Abstract
Infection by hepatitis B virus (HBV) genotype C is associated with a prolonged viremic phase, delayed hepatitis B e antigen (HBeAg) seroconversion, and an increased incidence of liver cirrhosis and hepatocellular carcinoma compared with genotype B infection. Genotype C is also associated with the more frequent emergence of core promoter mutations, which increase genome replication and are independently associated with poor clinical outcomes. We amplified full-length HBV genomes from serum samples from Chinese and U. S. patients with chronic HBV infection and transfected circularized genome pools or dimeric constructs of individual clones into Huh7 cells. The two genotypes could be differentiated by Western blot analysis due to the reactivities of M and L proteins toward a monoclonal pre-S2 antibody and slightly different S-protein mobilities. Great variability in replication capacity was observed for both genotypes. The A1762T/G1764A core promoter mutations were prevalent in genotype C isolates and correlated with increased replication capacity, while the A1752G/T mutation frequently found in genotype B isolates correlated with a low replication capacity. Importantly, most genotype C isolates with wild-type core promoter sequence replicated less efficiently than the corresponding genotype B isolates due to less efficient transcription of the 3.5-kb RNA. However, genotype C isolates often displayed more efficient virion secretion. We propose that the low intracellular levels of viral DNA and core protein of wild-type genotype C delay immune clearance and trigger the subsequent emergence of A1762T/G1764A core promoter mutations to upregulate replication; efficient virion secretion compensates for the low replication capacity to ensure the establishment of persistent infection by genotype C.
INTRODUCTION
The hepatitis B virus (HBV) is a hepatotropic enveloped DNA virus. The virion-associated relaxed circular DNA is converted to covalently closed circular DNA (cccDNA) in the nucleus of infected hepatocytes to serve as a template for viral gene expression and genome replication. The four genes are arranged on this 3.2-kb circular genome in the order of core, polymerase (P), surface, and X, with the P gene overlapping with the other three. Besides the core, P, small envelope (S), and hepatitis B X (HBx) proteins, the extra in-frame AUG codons upstream of the surface and core genes permit the expression of large (L) and middle (M) envelope proteins as well as the precore/core protein. The precore/core protein is processed by proteolytic cleavage and secreted as hepatitis B e antigen (HBeAg). The seven viral proteins are translated from five mRNAs generated from the cccDNA template: the 3.5-kb precore RNA for HBeAg, the slightly shorter 3.5-kb pregenomic (pg) RNA for core and P, and subgenomic RNAs of 2.4 kb for L protein, 2.1 kb for M/S proteins, and 0.7 kb for HBx. Transcription of the five RNAs is controlled by four promoters, with precore and pg RNAs being alternative products of the core promoter. Genome replication initiates with assembly of the cytoplasmic core protein into core particles. The P protein thus packaged converts the copackaged pg RNA into double-stranded DNA via a series of enzymatic reactions (11, 41). The core particles with double-stranded DNA genomes interact with envelope proteins anchored on the endoplasmic reticulum, leading to vesicle formation and virion release. Extra envelope proteins are secreted as noninfectious subviral particles, which constitute the bulk of hepatitis B surface antigen (HBsAg).
In Asia, HBV infection is transmitted primarily from HBeAg-positive mothers to their offspring at about the time of birth. This vertical mode of transmission, as opposed to horizontal transmission during adulthood, often induces chronic infection, leading to several decades of active virus replication. HBV is noncytopathic, and the first 1 to 2 decades of chronic infection are characterized by limited liver damage, despite high levels of virus replication. Such a state of immune tolerance is probably maintained by HBeAg, a secreted variant of the core protein (23). At the immune active phase that ensues, a cytotoxic T lymphocyte (CTL) response against viral proteins destroys infected hepatocytes, leading to hepatitis. However, the CTL response is often suboptimal and the immune clearance phase can drag on for decades. A critical turning point is HBeAg seroconversion (the disappearance of HBeAg from the blood, followed by detection of anti-HBe antibody), which is usually associated with a marked drop in viremia titer and normalization of alanine aminotransferase (ALT). To escape immune attack, HBV may switch off HBeAg expression by selecting for mutations in the precore region, especially the G1896A nonsense mutation (3, 43). Alternatively, HBeAg expression is reduced at the transcriptional level by point mutations in the core promoter, with the A1762T/G1764A double mutation being the most common (26). The double mutation also increases transcription of the pg RNA, thus augmenting HBV genome replication (2, 20, 31).
HBV isolates worldwide can be classified into eight genotypes on the basis of nucleotide sequence divergence of >8% (18, 25, 27, 35). Genotypes B and C are dominant in Asia. Numerous epidemiological studies suggest that genotype C is more pathogenic than genotype B due to a prolonged HBeAg-positive phase characterized by a high viral load and a higher prevalence of A1762T/G1764A core promoter mutations (6, 14). To understand the biological basis for their different behaviors, we amplified full-length HBV genomes from serum samples from a large number of patients with chronic HBV infection from China and the United States. The cloned genomes, with or without core promoter mutations, were transfected into Huh7 human hepatoma cells so as to examine their efficiencies of viral RNA transcription, protein expression, genome replication, and virion secretion.
MATERIALS AND METHODS
Serum samples and genotype determination.
Serum samples were collected at the Division of Gastroenterology, University of Michigan Medical Center (U.S. samples), or Huashan Hospital, Fudan University (Chinese samples), with informed consent from the patients. The protocols for sample collection were approved by the ethics committees at the respective hospitals. The subjects had no evidence of autoimmune hepatitis or markers of infection with hepatitis C virus, hepatitis D virus, or HIV and had not received antiviral therapies. They had been HBsAg positive for at least 6 months and were HBeAg positive with an HBV DNA titer of >10 4 copies/ml. Most Chinese subjects were incidentally identified as asymptomatic carriers with normal ALT levels. DNA was extracted from 200 μl of serum samples by a QIAamp DNA blood minikit (Qiagen), and HBV DNA was amplified by PCR. For the U.S. isolates, viral genotype as well as core promoter mutations were determined by line probe assay as previously described (5), and only isolates lacking the A1762T/G1764A core promoter mutations were used for amplification of the full-length genome. For the Chinese isolates, viral genotype was determined by sequencing of the envelope gene (positions 2815 to 3215 and 1 to 886), followed by phylogenetic analysis (33).
Generation of SphI dimers and circularized HBV genomes.
The full-length HBV genome was amplified from serum DNA by PCR (30 cycles for Chinese samples and 40 cycles for U.S. samples) using primers located in the precore region and High Fidelityplus polymerase (Roche) and inserted into the HindIII and SacI sites of the pUC18 vector as described previously (31). For the U.S. samples, the PCR clones were digested with SapI, and the 3.2-kb HBV genome was gel purified and ligated at a high insert/vector ratio with SapI-cut, dephosphorylated pUC18 DNA (the original SapI site was destroyed, while a new SapI site was introduced into the polylinker) to generate tandem dimers with junctions in the precore region (42). The dimeric DNA was then digested with SphI, and the 3.2-kb HBV genome thus purified was ligated with the SphI-cut, dephosphorylated pUC18 vector to generate tandem dimers ending at the SphI site (SphI dimers). For the Chinese samples, circularized genomes rather than SphI dimers were employed for transfection experiments. Briefly, pUC18 DNA with an altered SapI site was subjected to two rounds of digestion with HindIII and SacI, followed by digestion with SphI to minimize vector self-ligation (32). The vector DNA was ligated with HindIII/SacI-digested PCR products and transformed to MAX Efficiency DH5α competent cells (Invitrogen), followed by growth of the entire transformation product in 100 ml of LB broth containing ampicillin. Plasmid DNA, which represented clone pools, was extracted using a HiSpeed plasmid midikit (Qiagen). The midiprep DNA of the clone pools was used for sequencing of the core promoter and core gene. Prior to transfection, the HBV genome was released from the vector by digestion at 50°C with BspQI, an isoschizomer of SapI. The 3.2-kb HBV DNA was gel purified and treated with T4 DNA ligase at a concentration of 1 μg/ml to promote intramolecular ligation (genome circularization). The ligation product was extracted sequentially with phenol and chloroform-isoamyl alcohol and precipitated with ethanol in the presence of glycogen. The DNA concentration was determined by spectrometry and confirmed by running an aliquot in an agarose gel.
Transfection and analysis of genome replication and virion secretion.
Detailed procedures for transfection of Huh7 cells and subsequent detection of genome replication, protein expression, protein secretion, and virion secretion have been described previously (8, 9, 32, 44). Huh7 cells seeded at 6 × 105/wells in six-well plates were transfected with 1.5 μg of circularized HBV DNA or 1 μg of SphI dimer DNA using TransIT-LT1 reagent (Mirus). Cells and culture supernatant were harvested at day 5 posttransfection. Core particles were precipitated from 1/4th of the cell lysate by polyethylene glycol, followed by nuclease digestion, proteinase K digestion, and DNA extraction (31). Virions were immunoprecipitated from 1.5 ml of culture supernatant by a 1:1,000 dilution of horse polyclonal anti-HBs antibody (anti-ad/ay; Novus). Core particle- and virion-associated DNA was subjected to Southern blot analysis. The nearly full-length probe DNA was obtained from a cloned genome of genotype B or C by nested PCR amplification to eliminate the vector sequence and labeled with [32P]dCTP by random priming. A mixed probe of genotypes B and C at a 1:1 ratio was used when comparison was made between genotype B and genotype C isolates or clones. Unless stated otherwise, the blots were washed at 62°C with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS solution.
To further compare virion secretion efficiencies between isolates of genotypes B and C, we separated virions from naked core particles according to their different densities (31). Both types of particles were pelleted from culture supernatant by ultracentrifugation at 39,000 rpm for 18 h in a Sorvall rotor. The pellet was resuspended in a 0.33-g/ml CsCl2 solution in TEN buffer (10 mM Tris, 1 mM EDTA, 100 mM NaCl) and further centrifuged at 46,000 rpm for 72 h. Fractions of 450 μl were collected from the top, weighed, and dialyzed against TEN buffer solution. Samples were treated with DNase I and mung bean nuclease, followed by proteinase K digestion and DNA precipitation. DNA was separated in agarose gels for Southern blot analysis.
Analysis of protein expression and secretion.
Intracellular envelope proteins were detected by direct Western blot analysis (8, 9, 32). Briefly, 50 μg protein from cell lysate was separated overnight in an SDS-12% polyacrylamide gel and transferred to polyvinylidene difluoride membranes. The primary antibodies used for envelope protein detection were horse polyclonal anti-HBs (anti-ad/ay; Novus) at a 1:5,000 dilution, mouse monoclonal anti-pre-S2 (031-A; Virogen) at a 1:1,000 dilution, and rabbit anti-pre-S1 (R271; a kind gift from Camille Sureau, INTS, Paris, France) at a 1:2,000 dilution (8). The corresponding horseradish peroxidase (HRP)-conjugated secondary antibodies were rabbit antihorse at 1:100,000 (Abcam), antimouse at 1:40,000, and antirabbit at 1:100,000. For the loading control, the blots were incubated with a 1:10,000 dilution of mouse antibody against glyceraldehyde-3-phosphate dehydrogenase (GAPDH), followed by antimouse secondary antibody. Intracellular core protein was immunoprecipitated from 1/4th of the cell lysate with 1.3 μl of a polyclonal rabbit anti-core antibody (Dako) in a volume of 400 μl and revealed by Western blotting using the same antibody at a 1:2,000 dilution. Secreted HBsAg was detected by enzyme-linked immunosorbent assay (ELISA) kits from Abazyme using 5 to 10 μl of culture supernatant. Secreted HBeAg was measured by an ELISA kit (DiaSorin) using diluted samples corresponding to 0.7 μl of culture supernatant.
Northern blot analysis for HBV RNAs.
Huh7 cells were lysed at day 2 posttransfection using TRIzol (Invitrogen), and RNA was extracted according to the manufacturer's instructions. RNA (15 μg) was denatured at 58°C for 10 min and separated in a 1% agarose gel with morpholinepropanesulfonic acid and formaldehyde. After transfer, the blots were hybridized with a 32P-labeled, mixed HBV DNA probe of genotypes B and C as described for Southern blotting. The HBV probe was removed by boiling the blots in 2% SDS-0.1× SSC solution for 30 min, and the blots were further hybridized with 32P-labeled GAPDH probe. The probe DNA of 569 bp was generated by PCR amplification of RNA from Huh7 cells using sense primer 5′-CAGCAATGCCTCCTGCACCACC-3′ (nucleotides [nt] 543 to 564) and antisense primer 5′-CTTACTCCTTGGAGGCCATGTGG-3′ (nt 1111 to 1089).
RESULTS
Rationale.
Our goal was to compare the intrinsic protein expression, genome replication, and virion secretion capacities between genotype B (B2) and genotype C, which requires constructs for which transcription of the viral RNAs is driven by endogenous promoters and enhancers. Being mindful of possible variability within the same genotype, we analyzed a large number of genotype B and genotype C isolates from two countries. The Chinese carriers were relatively young. The U.S. samples were prescreened to eliminate those harboring the A1762T/G1764A mutations. Since the full-length HBV genome cloned to a vector is unable to generate the terminally redundant pg RNA, which is essential for genome replication, the U.S. clones were converted to tandem dimers via the unique SphI site. Considering possible quasispecies within the same sample and PCR errors (32), we converted two or more PCR clones into SphI dimers, which requires an intermediate step (see Materials and Methods). This procedure turned out to be extremely time-consuming. A simpler approach was taken during the subsequent study of Chinese samples: the full-length HBV genome was excised from the vector and circularized in vitro by treatment with T4 DNA ligase. Moreover, by expanding the entire transformation product (the clone pool) in liquid culture, we avoided the need to test several clones per serum sample.
High prevalence of A1752G mutation among Chinese genotype B isolates and its correlation with low replication capacity.
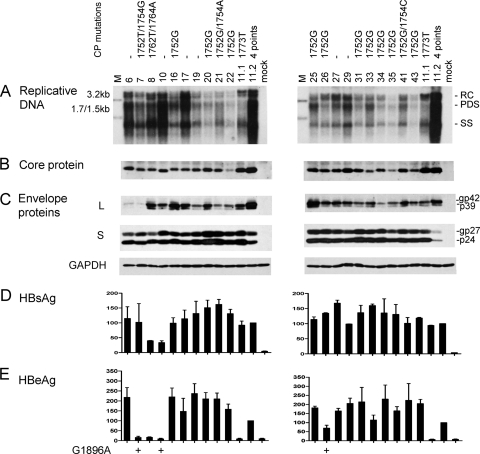
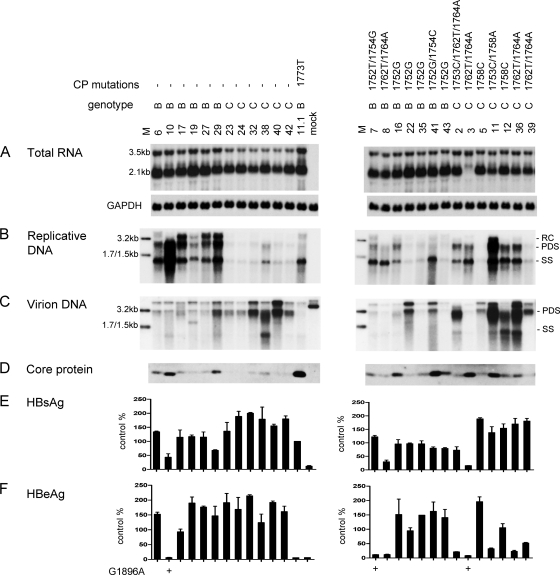
Full-length HBV clone pools were generated from 37 Chinese patients, 20 with genotype B infection and 17 with genotype C infection. Genotype B patients were significantly younger (25.2 ± 5.37 versus 34 ± 10.41 years) and were less likely to have elevated ALT levels (5/20 versus 7/17 patients). Figure 1 shows the functional characterization of the genotype B isolates, with U.S. genotype B clones 11.1 (low level of replication) and 11.2 (high level of replication) serving as positive controls. Six genotype B isolates showed wild-type core promoter sequence, and one had the classic A1762T/G1764A double mutation. The remaining 13 harbored the A1752G or A1752T mutation, and 3 of these also contained a point mutation at position 1754. Replicative DNA and virion DNA were detected by a genotype B probe, followed by washing under mild conditions to minimize detection bias. Replication capacity varied greatly among the 20 isolates (Fig. 1A), but the isolates replicating at the highest level (isolates 6, 10, and 17) contained wild-type core promoter sequence. In contrast, most isolates replicating at a low level (isolates 20, 22, 31, 34, 35, and 43) harbored the A1752G mutation. This is consistent with a previous report suggesting that the A1752G/T/C mutations reduce HBV DNA replication (24). Variability in intracellular core protein, L protein, as well as secreted HBsAg and HBeAg was also observed (Fig. 1B to E). The inability of isolates 7 and 10 to secrete HBeAg could be explained by the G1896A mutation in the precore region (Fig. 1E, left panel). Isolate 26 also harbored this mutation, although it continued to express a low level of HBeAg possibly due to the presence of the wild-type population as well.
Fig. 1.
Comparison of genome replication and protein expression/secretion among Chinese genotype B isolates. Huh7 cells were transfected with circularized HBV genome and harvested 5 days later. Clones 11.1 and 11.2 of genotype B were SphI dimers and were also transfected to serve as positive controls. The core promoter (CP) mutations of these isolates are shown. Clone 11.2 has four point mutations: G1764A/C1766T/T1767G/C1773T. (A) Southern blot analysis of intracellular replicative DNA using a genotype B probe. The blots were washed at 62°C with 2× SSC-0.1% SDS solution. Lanes M, 50 pg of 3.2-kb, 1.7-kb, and 1.5-kb HBV DNA as size markers. RC, relaxed circular; PDS, partially double stranded; SS, single stranded. (B) Immunoprecipitation-Western blot analysis of intracellular core protein. (C) Western blot analysis of intracellular L and S proteins, as well as GAPDH as a loading control. (D and E) ELISA for secreted HBsAg and HBeAg (averaged from 4 transfection experiments). The value from clone 11.2 was set at 100%. The presence of the G1896A mutation is indicated.
High prevalence of A1762T/G1764A mutations among Chinese genotype C isolates and their association with high replication capacity.
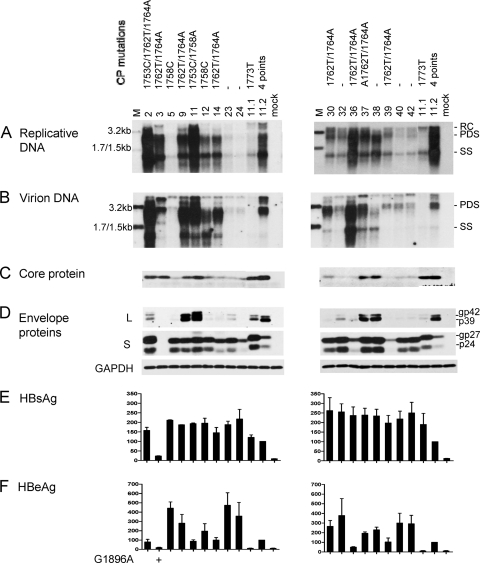
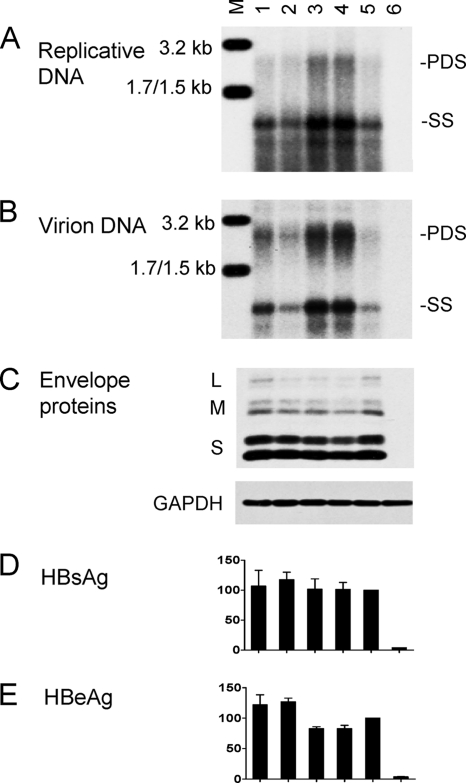
Six of the 17 genotype C isolates harbored wild-type core promoter sequence, and most displayed a low replication capacity, as revealed by a genotype C probe (Fig. 2A). No mutation was detected at position 1752 or 1754. Instead, the A1762T/G1764A mutations (together with T1753C for isolate 2) were detected in eight isolates, many of which (isolates 2, 3, 9, 36, and 37) showed a high replication capacity. Isolate 11 also showed a high replication capacity but harbored the T1753C/T1758A double mutation instead (Fig. 2A). The relevant core promoter mutations were introduced to clone 17.3 of genotype C to verify their contribution to the high-replication phenotype. We found that levels of replicative DNA and virion DNA were significantly improved by the A1762T/G1764A mutation but not further increased with the extra T1753C mutation (Fig. 3A and B; compare lanes 3 to 5). In contrast, the T1753C/T1758A double mutation did not increase genome replication (lane 2), suggesting that the high replication capacity of isolate 11 is caused by a sequence element(s) outside the core promoter.
Fig. 2.
Comparison of genome replication, virion secretion, and protein expression/secretion among Chinese genotype C isolates. Huh7 cells were transfected with circularized HBV genome and harvested 5 days later. Clones 11.1 and 11.2 of genotype B were also transfected to serve as positive controls. (A and B) Southern blot analysis of intracellular replicative DNA and virion-associated DNA using a genotype C probe. The blots were washed at 62°C with 2× SSC-0.1% SDS solution. Virions were immunoprecipitated with an anti-HBs antibody. Lanes M, HBV DNA markers of 3.2, 1.7, and 1.5 kb at 20 pg (A) and 10 pg (B). (C) Immunoprecipitation-Western blot analysis of intracellular core protein. (D) Western blot analysis of intracellular L and S proteins and GAPDH as a loading control. (E and F) ELISA for secreted HBsAg and HBeAg (averaged from 3 transfection experiments). Core promoter mutations and the G1896A mutation are indicated above panel A and below panel F, respectively.
Fig. 3.
Biological impacts of core promoter mutations introduced to a genotype C clone. Clone 17.3 and its site-directed mutants as SphI dimers were transfected to Huh7 cells. Cells and culture supernatant were harvested 5 days later. (A) Intracellular replicative DNA; (B) extracellular virion DNA; (C) intracellular envelope proteins and GAPDH; (D and E) secreted HBsAg and HBeAg based on three transfection experiments. The value for the parental construct was set at 100%. Lanes: M, HBV size markers; 1, T1758C; 2, T1753C/T1758A; 3, A1762T/G1764A; 4, T1753C/A1762T/G1764A; 5, parental construct; 6, mock-transfected cells.
The level of virion secretion among the Chinese genotype C isolates mostly mirrored that of genome replication (Fig. 2B). Variability in the expression of the L, S, and core proteins and secretion of HBsAg and HBeAg was observed (Fig. 2C to F). Overall, isolates with core promoter mutations displayed diminished HBeAg expression (Fig. 2F), and the A1762T/G1764A double mutation reduced HBeAg by about 20% when it was introduced into clone 17.3 (Fig. 3E). Isolate 3 was incapable of HBeAg expression due to the G1896A mutation (Fig. 2F).
Most genotype B isolates/clones with wild-type core promoter sequence displayed a higher replication capacity than corresponding genotype C isolates/clones.
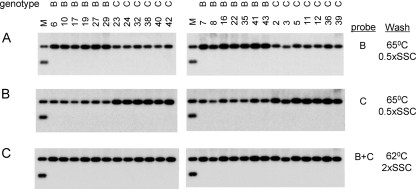
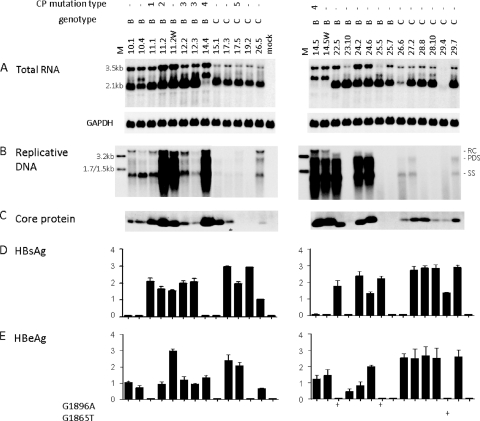
We found that a genotype B probe combined with a stringent washing condition (0.5× SSC-0.1% SDS at 65°C) diminished the detection sensitivity for genotype C isolates, whereas a genotype C probe followed by stringent washing favored detection of genotype C (Fig. 4 A and B). With the two probes mixed at a 1:1 ratio and a mild washing condition (2× SSC-0.1% SDS at 62°C), isolates of both genotypes were detected at comparable efficiencies (Fig. 4C). Using this hybridization condition, we found that the six Chinese genotype B isolates lacking core promoter mutations replicated at higher levels than the six genotype C isolates (Fig. 5B, left panel). Furthermore, of the nine U.S. genotype B clones and 10 genotype C clones lacking core promoter mutations, the genotype C clones uniformly showed a low replication capacity, whereas many genotype B clones had very efficient genome replication (especially clones 22.5, 24.2, and 24.6) (Fig. 6 B). For both Chinese and U.S. constructs, the higher replication capacity of genotype B correlated with more efficient transcription of the 3.5-kb RNA (Fig. 5A, left panel; Fig. 6A) and a higher level of core protein expression (Fig. 5D, left panel; Fig. 6C). Although clone 11.2 of genotype B with a high level of replication harbored G1764A/C1766T/T1767G/C1773T mutations, reverting these nucleotides into wild-type sequence did not markedly reduce 3.5-kb RNA or replicative DNA (clone 11.2W; Fig. 6A and B). Primer extension assay of clone 11.2W revealed no reduction in pg RNA but increased precore RNA (data not shown), which could explain the marked increase in HBeAg expression (Fig. 6E). Another clone of genotype B with a high level of replication, clone 14.5, harbored the T1754C/T1758G double mutation. Again, the corresponding revertant (clone 14.5W) did not show a marked reduction in the 3.5-kb RNA, replicative DNA, or core protein (Fig. 6A to C).
Fig. 4.
Effects of probe and washing conditions on detection of Chinese genotype B and C isolates. The 3.2-kb HBV genome was released from the pUC18 vector, and 500 pg was subjected to Southern blot analysis with genotype B probe, genotype C probe, or genotype B and C probes at a 1:1 ratio. The washing conditions are indicated. The washing buffer contained 0.1% SDS, in addition to 0.5× or 2× SSC.
Fig. 5.
Comparison of RNA transcription, genome replication, virion secretion, and viral protein expression/secretion between Chinese genotype B and genotype C isolates. Huh7 cells were transfected with circularized HBV genomes and harvested 2 days later (for RNA analysis) or 5 days later (for other analyses). (Left panels) Isolates lacking core promoter mutations, with genotype B clone 11.1 serving as a control; (right panels) isolates with the core promoter mutations indicated above. (A) Intracellular HBV RNA, with GAPDH serving as loading control. (B) Intracellular replicative DNA. (C) Virion DNA. Lanes M, 3.2-kb, 1.7-kb, and 1.5-kb HBV DNA markers at 100 pg (B) and 10 pg (C). For panels A to C, genotype B and C probes were used and washing was at 62°C with 2× SSC-0.1% SDS. (D) Immunoprecipitation-Western blot analysis of intracellular core protein. (E and F) ELISA for secreted HBsAg and HBeAg based on 3 transfection experiments, with values from clone 11.1 set at 100%.
Fig. 6.
Comparison of RNA transcription, genome replication, and protein expression/secretion between U.S. genotype B and genotype C clones. Huh7 cells were transfected with SphI dimers and harvested 5 days later. Clones 11.2W and 14.5W were derived from clones 11.2 and 14.5, respectively, by reverting the core promoter sequence. (A) Intracellular HBV RNA. (B) Replicative DNA. Lanes M, 100 pg of 3.2-kb, 1.7-kb, and 1.5-kb HBV DNA markers. (C) Immunoprecipitation-Western blot analysis of intracellular core protein. (D and E) ELISA for secreted HBsAg and HBeAg (based on 3 transfection experiments). The core promoter mutation types are as follows: 1, C1773T; 2, G1764A/C1766T/T1767G/C1773T; 3, A1752G; 4, T1754C/T1758G; 5, G1746A.
A1762T/G1764A mutants of genotype C may possess a higher replication capacity than A1752G mutants of genotype B.
Chinese genotype C isolates harboring core promoter mutations showed 3.5-kb RNA levels comparable to those of the core promoter mutants of genotype B (Fig. 5A, right panel). There was much greater variability in the replication capacity, but genotype C isolates showed comparable or even higher replication capacity in comparison to genotype B isolates (Fig. 5B, right panel). This finding could be explained by the ability of the A1762T/G1764A mutations, found in most genotype C isolates, to enhance genome replication (Fig. 3) and by the ability of the A1752G mutation, found in most genotype B isolates, to diminish genome replication (24).
Genotype C isolates often possess higher virion secretion capacity than genotype B isolates.
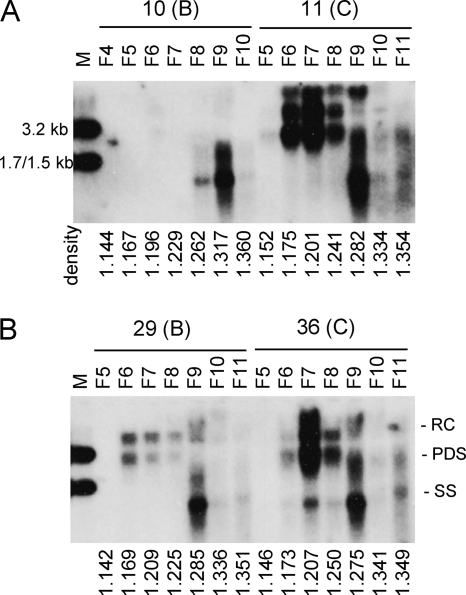
Although Chinese genotype B isolates with wild-type core promoter sequence showed higher intracellular levels of replicative DNA than the corresponding genotype C isolates (Fig. 5B, left panel), they produced less virion DNA (Fig. 5C, left panel). This result indicates a much higher virion secretion efficiency of genotype C isolates. The genotype C isolates with core promoter mutations displayed a higher level of replicative DNA but an even higher level of virion DNA than genotype B mutants (Fig. 5B and C, right panels; compare isolates 12 and 36 with isolates 7 and 8). To independently verify the virion secretion efficiency, we scaled up the transfection experiment and separated virions from naked core particles (which can be released from transfected Huh7 cells) by ultracentrifugation through a CsCl2 gradient. Such an analysis confirmed the more efficient virion secretion by isolate 36 of genotype C than isolate 29 of genotype B, despite comparable amounts of naked core particles (Fig. 7 B). Isolate 11 of genotype C also showed efficient virion secretion, in contrast to isolate 10 of genotype B (Fig. 7A).
Fig. 7.
Verification of different virion secretion capacities between genotype B and C isolates. (A) Comparison between isolates 10 and 11; (B) Comparison between isolates 29 and 36. Secreted particles were separated by ultracentrifugation through a CsCl2 gradient, and about 11 fractions were taken from the top. DNA was extracted from fractions 4 to 10 (F4 to F10) or 5 to 11 (F5 to F11) for Southern blot analysis using mixed genotype B and C probes. The density of each fraction is given.
Genotype-specific migration patterns and antibody affinities of envelope proteins.
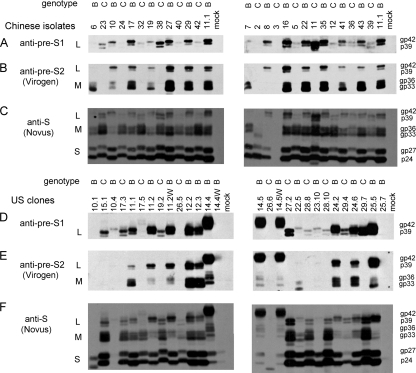
By running a small aliquot of cell lysate (50 μg of total proteins) overnight in a 12% separating gel of sufficient length (>10 cm), the S proteins of all the genotype B constructs displayed slightly accelerated mobility compared with those of genotype C constructs. Thus, by alternating constructs of the two genotypes in the same gel, a zigzag pattern was revealed by the horse polyclonal anti-HBs antibody (Fig. 8 C and F). Intriguingly, the L protein of genotype B manifested slower rather than faster mobility than that of genotype C, whether it was determined with this antibody (Fig. 8C and F) or with a rabbit anti-pre-S1 antibody (Fig. 8A and D). Furthermore, a monoclonal antibody (031-A; Virogen), which targets pre-S2 residues 13 to 18, recognized the L and M proteins of genotype B but not genotype C (Fig. 8B and E). Using a commercial ELISA kit (Abazyme), we found that the mean HBsAg levels (optical density at 450 nm) from 6 μl of culture supernatant were 0.91 ± 0.51 and 1.51 ± 0.52 for Chinese genotype B and C isolates, respectively (P < 0.05), and 1.06 ± 0.95 and 1.98 ± 1.15 for U.S. genotype B and C clones, respectively (P < 0.05). However, in earlier experiments with U.S. clones using the Auszyme kit from Abbott Laboratories, which has since been discontinued, we found consistently higher HBsAg secretion by genotype B clones (data not shown). On the basis of Northern blot analysis, on average, genotype B2 isolates or clones appeared to have higher levels of the 2.1-kb S transcript than genotype C2 constructs (Fig. 5A and 6A) (clones 27.2, 28.8, and 28.10 belong to the C1 subtype).
Fig. 8.
Differentiation of genotypes B and C by Western blot analysis of viral envelope proteins. About 50 μg of proteins from cell lysate was separated in a 12% polyacrylamide gel and reacted with three different antibodies. (A and D) Polyclonal pre-S1 antibody R271; (B and E) monoclonal pre-S2 antibody 031-A (Virogen); (C and F) polyclonal S antibody anti-ad/ay (Novus); (A to C) Chinese isolates; (D to F) U.S. clones.
For unknown reasons, the L and M proteins of U.S. clones 14.4, 14.5, and 14.5W showed retarded migration (Fig. 8D to F). These clones showed increased expression of the L protein but diminished expression of the S protein, which could explain the lack of HBsAg secretion (Fig. 6D) and might be a consequence of increased transcription of 2.4-kb subgenomic RNA but reduced transcription of the 2.1-kb RNA (Fig. 6A). Isolate 3 from China did not produce the 2.1-kb RNA (Fig. 5A) and was defective in envelope protein expression (Fig. 8A and C) and HBsAg secretion (Fig. 5E).
DISCUSSION
In the present study, we characterized the biological properties of a large number of HBV genotype B and C isolates from Chinese and U.S. chronic carriers. Genotyping was established by sequencing of the entire envelope gene for the Chinese samples and line probe assay of the S gene for the U.S. samples. Interestingly, the M and L proteins of none of the genotype C isolates could be detected in a Western blot by a monoclonal antibody targeting pre-S2 residues 13 to 18. Considering that pre-S2 residues 11 to 21 are TLQDPRVRALY for genotype B but ALLDPRVRGLY for genotype C (boldface indicates polymorphisms), the Q13 found in genotypes A, B, and D relative to L13 for genotype C is apparently responsible for the differential reactivity. Previously, Usuda and colleagues used the reactivity of serum HBsAg toward a panel of pre-S2 monoclonal antibodies in the ELISA format for HBV genotyping (45). Two monoclonal antibodies, targeting 7TFHQALLDP15 and 14DPRVRGLY21, respectively, were unable to recognize HBsAg of genotype B (45). Taken together, these results suggest that sequence variability at pre-S2 residues 11, 13, and 19 can be used to differentiate genotype B from genotype C through protein-based assays. Moreover, we demonstrated for the first time that the S protein of genotype B migrates slightly faster than that of genotype C, which, together with genotype-specific antibodies, should make Western blot analysis an alternative method to differentiate genotype B from genotype C.
Isolates belonging to the same HBV genotype can be further classified into subgenotypes on the basis of nucleotide sequence divergence of >4%. The B1 subgenotype is found exclusively in Japan, while the B2 subgenotype is prevalent in Taiwan, Hong Kong, and China (36, 37, 46, 49). Genotype C1 is found in Southeast Asia, Hong Kong, and southern China, whereas C2 is distributed in Japan, South Korea, Taiwan, and northern China (4, 12). On the basis of sequencing of the core promoter and precore/core gene, the Chinese HBV isolates, all collected in Shanghai, belong to the B2 and C2 subgenotypes. This is consistent with another report from Shanghai (51). The U.S. clones also belong to either the B2 or C2 subgenotype, except for clones 27.2, 28.8, and 28.10A, which are of the C1 subgenotype, as evidenced by the characteristic T1856/C1858 sequence. To overcome the effect of PCR errors on biological properties, we generated clone pools for the Chinese samples and characterized two independent clones for most U.S. samples. Bias in the Southern and Northern blot analyses was minimized by judicious selection of the probe and washing conditions.
The transfection experiments in Huh7 cells revealed marked variability in the replication capacities among constructs of the same genotype. The 12 Chinese genotype B isolates and the two U.S. genotype B clones harboring the A1752G mutation mostly displayed replication capacities lower than those of isolates with wild-type core promoter sequence. This is consistent with an earlier report revealing its association with low viremia titers in clinical samples and its downregulation of genome replication when introduced by site-directed mutagenesis (24). It is still unclear whether G1752 is present throughout infection or arises at a late stage of infection. Chinese genotype C isolates with A1762T/G1764A core promoter mutations mostly showed higher replication capacities than those with wild-type core promoter sequence, and introduction of these mutations into a genotype C clone increased intracellular replicative DNA and extracellular virion DNA. This is consistent with similar findings with other genotypes on the basis of site-directed mutagenesis (2, 20, 31). On the other hand, our mutational analysis revealed no major effect of the T1753C/T1758A, T1754C/T1758G, and G1764A/C1766T/T1767G/C1773T mutations on genome replication, despite a marked reduction of HBeAg expression by the last set of mutations. Therefore, the high replication capacities of clones 11.2 and 14.5 of genotype B and isolate 11 of genotype C are contributed by sequences outside the core promoter.
For both the Chinese and U.S. samples, genotype C constructs with wild-type core promoter sequence mostly displayed lower replication capacities than the corresponding genotype B constructs. Such a low replication capacity largely correlated with a low level of the core protein and diminished transcription of the 3.5-kb precore/pg RNAs. Previously, by transfecting Huh7 cells with terminally redundant HBV genomes, Sugiyama and colleagues also observed marked variability in replication capacity among genotype B and C clones lacking G1896A, A1762T, and G1764A mutations (38). However, two C2 clones and a B1 clone displayed the highest replication capacity. In this regard, Sugiyama and colleagues used nested PCR to generate 1.24 copies of the HBV genome, followed by selection of clones with a consensus sequence (38), which is different from our approach. The different findings could also be caused by biological differences between the Japanese and non-Japanese C2 and B2 isolates or simply clonal variability. Indeed, among our genotype B clones lacking core promoter mutations, some (clones 10.1, 10.4, 23.10, 25.5, and 25.7) replicated much less efficiently than others (clones 22.5, 24.2, and 24.6) (Fig. 6B).
A limitation of the present study is that the major findings were based on a single hepatoma cell line (Huh7) established from an Asian male. Since HBV genome replication in Huh7 cells is independent of a functional HBx protein (1), it will be important to repeat similar experiments in HepG2 cells, another human hepatoma cell line. Even with that, we cannot exclude the possibility that the levels of HBV genome replication and protein expression in hepatoma cell lines do not correlate with those of normal human hepatocytes due to the altered expression profiles of transcription factors required for HBV RNA production. Assuming that the main findings in Huh7 cells can be extrapolated to in vivo HBV infection, the overall lower replication capacity of the wild-type genotype C isolates than the corresponding genotype B isolates may underlie some of the key clinical differences between these two genotypes. Genotype C patients are more likely to be HBeAg positive than genotype B patients of the same age group due to a decade delay in HBeAg seroconversion (5, 16, 28, 48). The prevalence of genotype C relative to genotype B increases as the infection progresses from asymptomatic carriage status through chronic hepatitis to liver fibrosis/cirrhosis (7, 15, 40, 46). On the other hand, genotype B is more likely implicated in fulminant hepatitis and acute-on-chronic liver failure than genotype C (13, 30, 34). In Taiwan and China, genotype B (B2) infection might be associated with earlier hepatocellular carcinoma (HCC) development (15, 47). These findings can be explained if the higher levels of genome replication and core protein expression by genotype B2 trigger an earlier transition from the immune tolerance phase to the immune active phase, thus accelerating HBeAg seroconversion and viral clearance and diminishing the lifelong risk for HCC. On the other hand, the more vigorous though short-lived immune clearance phase may trigger fulminant hepatitis and earlier onset of HCC, if it does occur. Although the core protein sequence may also affect the magnitude of the CTL response, genotype B2 has a genotype C-like core gene due to a past recombination event (36, 37).
The different replication capacities between wild-type genotype B and genotype C may also shed light on the much higher prevalence of A1762T/G1764A core promoter mutations in genotype C (29, 48). The prevalence of the two point mutations is C1 > C2 > B2 > B1 (4, 36, 46, 47). The higher rate of A1762T/G1764A mutations in C1 than C2 may be due to the inability of C1 to develop the G1896A mutation (4, 22, 46, 47). C1 has C1858 instead of T1858; thus, a G1896A mutation will disrupt a preexisting C-G base pair in the ε signal of pg RNA to impair DNA replication (21). With reduced opportunity to abolish HBeAg expression by the precore mutation, C1 may primarily employ the core promoter mutations to downregulate HBeAg expression. The other effect of the A1762T/G1764A mutations is the upregulation of HBV genome replication. We hypothesize that the low replication capacity of the wild-type genotype C triggers emergence of the A1762T/G1764A mutations during the immune clearance phase to maximize viral survival. Cumulative evidence suggests that the increased pathogenicity of genotype C, including HCC risk, is due to its high prevalence of A1762T/G1764A core promoter mutations (49, 50). In fact, these mutations have been used as an independent marker for HCC development (19).
A low replication capacity, if it is not compensated for by more efficient virion secretion or better infectivity, will slow virus spread during the onset of infection. Such low kinetics of virus spread may give the host immune system windows of opportunity to contain the infection at its infancy, thus preventing viral persistence. However, study of the Chinese genotype C isolates revealed that they had much more efficient virion secretion than the genotype B isolates. This finding, based on immunoprecipitation with a polyclonal anti-S antibody, was confirmed for some isolates by the ultracentrifugation method. A low replication capacity combined with very efficient virion secretion could enable rapid spread of wild-type genotype C isolates without provoking a strong immune response, thus predisposing the individual to persistent infection. Indeed, in adulthood, acute infection with genotype C is more likely to progress to chronic infection than genotype B infection (13, 51). The doubling time of HBV DNA was 3.4 days for genotype A but only 1.9 days for genotype C in experimentally infected chimpanzees (17). The 50% chimpanzee infectious doses were found by others to be 169 copies of HBV DNA for genotype A, 78 copies for genotype D, and only 3 copies for genotype C (10). HBV can infect severe combined immunodeficiency mice transgenic for urokinase-type plasminogen activator (uPA-SCID mice) and repopulated with human hepatocytes. In this small-animal model, genotype C also showed a faster rise of viremia titer than genotypes A and B (38, 39). Whether these in vivo findings are the outcome of efficient virion secretion or infectivity remain to be clarified.
ACKNOWLEDGMENTS
This work was supported by Research Scholar Grant 06-059-01-MBC from the American Cancer Society; grants CA133976, CA109733, AA08169, and AA19072 from the NIH; and National Science and Technology Major Projects of China grants 2008ZX10002-006 and 2009ZX10603.
Footnotes
Published ahead of print on 20 July 2011.
REFERENCES
- 1. Blum H. E., et al. 1992. Hepatitis B virus X protein is not central to the viral life cycle in vitro. J. Virol. 66:1223–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Buckwold V. E., Xu Z., Chen M., Yen T. S., Ou J. H. 1996. Effects of a naturally occurring mutation in the hepatitis B virus basal core promoter on precore gene expression and viral replication. J. Virol. 70:5845–5851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carman W. F., et al. 1989. Mutation preventing formation of hepatitis B e antigen in patients with chronic hepatitis B infection. Lancet ii:588–591 [DOI] [PubMed] [Google Scholar]
- 4. Chan H. L., et al. 2005. Epidemiological and virological characteristics of 2 subgroups of hepatitis B virus genotype C. J. Infect. Dis. 191:2022–2032 [DOI] [PubMed] [Google Scholar]
- 5. Chu C. J., Hussain M., Lok A. S. 2002. Hepatitis B virus genotype B is associated with earlier HBeAg seroconversion compared with hepatitis B virus genotype C. Gastroenterology 122:1756–1762 [DOI] [PubMed] [Google Scholar]
- 6. Chu C. J., Lok A. S. 2002. Clinical significance of hepatitis B virus genotypes. Hepatology 35:1274–1276 [DOI] [PubMed] [Google Scholar]
- 7. Chu C. M., Liaw Y. F. 2005. Genotype C hepatitis B virus infection is associated with a higher risk of reactivation of hepatitis B and progression to cirrhosis than genotype B: a longitudinal study of hepatitis B e antigen-positive patients with normal aminotransferase levels at baseline. J. Hepatol. 43:411–417 [DOI] [PubMed] [Google Scholar]
- 8. Garcia T., et al. 2009. Drastic reduction in the production of subviral particles does not impair hepatitis B virus virion secretion. J. Virol. 83:11152–11165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guarnieri M., et al. 2006. Point mutations upstream of hepatitis B virus core gene affect DNA replication at the step of core protein expression. J. Virol. 80:587–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hsia C. C., Purcell R. H., Farshid M., Lachenbruch P. A., Yu M. Y. 2006. Quantification of hepatitis B virus genomes and infectivity in human serum samples. Transfusion 46:1829–1835 [DOI] [PubMed] [Google Scholar]
- 11. Hu J., Boyer M. 2006. Hepatitis B virus reverse transcriptase and epsilon RNA sequences required for specific interaction in vitro. J. Virol. 80:2141–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huy T. T., et al. 2004. Genotype C of hepatitis B virus can be classified into at least two subgroups. J. Gen. Virol. 85:283–292 [DOI] [PubMed] [Google Scholar]
- 13. Imamura T., et al. 2003. Distribution of hepatitis B viral genotypes and mutations in the core promoter and precore regions in acute forms of liver disease in patients from Chiba, Japan. Gut 52:1630–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kao J. H. 2002. Hepatitis B viral genotypes: clinical relevance and molecular characteristics. J. Gastroenterol. Hepatol. 17:643–650 [DOI] [PubMed] [Google Scholar]
- 15. Kao J. H., Chen P. J., Lai M. Y., Chen D. S. 2000. Hepatitis B genotypes correlate with clinical outcomes in patients with chronic hepatitis B. Gastroenterology 118:554–559 [DOI] [PubMed] [Google Scholar]
- 16. Kao J. H., Chen P. J., Lai M. Y., Chen D. S. 2004. Hepatitis B virus genotypes and spontaneous hepatitis B e antigen seroconversion in Taiwanese hepatitis B carriers. J. Med. Virol. 72:363–369 [DOI] [PubMed] [Google Scholar]
- 17. Komiya Y., et al. 2008. Minimum infectious dose of hepatitis B virus in chimpanzees and difference in the dynamics of viremia between genotype A and genotype C. Transfusion 48:286–294 [DOI] [PubMed] [Google Scholar]
- 18. Kramvis A., et al. 2008. Relationship of serological subtype, basic core promoter and precore mutations to genotypes/subgenotypes of hepatitis B virus. J. Med. Virol. 80:27–46 [DOI] [PubMed] [Google Scholar]
- 19. Kuang S. Y., et al. 2004. Specific mutations of hepatitis B virus in plasma predict liver cancer development. Proc. Natl. Acad. Sci. U. S. A. 101:3575–3580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li J., Buckwold V. E., Hon M. W., Ou J. H. 1999. Mechanism of suppression of hepatitis B virus precore RNA transcription by a frequent double mutation. J. Virol. 73:1239–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li J. S., et al. 1993. Hepatitis B virus genotype A rarely circulates as an HBe-minus mutant: possible contribution of a single nucleotide in the precore region. J. Virol. 67:5402–5410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lok A. S., Akarca U., Greene S. 1994. Mutations in the pre-core region of hepatitis B virus serve to enhance the stability of the secondary structure of the pre-genome encapsidation signal. Proc. Natl. Acad. Sci. U. S. A. 91:4077–4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Milich D., Liang T. J. 2003. Exploring the biological basis of Hepatitis B e antigen in hepatitis B virus infection. Hepatology 38:1075–1086 [DOI] [PubMed] [Google Scholar]
- 24. Ng L. F., et al. 2005. Host heterogeneous ribonucleoprotein K (hnRNP K) as a potential target to suppress hepatitis B virus replication. PLoS Med. 2:e163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Norder H., et al. 2004. Genetic diversity of hepatitis B virus strains derived worldwide: genotypes, subgenotypes, and HBsAg subtypes. Intervirology 47:289–309 [DOI] [PubMed] [Google Scholar]
- 26. Okamoto H., et al. 1994. Hepatitis B virus with mutations in the core promoter for an e antigen-negative phenotype in carriers with antibody to e antigen. J. Virol. 68:8102–8110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Okamoto H., et al. 1988. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J. Gen. Virol. 69 (Pt 10):2575–2583 [DOI] [PubMed] [Google Scholar]
- 28. Orito E., et al. 2001. Geographic distribution of hepatitis B virus (HBV) genotype in patients with chronic HBV infection in Japan. Hepatology 34:590–594 [DOI] [PubMed] [Google Scholar]
- 29. Orito E., et al. 2001. A case-control study for clinical and molecular biological differences between hepatitis B viruses of genotypes B and C. Japan HBV Genotype Research Group. Hepatology 33:218–223 [DOI] [PubMed] [Google Scholar]
- 30. Ozasa A., et al. 2006. Influence of genotypes and precore mutations on fulminant or chronic outcome of acute hepatitis B virus infection. Hepatology 44:326–334 [DOI] [PubMed] [Google Scholar]
- 31. Parekh S., et al. 2003. Genome replication, virion secretion, and e antigen expression of naturally occurring hepatitis B virus core promoter mutants. J. Virol. 77:6601–6612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Qin Y., et al. 2011. Improved method for rapid and efficient determination of genome replication and protein expression of clinical hepatitis B virus isolates. J. Clin. Microbiol. 49:1226–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Qin Y., et al. 2009. Prevalence of basal core promoter and precore mutations in Chinese chronic hepatitis B patients and correlation with serum HBeAg titers. J. Med. Virol. 81:807–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ren X., et al. 2010. Hepatitis B virus genotype and basal core promoter/precore mutations are associated with hepatitis B-related acute-on-chronic liver failure without pre-existing liver cirrhosis. J. Viral Hepat. 17:887–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schaefer S. 2005. Hepatitis B virus: significance of genotypes. J. Viral Hepat. 12:111–124 [DOI] [PubMed] [Google Scholar]
- 36. Sugauchi F., et al. 2003. Epidemiologic and virologic characteristics of hepatitis B virus genotype B having the recombination with genotype C. Gastroenterology 124:925–932 [DOI] [PubMed] [Google Scholar]
- 37. Sugauchi F., et al. 2002. Hepatitis B virus of genotype B with or without recombination with genotype C over the precore region plus the core gene. J. Virol. 76:5985–5992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sugiyama M., et al. 2006. Influence of hepatitis B virus genotypes on the intra- and extracellular expression of viral DNA and antigens. Hepatology 44:915–924 [DOI] [PubMed] [Google Scholar]
- 39. Sugiyama M., et al. 2009. Direct cytopathic effects of particular hepatitis B virus genotypes in severe combined immunodeficiency transgenic with urokinase-type plasminogen activator mouse with human hepatocytes. Gastroenterology 136:652–662 [DOI] [PubMed] [Google Scholar]
- 40. Sumi H., et al. 2003. Influence of hepatitis B virus genotypes on the progression of chronic type B liver disease. Hepatology 37:19–26 [DOI] [PubMed] [Google Scholar]
- 41. Tavis J. E., Ganem D. 1996. Evidence for activation of the hepatitis B virus polymerase by binding of its RNA template. J. Virol. 70:5741–5750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tong S. P., Li J. S., Vitvitski L., Benjelloun S., Trepo C. 1991. Rapid screening for bacterial colonies harbouring tandem hepatitis B virus sequences by an oligonucleotide probe. J. Virol. Methods 32:109–114 [DOI] [PubMed] [Google Scholar]
- 43. Tong S. P., Li J. S., Vitvitski L., Trepo C. 1990. Active hepatitis B virus replication in the presence of anti-HBe is associated with viral variants containing an inactive pre-C region. Virology 176:596–603 [DOI] [PubMed] [Google Scholar]
- 44. Tsai A., et al. 2009. Chimeric constructs between two hepatitis B virus genomes confirm transcriptional impact of core promoter mutations and reveal multiple effects of core gene mutations. Virology 387:364–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Usuda S., et al. 1999. Serological detection of hepatitis B virus genotypes by ELISA with monoclonal antibodies to type-specific epitopes in the preS2-region product. J. Virol. Methods 80:97–112 [DOI] [PubMed] [Google Scholar]
- 46. Wang Z., et al. 2007. Clinical and virological characteristics of hepatitis B virus subgenotypes Ba, C1, and C2 in China. J. Clin. Microbiol. 45:1491–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yuan J., et al. 2007. Hepatitis B virus (HBV) genotypes/subgenotypes in China: mutations in core promoter and precore/core and their clinical implications. J. Clin. Virol. 39:87–93 [DOI] [PubMed] [Google Scholar]
- 48. Yuen M. F., et al. 2003. Significance of hepatitis B genotype in acute exacerbation, HBeAg seroconversion, cirrhosis-related complications, and hepatocellular carcinoma. Hepatology 37:562–567 [DOI] [PubMed] [Google Scholar]
- 49. Yuen M. F., et al. 2004. Role of hepatitis B virus genotypes Ba and C, core promoter and precore mutations on hepatocellular carcinoma: a case control study. Carcinogenesis 25:1593–1598 [DOI] [PubMed] [Google Scholar]
- 50. Yuen M. F., et al. 2008. Risk for hepatocellular carcinoma with respect to hepatitis B virus genotypes B/C, specific mutations of enhancer II/core promoter/precore regions and HBV DNA levels. Gut 57:98–102 [DOI] [PubMed] [Google Scholar]
- 51. Zhang H. W., et al. 2008. Risk factors for acute hepatitis B and its progression to chronic hepatitis in Shanghai, China. Gut 57:1713–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]