Abstract
Dimerization of HIV protease is essential for the acquisition of protease's proteolytic activity. We previously identified a group of HIV protease dimerization inhibitors, including darunavir (DRV). In the present work, we examine whether loss of DRV's protease dimerization inhibition activity is associated with HIV development of DRV resistance. Single amino acid substitutions, including I3A, L5A, R8A/Q, L24A, T26A, D29N, R87K, T96A, L97A, and F99A, disrupted protease dimerization, as examined using an intermolecular fluorescence resonance energy transfer (FRET)-based HIV expression assay. All recombinant HIVNL4-3-based clones with such a protease dimerization-disrupting substitution failed to replicate. A highly DRV-resistant in vitro-selected HIV variant and clinical HIV strains isolated from AIDS patients failing to respond to DRV-containing antiviral regimens typically had the V32I, L33F, I54M, and I84V substitutions in common in protease. None of up to 3 of the 4 substitutions affected DRV's protease dimerization inhibition, which was significantly compromised by the four combined substitutions. Recombinant infectious clones containing up to 3 of the 4 substitutions remained sensitive to DRV, while a clonal HIV variant with all 4 substitutions proved highly resistant to DRV with a 205-fold 50% effective concentration (EC50) difference compared to HIVNL4-3. The present data suggest that the loss of DRV activity to inhibit protease dimerization represents a novel mechanism contributing to HIV resistance to DRV. The finding that 4 substitutions in PR are required for significant loss of DRV's protease dimerization inhibition should at least partially explain the reason DRV has a high genetic barrier against HIV's acquisition of DRV resistance.
INTRODUCTION
Currently available combination therapy or highly active antiretroviral therapy (HAART) for human immunodeficiency virus type 1 (HIV) infection and AIDS has been shown to potently suppress the replication of HIV and extend the life expectancy of HIV-infected individuals (32, 34). Recent analyses have revealed that life expectancy in HIV-infected patients treated with HAART has significantly increased, that mortality rates for HIV-infected persons have recently become close to that of general population, and that the appearance of the current first-line antiretroviral therapy with boosted protease inhibitor (PI)-based regimens has made the development of HIV resistance relatively less likely (2, 7, 18, 39). However, the ability to provide effective long-term antiretroviral therapy for HIV infection remains a complex issue since many of those who initially achieved favorable viral suppression to undetectable levels still suffer treatment failure (12, 18, 29).
Dimerization of HIV protease (PR) subunits is an essential process for the acquisition of proteolytic activity of HIV PR, which plays a critical role in the maturation and replication of the virus (28, 40). Thus, inhibition of PR dimerization by chemical reagents is likely to abolish proteolytic activity and intervene in HIV replication. We have recently developed an intermolecular fluorescence resonance energy transfer (FRET)-based HIV-expression assay that employs cyan fluorescent protein (CFP)- and yellow fluorescent protein (YFP)-tagged HIV PR monomers to detect and quantify PR dimerization (26). Using this assay, we identified a group of nonpeptidyl small molecule inhibitors of HIV PR dimerization. These inhibitors, including darunavir (DRV) and tipranavir (TPV) as well as a series of potent experimental antiretroviral agents such as TMC126 (41), blocked PR dimerization at concentrations of as low as 0.01 μM and potently blocked HIV replication in vitro (26).
DRV contains a structure-based and designed privileged nonpeptidic P2 ligand, 3(R),3a(S),6a(R)-bis-tetrahydrofuranylurethane (bis-THF) (14, 15, 27), which potently inhibits the enzymatic activity and dimerization of HIV PR (26) and has a high-level genetic barrier against HIV development of resistance to DRV (9, 10). Nevertheless, we have witnessed that HIV acquires significant levels of resistance against DRV among HIV-infected individuals who have received long-term combination chemotherapy (33, 38). Indeed, a variety of amino acid substitutions that are potentially related to HIV resistance to DRV have been reported (9, 24, 33, 38). Thus, the elucidation of the mechanism of the development of HIV drug resistance represents an urgent subject in the research area of HIV-1 infection/AIDS and therapy.
MATERIALS AND METHODS
Cells, viruses, and antiviral agents.
MT-4 cells were grown in RPMI 1640-based culture medium, and 293T and COS7 cells were propagated in Dulbecco's modified Eagle's medium. These media were supplemented with 10% fetal calf serum (FCS; PAA Laboratories GmbH, Linz, Austria) plus 50 U of penicillin and 50 μg of kanamycin per ml. The following HIV strains were used for the determination of 50% effective concentrations (EC50s) against DRV and to construct plasmids for use in the FRET-based HIV expression assay, including HIVNL4-3 and HIV8MIXP51. Three recombinant clinical HIV isolates (rCLHIVF16, rCLHIVT45, and rCLHIVT48) used in this study were produced using recombinant HIVNL4-3-based infectious molecular clones generated by ligating patient-derived amplicons encompassing approximately 200 nucleotides of 3′ Gag (beginning at the unique ApaI restriction site), the entire protease, and the first 72 nucleotides of reverse transcriptase (RT) using the expression vector pNLPFB (a generous gift from Tomozumi Imamichi of the National Institute of Infectious Diseases and Allergy). The four clinical HIV isolates examined in the present study were chosen from 32 isolates that had been obtained from multi-PI-treated patients whose protease genotype contained prototypical patterns of PI resistance. The median duration of continuous PI treatment was 7.5 years (range, 6 to 10 years). The median number of PIs received (excluding the use of ritonavir for pharmacokinetic boosting) was 5 (range, 4 to 8). According to the PhenoSense assay, two isolates (rCLHIVT45 and rCLHIVF16) had high-level resistance to all PIs, including >90-fold decreased susceptibility to DRV and >8-fold decreased susceptibility to TPV, and one isolate (rCLHIVT48) had intermediate resistance to DRV and TPV and high-level resistance to the remaining PIs (see Table 2).
Table 2.
Amino acid substitutions associated with DRV resistance
| DRV resistance-associated amino acid substitutions identifieda | Variantb | EC50, μM (fold change)c |
|---|---|---|
| L10I, I15V, K20R, L24I, V32I, L33F, M36I, M46L, I54 M, L63P, K70Q, V82I, I84V, L89 M | HIV8MIXP51 | >1 (>333) |
| L10F, V11I, I13V, L19Q, K20 M, V32I, L33F, E35A, M36I, M46I, I47V, I54 M, R57K, I62V, L63P, I64V, G73T, T74A, I84V, L89V, L90 M | rCLHIVF16 | 0.30 (97) |
| L10F, V11I, T12P, I13V, I15V, L19P, K20T, V32I, L33F, E35G, M36I, I54V, I62V, L63P, K70T, A71I, G73S, P79A, I84V, L89V, L90 M | rCLHIVT45 | 0.33 (105) |
| L10I, I13V, I15V, L19V, L24I, V32I, L33F, K43E, M46L, I54L, D60E, L63P, A71V, I72V, V82A, I84V | rCLHIVT48 | 0.17 (54) |
| V11I, V32I, L33F, I47V, I50V, I54L/M, G73S, L76V, I84V, L89V | ||
| V32I, I50V, I54L, I54 M, L76V, V82F |
Amino acid substitutions identified in the protease-encoding region of HIV are shown. The amino acid substitutions shown in the second row from the bottom were reported by De Meyer et al. (9) and Mitsuya et al. (33). Those in the bottom row were reported by Van Marck et al. (38) and were reported to have the greatest impact on HIV-1 resistance to DRV. Those for HIV8MIXP51 were reported by Koh et al. (24).
Three infectious clones (rCLHIVF16, rCLHIVT45, and rCLHIVT48) were derived from clinical strains isolated from patients who failed to respond to DRV-containing regimens.
Shown are EC50s of DRV for each infectious clone. Values in parentheses represent fold changes of EC50s of DRV compared to EC50s against a wild-type clinical strain, HIV-1ERS104pre. All assays were conducted in triplicate, and the mean values are shown. The EC50s of ritonavir and lopinavir against HIV8MIXP51, rCLHIVF16, rCLHIVT45, and rCLHIVT48 were all >1 μM.
DRV was synthesized by A. K. Ghosh as described previously (16, 27). GRL-0216 (37), GRL-98065 (1), and TMC-126 (41) were synthesized in a convergent manner by coupling an optically active P2 ligand and an (R)-hydroxyethylamino sulfonamide isostere (17). TPV was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health.
Generation of FRET-based HIV expression system.
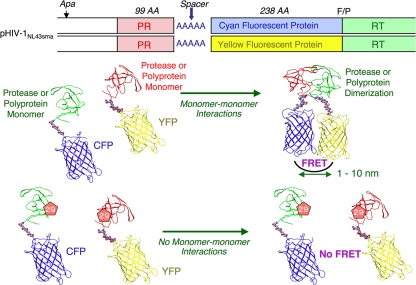
Cyan fluorescent protein (CFP)- and yellow fluorescent protein (YFP)-tagged HIV PR constructs were generated using BD Creator DNA cloning kits (BD Biosciences, San Jose, CA). The basic concepts of the intermolecular FRET-based HIV expression assay (FRET-HIV assay) are illustrated in Fig. 1. In brief, XhoI and HindIII fragments from the pCR-XL-TOPO vector containing the HIV PR-encoding gene excised from pHIVNL4-3 were inserted into pDNR-1r, the donor vector, which had been digested with XhoI and HindIII. In the transfer of the PR gene from the donor vector into pLP-CFP/YFP-C1 (acceptor vector), the Cre-loxP site-specific recombination method was used according to the manufacturer's instructions. Using Cre-recombinase with the loxP site, the PR gene from pDNR-1r was inserted into pLP-CFP-C1 or pLP-YFP-C1 through Cre-mediated recombination (19), generating a plasmid expressing CFP-tagged wild-type PR (PRWT) and one expressing YFP-tagged PRWT, with which HIV PR was successfully expressed as a fusion protein with CFP and YFP tagged at the C terminus, respectively. Western blot assay using anti-green fluorescent protein-specific rabbit polyclonal antibodies revealed that PR was correctly tagged with CFP or YFP (26).
Fig. 1.
FRET-based HIV expression system. Plasmids encoding full-length molecular infectious HIV (HIVNL4-3) clones that produce CFP- or YFP-tagged PR were prepared using the PCR-mediated recombination method as described in Materials and Methods. A linker consisting of five alanines was inserted between PR and the fluorescent protein. A phenylalanine-proline site (F/P) that HIV PR cleaves was introduced between the fluorescent protein and RT. Shown are structural representations of PR monomers and dimer in association with the linker atoms and fluorescent proteins. FRET occurs when the two fluorescent proteins become 1 to 10 nm apart. If an agent that is capable of inhibiting the dimerization of PR monomer subunits is present when the CFP- and YFP-tagged PR monomers are produced within the cell upon cotransfection, no FRET occurs. If certain amino acid substitutions (AA) such as D29N (shown below) are introduced, PR subunits do not get dimerized and no FRET occurs.
For the generation of full-length molecular infectious clones containing CFP- or YFP-tagged PR, the PCR-mediated recombination (PMR) method was used (11). To this end, we amplified an upstream proviral DNA fragment containing an ApaI site and HIV PR (excised from pHIVNL4-3) with primer pair Apa-PRO-F (5′-TTG CAG GGC CCC TAG GAA AAA GG-3′) plus PR-5Ala-R (5′-GGC TGC TGC GGC AGC AAA ATT TAA AGT GCA GCC AAT CT-3′), a middle proviral DNA fragment containing CFP (excised from pCFP-C1) or YFP (excised from pYFP-C1) (Clontech, Mountain View, CA) with primer pair CFPYFP-5Ala-F (5′-GCT GCC GCA GCA GCC GTG AGC AAG GGC GAG GAG CTG-3′) plus CFPYFP-FP-R (5′-ACT AAT GGG AAA CTT GTA CAG CTC GTC CAT GCC G-3′), and a downstream proviral DNA fragment containing the 5′-DNA fragment of RT and an SmaI site from pHIVNLSma (13, 25), which had been created to have an SmaI site by changing two nucleotides (2590 and 2593) of pHIVNL4-3, with primer pair FRT-F (5′-TTT CCC ATT AGT CCT ATT GAG ACT GTA-3′) plus NL4-3-RT263-R (5′-CCA GAA ATC TTG AGT TCT CTT ATT-3′). A linker consisting of five alanines was inserted between the PR and fluorescent protein. The phenylalanine-proline site that HIV PR cleaves was also introduced between the fluorescent protein and RT. Thus, the three DNA fragments obtained were subsequently joined by using the PMR reaction performed under the standard condition for ExTaq polymerase (Takara Bio, Inc., Otsu, Japan) with 10 pmol of Apa-PRO-F (5′-TTG CAG GGC CCC TAG GAA AAA GG-3′) and NL4-3-RT263-R (5′-CCA GAA ATC TTG AGT TCT CTT ATT-3′) and the three DNA fragments (100 ng each) in a 20-μl reaction solution. Thermal cycling was carried out at 94°C for 3 min, followed by 35 cycles of 94°C for 50 s, 53°C for 50 s, and 72°C for 2 min, and finally 72°C for 15 min. The amplified PCR products were cloned into pCR-XL-TOPO vector according to the manufacturer's instructions (Gateway cloning system; Invitrogen). PCR products were generated with pCR-XL-TOPO vector as templates, followed by digestion by both ApaI and SmaI, and the ApaI-SmaI fragment was introduced into pHIVNLSma (13), generating pHIV-PRWTCFP and pHIV-PRWTYFP, respectively.
Analysis of inter- and intramolecular interactions of PR subunits.
Analysis of inter- and intramolecular interactions of PR subunits was conducted by examining the crystal structure of DRV with HIV PR (Protein Data Bank identification no. [PDB ID no.] 2IEN). Hydrogens were added and minimized using the OPLS2005 force field with constraints on heavy atom positions. The calculation was performed using MacroModel 9.1 from Schrödinger, LLC. Hydrogen bonds were assigned when the following distance and angle cutoff was satisfied: 3.0 Å for H-A distance, with a D-H-A angle of >90° and an H-A-B angle of >60°, where H is the hydrogen, A is the acceptor, D is the donor, and B is a neighbor atom bonded to the acceptor. The representative distance between the termini of two monomers was determined by analyzing the PR-DRV crystal structure (PDB ID no. 2IEN). The distance between the α carbons at the N termini and C termini is around 0.5 nm, whereas the distance between the α carbons of the N termini ends of two monomers is around 1.8 nm.
FRET procedure.
COS7 cells plated on an EZ view cover glass bottom culture plate (Iwaki, Tokyo) were transfected with the indicated plasmid constructs, using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions in the presence of various concentrations of each compound, cultured for 72 h, and analyzed under a Fluoview FV500 confocal laser scanning microscope (Olympus Optical Corp., Tokyo) at room temperature. When the effect of each compound was analyzed by FRET, test compounds were added to the culture medium simultaneously with plasmid transfection. The results of FRET were determined by quenching of CFP (donor) fluorescence and an increase in YFP (acceptor) fluorescence (sensitized emission), because part of the energy of CFP is transferred to YFP instead of being emitted. This phenomenon can be measured by bleaching YFP, which should result in an increase in CFP fluorescence. This technique, also known as acceptor photobleaching, is a well-established method of determining the occurrence of FRET (5, 6, 35, 36). Dequenching of the donor CFP by selective photobleaching of the acceptor YFP was performed by first obtaining YFP and CFP images at the same focal plane, followed by illuminating the same image for 3 min at a wavelength of 488 nm with a laser power set at the maximum intensity to bleach YFP and then recapturing the same CFP and YFP images. The changes in the CFP and YFP fluorescence intensity in the images of selected regions were examined and quantified using the Olympus FV500 Image software system (Olympus Optical Corp.). Background values were obtained from the regions where no cells were present and were subtracted from the values for the cells examined in all calculations. For each chimeric protein, the data were obtained from at least three independent experiments. Digitized image data obtained from the experiment were prepared for presentation using Photoshop 6.0 (Adobe Systems, Mountain View, CA). Ratios of intensities of CFP fluorescence after photobleaching to CFP fluorescence prior to photobleaching (CFPA/B ratios) were determined. It is well established that CFPA/B ratios of >1.0 indicate that association of CFP- and YFP-tagged proteins occurred and were interpreted to indicate that the dimerization of PR subunits occurred. CFPA/B ratios of <1 indicated that the association of the two subunits did not occur and were interpreted to indicate that PR dimerization was inhibited. The difference in the CFPA/B ratios determined in the presence or absence of test drugs was evaluated using the nonparametric Mann-Whitney U statistic test.
Replication kinetics of various NL-PRmutantYFP strains.
MT-4 cells (105) were exposed to each infectious HIV-PRYFP clone (100 ng of p24 Gag protein/ml) for 6 h, washed twice with phosphate-buffered saline (PBS), and cultured in 7 ml of complete medium with some modification as described previously (3, 13). Culture supernatants (50 μl) were harvested every other day, and virus replication was monitored by the amounts of p24 Gag produced in the culture supernatants.
RESULTS
DRV inhibits the dimerization of HIV PR expressed as a single protein.
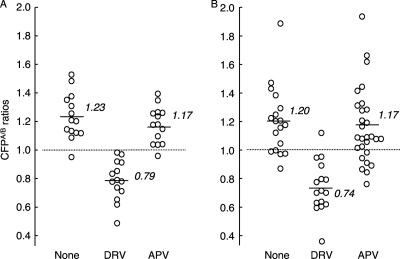
The basic concepts of the intermolecular FRET-based HIV-expression assay (FRET-HIV expression assay) to assess PR dimerization are illustrated in Fig. 1 (26). Using the FRET-based HIV expression assay, we previously identified a group of PR dimerization inhibitors (PDIs), including DRV and TPV, although other conventional PR inhibitors (PIs), such as amprenavir (APV), failed to block dimerization (Fig. 2A) (26). In the FRET-based HIV expression assay, YFP- or CFP-tagged PR should be primarily expressed as a part of Pr160gag-pol polyprotein, and it was assumed that DRV blocks the dimerization of the PR subunit within the polyprotein. Thus, it remained to be determined whether DRV also blocks the dimerization of PR in the form of a single PR molecule. We, therefore, generated a pair of plasmids encoding wild-type HIVNL4-3 PR tagged with YFP and CFP in the 3′ terminus (pPRWTYFP and pPRWTCFP, respectively), transfected COS7 cells with the pair, and determined whether DRV blocked the dimerization of PRWTYFP and PRWTCFP. As shown in Fig. 2B, the average value of CFPA/B ratios obtained in the absence of drug was 1.20 ± 0.24, which indicated that the dimerization between PRWTYFP and PRWTCFP occurred. The average value of the ratios determined in the presence of 1 μM DRV was 0.74 ± 0.18 (P = 0.000003), signifying that DRV clearly blocked the dimerization of PRWTYFP and PRWTCFP, while the value with APV was 1.17 ± 0.27 (P = 0.60), indicating that APV failed to block the dimerization, in line with our previous data (26). These results strongly suggest that DRV blocks dimerization of the PR monomer subunit in the form of Pr160gag-pol polyprotein as well as in the form of a single molecule.
Fig. 2.
DRV blocks the dimerization of both pHIV-PRWT-encoded PR and pPRWT-encoded PR. (A) COS7 cells were cotransfected with pHIV-PRWTCFP plus pHIV-PRWTYFP in the absence or presence of 1 μM DRV or APV. On day 3 after transfection, CFPA/B ratios were determined using an FV500 confocal laser microscope. When the average value of CFPA/B ratios was greater than 1.0, it was judged that the dimerization of PR occurred, whereas when it was less than 1.0, it was judged that the dimerization did not occur. (B) COS7 cells were cotransfected with a pair of wild-type PR-expressing plasmids (pPRWTCFP plus pPRWTYFP) in the absence or presence of 1 μM DRV or APV, and CFPA/B ratios were determined as described above. Note that DRV inhibited the dimerization of PR when it was expressed as HIV virions and virion-free PR. The results of statistical evaluation of the changes in the CFPA/B ratios, determined in the presence or absence of DRV or APV, using the nonparametric Mann-Whitney U test, are as follows. (A) For the CFPA/B ratios in the absence of drug (CFPA/BNo Drug) versus the CFPA/B ratios in the presence of 1.0 μM DRV (CFPA/B1.0 DRV), P = 0.00001, and for CFPA/BNo Drug versus CFPA/B1.0 APV, P = 0.42. (B) For CFPA/BNo Drug versus CFPA/B1.0 DRV, P = 0.000003, and for CFPA/BNo Drug versus CFPA/B1.0 APV, P = 0.60.
Dimerization profiles of single PR mutants in the presence of DRV.
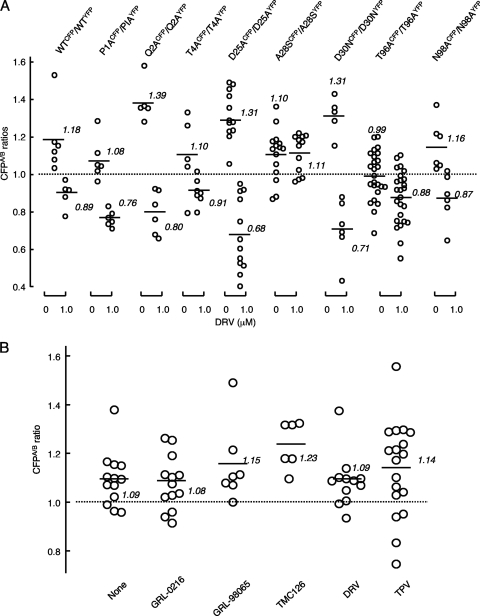
Certain amino acids in the termini and active site interfaces, both of which are critical for the dimerization of PR monomer subunits (28, 40), do not significantly affect the dimerization process of PR. Such amino acids include Pro-1, Gln-2, Thr-4, Asp-25, Ala-28, Asp-30, Thr-96, and Asn-98 (26). It is assumed that DRV blocks PR dimerization by binding to a certain structural domain or domains within or in the proximity of either or both of the two interfaces (4, 22, 23). We, therefore, examined whether amino acid substitutions at positions 1, 3, 5, 25, 28, 30, 96, and 98, which allow PR to dimerize, affected the PR dimerization disruption by DRV. We reasoned that if any of the amino acid substitutions at these positions would affect PR dimerization inhibition by DRV, such amino acids could possibly be associated with the binding of DRV to the PR subunit. However, 1 μM DRV effectively blocked the dimerization of all of the mutated PR species, except that of the species with the A28S substitution (Fig. 3 A). These data suggest that all amino acid residues examined except A28S were not associated with the binding of DRV to the PR monomer subunit.
Fig. 3.
Dimerization profiles of single PR mutants in the presence of DRV. (A) COS-7 cells were cotransfected with pHIV-PRWTCFP plus pHIV-PRWTYFP (shown as WTCFP/WTYFP) or mutated pairs such as pHIV-PRP1ACFP plus pHIV-PRP1AYFP (shown as P1ACFP/P1AYFP) in the absence or presence of 1 μM DRV. On day 3 after transfection, CFPA/B ratios were determined. (B) COS7 cells were cotransfected with plasmid pair pHIV-PRA28SCFP and pHIV-PRA28SYFP in the absence or presence of an agent (1 μM GRL-0216, DRV, GRL-98065, TPV, or TMC126), and CFPA/B ratios were determined as described above. (A) The statistical evaluation of all the changes in the CFPA/B ratios determined in the presence or absence of DRV using the nonparametric Mann-Whitney U test, gave P values ranging 0.000037 to 0.044, except for the P value for the pair A28SCFP and A28SYFP, which was 0.57. (B) The differences between the CFPA/B ratios in the absence of drug (CFPA/BNo Drug) and the CFPA/B ratios in the presence of 1.0 μM DRV (CFPA/B1.0 DRV) were statistically insignificant, indicating that all of the agents examined failed to block the dimerization of A28SCFP/A28SYFP.
We have previously shown that, in addition to DRV and TPV, the three compounds GRL-0216 (37), GRL-98065 (1), and TMC126 (41) effectively blocked PR dimerization in the FRET-based HIV expression assay (26). Since the structures of these five compounds differ from each other, it was thought that the binding profiles of each compound also differed. We, therefore, examined if the four compounds other than DRV disrupted the dimerization of the A28S-carrying PR subunit. As shown in Fig. 3B, all four compounds failed to block protease dimerization, suggesting that Ala-28 is likely involved directly or indirectly in the binding of all four compounds to the PR monomer subunit.
Replication kinetics of HIV variants with failed PR dimerization.
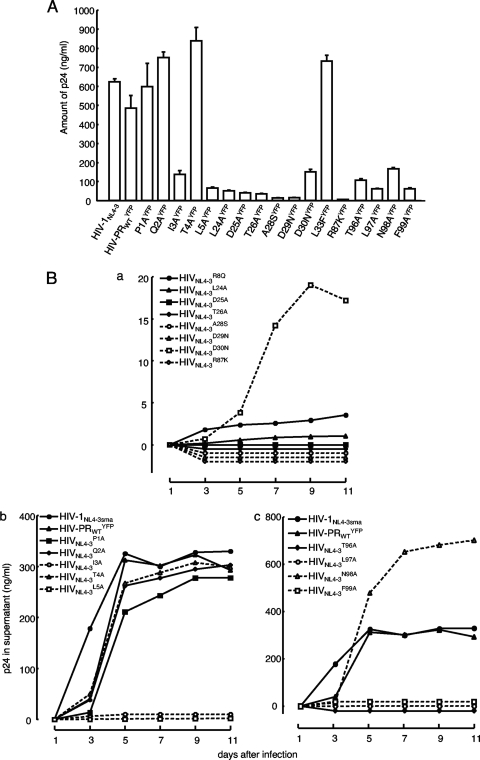
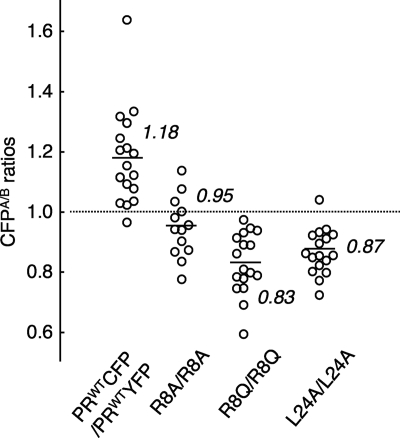
The failure of PR dimerization should completely block or significantly compromise the replication of HIV. In order to confirm that the observed dimerization failure elicited by a single amino acid substitution (26) causes replication failure of HIV, we generated a panel of HIV variants carrying YFP-tagged PR with a single amino acid substitution and examined the replicative capability of each variant. When the amount of p24 antigen produced into culture medium following transfection of COS7 cells with each plasmid was quantified, an HIV variant containing I3A, L5A, L24A, D25A, T26A, A28S, D29N, R87K, T96A, L97A, or F99A produced no or a significantly small amount of p24 (Fig. 4 A). Among these variants, we have previously shown that I3A, L5A, T26A, D29N, R87K, T96A, L97A, and F99A disrupted PR dimerization (26). Figure 5 shows that R8A (P = 0.000099), R8Q (P = 0.000084), and L24A (P = 0.0000014) also disrupted PR dimerization, as examined in the FRET-based HIV expression assay (Fig. 5). When fresh MT-4 cells were exposed to each cell-free culture supernatant of the transfected COS7 cells, as described above, no further replication was seen over the 11-day period of the culture (Fig. 4B). The HIV variant carrying A28S also failed to replicate; however, this failure was explained by an observation that protease with A28S has almost no enzymatic activity, as reported by Hong et al. (20, 21). In contrast, recombinant HIV clones containing either of P1A, Q2A, T4A, D30N, or N98A continued to replicate (Fig. 4B).
Fig. 4.
Replication kinetics of HIV-PRYFP with wild-type or mutated PR. (A) 293T cells were transfected with pHIV-PRWTYFP or mutated pHIV-PRYFP (if pHIV-PRP1AYFP was used, it is shown as P1AYFP), and the amounts of p24 Gag in the culture supernatants were determined 48 h after transfection. (B) MT-4 cells (105) were exposed to the harvested supernatant of each infectious HIV-PRYFP clone shown in panel A (100 ng of p24 Gag protein/ml) for 6 h, washed twice with phosphate-buffered saline (PBS), and further cultured in 7 ml of complete medium. Culture supernatants (50 μl) were harvested every other day, and virus replication was monitored by the amounts of p24 Gag produced in the culture supernatants. Replication kinetics of various HIV-PRYFP mutants are shown over 11 days. In subpanels a, b, and c, the replication kinetics of infectious clones carrying mutations in the active site, N ternimus, and C terminus, respectively, are shown. Note that recombinant HIV clones, whose replication rates were relatively poor, are illustrated in subpanel a. The experiments that generated data in subpanels a and b were performed on the same occasion. Thus, two controls (HIV-1NL4-3sma and HIV-PRWTYFP) in subpanel b serve as controls in subpanel a as well.
Fig. 5.
Dimerization inhibition profiles of selected HIV-1 PR mutants. COS7 cells were cotransfected with a pair of HIV-PRCFP and HIV-PRYFP strains either wild type or carrying single amino acid (AA) substitutions, such as the R8A, R8Q, or L24A, in the absence of drug. The CFPA/B ratios were determined at the conclusion of the 3-day period of culture. The differences between the CFPA/B ratios of the WT and the CFPA/B ratios of the mutant had P values of 0.000099 for R8A, 0.000084 for R8Q, and 0.0000014 for L24A.
DRV resistance profiles of PR species carrying DRV-resistance-associated amino acid substitutions.
Using the standardized protocol for selection of drug-resistant HIV variants, we along with others have experienced difficulty in selecting DRV-resistant HIV variants in vitro (8, 24). The emergence of DRV-resistant HIV variants was substantially slower than that of variants resistant to other FDA-approved PIs when a single HIV strain was employed as a starting viral strain (8, 24). In this respect, we have recently succeeded in selecting a highly DRV-resistant HIV variant by using a mixture of 8 highly multi-PI-resistant, DRV-susceptible clinical HIV strains (HIV8MIX strains HIVA, HIVB, HIVC, HIVG, HIVTM, HIVMM, HIVJSL, and HIVSS), which were originally isolated from patients with AIDS, who had failed then-existing anti-HIV regimens after receiving 9 to 11 anti-HIV drugs over the previous 32 to 83 months in the late 1990s and contained 9 to 14 amino acid substitutions in the PR-encoding region (42). By passage 39 in the selection with DRV, HIV8MIX (HIV8MIXP39) became highly resistant to DRV, with an EC50 ∼333-fold greater than that against HIVNL4-3. HIV8MIX at passage 39 (HIV8MIXP39) was capable of replicating in the presence of 1 μM DRV with a replication fitness comparable to that of HIVNL4-3 (24). HIV8MIX at passage 51 (HIV8MIXP51), which was also capable of replicating in the presence of 5 μM DRV, was found to contain the following 14 mutations: L10I, I15V, K20R, L24I, V32I, L33F, M36I, M46L, I54M, L63P, K70Q, V82I, I84V, and L89M (24). As illustrated in Table 1, when HIV8MIXP51 was propagated in the presence of 0.1 and 1.0 μM DRV in CD4+ MT-4 cells, the virus replicated comparably to HIVNL4-3 during the 9-day period of culture, while HIVNL4-3 completely failed to replicate in the presence of 0.1 or 1.0 μM DRV, as examined according to the amounts of Gag protein produced in the culture supernatant, indicating that HIV8MIXP51 had acquired a high-level resistance against DRV, while it maintained its robust replication fitness.
Table 1.
HIV DRV-resistant strain HIV8MIXP51 is capable of replicating in the presence of DRVa
| Virus | DRV (μM) | Replication (ng/ml) at day postexposure: |
||||
|---|---|---|---|---|---|---|
| 1 | 3 | 5 | 7 | 9 | ||
| HIVNL4-3 (WT) | 0 | 0 | 33 ± 9.9 | 955 ± 9.9 | 993 ± 57 | 1152 ± 127 |
| 0.1 | 0 | 0 | 0 | 0 | 0 | |
| 1.0 | 0 | 0 | 0 | 0 | 0 | |
| HIV8MIXP51 | 0 | 0 | 701 ± 45 | 734 ± 68 | 771 ± 19 | 877 ± 88 |
| 0.1 | 1.5 ± 2.1 | 590 ± 103 | 682 ± 199 | 729 ± 3 | 909 ± 178 | |
| 1.0 | 0.5 ± 0.7 | 270 ± 10 | 886 ± 117 | 936 ± 18 | 1,201 ± 170 | |
CD4+ MT-4 cells were exposed to HIVNL4-3 or HIV8MIXP51 (a highly DRV-resistant HIV variant derived from the mixture of 8 highly-PI-resistant clinical HIV isolates exposed to increasing concentrations of DRV up to 1 μM) (24), cultured in the absence or presence of 0.1 or 1.0 μM DRV. Viral replication was monitored by the amounts of p24 Gag protein (ng/ml) produced in the culture supernatant.
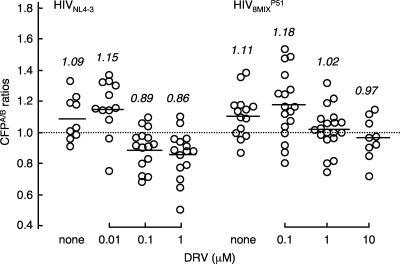
We, therefore, asked if the dimerization of the PR of HIV8MIXP51 was blocked by DRV, exploiting the FRET-based HIV expression system by using a pair of newly generated plasmids encoding a full-length molecular infectious HIV clone containing CFP- or YFP-tagged PR with all 14 amino acid substitutions. As shown in Fig. 6, DRV significantly blocked the dimerization of the wild-type PR of HIVNL4-3 at concentrations of 0.1 and 1 μM. However, DRV failed to block the dimerization of the PR of HIV8MIXP51 at 0.1 μM (P = 0.42). These data suggested that all amino acid substitutions present in the PR of HIV8MIXP51 or subsets of them were associated with the HIV8MIXP51 strain's acquisition of DRV resistance.
Fig. 6.
DRV fails to inhibit the dimerization of the protease of a highly DRV-resistant HIV8MIXP51 variant. COS7 cells were transfected with a pair of plasmids encoding a full-length molecular infectious HIV-1 clone (HIV8MIXP51) containing CFP- or YFP-tagged PR with 14 amino acid substitutions (L10I, I15V, K20R, L24I, V32I, L33F, M36I, M46L, I54M, L63P, K70Q, V82I, I84V, and L89M) in the presence or absence of 0.1, 1, or 10 μM DRV. On day 3 after transfection, CFPA/B ratios were determined as described in the legend to Fig. 2. HIVNL4-3 served as a reference. Note that 0.1 and 1 μM DRV failed to block the dimerization of the protease of HIV8MIXP51, while the same concentration of DRV blocked protease dimerization in HIVNL4-3. The differences between the CFPA/B ratios in the absence of drug (CFPA/BNo Drug) and the CFPA/B ratios in the presence of 0.01 μM DRV (CFPA/B0.01 DRV), between the CFPA/B ratios in the presence of 0.01 μM DRV (CFPA/B0.01 DRV) and 0.1 μM DRV (CFPA/B0.1 DRV), and between the CFPA/B ratios in the presence of 0.1 μM DRV (CFPA/B0.1 DRV) and 1.0 μM DRV (CFPA/B1.0 DRV) had P values of 0.32, 0.0025, and 0.34 for HIVNL4-3, respectively. The differences between the CFPA/BNo Drug and the CFPA/B0.1 DRV, between the CFPA/B0.1 DRV and CFPA/B1.0 DRV, and between the CFPA/B1.0 DRV and the CFPA/B10.0 DRV had P values of 0.42, 0.022, and 0.26, respectively, for HIV8MIXP51.
Effects of V32I, L33F, I54M/L, and/or I84V substitutions on HIV susceptibility to DRV and PR dimerization inhibition by DRV.
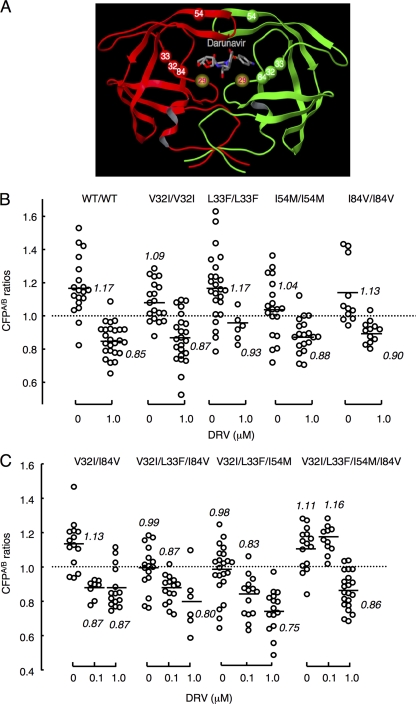
When we examined the sequence of the PR-encoding gene in HIV8MIXP51 and three clinical HIV variants isolated from individuals with AIDS who did not respond to DRV-containing antiviral regimens (Table 2) (33), four amino acid substitutions (V32I, L33F, I54M, and I84V) were found to be mostly in common and thought to be relatively unique in such DRV-resistant HIV variants. The locations of the four amino acid substitutions are illustrated in Fig. 7A. This notion was further confirmed when we examined reports by others (9, 24, 33, 38) regarding the sequence of the PR-encoding region of DRV-resistant variants, as illustrated in Table 2. We consequently examined whether the notion described above was plausible by incorporating one of the four amino acid substitutions or subsets of them.
Fig. 7.
Amino acid changes conferring DRV resistance on HIV. (A) Locations of amino acid substitutions V32I, L33A/F, I54M, and I84V associated with HIV's DRV resistance. The location of Asp29 (D29), which is known to be an essential amino acid for dimerization, is also shown. (B) Profiles of DRV's dimerization inhibition of PR carrying a single amino acid substitution. COS7 cells were cotransfected with a pair of HIV-PRCFP and HIV-PRYFP variants carrying wild-type PR or a single amino acid substitution such as V32I, L33F, I54M, or I84V, each of which was found to be associated with the development of HIV resistance to DRV, in the presence of 1 μM DRV, further cultured, and the CFPA/B ratios were determined. Note that none of the amino acid substitutions introduced blocked the dimerization of PR. The statistical evaluation of all the changes in the CFPA/B ratios determined in the presence or absence of DRV, conducted using the nonparametric Mann-Whitney U test, showed P values ranging from 0.00000034 (3.4E−07) to 0.0026. (C) Profiles of DRV's dimerization inhibition of PR carrying combined amino acid substitutions. COS7 cells were cotransfected with a pair of HIV-PRCFP and HIV-PRYFP variants carrying combined amino acid substitutions such as V32I and I84V, V32I, L33F, and I84V, V32I, L33F, and I54M, or V32I, L33F, I54M, and I84V. The COS7 cells were further cultured in the continuous presence of 0, 0.1, and 1 μM DRV, and the CFPA/B ratios were determined at the conclusion of the 3-day period of culture. The differences between the CFPA/B ratios in the absence of drug (CFPA/BNo Drug) and the CFPA/B ratios in the presence of 0.1 μM DRV (CFPA/B0.1 DRV) and between the CFPA/B ratios in the presence of 0.1 μM DRV (CFPA/B0.1 DRV) and 1.0 μM DRV (CFPA/B1.0 DRV) had P values of 0.0015 and 0.42 for V32I and I84V, 0.0047 and 0.15 for V32I, L33F, and I84V, 0.033 and 0.07 for V32I, L33F, and I54M, and 0.3 and 0.0000073 for V32I, L33F, I54M, and I84V, respectively.
When we introduced each of the four substitutions into the wild-type strain, HIVNL4-3, there was no increase observed in the EC50s of DRV against such infectious recombinant clones, with resistance ranging from 0.07- to 1.0-fold, as shown in Table 3. Introduction of the combinations of two or three amino acid substitutions did not increase the EC50s of DRV against such clones either, with resistance ranging from 0.09- to 0.9-fold. However, when we introduced all four amino acid substitutions (V32I, L33F, I54M, and I84V), into HIVNL4-3, generating HIVNL4-3V32I/L33F/I54M/I84V, the EC50 of DRV against HIVNL4-3V32I/L33F/I54M/I84V was as high as 0.64 ± 0.02 μM with resistance of 205-fold (Table 3). These data suggested that the four amino acid substitutions are associated with the high-level resistance to DRV seen in HIV8MIXP51.
Table 3.
The four amino acid substitutions V32I, L33F, I54M, and I84V confer on HIVNL4-3 variants high-level resistance to DRVa
| Wild-type strain or recombinant HIV variant | Amino acid substitution(s) in PR | Mean EC50 ± SD (μM) | Fold resistance |
|---|---|---|---|
| HIV-1NL4-3 | None (wild type) | 0.0031 ± 0.0002 | 1 |
| HIV-1NL4-3V32I | V32I | 0.00022 ± 0.00006 | 0.07 |
| HIV-1NL4-3L33F | L33F | 0.0028 ± 0.0008 | 0.9 |
| HIV-1NL4-3I54M | I54M | 0.0026 ± 0.0001 | 0.8 |
| HIV-1NL4-3I84V | I84V | 0.0035 ± 0.0001 | 1 |
| HIV-1NL4-3V32I/I54V | V32I, I54M | 0.0017 ± 0.0002 | 0.5 |
| HIV-1NL4-3V32I/I84V | V32I, I84V | 0.00028 ± 0.00008 | 0.09 |
| HIV-1NL4-3V32I/L33F/I54V | V32I, L33F, I54M | 0.0019 ± 0.0006 | 0.6 |
| HIV-1NL4-3V32I/L33F/I84V | V32I, L33F, I84V | 0.0030 ± 0.0004 | 0.9 |
| HIV-1NL4-3V32I/L33F/I54MV/82Ab | V32I, L33F, I54M, V82A | ||
| HIV-1NL4-3V32I/L33F/I54 M/V82I | V32I, L33F, I54M, V82I | 0.034 ± 0.018 | 11 |
| HIV-1NL4-3V32I/L33F/I54V/I84V | V32I, L33F, I54M, I84V | 0.64 ± 0.02 | 205 |
The data shown represent mean values derived from the results of three independent experiments conducted in triplicate. The EC50s were determined by employing MT-4 cells exposed to each infectious recombinant HIV-1 clone (50 TCID50) and cultured in the presence of various concentrations of DRV, using the inhibition of p24 Gag protein production by DRV by 50% as an endpoint.
Note that there are no EC50 or fold resistance values for HIV-1NL4-3V32I/L33F/I54MV/82A because this variant was replication incompetent.
We next examined the effects of one of the four amino acid substitutions or subsets of them on PR dimerization inhibition by DRV using the FRET-based HIV expression assay. Figure 7B shows that each single mutation of the four mutations allowed PR to undergo dimerization, and 1.0 μM DRV effectively blocked the dimerization (P values ranging from 3.4E−07 to 0.0026). We next determined the effects of various combinations of the four amino acid mutations on dimerization inhibition by DRV (Fig. 7C). Two mutations such as V32I and I84V still allowed PR to undergo dimerization, and DRV effectively blocked the dimerization at 0.1 and 1 μM. When combinations of three amino acid substitutions such as V32I, L33F, and I84V and V32I, L33F, and I54M were introduced, the mean CFPA/B ratios were close to 1.0, the threshold for indication of the occurrence of dimerization, with 0.99 and 0.98, respectively, in the absence of DRV, suggesting that possibly such groups of three substitutions somewhat compromised PR dimerization, although DRV still significantly blocked the dimerization at 0.1 and 1 μM, giving mean ratios ranging from 0.75 to 0.87. PR dimerization still occurred with all four substitutions, giving a mean CFPA/B ratio of 1.11; however, 0.1 μM DRV clearly failed the dimerization, giving a ratio of 1.16 (P = 0.3), while 1.0 μM DRV blocked dimerization (P = 0.00000073) (Fig. 7C).
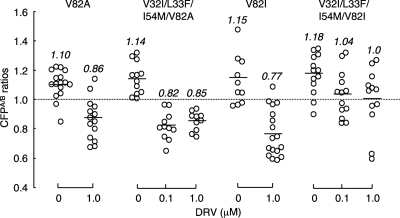
V82I, not V82A, contributes to the loss of DRV's activity to inhibit PR dimerization.
Since V82A and V82I were seen in rCLHIVT48 and HIV8MIXP51, respectively (Table 2), we examined whether V82A and V82I had effects on the loss of DRV's protease dimerization inhibition activity. Figure 8 shows that with V82A alone, PR dimerization occurred and 1 μM DRV effectively blocked the dimerization. When V82A was combined with the three amino acid substitutions V32I, L33F, and I54M, PR dimerization still occurred, which DRV effectively blocked at both 0.1 and 1 μM, suggesting that V82A substitution plays no significant role in conferring on HIV reduced sensitivity to the PR dimerization inhibition by DRV. We also examined the effects of the V82I substitution, which was identified in HIV8MIXP51, with or without the three amino acid substitutions, on inhibition of PR dimerization by DRV. Interestingly, the four combined amino acid substitutions V32I, L33F, I54M, and V82I abrogated DRV's PR dimerization inhibition activity at both 0.1 and 1.0 μM (Fig. 8), strongly suggesting that V82I with other three amino acid substitutions contributes to the loss of DRV's activity to inhibit PR dimerization.
Fig. 8.
Effects of V82A and V82I substitutions on DRV's activity to inhibit PR dimerization. COS7 cells were cotransfected with the pHIV-PRCFP and pHIV-PRYFP pair of plasmids carrying a single V82A or V82I substitution or four combined mutations (V32I, L33F, and I54M plus V82A or V82I). The COS7 cells were further cultured in the continuous presence of 0.1 and 1 μM DRV, and the CFPA/B ratios were determined at the conclusion of the 3-day period of culture. Note that combined with other three substitutions V32I, L33F, and I54M, V82A did not have a significant effect on DRV's dimerization inhibition activity, while V82I compromised the dimerization inhibition of DRV at 0.1 and 1.0 μM. The differences between the CFPA/B ratios in the absence of drug (CFPA/BNo Drug) and the CFPA/B ratios in the presence of 1.0 μM DRV (CFPA/B1.0 DRV) had P values of 0.00017 for V82A and 0.0027 for V82I. The differences between the CFPA/B ratios in the absence of drug (CFPA/BNo Drug) and the CFPA/B ratios in the presence of 0.1 μM DRV (CFPA/B0.1 DRV) and between the CFPA/B ratios in the presence of 0.1 μM DRV (CFPA/B0.1 DRV) and 1.0 μM DRV (CFPA/B1.0 DRV) were 0.000055 and 0.38 for V32I, L33F, I54M, and V82A and 0.026 and 0.91 for V32I, L33F, I54M, and V82I, respectively.
DISCUSSION
In the FRET-based HIV expression assay we previously reported, YFP- or CFP-containing PR should be initially expressed as a part of Pr160gag-pol polyprotein, and it was assumed that PDIs block the dimerization of PR monomer subunit within the polyprotein. In the present study, we examined whether one of the PDIs, DRV, blocked the dimerization of HIV PR expressed as a single protease molecule. As expected, DRV effectively blocked the dimerization (Fig. 2B), indicating that PDIs can block the dimerization of the PR monomers in the form of polyprotein as well as after autolysis (30, 31). The dimerization of two identical PR monomers is required for the acquisition of PR catalytic activity (28, 40), and the failure of PR dimerization should result in the loss of viral replication or compromised viral replication.
In our previous study (26) and in the present study, we demonstrated that a single amino acid substitution, such as I3A, L5A, R8Q, L24A, T26A, D29N, R87K, T96A, L97A, or F99A, effectively disrupts PR dimerization, as examined in the FRET-based HIV expression assay. In the present work, we constructed plasmids containing HIVNL4-3-based recombinant clones with one of those amino acid substitutions and examined if such clones replicated in MT-4 cells. To this end, we attempted to propagate such recombinant clones in MT-4 cells using the supernatants obtained through transfection of 293T cells; however, all clones failed to replicate (Fig. 4B). In contrast, similarly generated recombinant clones containing a single amino acid substitution such as P1A, Q2A, T4A, D30N, and N98A (26), none of which disrupted PR dimerization, as examined with the FRET-based HIV expression assay (26), continued to replicate in MT-4 cells (Fig. 4B). These data further confirmed the validity of the results obtained in the FRET-based HIV expression assay.
It is of note that HIV-1 protease containing A28S has virtually no enzymatic activity, as previously published by Hong et al. (20), and as expected, recombinant HIV-1NL4-3 with A28S (HIVNL4-3A28S) was totally replication incompetent, as shown in Fig. 4B. The D25A substitution is also known to render HIV-1 protease virtually enzymatically inactive; however, that substitution does not disrupt the dimerization of D25A protease as we previously published (26). Yet, 1 μM DRV clearly blocks the dimerization of D25A protease (Fig. 3A), suggesting that D25 is not significantly associated with the putative DRV binding site in monomer subunit. These observations have led us to conclude that A28 is likely involved directly or indirectly in binding of DRV to the protease monomer subunit.
In recent clinical studies, a set of mutations, including V32I, L33F, I47V, I54L, and L89V has been identified in HIV strains isolated from patients failing DRV-containing regimens and has been associated with a diminished virological response to a regimen containing DRV boosted with ritonavir (DRV/r) (9, 33). More recently, Van Marck et al. reported that a set of amino acid substitutions (V32I, I50V, I54L/M, L76V, and V82F) in the PR-encoding region of HIV might be associated with the failure of HIV-1 infected individuals receiving DRV-containing antiviral regimens (38). We have recently selected a highly DRV-resistant HIV variant (HIV8MIXP51) by using a mixture of eight highly multi-PI-resistant (but DRV-sensitive) clinical HIV strains, isolated from those who had received various PIs and failed to respond to PI (not DRV)-containing regimens, by propagating the mixture in the presence of increasing concentrations of DRV in phytohemagglutinin-activated peripheral blood mononuclear cells (PHA-PBMCs) followed by MT-4 cells. HIV8MIXP51 proved to have an EC50 of DRV ∼333-fold greater than that against a wild-type HIV strain. HIV8MIXP51 was highly resistant to amprenavir, indinavir, nelfinavir, ritonavir, lopinavir, and atazanavir (all EC50s of >1 μM) and moderately resistant to saquinavir and TPV (EC50s 33- and 18-fold greater, respectively) and replicated as rapidly as the wild-type HIVNL4-3 strain in the presence of 1 μM DRV (Table 1). The amino acid substitutions identified in HIV8MIXP51 were L10I, I15V, K20R, L24I, V32I, L33F, M36I, M46L, I54M, L63P, K70Q, V82I, I84V, and L89M (24). When we determined the amino acid sequences of three highly DRV-resistant clinical HIV strains, rCLHIVF16, rCLHIVT45, and rCLHIVT48, which had EC50s of DRV of 0.30 μM (97-fold), 0.33 μM (105-fold), and 0.33 μM (105-fold), respectively, we recognized that all three clinical isolates contained V32I, L33F, I54V, and I84V in common.
Thus, we hypothesized that PR dimerization inhibition by DRV might contribute to the antiviral activity of DRV and that the loss of PR dimerization inhibition by DRV might be associated with the decreased antiviral activity of DRV. When we introduced single, double, or triple amino acid substitutions of the four substitutions into pHIV-PRWTCFP and pHIV-PRWTYFP and cotransfected COS7 cells with such two plasmids, the dimerization of HIV-PRWTCFP and HIV-PRWTYFP clearly occurred in the absence of DRV, giving CFPA/B ratios ranging from 1.04 to 1.17 (Fig. 7B). DRV at 1 μM effectively blocked the dimerization of the PR containing a single one of the four amino acid substitutions (Fig. 7B), giving average CFPA/B ratios ranging from 0.88 to 0.93. DRV at 0.1 and 1.0 μM also blocked dimerization when two amino acid substitutions (V32I and I84V) and three substitutions (V32I, L33F, and I84V or V32I, L33F, and I54M) were introduced. However, when all four combined substitutions were introduced, DRV at 0.1 μM failed to block the dimerization (Fig. 7C), suggesting that the combination of the four substitutions compromised the activity of DRV to block the dimerization, presumably through altering the conformation of the monomer's putative binding site for DRV.
Considering that (i) the four amino acid substitutions in PR V32I, L33F, I54M, and I84V, were identified in the highly DRV-resistant HIV variant (HIV8MIXP51) and various DRV-resistant clinical HIV strains, such as rCLHIVF16, rCLHIVT45, and rCLHIVT48, isolated from individuals failing DRV-containing regimens (9, 33); that (ii) multi-PI-resistant, but DRV-sensitive HIV variants (24) do not contain the combination of the four substitutions; and (iii) that with the four substitutions combined, the activity of DRV to block PR dimerization is compromised, it is strongly suggested that the loss of PR dimerization activity is associated with the acquisition of resistance of HIV against DRV. More critically, the fact that the emergence of the four combined mutations is often seen in individuals failing DRV-containing regimens strongly suggests that the protein dimerization inhibition activity of DRV is in operation for the drug to exert its anti-HIV activity in a clinical setting. Our finding that four combined amino acid substitutions are required for the loss of PR dimerization inhibition activity of DRV should explain at least in part the reason why DRV has a high level of genetic barrier to HIV acquisition of DRV resistance. It is noteworthy that the crystallographic data of HIV PR monomer complexed with DRV are as yet unavailable; however, if the results of such structural analysis are obtained, it should help our understanding of the dynamics of the dimerization process of HIV PR, the mechanisms of the activity of DRV and other PDIs to block the dimerization, and the mechanisms of the emergence of HIV resistance against PDIs.
ACKNOWLEDGMENTS
This work was supported in part by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health, in part by a grant for global education and research centers aimed at the control of AIDS (Global Center of Excellence supported by Monbu-Kagakusho), Promotion of AIDS Research from the Ministry of Health, Welfare, and Labor of Japan, by a grant to the Cooperative Research Project on Clinical and Epidemiological Studies of Emerging and Reemerging Infectious Diseases (Renkei Jigyo no. 78, Kumamoto University) of Monbu-Kagakusho, and by a grant from the National Institutes of Health (GM53386 to A.K.G.).
Footnotes
Published ahead of print on 3 August 2011.
REFERENCES
- 1. Amano M., et al. 2007. A novel bis-tetrahydrofuranylurethane-containing nonpeptidic protease inhibitor (PI) GRL-98065 potent against multi-PI-resistant HIV in vitro. Antimicrob. Agents Chemother. 51:2143–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Antiretroviral Therapy Cohort Collaboration 2008. Life expectancy of individuals on combination antiretroviral therapy in high-income countries: a collaborative analysis of 14 cohort studies. Lancet 372:293–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aoki M., et al. 2009. Non-cleavage site Gag mutations in amprenavir-resistant human immunodeficiency virus type 1 (HIV-1) predispose HIV-1 to rapid acquisition of amprenavir resistance but delay development of resistance to other protease inhibitors. J. Virol. 83:3059–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Babe L. M., Rose J., Craik C. S. 1992. Synthetic “interface” peptides alter dimeric assembly of the HIV 1 and 2 proteases. Protein Sci. 1:1244–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bastiaens P. I., Jovin T. M. 1996. Microspectroscopic imaging tracks the intracellular processing of a signal transduction protein: fluorescent-labeled protein kinase C beta I. Proc. Natl. Acad. Sci. U. S. A. 93:8407–8412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bastiaens P. I., Majoul I. V., Verveer P. J., Soling H. D., Jovin T. M. 1996. Imaging the intracellular trafficking and state of the AB5 quaternary structure of cholera toxin. EMBO J. 15:4246–4253 [PMC free article] [PubMed] [Google Scholar]
- 7. Bhaskaran K., et al. 2008. Changes in the risk of death after HIV seroconversion compared with mortality in the general population. JAMA 300:51–59 [DOI] [PubMed] [Google Scholar]
- 8. De Meyer S., et al. 2005. TMC114, a novel human immunodeficiency virus type 1 protease inhibitor active against protease inhibitor-resistant viruses, including a broad range of clinical isolates. Antimicrob. Agents Chemother. 49:2314–2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de Meyer S., et al. 2008. Resistance profile of darunavir: combined 24-week results from the POWER trials. AIDS Res. Hum. Retroviruses 24:379–388 [DOI] [PubMed] [Google Scholar]
- 10. Dierynck I., et al. 2007. Binding kinetics of darunavir to human immunodeficiency virus type 1 protease explain the potent antiviral activity and high genetic barrier. J. Virol. 81:13845–13851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fang G., Weiser B., Visosky A., Moran T., Burger H. 1999. PCR-mediated recombination: a general method applied to construct chimeric infectious molecular clones of plasma-derived HIV-1 RNA. Nat. Med. 5:239–242 [DOI] [PubMed] [Google Scholar]
- 12. Ferrer E., et al. 2003. Genotype and phenotype at baseline and at failure in human immunodeficiency virus-infected antiretroviral-naive patients in a randomized trial comparing zidovudine and lamivudine plus nelfinavir or nevirapine. J. Infect. Dis. 187:687–690 [DOI] [PubMed] [Google Scholar]
- 13. Gatanaga H., et al. 2002. Amino acid substitutions in Gag protein at non-cleavage sites are indispensable for the development of a high multitude of HIV-1 resistance against protease inhibitors. J. Biol. Chem. 277:5952–5961 [DOI] [PubMed] [Google Scholar]
- 14. Ghosh A. K., et al. 1998. Potent HIV protease inhibitors incorporating high-affinity P2-ligands and (R)-(hydroxyethylamino)sulfonamide isostere. Bioorg. Med. Chem. Lett. 8:687–690 [DOI] [PubMed] [Google Scholar]
- 15. Ghosh A. K., et al. 1998. Structure based design: novel spirocyclic ethers as nonpeptidal P2-ligands for HIV protease inhibitors. Bioorg. Med. Chem. Lett. 8:979–982 [DOI] [PubMed] [Google Scholar]
- 16. Ghosh A. K., Leshchenko S., Noetzel M. 2004. Stereoselective photochemical 1,3-dioxolane addition to 5-alkoxymethyl-2(5H)-furanone: synthesis of bis-tetrahydrofuranyl ligand for HIV protease inhibitor UIC-94017 (TMC-114). J. Org Chem. 69:7822–7829 [DOI] [PubMed] [Google Scholar]
- 17. Ghosh A. K., Pretzer E., Cho H., Hussain K. A., Duzgunes N. 2002. Antiviral activity of UIC-PI, a novel inhibitor of the human immunodeficiency virus type 1 protease. Antiviral Res. 54:29–36 [DOI] [PubMed] [Google Scholar]
- 18. Gupta R., Hill A., Sawyer A. W., Pillay D. 2008. Emergence of drug resistance in HIV type 1-infected patients after receipt of first-line highly active antiretroviral therapy: a systematic review of clinical trials. Clin. Infect. Dis. 47:712–722 [DOI] [PubMed] [Google Scholar]
- 19. Hoess R. H., Abremski K. 1984. Interaction of the bacteriophage P1 recombinase Cre with the recombining site loxP. Proc. Natl. Acad. Sci. U. S. A. 81:1026–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hong L., Hartsuck J. A., Foundling S., Ermolieff J., Tang J. 1998. Active-site mobility in human immunodeficiency virus, type 1, protease as demonstrated by crystal structure of A28S mutant. Protein Sci. 7:300–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hong L., Zhang X. C., Hartsuck J. A., Tang J. 2000. Crystal structure of an in vivo HIV-1 protease mutant in complex with saquinavir: insights into the mechanisms of drug resistance. Protein Sci. 9:1898–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ishima R., et al. 2001. Folded monomer of HIV-1 protease. J. Biol. Chem. 276:49110–49116 [DOI] [PubMed] [Google Scholar]
- 23. Ishima R., Torchia D. A., Lynch S. M., Gronenborn A. M., Louis J. M. 2003. Solution structure of the mature HIV-1 protease monomer: insight into the tertiary fold and stability of a precursor. J. Biol. Chem. 278:43311–43319 [DOI] [PubMed] [Google Scholar]
- 24. Koh Y., et al. 2010. In vitro selection of highly darunavir-resistant and replication-competent HIV-1 variants by using a mixture of clinical HIV-1 isolates resistant to multiple conventional protease inhibitors. J. Virol. 84:11961–11969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koh Y., et al. 2009. GRL-02031, a novel nonpeptidic protease inhibitor (PI) containing a stereochemically defined fused cyclopentanyltetrahydrofuran potent against multi-PI-resistant human immunodeficiency virus type 1 in vitro. Antimicrob. Agents Chemother. 53:997–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Koh Y., et al. 2007. Potent inhibition of HIV-1 replication by novel non-peptidyl small molecule inhibitors of protease dimerization. J. Biol. Chem. 282:28709–28720 [DOI] [PubMed] [Google Scholar]
- 27. Koh Y., et al. 2003. Novel bis-tetrahydrofuranylurethane-containing nonpeptidic protease inhibitor (PI) UIC-94017 (TMC114) with potent activity against multi-PI-resistant human immunodeficiency virus in vitro. Antimicrob. Agents Chemother. 47:3123–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kohl N. E., et al. 1988. Active human immunodeficiency virus protease is required for viral infectivity. Proc. Natl. Acad. Sci. U. S. A. 85:4686–4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Little S. J., et al. 2002. Antiretroviral-drug resistance among patients recently infected with HIV. N. Engl. J. Med. 347:385–394 [DOI] [PubMed] [Google Scholar]
- 30. Louis J. M., Ishima R., Torchia D. A., Weber I. T. 2007. HIV-1 protease: structure, dynamics, and inhibition. Adv. Pharmacol. 55:261–298 [DOI] [PubMed] [Google Scholar]
- 31. Louis J. M., Weber I. T., Tozser J., Clore G. M., Gronenborn A. M. 2000. HIV-1 protease: maturation, enzyme specificity, and drug resistance. Adv. Pharmacol. 49:111–146 [DOI] [PubMed] [Google Scholar]
- 32. Mitsuya H., Erickson J. 1999. Discovery and development of antiretroviral therapeutics for HIV infection, p. 751–780 In Merigan T. C., Bartlet J. G., Bolognesi D. (ed.), Textbook of AIDS Medicine. Williams & Wilkins, Baltimore, MD [Google Scholar]
- 33. Mitsuya Y., Liu T. F., Rhee S. Y., Fessel W. J., Shafer R. W. 2007. Prevalence of darunavir resistance-associated mutations: patterns of occurrence and association with past treatment. J. Infect. Dis. 196:1177–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Murphy E. L., et al. 2001. Highly active antiretroviral therapy decreases mortality and morbidity in patients with advanced HIV disease. Ann. Intern. Med. 135:17–26 [DOI] [PubMed] [Google Scholar]
- 35. Sekar R. B., Periasamy A. 2003. Fluorescence resonance energy transfer (FRET) microscopy imaging of live cell protein localizations. J. Cell Biol. 160:629–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Szczesna-Skorupa E., Mallah B., Kemper B. 2003. Fluorescence resonance energy transfer analysis of cytochromes P450 2C2 and 2E1 molecular interactions in living cells. J. Biol. Chem. 278:31269–31276 [DOI] [PubMed] [Google Scholar]
- 37. Tojo Y., et al. 2010. Novel protease inhibitors (PIs) containing macrocyclic components and 3(R),3a(S),6a(R)-bis-tetrahydrofuranylurethane that are potent against multi-PI-resistant HIV-1 variants in vitro. Antimicrob. Agents Chemother. 54:3460–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Van Marck H., et al. 2009. The impact of individual human immunodeficiency virus type 1 protease mutations on drug susceptibility is highly influenced by complex interactions with the background protease sequence. J. Virol. 83:9512–9520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Walensky R. P., et al. 2006. The survival benefits of AIDS treatment in the United States. J. Infect. Dis. 194:11–19 [DOI] [PubMed] [Google Scholar]
- 40. Wlodawer A., et al. 1989. Conserved folding in retroviral proteases: crystal structure of a synthetic HIV-1 protease. Science 245:616–621 [DOI] [PubMed] [Google Scholar]
- 41. Yoshimura K., et al. 2002. A potent human immunodeficiency virus type 1 protease inhibitor, UIC-94003 (TMC-126), and selection of a novel (A28S) mutation in the protease active site. J. Virol. 76:1349–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yoshimura K., et al. 1999. JE-2147: a dipeptide protease inhibitor (PI) that potently inhibits multi-PI-resistant HIV-1. Proc. Natl. Acad. Sci. U. S. A. 96:8675–8680 [DOI] [PMC free article] [PubMed] [Google Scholar]