania. Western Tanzania is home to an estimated 2,700 to 2,800 wild-living chimpanzees (26, 32), only about 25% of whom live in the protected areas of Gombe and Mahale Mountains (Fig. 1B). An estimated 2,600 live in the Greater Mahale Ecosystem, which includes Mahale Mountains National Park and the regions of Masito and Ugalla. The latter areas encompass some of the most arid habitats in which chimpanzees have been found (Fig. 1C). Apes living in Masito and Ugalla have adapted to exceptional living conditions (Fig. 1D). Such “savanna chimpanzees” occupy much larger home ranges, live at 10- to 50-fold lower population densities, and face more extreme seasonal changes than forest chimpanzees (4, 23–25). Because of their challenging environment, savanna chimpanzees are also predicted to differ in aspects of their behavior and social structure from forest chimpanzees (24, 25). Finally, they are often used as a model system for the behavioral ecology of early hominids, who are believed to have evolved in similar habitats (24, 34). Here, we report the first survey of savanna chimpanzees for SIVcpz infection.
MATERIALS AND METHODS
Study site.
The Ugalla study area lies in the northeastern portion of the Greater Mahale Ecosystem, about 80 km inland from Lake Tanganyika, and includes the study sites of the Issa Valley and Shangwa River (Fig. 1B). Ugalla itself is a 3,300-km2 region of broad valleys broken up by steep mountains and flat hilltop plateaus 900 to 1,800 m in elevation. The boundaries of this area are (i) the Uvinza-Mpanda Road to the west, (ii) the Malagarasi River to the north, (iii) the Ugalla River to the east, and (iv) a decline in elevation to the south (Fig. 1B). Miombo woodland, named for the dominant tree genera of Brachystegia and Julbernardia, dominates the landscape, interspersed with grassland, riverine forest strips, and small patches of swamp. Only 1.7% of the region is evergreen forest, located principally in the thin riverine strips (Fig. 1C). There is a rainy season (October to April) and a dry season (May to September), with dry months defined as having less than 100 mm of rainfall. Rainfall averages less than 1,000 mm per annum (13), and temperatures range from 11 to 35°C.
Sample collection.
Chimpanzees in the Ugalla area are not habituated. In the Issa Valley, they were tracked using an acoustic system that monitored nighttime vocalizations to identify the location of nesting sites (30). At the Shangwa River site, they were tracked by an experienced field assistant familiar with the area. Most fecal samples were collected early in the morning, after the chimpanzees had left their night nests. Samples were also collected during biweekly walked line transects or when encountered opportunistically. Fecal samples (∼10 to 20 g) were placed into 50-ml conical tubes and mixed with an equal volume of RNAlater (Ambion). Tubes were labeled with a sample code and the Global Positioning System (GPS) coordinates of the collection site. Samples were stored at ambient temperature until transported to the Jane Goodall Institute in Kigoma, Tanzania, where they were stored at −20°C. Samples were shipped to the University of Alabama at Birmingham at ambient temperature and stored at −80°C thereafter.
SIVcpz antibody detection.
All fecal samples were screened for the presence of SIVcpz-specific antibodies using an enhanced chemiluminescence Western blot approach as previously reported (15, 16, 41). Sample integrity was confirmed using an IgG control.
Viral RNA extraction and amplification.
Fecal RNA was extracted from SIVcpz antibody-positive samples using an RNAqueous Midi Kit (Ambion) and subjected to reverse transcription-PCR (RT-PCR) amplification using SIVcpz-specific pol, gp41, and gp41/nef consensus primers as previously described (15, 16, 37, 41, 42). All amplicons were gel purified and sequenced directly.
Individual identification.
All samples were subjected to mitochondrial DNA (mtDNA) and microsatellite analyses to confirm their species origin and determine the number of sampled individuals. Fecal DNA was extracted as described previously (15, 16, 37, 41, 42). A 419-bp fragment of the mitochondrial D-loop region was amplified, sequenced, and subjected to phylogenetic analysis (39) to identify distinct mitochondrial haplotypes (see Fig. S1 in the supplemental material). All samples were genotyped at eight autosomal microsatellite loci (5, 15, 16, 37, 50), with amplification products analyzed on an automated sequencer (Applied Biosystems) and sized using the GeneMapper, version 4.0, program (Applied Biosystems). For individual identification, samples were first grouped by mitochondrial DNA haplotypes. Within each haplotype, samples were then grouped by microsatellite genotypes at eight loci (see Table S1 in the supplemental material). Due to the prolonged storage at ambient temperatures, we suspected partial sample degradation. To guard for potential allelic dropout, we thus allowed allelic mismatches at up to four loci, but only if they represented a missing allele. This genotyping approach is rather conservative and may have resulted in an underestimation of the number of individuals screened. Gender was determined by amplifying a region of the amelogenin gene that contains a 6-bp insertion in the Y chromosome but not the X chromosome. Samples with evidence of DNA admixture (multiple peaks for the same locus or double peaks in the mtDNA sequence) were discarded.
SIVcpz prevalence determination.
The prevalence of SIVcpz infection was estimated on the basis of the proportion of infected individuals determined by microsatellite analyses. For each chimpanzee, the probability that it would be detected as being infected, if it was truly infected, was calculated by taking into consideration the number of specimens analyzed as well as the sensitivity of the fecal antibody detection test, previously estimated to be 0.92 (16). SIVcpz prevalence was then estimated as the proportion of chimpanzees identified to be infected, but corrected for this sensitivity; 95% confidence limits were calculated on the basis of binomial sampling.
Full-length genome amplification.
Full-length SIVcpz sequences were generated by amplifying partially overlapping subgenomic fragments from fecal RNA as described previously (40, 47, 48). Amplicons were gel purified and sequenced directly. Chromatograms were examined for positions of base mixtures using the Sequencher (version 4.7) program (Gene Codes Corporation, Ann Arbor, MI). Nucleotide differences between adjoining fragments in regions of sequence overlap were recorded. Ambiguous sites were resolved as previously reported (48). Briefly, for positions that did not affect the corresponding amino acid, the predominant nucleotide (the one with the highest amplitude in the sequence chromatogram or the most frequent nucleotide in repeat sequencing reactions) was chosen. For positions that affected the corresponding amino acid, the base that encoded the more common amino acid residue in alignments of existing SIVcpz and HIV-1 protein sequences was selected. In the absence of an apparent common amino acid (e.g., in hypervariable protein regions), the nucleotide with the highest amplitude in the sequence chromatograms was selected. Using these criteria, all ambiguous sites were resolved, except for two positions in the long terminal repeat (LTR) region of UG31.
Distance plots.
Distance plots comparing the newly derived SIVcpzUG38, SIVcpzUG31, and SIVcpzTAN5 sequences to each other and to the SIVcpzANT reference strain were generated using SimPlot (version 3.5.1) software (21). Pairwise distance values were calculated for a window of 300 bp moved in 20-base increments along the alignment. Regions that could not be unambiguously aligned were removed from the analysis.
Phylogenetic analyses.
Partial pol (HXB2 coordinates 4107 to 4775) and gp41/nef (8358 to 9054) sequences from the newly characterized Ugalla viruses were aligned to P. t. schweinfurthii SIVcpz (SIVcpzPts) reference sequences from Gombe and the Democratic Republic of the Congo by using the CLUSTAL W program (18). GenBank accession numbers are as indicated for the following SIVcpzPts strains: ANT, U42720; TAN1, AF447763; TAN2, DQ374657; TAN3, DQ374658; TAN5, FJ895394 and FJ895382; TAN6, FJ895395 and FJ895383; TAN8, FJ895403 and FJ895389; TAN9, FJ895405; TAN10, FJ895398 and FJ895386; TAN11, FJ895399 and FJ895387; TAN12, FJ895400 and FJ895388; TAN13, FJ895393 and FJ895381; TAN14, FJ895397 and FJ895385; TAN15, FJ895404; TAN16, FJ895384; TAN18, FJ895396; TAN19, FJ895402; TAN20, FJ895401 and FJ895390; TAN21, FJ895392. Regions of the sequences that could not be unambiguously aligned were removed from further analyses. For SIVcpzUG38, SIVcpzUG31, and SIVcpzTAN5, deduced Gag, Pol, Vif, and Env sequences were aligned with the corresponding protein sequences in HIV-1, P. t. troglodytes SIVcpz (SIVcpzPtt), and SIVcpzPts reference strains (GenBank accession numbers for HIV-1 strains were K03455 for group M HXB2, AJ006022 for group N YBF30, L20587 for group O ANT70, and GU1115555 for group P RB168; GenBank accession numbers for SIVcpzPtt strains were X52154 for GAB1, AF115393 for CAM3, AJ271369 for CAM5, AY169968 for CAM13, AF103818 for US, DQ373063 for MB66, DQ373064 for LB7, DQ373065 EK505, and DQ373066 for MT145; and the GenBank accession number for SIV gorilla [SIVgor] strain CP684 was FJ424871). Gag/Pol and Pol/Vif protein overlaps were removed from the N and C termini of the deduced Pol protein sequences. In addition, the concatenated Pol and Vif alignment was divided into two regions around a previously reported recombination breakpoint between the SIVcpzCAM3/CAM5/US and HIV-1 group N lineages (8, 45). Appropriate evolutionary models for phylogenetic analyses were selected using the ModelTest (33) and ProtTest (1) programs. Phylogenetic trees were constructed using maximum likelihood (12) and Bayesian (36) methods, the latter with a 25% burn-in and using as a convergence criterion an average standard deviation of partition frequencies of <0.01.
Nucleotide sequence accession numbers.
New SIVcpzPts sequences have been deposited in GenBank under accession numbers JN054704 to JN054707 and JN091690 to JN091702; the new P. t. schweinfurthii mitochondrial D-loop sequences are available under accession numbers JN091703 and JN091704.
RESULTS
Demographics of study population.
To examine whether the chimpanzees of Gombe were the only apes in Tanzania with endemic SIVcpz infection, we conducted a noninvasive survey of savanna chimpanzees in the Ugalla region approximately 135 km to the south of Gombe (Fig. 1B). Fecal specimens were collected between February 2009 and March 2010 at two locations: 375 samples were obtained in an 85-km2 area of the Issa Valley, while an additional 6 samples were collected at a single site near the Shangwa River approximately 20 km southwest of the Issa Valley base camp (Fig. 1B). To estimate the number of sampled individuals, fecal samples were subjected to mitochondrial and microsatellite analyses. Of 381 samples, 44 (43 from the Issa Valley site and 1 from the Shangwa River site) were not usable because they either were not of chimpanzee origin (n = 2), represented fecal admixtures (n = 23), or were too degraded for further analysis (n = 19). The remaining 337 fecal samples were genotyped at eight microsatellite loci, which identified a total of 72 individuals, 67 in the Issa Valley and 5 at the Shangwa River site (Table 1; see Table S1 in the supplemental material). It should be noted that this represents a minimum estimate, since up to four allelic mismatches were permitted to exclude possible allelic dropout due to specimen degradation.
Table 1.
SIVcpz infection in a community of savanna chimpanzees
| Chimp identifiera | Sexb | No. of samples | Collection date(s) (mo/day/yr) | mtDNA haplotypec | Microsatellite locid |
SIVcpz fecal WBe | SIVcpz viral RNAf | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D18s536 | D4s243 | D10s676 | D9s922 | D2s1326 | D2s1333 | D4s1627 | D9s905 | |||||||
| Ch-01 | F | 1 | 6/12/09 | GM10 | 135/139 | 194/198 | 179/183 | 282/286 | 259/267 | NAg | NA | 293/293 | Pos | |
| Ch-02 | M | 1 | 12/19/09 | GM10 | 139/159 | 218/230 | 183/187 | 270/282 | 251/263 | 321/325 | 221/221 | 281/293 | Neg | |
| Ch-03 | M | 12 | 11/2/09–3/5/10 | GM10 | 139/175 | 218/230 | 179/183 | 286/286 | 251/251 | 317/321 | 221/233 | 285/293 | Pos* | pol,gp41, gp41/nef |
| Ch-04 | F | 7 | 7/23/09–11/6/09 | GM10 | 159/175 | 218/230 | 175/183 | 286/298 | 251/263 | 317/325 | 221/233 | 285/293 | Neg | |
| Ch-05 | F | 8 | 6/27/09–11/20/09 | GM10 | 159/159 | 198/218 | 183/183 | 290/298 | 247/251 | 317/321 | 221/221 | 285/285 | Neg | |
| Ch-06 | F | 6 | 3/06/09–11/20/09 | GM10 | 159/175 | 202/218 | 179/183 | 270/286 | 231/263 | 325/337 | 221/221 | 293/293 | Pos | |
| Ch-07 | Unk | 1 | NA | GM10 | 159/159 | 198/198 | 183/183 | NA | 239/251 | NA | NA | NA | Neg | |
| Ch-08 | M | 7 | 2/7/09–3/1/10 | GM7 | 135/175 | 202/230 | 183/195 | 270/302 | 255/263 | 321/337 | 221/221 | 293/293 | Neg | |
| Ch-09 | F | 5 | 6/26/09–3/1/10 | GM7 | 135/175 | 194/230 | 183/195 | 286/302 | 243/255 | 321/325 | 221/229 | 293/293 | Neg | |
| Ch-10 | Unk | 1 | 12/7/09 | GM7 | 139/139 | 198/234 | 179/183 | 286/290 | 231/231 | NA | 221/225 | 281/293 | Pos | |
| Ch-11 | Unk | 2 | 11/06/09–11/13/09 | GM7 | 139/139 | 202/230 | 179/183 | 270/286 | 263/263 | 321/321 | 221/225 | 269/293 | Neg | |
| Ch-12 | M | 5 | 10/11/09–11/20/09 | GM7 | 139/159 | 194/198 | 179/183 | 270/302 | 251/263 | 321/337 | 221/233 | 289/293 | Neg | |
| Ch-13 | F | 8 | 7/02/09–2/15/10 | GM7 | 139/159 | 194/202 | 175/187 | 302/302 | 255/263 | 321/321 | 221/233 | 285/293 | Neg | |
| Ch-14 | F | 7 | 10/11/09–12/15/09 | GM7 | 139/159 | 198/234 | 179/183 | 290/302 | 263/263 | 321/337 | 221/221 | 293/293 | Neg | |
| Ch-15 | M | 3 | 8/14/09–10/19/09 | GM7 | 139/159 | 202/230 | 179/183 | 282/290 | 231/263 | 321/337 | 221/221 | 281/293 | Pos | pol, gp41, gp41/nef |
| Ch-16 | F | 1 | 12/15/09 | GM7 | 139/159 | 198/230 | 175/175 | 290/298 | 251/263 | NA | 209/233 | 285/293 | Neg | |
| Ch-17 | F | 8 | 6/12/09–12/15/09 | GM7 | 139/159 | 230/230 | 175/183 | 286/290 | 231/251 | 321/321 | 221/221 | 269/293 | Pos | gp41, gp41/nef |
| Ch-18 | F | 7 | 6/12/09–12/30/09 | GM7 | 139/171 | 230/234 | 175/179 | 286/290 | 263/263 | 321/325 | 221/225 | 285/293 | Pos | pFL |
| Ch-19 | M | 3 | 6/12/09–11/14/09 | GM7 | 139/175 | 202/230 | 183/183 | 286/290 | 251/255 | 317/321 | 221/233 | 281/293 | Neg | |
| Ch-20 | F | 10 | 7/2/09–2/15/10 | GM7 | 151/159 | 194/218 | 175/179 | 282/290 | 251/255 | 317/317 | 233/233 | 285/293 | Neg | |
| Ch-21 | M | 5 | 7/2/09–2/22/10 | GM7 | 159/159 | 194/230 | 175/183 | 282/298 | 251/255 | 317/317 | 221/233 | 281/285 | Neg | |
| Ch-22 | M | 9 | 7/2/09–3/5/10 | GM7 | 159/175 | 202/202 | 175/183 | 298/302 | 251/263 | 317/321 | 221/221 | 285/293 | Neg | |
| Ch-23 | M | 2 | 11/2/09–2/14/10 | GM7 | 159/175 | 202/218 | 183/187 | 286/302 | 251/263 | 321/321 | 221/221 | 293/293 | Neg | |
| Ch-24 | Unk | 1 | 2/11/10 | GM7 | 159/159 | 202/202 | NA | 262/262 | NA | NA | 221/221 | NA | Pos | |
| Ch-25 | M | 6 | 8/2/09–11/14/09 | GM8 | 159/159 | 194/230 | 179/183 | 270/270 | 251/255 | 321/325 | 221/221 | 293/293 | Pos | pol,gp41, gp41/nef |
| Ch-26 | M | 9 | 7/2/09–11/1/09 | GM8 | 159/159 | 202/234 | 183/191 | 298/302 | 251/255 | 329/337 | 221/221 | 293/293 | Pos | pol, gp41, gp41/nef |
| Ch-27 | Unk | 1 | 11/13/09 | GM8 | 139/139 | 202/234 | 179/179 | 286/286 | 251/267 | NA | 221/233 | 281/285 | Neg | |
| Ch-28 | F | 4 | 8/1/09–8/14/09 | MH32 | 135/139 | 194/198 | 179/183 | 282/286 | 259/267 | 317/321 | 233/233 | 293/293 | Neg | |
| Ch-29 | M | 5 | 7/27/09–11/6/09 | MH32 | 139/159 | 194/230 | 179/183 | 282/286 | 247/255 | 321/321 | 221/221 | 293/293 | Neg | |
| Ch-30 | M | 1 | 12/15/09 | MH32 | 139/139 | 198/230 | 175/179 | 282/290 | 251/263 | 321/321 | 209/221 | 281/293 | Neg | |
| Ch-31 | F | 1 | 11/4/09 | MH32 | 135/139 | NA | 183/183 | NA | 255/255 | NA | 221/221 | 293/293 | Pos | |
| Ch-32 | M | 9 | 7/27/09–12/15/09 | MH32 | 139/139 | 230/230 | 179/187 | 282/286 | 247/251 | 321/325 | 221/221 | 281/293 | Pos | |
| Ch-33 | F | 2 | 10/31/09–12/30/09 | MH32 | 139/159 | 194/234 | 175/195 | 286/298 | 259/263 | 321/325 | 221/225 | 285/293 | Neg | |
| Ch-34 | M | 5 | 6/12/09–12/15/09 | MH32 | 139/159 | 194/198 | 183/195 | 270/270 | 251/255 | 325/337 | 221/221 | 293/293 | Neg | |
| Ch-35 | M | 1 | 10/1/09 | MH32 | 139/159 | 198/202 | 183/183 | 290/290 | 231/231 | 321/337 | 221/221 | 285/293 | Neg | |
| Ch-36 | F | 12 | 8/2/09–2/12/10 | MH32 | 139/159 | 198/202 | 175/195 | 270/290 | 251/255 | 337/337 | 221/221 | 293/293 | Neg | |
| Ch-37 | F | 9 | 8/2/09–2/1/10 | MH32 | 139/159 | 198/218 | 175/179 | 286/306 | 243/263 | 321/321 | 221/225 | 285/293 | Neg | |
| Ch-38 | F | 6 | 3/19/09–10/11/09 | MH32 | 139/159 | 198/230 | 175/175 | 290/298 | 251/263 | 301/321 | 209/233 | 285/293 | Neg | |
| Ch-39 | M | 1 | 10/31/09 | MH32 | 139/159 | 198/234 | 179/183 | 290/302 | 263/263 | 321/337 | 221/221 | 293/293 | Pos | |
| Ch-40 | M | 9 | 8/2/09–12/30/09 | MH32 | 139/159 | 202/230 | 179/179 | 286/306 | 255/255 | 325/337 | 221/221 | 281/293 | Pos | gp41, gp41/nef |
| Ch-41 | M | 5 | 7/27/09–11/14/09 | MH32 | 139/159 | 202/234 | 179/179 | 282/286 | 251/255 | 321/325 | 221/221 | 281/293 | Pos | |
| Ch-42 | M | 4 | 6/12/09–12/30/09 | MH32 | 139/159 | 218/234 | 175/179 | 286/286 | 259/263 | 321/321 | 221/225 | 285/293 | Neg | |
| Ch-43 | F | 1 | 8/14/09 | MH32 | 139/159 | 230/230 | 175/183 | 286/290 | 231/251 | 321/321 | 221/221 | 293/293 | Pos | |
| Ch-44 | M | 2 | 6/12/09 | MH32 | 139/159 | 230/234 | 175/183 | 286/298 | 231/259 | 321/321 | 221/221 | 281/293 | Pos | pol,gp41, gp41/nef |
| Ch-45 | F | 3 | 6/12/09–11/6/09 | MH32 | 139/175 | 194/230 | 179/179 | 270/286 | 263/263 | NA | 221/221 | 293/293 | Neg | |
| Ch-46 | M | 1 | 10/4/09 | MH32 | 159/175 | 194/202 | 175/183 | 270/290 | 255/259 | 317/321 | 221/221 | 293/293 | Neg | |
| Ch-47 | F | 3 | 6/12/09-10/12/09 | MH32 | 159/159 | 194/202 | 179/183 | 286/286 | 255/263 | 321/325 | 221/221 | 285/293 | Neg | |
| Ch-48 | F | 3 | 10/11/09-10/12/09 | MH32 | 159/171 | 194/202 | 175/179 | 270/286 | 251/263 | 325/325 | 221/221 | 281/285 | Pos | |
| Ch-49 | F | 7 | 6/12/09-11/6/09 | MH32 | 159/175 | 194/194 | 179/195 | 286/286 | 251/263 | 321/325 | 225/233 | 285/285 | Neg | |
| Ch-50 | Unk | 1 | 12/15/09 | MH32 | 159/175 | 218/230 | 175/183 | 286/298 | 251/263 | 317/325 | 221/233 | 285/293 | Neg | |
| Ch-51 | F | 4 | 7/27/09-11/5/09 | MH32 | 171/175 | 194/202 | 175/175 | 286/290 | 231/259 | 317/317 | 221/221 | 293/293 | Neg | |
| Ch-52 | F | 1 | 11/6/09 | MH32 | 139/159 | 198/198 | 175/199 | 290/290 | NA | NA | NA | 285/293 | Neg | |
| Ch-53 | F | 9 | 6/12/09-11/13/09 | MH37 | 159/171 | 230/230 | 183/183 | 290/298 | 251/255 | 309/337 | 221/221 | 269/269 | Neg | |
| Ch-54 | M | 6 | 10/12/09-12/15/09 | UG135 | 151/159 | 198/202 | 175/183 | 270/286 | 231/255 | 321/325 | 221/221 | 293/293 | Neg | |
| Ch-55 | F | 1 | 12/16/09 | UG59 | 131/159 | 230/230 | 179/179 | NA | 251/255 | NA | 221/221 | 293/293 | Neg | |
| Ch-56 | F | 3 | 11/2/09-2/22/10 | UG59 | 139/159 | 194/230 | 175/195 | 286/298 | 259/263 | 317/337 | 217/221 | 281/293 | Neg | |
| Ch-57 | M | 13 | 3/7/09-2/22/10 | UG59 | 139/159 | 198/202 | 183/183 | 290/290 | 231/263 | 321/337 | 221/221 | 285/293 | Neg | |
| Ch-58 | F | 7 | 6/14/09-2/23/10 | UG59 | 139/159 | 202/202 | 175/183 | 286/290 | 231/263 | 337/337 | 221/221 | 269/293 | Pos | FL |
| Ch-59 | F | 12 | 8/14/09-2/12/10 | UG59 | 139/159 | 202/230 | 179/179 | 286/286 | 255/263 | 317/321 | 221/221 | 285/293 | Neg | |
| Ch-60 | Unk | 1 | 10/31/09 | UG59 | 139/159 | 202/230 | 175/183 | 270/290 | 231/231 | NA | 221/221 | 269/269 | Neg | |
| Ch-61 | Unk | 1 | 11/2/09 | UG59 | 139/159 | 230/230 | 175/183 | 270/286 | 251/263 | NA | 221/233 | 269/285 | Pos | |
| Ch-62 | M | 2 | 7/30/09-8/9/09 | UG59 | 139/179 | 198/198 | 183/183 | 270/290 | 263/263 | 321/325 | 221/233 | 285/293 | Neg | |
| Ch-63 | F | 13 | 3/7/09-2/15/10 | UG59 | 159/175 | 194/230 | 179/179 | 270/286 | 255/263 | 321/337 | 221/221 | 285/293 | Neg | |
| Ch-64 | F | 10 | 3/7/09-2/22/10 | UG59 | 139/159 | 230/234 | 175/179 | 270/286 | 251/263 | 321/325 | 221/221 | 269/281 | Pos* | gp41, gp41/nef |
| Ch-65 | M | 1 | 10/26/09 | UG59 | 159/159 | 194/230 | 175/183 | 282/298 | 251/255 | 317/317 | 221/233 | 281/285 | Neg | |
| Ch-66 | M | 12 | 2/24/09-2/23/10 | UG59 | 159/175 | 198/202 | 183/183 | 282/290 | 259/263 | 321/325 | 229/233 | 269/293 | Neg | |
| Ch-67 | F | 5 | 3/9/09-2/22/10 | UG59 | 175/179 | 194/198 | 183/191 | 270/290 | 263/263 | 321/321 | 233/233 | 269/293 | Neg | |
Non-boldface type, uninfected; boldface type, infected.
F, female; M, male; Unk, unknown.
MH and GM haplotypes have previously been reported (15, 20, 37); UG haplotypes have been deposited in GenBank (accession numbers JN091703 and JN091704).
Consensus genotypes are shown.
WB, Western blot; Pos, positive; Neg, negative; asterisks, the two individuals which became newly infected during our survey.
FL, full-length genome; pFL, partial full-length genome.
NA, not available.
Members of individual chimpanzee communities range together over a circumscribed geographic area, usually traveling in small temporary parties. Since many of the Issa Valley chimpanzees were sampled on more than one occasion (range, 1 to 13), their movement in space and time could be examined. These analyses documented associations among 64 of the 67 individuals, indicating that they shared the same social network as well as habitat. The remaining three, who were sampled only once, lacked such association but were identified in the same geographic area. It is thus likely that all 67 Issa Valley chimpanzees, including 31 females, 27 males, and 9 individuals whose sex could not be determined, comprised members of a single community. In contrast, the four males and one female sampled at the Shangwa River site did not share any co-nesting associations with the Issa Valley chimpanzees and thus do not seem to belong to the Issa Valley community (see Table S1 in the supplemental material).
Chimpanzee population density and home range in Ugalla.
Chimpanzees build nightly sleeping nests, and together with estimates of sighting distances, nest decay rates, the proportion of nests reused, and the proportion of infants (who do not build independent nests), the resulting counts can be used to generate estimates of population density (17, 31). Previous nest surveys using these methods within the Issa Valley study area of 85 km2 yielded a density estimate of 0.14 individual/km2, but surveys of nearby areas yielded estimates as low as 0.03/km2 (28). When the size of a community is known, an estimate of home-range size can be obtained by the reciprocal of the population density; e.g., at a density of 0.14 individual/km2 in Issa Valley, a community of 67 (see above) would occupy a minimum range of 478 km2, assuming no overlap in nesting areas with neighboring communities. In reality, calculating home-range sizes from nest-based densities underestimates the actual home range because (i) nesting areas may overlap and (ii) individuals may make excursions during the day and then return toward the center of their range to sleep.
High prevalence of SIVcpz infection in Ugalla.
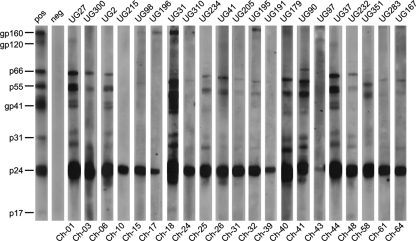
To determine whether the chimpanzees of Ugalla were SIVcpz infected, we tested all genotyped fecal samples for the presence of antibodies using a highly sensitive and specific Western blot approach (16). None of the five Shangwa River site samples were SIVcpz positive (Table 1; see Table S1 in the supplemental material); however, 96 fecal specimens from 21 Issa Valley chimpanzees exhibited clear evidence of SIVcpz infection (Fig. 2). Importantly, 14 of these individuals were sampled on more than one occasion, and except for two newly infected apes (see below), all specimens were fecal antibody positive, consistent with the fact that SIVcpz causes a persistent life-long infection. Thus, the prevalence of SIVcpz infection in the Issa Valley community was 31%, with 95% confidence limits ranging from 21% to 44%. Combining the individual test sensitivity (0.92) with the number of repeat samples per chimpanzee (from 1 to 13) increased the average overall sensitivity to 0.98, which raised the prevalence estimate only marginally, to 32%. There was no evidence of SIVcpz infection at the Shangwa River site; however, because only five chimpanzees were sampled there only once, the prevalence at the Shangwa River site may not really be different from that for Issa Valley, especially since the five chimpanzees likely represent only a subset of a larger community. Thus, the possibility of SIVcpz infection in the Sangwa River area cannot be excluded.
Fig. 2.
Evidence of SIVcpz infection in Ugalla. Chimpanzee fecal samples were tested for SIVcpz-specific antibodies using an enhanced chemiluminescent Western blot approach and HIV-1 antigen-containing strips. Samples are numbered, with the corresponding individual denoted on the bottom (also see Table S1 in the supplemental material). Molecular weights of HIV-1 specific proteins are indicated. The banding patterns of plasma from an HIV-1-infected patient (positive [pos] at a 1:10,000 dilution) and an uninfected human control (neg) are shown.
Continuing spread of SIVcpz in Ugalla.
Repeat sampling of a subset of individuals also provided an opportunity to look for incident infections. This analysis identified two chimpanzees, one male (Ch-03) and one female (Ch-64), who became newly infected over the 14-month observation period (Table 2). Ch-03 was negative for SIVcpz infection on two occasions in November 2009; however, a sample taken 3 months later was both antibody and viral RNA positive. Samples from two subsequent time points were also antibody and/or viral RNA positive. Similarly, Ch-64 was SIVcpz negative in March 2009, but a sample taken 3 months later was antibody and viral RNA positive. Samples obtained at six subsequent time points were also antibody positive (Tables 2; see Table S1 in the supplemental material). Since each of these two individuals had at least two early negative samples, the chance of misdiagnosis due to false-negative testing is less than 1%. Interestingly, Ch-03 and Ch-64 were infected with nearly identical viruses (see below). This suggests either that one transmitted SIVcpz to the other or that both became infected from a common source. Irrespective of the circumstances, the finding of two incident infections within a 14-month time frame indicates that SIVcpz continues to spread in the Ugalla area.
Table 2.
SIVcpz incident infections
| Chimp identifiera | Sexb | UG sample code | Collection date (day-mo-yr) | mtDNA haplotypec | Microsatellite locid: |
SIVcpz fecal WBe | SIVcpz viral RNA | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D18s536 | D4s243 | D10s676 | D9s922 | D2s1326 | D2s1333 | D4s1627 | D9s905 | |||||||
| Ch-03 | M | 253 | 2-Nov-09 | GM10 | 139/175 | 218/230 | 179/183 | 286/286 | 251/251 | 317/321 | 221/233 | 285/293 | Neg | |
| 284 | 2-Nov-09 | GM10 | 139/175 | 218/230 | 179/183 | 286/286 | 251/251 | 221/233 | 285/— | Neg | ||||
| 211 | 4-Nov-09 | GM10 | 139/175 | 218/230 | 179/— | 286/286 | 251/251 | 221/233 | 285/293 | Neg | Neg | |||
| M | 305 | 15-Feb-10 | GM10 | 139/175 | 218/230 | 179/183 | 286/286 | 251/251 | 317/321 | 221/233 | 285/293 | Pos | gp41 | |
| 323 | 15-Feb-10 | GM10 | 139/175 | 218/230 | 179/183 | 286/286 | 251/251 | 221/233 | 285/293 | Pos | ||||
| 298 | 22-Feb-10 | GM10 | 139/175 | 218/230 | 179/183 | 286/286 | 251/251 | 317/321 | 221/233 | 285/293 | Pos | pol | ||
| 300 | 22-Feb-10 | GM10 | 139/175 | 218/230 | 179/183 | 286/286 | 251/251 | 317/321 | 221/233 | 285/293 | Pos | pol, gp41, gp41/nef | ||
| 306 | 22-Feb-10 | GM10 | 139/175 | 218/230 | 179/183 | 286/286 | 251/251 | 317/321 | 221/233 | 285/293 | Pos | pol, gp41 | ||
| 336 | 22-Feb-10 | GM10 | 139/175 | 218/230 | 179/183 | 286/286 | 251/251 | 317/321 | 221/233 | 285/293 | Pos | |||
| 341 | 22-Feb-10 | GM10 | 139/175 | 218/230 | 179/183 | 286/286 | 251/251 | 221/233 | 285/293 | Pos | ||||
| 334 | 5-Mar-10 | GM10 | 139/— | 218/230 | 179/— | 286/286 | 251/251 | 221/233 | 285/293 | Pos | ||||
| 307 | NAf | GM10 | 139/175 | 218/230 | 179/183 | 286/286 | 251/251 | 221/233 | 285/293 | Pos | ||||
| Ch-64 | F | 11 | 7-Mar-09 | UG59 | —/159 | 230/234 | 175/— | 270/286 | 251/263 | 321/325 | 221/221 | 269/281 | Neg | |
| F | 17 | 7-Mar-09 | UG59 | —/159 | 230/234 | 175/— | 270/286 | 251/263 | 221/221 | 269/281 | Neg | Neg | ||
| F | 59 | 30-Jul-09 | UG59 | —/159 | 230/234 | 175/— | 270/286 | 251/263 | 321/325 | 221/221 | 269/281 | Pos | gp41, gp41/nef | |
| 286 | 2-Nov-09 | UG59 | —/159 | 230/234 | 175/— | 270/286 | 251/263 | 221/221 | 269/281 | Pos | ||||
| 206 | 4-Nov-09 | UG59 | —/159 | 230/234 | 175/— | 270/286 | 251/263 | 321/325 | 221/221 | 269/281 | Pos | |||
| 177 | 20-Dec-09 | UG59 | —/159 | 230/234 | 175/— | —/286 | 251/263 | 221/221 | 269/281 | Pos | ||||
| 167 | 29-Dec-09 | UG59 | 139/159 | 230/234 | 175/179 | 270/286 | 251/263 | 221/221 | 269/281 | Pos | ||||
| 317 | 15-Feb-10 | UG59 | —/159 | 230/234 | 175/— | 270/286 | 251/263 | 321/325 | 221/221 | 269/281 | Pos | |||
| 308 | 22-Feb-10 | UG59 | —/159 | 230/234 | 175/— | 270/286 | 251/263 | 221/221 | 269/281 | Pos | ||||
| 309 | 22-Feb-10 | UG59 | —/159 | 230/234 | 175/— | 270/286 | 251/263 | 321/325 | 221/221 | 269/281 | Pos | |||
Non-boldface type, uninfected; boldface type, infected.
F, female; M, male.
MH and GM haplotypes have previously been reported (15, 20, 37); UG haplotypes have been deposited in GenBank (accession numbers JN091703 and JN091704).
Microsatellite loci are indicated. —, alleles that did not amplify; empty cells, loci that failed to amplify.
WB, Western blot; Pos, positive; Neg, negative.
NA, not available.
Phylogeography of Ugalla SIVcpz.
To characterize the newly identified SIVcpz infections, we used consensus primers to amplify subgenomic pol and gp41/nef viral fragments (15, 16, 37, 41). Using all available samples as well as up to eight different RT-PCR attempts, we were able to detect SIVcpz sequences in 10 of the 21 infected individuals. Amplified regions included 231 to 669 bp of pol, 239 to 278 bp of gp41, and 545 to 791 bp of gp41/nef (Table 1; see Table S1 in the supplemental material). Our failure to amplify viral RNA from the remaining 11 infected individuals was most likely caused by viral RNA degradation due to extended sample storage at ambient temperatures.
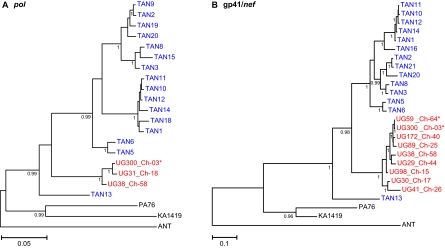
To determine the evolutionary relationships of the Ugalla viruses to each other and to previously characterized SIVcpz strains, we constructed phylogenetic trees from subgenomic gp41/nef and pol sequences (Fig. 3). These analyses showed that all 10 Ugalla viruses clustered together, forming a monophyletic lineage within the clade of SIVcpz from P. t. schweinfurthii apes (SIVcpzPts). The closest relatives of the Ugalla viruses were SIVcpz strains from Gombe National Park (Fig. 1B). As shown in Fig. 3, all Ugalla viruses fell within the radiation of Gombe SIVcpz, forming a sister clade to the main Gombe group. Interestingly, one Gombe virus (SIVcpzTAN13) previously identified in a young female who was first observed in Gombe in 1993 and is believed to be the daughter of an immigrant (37) formed an outgroup to both the Ugalla and the main Gombe clades. This divergent lineage, which was observed in both pol and gp41/nef trees, may thus represent an independent introduction of SIVcpz into Gombe, suggesting that some (or all) of the Ugalla and Gombe lineages originated in chimpanzee communities that have not yet been identified or may no longer exist. Alternatively, SIVcpz may have been introduced into Gombe a long time ago, and the Ugalla lineage could have originated there. Although current data are not sufficient to trace the origin of either the Gombe or Ugalla viruses, they indicate that these now separated communities must have been connected, either directly or indirectly, at some point in the past.
Fig. 3.
Phylogeny of SIVcpz from Ugalla. Maximum likelihood trees (12) were constructed from partial pol (HXB2 coordinates 4107 to 4775) (A) and gp41/nef (HXB2 coordinates 8358 to 9054) (B) sequences. Newly characterized SIVcpz strains from Ugalla are highlighted in red; asterisks denote two incident infections. Previously characterized viruses from Gombe and the Democratic Republic of the Congo are shown in blue and black, respectively. Numbers indicate posterior probability values for the clustering of the clade immediately to the right (only values above 0.95 are shown). The scale bars represent 0.05 (A) and 0.1 (B) substitutions per site.
Analysis of Ugalla SIVcpz genomes.
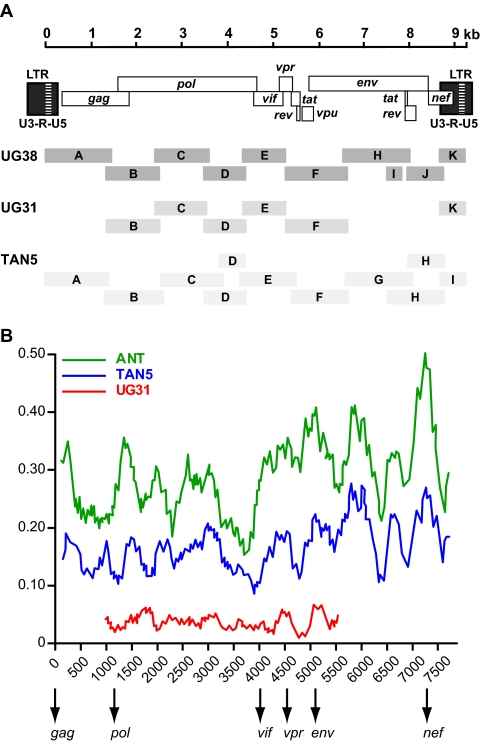
To examine the evolutionary relationship of the Ugalla viruses over the entire length of their genomes, we amplified one full-length (UG38) and one partial (UG31) SIVcpz strain from fecal samples with high RNA loads (Fig. 4A). In addition, we amplified the genome of SIVcpzTAN5, a Gombe virus representing a lineage not previously characterized. Fecal RNA was extracted and subjected to RT-PCR analysis using consensus as well as strain-specific primers. For UG38 and TAN5, this approach yielded 10 to 11 partially overlapping fragments, which in each case spanned the entire proviral genome (Fig. 4A). Amplification of UG31 was more difficult. Despite repeated RT-PCR attempts, only seven fragments spanning parts of gag, pol, and the accessory gene region could be amplified (Fig. 4A). All amplicons were sequenced directly and concatenated to generate fecal consensus sequences. As expected, these contained a limited number of ambiguous sites, which were resolved as previously described (48).
Fig. 4.
Molecular characterization of full-length SIVcpz proviruses. (A) Individual RT-PCR amplicons (shaded boxes) of SIVcpzUG38, SIVcpzUG31, and SIVcpzTAN5 are shown in relation to the SIVcpz genome (top). All fragments are drawn to scale, with nucleotide sequences numbered starting at the beginning of the R region in the 5′ LTR (see scale bar). (B) Distance plots illustrating the extent of nucleotide sequence divergence between SIVcpzUG38 and SIVcpzUG31 (red), SIVcpzTAN5 (blue), and SIVcpzANT (green). Differences were calculated for windows of 300 bp moved in steps of 20 bp along the alignment; values are plotted at the midpoint of the window. Arrows indicate the start of the gag, pol, vif, vpr, env, and nef reading frames.
The concatenated sequences of UG38, UG31, and TAN5 were 9,283 bp, 5,783 bp, and 9,893 bp in length, respectively, and all viral genes within these sequences contained intact open reading frames. The new sequences allowed us to examine previously identified SIVcpzPts signatures (40). Both Ugalla viruses and TAN5 contained insertions of 4 to 5 amino acids (aa) within the conserved PPLP domain at the C terminus of Vif. Moreover, their Vpr proteins were 89 to 90 amino acids in length and, thus, about 10 residues shorter than the Vpr proteins typically found in SIVcpz from central chimpanzees (SIVcpzPtt). Finally, UG38, UG31, and TAN5 contained a 5-amino-acid deletion in the C terminus of Nef and an 8- to 11-amino acid insertion in the ectodomain of gp41 compared to SIVcpzPtt strains. Thus, the newly sequenced Ugalla viruses as well as TAN5 contained signatures typical of SIVcpzPts strains (40).
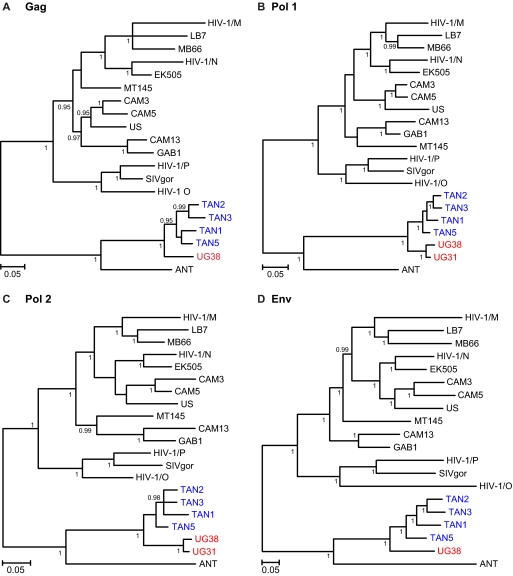
To determine the relationships of UG38, UG31, and TAN5 to other SIVcpz strains, we performed distance and phylogenetic tree analyses. Figure 4B illustrates the distances between UG38 and each of UG31, TAN5, and ANT (the first described SIVcpzPts strain) across the length of their genomes. As expected, UG38 and UG31 were most similar in all genomic regions, while TAN5 and, in particular, ANT were more distant. We also constructed trees from Gag (420 aa), N-terminal Pol (630 aa), C-terminal Pol/Vif (453 aa), and Env (729 aa) protein sequences (Fig. 5). These analyses confirmed the relationships derived from the partial pol and gp41/nef sequences and revealed no evidence for recombination between the major SIVcpzPts lineages. Thus, the available Ugalla and Gombe viruses exhibited phylogeographic clustering, forming two sister clades within the SIVcpzPts radiation.
Fig. 5.
Evolutionary relationships of Ugalla SIVcpz derived from full-length genome sequences. Trees were inferred by the Bayesian method (36) from amino acid sequence alignments of the major proteins, Gag (420 aa), N-terminal Pol (630 aa), C-terminal Pol/Vif (453 aa), and Env (729 aa); the Pol protein was separated into two fragments at a point where a recombination breakpoint was previously identified in HIV-1 group N (8, 45). SIVcpz strains from Ugalla and Gombe are highlighted in red and blue, respectively; HIV-1 group M to P, SIVgor, SIVcpzPtt, and SIVcpzPts reference sequences are shown in black. Numbers on internal branches indicate posterior probabilities (only values of 0.95 or greater are shown). Branches with posterior probability values of less than 0.5 were collapsed. The scale bars represent 0.05 amino acid replacement per site.
DISCUSSION
Studies of SIVcpz in Tanzania began in 2000, when infection was first identified in Gombe National Park (42). This discovery prompted surveys of other chimpanzee communities in East Africa, including the M group in Mahale (Fig. 1B), none of which uncovered additional infections (44). In contrast, surveys of wild apes in the Democratic Republic of the Congo revealed SIVcpz infection at multiple different field sites (19, 43). Thus, the presence of SIVcpz in Gombe seemed to represent an isolated case, possibly the remnant of a previously more widespread infection that had largely gone extinct. To test this hypothesis, we conducted a survey in the Masito-Ugalla region of the Greater Mahale Ecosystem, which comprises one of the largest remaining chimpanzee habitats in western Tanzania. There were two reasons for selecting this particular area. First, we wished to test populations that were less geographically confined than those in Gombe and Mahale Mountains National Parks (22, 25, 26, 32). Second, we were intrigued by the ecology of savanna chimpanzees, which provided an opportunity to study a low-density ape population. Testing fecal samples from just two collection sites yielded a number of unexpected results. First, we found that the 67 Issa Valley chimpanzees all shared the same home range, indicating a community size much larger than that of other known savanna chimpanzees (4, 34, 46). Second, we discovered SIVcpz infection among these apes, indicating that there was, in fact, a second area of SIVcpz endemicity in Tanzania. Third, we found that 21 of 67 savanna chimpanzees were infected, indicating that even low-density ape populations can harbor SIVcpz at high prevalence rates. Finally, we documented two incident SIVcpz infections in Issa Valley, indicating ongoing virus spread among chimpanzees in this area.
Nearly one-third of chimpanzees sampled within an 85-km2 area of the Issa Valley harbored SIVcpz. This rate of infection is at the upper end of prevalence rates previously determined for wild-living populations (16, 19, 37, 41, 49), with only the Kalande community in Gombe consistently exhibiting a larger proportion of infected individuals (37, 41). Previous surveys estimated the population density of chimpanzees in the Ugalla region to be 0.08 individual/km2, with a slightly higher density in the Issa Valley (0.14 individual/km2) (26). Thus, these savanna chimpanzees live at 20- to 50-fold lower population densities than the forest chimpanzees in Gombe and Mahale (25). For SIVcpz to be able to spread and persist in a population, the basic reproductive number, R0, must be greater than 1. For sexually transmitted diseases, R0 is the product of the transmission probability per partnership (β), the rate of partner change (c), and the duration of infectiousness (D), i.e., R0 = βcD (for a review, see reference 2). Although population density does not directly factor into this equation, it might be expected to influence the number of sexual contacts and partner turnover. In fact, a recent study in beetles demonstrated that the spread of a sexually transmitted disease (a parasitic mite) was more dependent on the population density of infected individuals than their proportion within the population (38). Importantly, our data indicate that the low population density in Ugalla has not impeded the spread of SIVcpz among these savanna chimpanzees.
The finding of 67 individuals within the Issa Valley study area, 64 of whom were interconnected by joint nesting episodes or diurnal associations, was surprising. Other known savanna chimpanzee communities are either much smaller (28 to 35 individuals) (4, 34) or from sites with appreciably greater population densities (0.2 to 0.7/km2) (14, 46). The significance of such a large community (a minimum of 67 individuals) living at such a low density (less than 0.14 individual/km2) is that the implied home range is enormous, i.e., greater than 478 km2, which is nearly twice the size of the largest similarly estimated home range published to date (4). Such a vast home range would be expected to have major consequences for within- and between-community social behaviors, including those affecting SIVcpz transmission (24, 25). Nonetheless, the high SIVcpz prevalence implies that the number of contacts between different community members is similar to that of forest chimpanzees. Additional research will be necessary to resolve this seeming paradox.
Molecular characterization of the newly identified viruses revealed a close genetic relationship of Ugalla and Gombe viruses (Fig. 3 and 5). Viruses from each community formed well-defined lineages in phylogenetic trees, with one Gombe strain outflanking both the main Gombe and Ugalla clades (Fig. 3). The SIVcpz strains from Gombe and Ugalla share a recent common ancestor, but the historical links between these communities are unclear. Since the 1940s, human settlements and deforestation have significantly reduced chimpanzee habitat between Gombe and Ugalla (35), and the original distribution of chimpanzees in this area is not known. The Malagarasi River forms the northern border of the Ugalla region and could represent a biogeographic barrier. Only two isolated chimpanzee communities occur between Gombe and this river, i.e., Kwitanga 15 km east of Gombe and Lilanshimba on the north side of the Malagarasi (Fig. 1B) (29, 35). Our finding of related viruses in Gombe and Ugalla thus suggests either that chimpanzees were previously able to cross the Malagarasi River or that SIVcpz was introduced into Gombe and Ugalla from the west shore of Lake Tanganyika by two separate routes, i.e., from the north through Burundi to Gombe and from the south through Zambia to the Mahale/Masito/Ugalla ecosystem. Among these, the former explanation seems more likely.
Land surveys between Mahale and Ugalla have provided ample evidence for the presence of chimpanzees, suggesting that large parts of this vast area are suitable chimpanzee habitats (Fig. 1B). Although human encroachment, including the Mishamo Refugee Settlement (measuring 55 km by 23 km) between Mahale and Ugalla, may hinder chimpanzee movement, no major geographical barriers exist that would prevent virus spread within the Mahale and Ugalla region. We have previously failed to detect SIVcpz in Mahale Mountains National Park, but our study was limited to only 20 individuals from one particular community (44). The Greater Mahale Ecosystem—which includes Ugalla—is believed to provide habitat for approximately 2,600 chimpanzees (26). It will thus be critical to determine to what extent SIVcpz has penetrated this population. In this context, the pathogenic potential of SIVcpz has to be considered. While chimpanzee density in Ugalla does not seem to have changed much since the 1960s (26), data from the Kalande community in Gombe would suggest that the impact of SIVcpz on chimpanzee population growth has a considerable lag time (37). It is thus possible that any detrimental effect of SIVcpz on these savanna chimpanzees may have not yet manifested itself.
In summary, we report here that SIVcpz infection in Tanzania is more widespread than previously appreciated. We also provide evidence that a large number of some of the most endangered, yet largely unstudied, ape populations in Tanzania are at risk of SIVcpz infection. Given the value of these communities to different disciplines, it will be critical to determine whether and to what extent other savanna chimpanzees across western Tanzania harbor SIVcpz, in order to be able to gauge what impact this infection might have on their long-term survival.
Supplementary Material
ACKNOWLEDGMENTS
We thank Shadrack Lucas, Busoti Juma, Ndai Sammwely, Abdalla Kanimba, Halufani Mulalelwa, Abdallah Moshi, and Moshi Rajabu for field assistance and collecting fecal samples from wild chimpanzees; Iddi Lipende and Anthony Collins for logistical support; the Tanzania Commission for Science and Technology and the Tanzania Wildlife Research Institute for permission to conduct research in Ugalla; Joel Robertson, Matthias Kraus, and Maria Salazar for technical assistance; James Holland Jones for helpful discussions; and Jamie C. White for artwork and manuscript preparation.
This work was supported by grants from the National Institutes of Health (R01 AI50529, R01 AI58715), the UAB Center for AIDS Research (P30 AI 27767), the National Science Foundation (DDIG), the Margot Marsh Biodiversity Foundation, the Department of Anthropology at the University of Texas at San Antonio, the Ruggles-Gates Fund for Biological Anthropology, the University of California in San Diego (UCSD COR), the Carnegie Trust for Universities of Scotland, the Harold Hyam Wingate Foundation, the L. S. B. Leakey Foundation, the International Primatological Society, and the Wenner Gren Foundation. R.S.R. was funded by a Howard Hughes Medical Institute Med-into-Grad Fellowship.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 20 July 2011.
REFERENCES
- 1. Abascal F., Zardoya R., Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105 [DOI] [PubMed] [Google Scholar]
- 2. Anderson R. M., Garnett G. P. 2000. Mathematical models of the transmission and control of sexually transmitted diseases. Sex. Transm. Dis. 27:636–643 [DOI] [PubMed] [Google Scholar]
- 3. Bailes E., et al. 2003. Hybrid origin of SIV in chimpanzees. Science 300:1713. [DOI] [PubMed] [Google Scholar]
- 4. Baldwin P. J., McGrew W. C., Tutin C. E. G. 1982. Wide-ranging chimpanzees at Mt. Assirik, Senegal. Int. J. Primatol. 3:367–385 [DOI] [PubMed] [Google Scholar]
- 5. Constable J. L., Ashley M. V., Goodall J., Pusey A. E. 2001. Noninvasive paternity assignment in Gombe chimpanzees. Mol. Ecol. 10:1279–1300 [DOI] [PubMed] [Google Scholar]
- 6. Etienne L., et al. 2011. Characterization of a new simian immunodeficiency virus strain in a naturally infected Pan troglodytes troglodytes chimpanzee with AIDS related symptoms. Retrovirology 8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gagneux P., et al. 1999. Mitochondrial sequences show diverse evolutionary histories of African hominoids. Proc. Natl. Acad. Sci. U. S. A. 96:5077–5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gao F., et al. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436–441 [DOI] [PubMed] [Google Scholar]
- 9. Gonder M. K., et al. 2011. Evidence from Cameroon reveals differences in the genetic structure and histories of chimpanzee populations. Proc. Natl. Acad. Sci. U. S. A. 108:4766–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gray R. H., et al. 2001. Probability of HIV-1 transmission per coital act in monogamous, heterosexual, HIV-1-discordant couples in Rakai, Uganda. Lancet 357:1149–1153 [DOI] [PubMed] [Google Scholar]
- 11. Groves C. 2001. Primate taxonomy. Smithsonian Institution Press, Washington, DC [Google Scholar]
- 12. Guindon S., et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321 [DOI] [PubMed] [Google Scholar]
- 13. Hernandez-Aguilar R. A. 2009. Chimpanzee nest distribution and site reuse in a dry habitat: implications for early hominin ranging. J. Hum. Evol. 57:350–364 [DOI] [PubMed] [Google Scholar]
- 14. Kano T. 1971. The chimpanzees of Filabanga, Western Tanzania. Primates 12:229–246 [Google Scholar]
- 15. Keele B. F., et al. 2009. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature 460:515–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Keele B. F., et al. 2006. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science 313:523–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kouakou C. Y., Boesch C., Kuehl H. 2009. Estimating chimpanzee population size with nest counts: validating methods in Tai National Park. Am. J. Primatol. 71:447–457 [DOI] [PubMed] [Google Scholar]
- 18. Larkin M. A., et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 19. Li Y., et al. 2010. Molecular epidemiology of SIV in eastern chimpanzees and gorillas, abstr. 440. Abstr. 7th Conf. Retroviruses Opportunistic Infect. http://www.retroconference.org/2010/PDFs/440.pdf [Google Scholar]
- 20. Liu W., et al. 2008. Molecular ecology and natural history of simian foamy virus infection in wild-living chimpanzees. PLoS Pathog. 4:e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lole K. S., et al. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Massawe E. T. 1992. Assessment of the status of chimpanzee populations in Western Tanzania. Afr. Study Monogr. 13:35–55 http://jambo.africa.kyoto-u.ac.jp/kiroku/asm_normal/abstracts/pdf/ASM%20%20Vol.13%20No.1%201992/Edeus%20T.%20MASSAWE.pdf [Google Scholar]
- 23. McGrew W. C., Baldwin P. J., Tutin C. E. G. 1981. Chimpanzees in a hot, dry, and open habitat: Mt. Assirik, Senegal, West Africa. J. Hum. Evol. 10:227–244 [Google Scholar]
- 24. Moore J. 1996. Savanna chimpanzees, referential models and the last common ancestor, p. 275–292In McGrew W. C., Marchant L. F., Nishida T. (ed.), Great ape societies. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 25. Moore J. 1992. “Savanna” chimpanzees, p. 99–118In Nishida T., McGrew W. C., Marler P., Pickford M., de Waal F. B. M. (ed.), Topics in primatology, vol. I Human origins. University of Tokyo Press, Tokyo, Japan [Google Scholar]
- 26. Moyer D., et al. 2006. Surveys of chimpanzees and other biodiversity in Western Tanzania. Report to the U.S. Fish and Wildlife Service. The Jane Goodall Institute, Wildlife Conservation Society, University of California at San Diego, La Jolla, CA: http://ugalla.ucsd.edu/objetos/herwcsta.pdf [Google Scholar]
- 27. Oates J. F., et al. 2008, posting date Pan troglodytes. In IUCN 2010. IUCN red list of threatened species, version 2010.4. http://www.iucnredlist.org/apps/redlist/details/15933/0
- 28. Ogawa H., Idani G., Moore J., Pintea L., Hernandez-Aguilar A. 2007. Sleeping parties and nest distribution of chimpanzees in the savanna woodland, Ugalla, Tanzania. Int. J. Primatol. 28:1397–1412 [Google Scholar]
- 29. Ogawa H., Sakamaki T., Idani G. 2006. The influence of Congolese refugees on chimpanzees in the Lilanshimba area, Tanzania. Pan Africa News 13:21–22 http://mahale.main.jp/PAN/13_2/13(2)_04.html [Google Scholar]
- 30. Piel A., Moore J. 2010. Monitoring movements: tracking unhabituated chimpanzees in Ugalla, Western Tanzania using a novel method—real-time acoustic localization, abstr. 656. Int. Primatol. Soc. XXIII Congr. http://primate-society.com/ips/public/ips_program/IPS10-656.pdf
- 31. Plumptre A. J., Reynolds V. 1997. Nesting behavior of chimpanzees: implications for censuses. Int. J. Primatol. 18:475–485 [Google Scholar]
- 32. Plumptre A. J., et al. 2010. Eastern chimpanzee (Pan troglodytes schweinfurthii): status survey and conservation action plan 2010-2020. IUCN, Gland, Switzerland: http://www.iucn.org/knowledge/publications_doc/publications/?5481/East-African-chimpanzee-Pan-troglodytes-schweinfurthii-status-survey-and-conservation-action-plan-2010-2020 [Google Scholar]
- 33. Posada D., Crandall K. A. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818 [DOI] [PubMed] [Google Scholar]
- 34. Pruetz J. D., Bertolani P. 2009. Chimpanzee (Pan troglodytes verus) behavioral responses to stresses associated with living in a savanna-mosaic environment: implications for hominin adaptation to open habitats. PaleoAnthropology 2009:252–262 [Google Scholar]
- 35. Pusey A. E., Pintea L., Wilson M. L., Kamenya S., Goodall J. 2007. The contribution of long-term research at Gombe National Park to chimpanzee conservation. Conserv. Biol. 21:623–634 [DOI] [PubMed] [Google Scholar]
- 36. Ronquist F., Huelsenbeck J. P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574 [DOI] [PubMed] [Google Scholar]
- 37. Rudicell R. S., et al. 2010. Impact of simian immunodeficiency virus infection on chimpanzee population dynamics. PLoS Pathog. 6:e1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ryder J. J., Webberley K. M., Boots M., Knell R. J. 2005. Measuring the transmission dynamics of a sexually transmitted disease. Proc. Natl. Acad. Sci. U. S. A. 102:15140–15143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 40. Santiago M. L., et al. 2003. Amplification of a complete simian immunodeficiency virus genome from fecal RNA of a wild chimpanzee. J. Virol. 77:2233–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Santiago M. L., et al. 2003. Foci of endemic simian immunodeficiency virus infection in wild-living eastern chimpanzees (Pan troglodytes schweinfurthii). J. Virol. 77:7545–7562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Santiago M. L., et al. 2002. SIVcpz in wild chimpanzees. Science 295:465. [DOI] [PubMed] [Google Scholar]
- 43. Sharp P. M., Hahn B. H. Origins of HIV and the AIDS pandemic. Cold Spring Harb. Perspect. Biol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sharp P. M., Shaw G. M., Hahn B. H. 2005. Simian immunodeficiency virus infection of chimpanzees. J. Virol. 79:3891–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Simon F., et al. 1998. Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat. Med. 4:1032–1037 [DOI] [PubMed] [Google Scholar]
- 46. Suzuki A. 1969. An ecological study of chimpanzees in a savanna woodland. Primates 10:103–148 [Google Scholar]
- 47. Takehisa J., et al. 2009. Origin and biology of simian immunodeficiency virus in wild-living western gorillas. J. Virol. 83:1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Takehisa J., et al. 2007. Generation of infectious molecular clones of simian immunodeficiency virus from fecal consensus sequences of wild chimpanzees. J. Virol. 81:7463–7475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Van Heuverswyn F., et al. 2007. Genetic diversity and phylogeographic clustering of SIVcpzPtt in wild chimpanzees in Cameroon. Virology 368:155–171 [DOI] [PubMed] [Google Scholar]
- 50. Wroblewski E. E., et al. 2009. Male dominance rank and reproductive success in chimpanzees, Pan troglodytes schweinfurthii. Anim. Behav. 77:873–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.