Abstract
Viruses encode RNA silencing suppressors to counteract host antiviral silencing. In this study, we analyzed the suppressors encoded by potato virus M (PVM), a member of the genus Carlavirus. In the conventional green fluorescent protein transient coexpression assay, the cysteine-rich protein (CRP) of PVM inhibited both local and systemic silencing, whereas the triple gene block protein 1 (TGBp1) showed suppressor activity only on systemic silencing. Furthermore, to elucidate the roles of these two suppressors during an active viral infection, we performed PVX vector-based assays and viral movement complementation assays. CRP increased the accumulation of viral RNA at the single-cell level and also enhanced viral cell-to-cell movement by inhibiting RNA silencing. However, TGBp1 facilitated viral movement but did not affect viral accumulation in protoplasts. These data suggest that CRP inhibits RNA silencing primarily at the viral replication step, whereas TGBp1 is a suppressor that acts at the viral movement step. Thus, our findings demonstrate a sophisticated viral infection strategy that suppresses host antiviral silencing at two different steps via two mechanistically distinct suppressors. This study is also the first report of the RNA silencing suppressor in the genus Carlavirus.
INTRODUCTION
Viruses are obligate intracellular pathogens that manipulate and exploit the molecular mechanism of the host to survive in a hostile cellular environment. The presence of viruses and their propagation induces diverse mechanisms in the host for combating viral infection at both the single-cell and the whole-organism levels. In plants, one of the most important defense mechanisms against viruses is RNA silencing, which is a sequence-specific RNA degradation process regulated by small RNA molecules (11, 12). Since RNA viruses go through single-stranded RNA or double-stranded RNA (dsRNA) stages at a given point in their replication, they are both active initiators and targets of host RNA silencing. When plant RNA viruses invade host cells, dsRNAs derived from their replication intermediates or highly structured genomic RNAs are recognized by plant dicer-like nucleases and processed into small interfering RNAs (siRNAs) of 21 to 24 nucleotides in length. The siRNAs are then recruited to a multiprotein effector complex called the RNA-induced silencing complex (RISC), which includes the slicer endonuclease Argonaute and subsequently mediates the cleavage of cognate viral RNAs. RNA silencing is a non-cell-autonomous event in higher plants and, once RNA silencing is induced in the initial cell, it spreads over the whole organism through the vasculature and from cell to cell presumably via plasmodesmata, mediated by mobile small RNA signals (3, 14, 22, 36, 48, 51). The cell-to-cell and long-distance movement of virus-derived small RNA signals likely serves to immunize surrounding naive cells ahead of the infection front. In fact, in addition to the reduction in viral RNAs at the initial infection site, the plants exhibiting a “recovery” phenomenon present a complete loss of viral disease symptoms and virus accumulation, and they become resistant to secondary infection by the homologous virus in upper systemically infected leaves as a result of highly effective antiviral silencing (15, 34, 39).
As a counterattack against host antiviral silencing, viruses have evolved RNA silencing suppressors (1, 28). To date, a number of RNA silencing suppressors have been identified in plant viruses. These suppressors share no significant sequence similarity and target different steps of the host silencing pathway. Tombusvirus p19 binds siRNAs to interfere with their incorporation into the RISC (27, 44), and many other viral suppressors bind dsRNAs (26, 33). Alternatively, some silencing suppressors interact with the protein components of the RISC. The 2b protein of cucumber mosaic virus, P0 of poleroviruses, p38 of turnip crinkle virus (TCV), and P1 of sweet potato mild mottle virus inhibit RISC activity by quenching Argonaute (2, 4, 6, 17, 56). However, except for these specific proteins, the targets of viral suppressors and the molecular mechanisms by which they suppress RNA silencing remain unclear.
Although the molecular characterization of the target and machinery of RNA silencing suppression is important, it is also important to elucidate what roles viral suppressors play in infection by individual viruses. In fact, many suppressors reported at present have been identified through a transient coexpression assay using reporter genes, such as green fluorescent protein (GFP) in Nicotiana benthamiana, and the rest were discovered as suppressors of systemic silencing using a similar coexpression assay or by grafting experiments in reporter gene-transgenic Nicotiana plants. However, many of these suppressors are primarily known as “pathogenicity determinants” or “long-distance movement factors” involved in symptom development, viral replication, or viral movement during systemic infection (20, 43). The potyviral HC-Pro, a multifunctional proteinase, is required for long-distance movement and the maintenance of genome replication, and site-directed mutagenesis experiments revealed that these functions are involved in silencing suppressor activity (9, 23, 24). HC-Pro also enhances synergistic viral disease symptoms by activating the replication of heterologous viruses (38). In addition, inactivation of the p19 protein in tomato bushy stunt virus (TBSV) has an attenuating effect on lethal apical necrosis in N. benthamiana (40) and also prevents systemic viral spread in spinach plants (41). Because these findings imply divergent roles for these suppressors in viral infections, it is essential to determine the influences and roles of the individual suppressors in actual viral infection processes to comprehensively understand the strategy that viruses use to invade host plants.
Potato virus M (PVM) belongs to the genus Carlavirus of the family Flexiviridae, which has a monopartite, single-stranded, positive-sense RNA genome containing six open reading frames (ORFs) (see Fig. 1A) (55). ORF1 encodes a viral replicase. Three overlapping ORFs, ORF2 to ORF4, encode movement proteins named triple gene block proteins 1, 2, and 3 (TGBp1, -2, and -3), and ORF5 encodes a coat protein (CP). The product of ORF6 at the 3′ proximal end is a cysteine-rich protein (CRP) that possesses nucleic acid-binding activity (18). Although silencing suppressors have not yet been identified in the genus Carlavirus, TGBp1 and CRP encoded by members of other genera have been reported to inhibit RNA silencing. The TGBp1 of potexviruses (genus Potexvirus, family Flexiviridae) has been shown to inhibit both local and systemic silencing in GFP coexpression assays (5, 42, 49). CRPs encoded by genetically diverse plant RNA viruses, including members of the genera Tobravirus, Hordeivirus, Pecluvirus, and Vitivirus, all of which contain a characteristic sequence motif consisting of conserved cysteine residues, have been demonstrated to function as silencing suppressors (1, 13, 16, 53, 57).
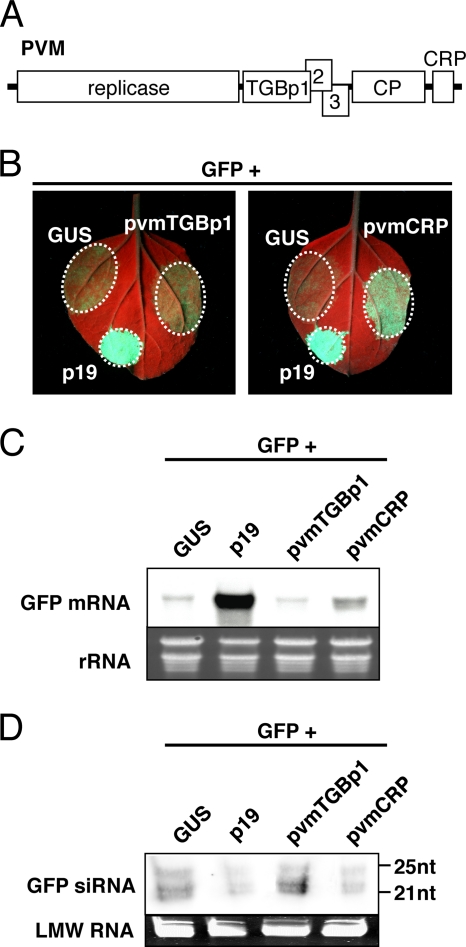
Fig. 1.
Suppressor activity of pvmTGBp1 and pvmCRP on sense transgene-induced local RNA silencing. (A) Schematic representation of the genome of potato virus M showing ORFs. (B) GFP fluorescence images of N. benthamiana leaves infiltrated with Agrobacterium mixtures containing a vector expressing GFP and GUS (left upper patches of each panel), p19 (left lower patches), pvmTGBp1 (right patch of left panel), or pvmCRP (right patch of right panel). Photographs were taken under UV light at 4 dpi. (C and D) Northern blot analysis of GFP mRNA (C) and siRNAs (D) extracted from the infiltrated patches shown in panel B. Ethidium bromide-stained rRNA or low-molecular-weight RNA (LMW RNA) is shown below each panel as a loading control. Molecular size markers are indicated on the right.
In the present study, we analyzed the silencing suppression activities of the TGBp1 and CRP encoded by PVM. In a GFP transient-coexpression assay, only CRP showed suppressive activity on local silencing. However, both TGBp1 and CRP inhibited the systemic spread of GFP silencing. Furthermore, we performed PVX vector-based assays to examine the effect of these two suppressors on an active viral infection. These analyses revealed that CRP can increase accumulation of viral RNA at the single-cell level, whereas TGBp1 promotes viral cell-to-cell movement by inhibiting RNA silencing. Taken together, our results suggest that PVM uses two suppressors that utilize different mechanisms to inhibit host antiviral silencing at different infection steps, thereby achieving successful PVM infection.
MATERIALS AND METHODS
Construction of binary plasmids and PVX recombinants.
The binary plasmids for expressing GFP and β-glucuronidase (GUS) were described previously (42). The plasmid pBin19, which contains the full-length cDNA of TBSV p19, was kindly provided by D. C. Baulcombe (University of Cambridge, Cambridge, United Kingdom). To construct the plasmids expressing pvmTGBp1 and pvmCRP, full-length cDNA was amplified by reverse transcription-PCR (RT-PCR; using 5′- and 3′-specific primers containing suitable restriction sites) from total RNA extracted from plants infected with PVM (MAFF 307027; National Institute of Agribiological Sciences GenBank) and cloned into the binary vector pBI121 using BamHI and SacI sites. The pvmCRP mutants CRPmBM and CRPmZF are derivatives of the pvmCRP expression plasmid, into which mutations into the basic motif and zinc-finger motif of pvmCRP, respectively, were incorporated by basic site-directed mutagenesis PCR (see Fig. 2A). p25 mutants, p25A104V and p25T117A, which contain single amino acid changes at the 104th and 117th residues of p25, respectively, were generated from the p25 expression plasmid (42) by site-directed mutagenesis PCR.
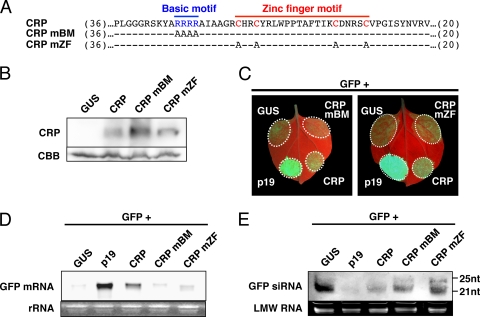
Fig. 2.
Suppressor activity of pvmCRP mutants on sense transgene-induced local RNA silencing. (A) Schematic representation of the site-directed mutagenesis of pvmCRP. The positions of the basic motif (in blue letters) and the zinc-finger motif (in red letters) are shown. The mutants CRPmBM and CRPmZF harbor alanine substitutions in each motif, and the dashes indicate residues identical to the wild type. Numbers in parentheses indicate amino acid residues not shown here. (B) Immunoblot analysis of pvmCRP and its mutants. Total protein was extracted from the leaves infiltrated with Agrobacterium mixtures containing a vector expressing p19 and GUS, tagged CRP, CRPmBM, or CRPmZF at 3 dpi. Coomassie brilliant blue-stained total protein (CBB) is shown as a loading control. (C) GFP fluorescence images of N. benthamiana leaves infiltrated with Agrobacterium mixtures containing a vector expressing GFP and GUS (left upper patches of each panel), p19 (left lower patches), wild-type CRP (right lower patches) CRPmBM (right upper patch of left panel), or CRPmZF (right upper patch of right panel). The photographs were taken under UV light at 4 dpi. (D and E) Northern blot analysis of GFP mRNA (D) and siRNAs (E) extracted from the infiltrated patches shown in panel C. Ethidium bromide-stained rRNA or low-molecular-weight RNA (LMW RNA) is shown below each panel as a loading control. Molecular size markers are indicated on the right.
The infectious cDNA clone of PVX carrying GFP (PVX-GFP) was described before (10) and was kindly provided by D. C. Baulcombe. The infectious cDNA clone PVXΔp25-GFP was generated from PVX-GFP, as described previously (5). To construct other PVX recombinants (PVX-pvmTGBp1, PVX-pvmCRP, PVX-p19, PVXΔp25-pvmTGBp1, PVXΔp25-pvmCRP, and PVXΔp25-p19), the full-length cDNA of each protein was PCR amplified from the above-mentioned expression plasmids and substituted for the GFP sequence in the PVX-GFP or PVXΔp25-GFP.
Plant material and agroinfiltration.
Plants were maintained in growth chambers at 20 to 25°C throughout the assays. Agrobacterium tumefaciens infiltration was conducted as described previously (42). Bacterial cells were harvested and resuspended in infiltration buffer to a final optical density at 600 nm of 1.0. In coexpression assays, equal volumes of each suspension were mixed before infiltration. For the viral movement assay depicted in Fig. 6B, an Agrobacterium culture carrying the binary plasmid of each suppressor protein was infiltrated, and then PVX-GFP was inoculated mechanically with an extract of a virus-infected N. benthamiana plant. For the viral movement complementation assay (VMCA) (Fig. 6D), an Agrobacterium culture carrying the PVXΔp25-GFP binary plasmid was diluted 10,000-fold and mixed 1:1:1 with two Agrobacterium cultures carrying the binary plasmid of the p25 mutant and of each suppressor protein, respectively. For inoculation of the PVX recombinants (see Fig. 4), carborundum-dusted leaves of 3- to 4-week-old N. benthamiana plants were inoculated with 1 μg of each plasmid DNA.
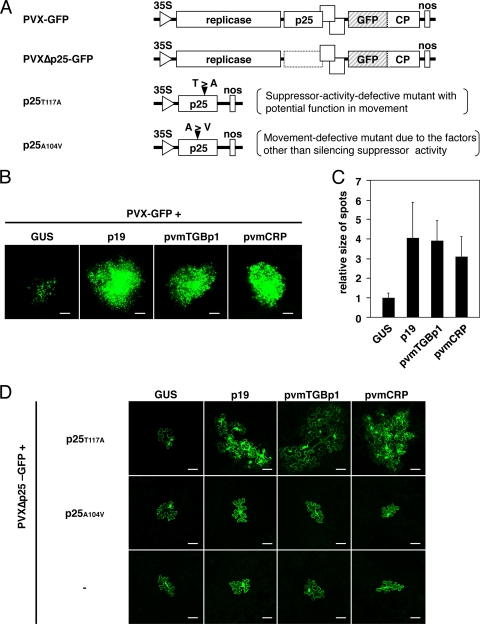
Fig. 6.
Effects of pvmTGBp1 and pvmCRP on viral cell-to-cell movement. (A) Schematic representation of recombinant PVX variants and mutant derivatives of PVX p25. PVXΔp25-GFP consists of an infectious PVX cDNA clone that expresses GFP from the CP promoter and harbors stop codons and a deletion in the p25 ORF (broken box). For the p25 mutants, p25T117A and p25A104V, the amino acid substitutions are shown above each ORF (solid triangles). (B) Fluorescence microscopic examination of cell clusters expressing GFP in N. benthamiana leaves cointroduced with PVX-GFP and either GUS, p19, pvmTGBp1, or pvmCRP. Photographs were taken at 6 dpi. Scale bars, 500 μm. (C) Quantification of the size of the green fluorescent spots derived from PVX-GFP. Twenty spots, as shown in panel B, were selected randomly from at least three leaves of two independent replicates for each expression construct (GUS, p19, pvmTGBp1, or pvmCRP), and the fluorescent areas were measured using ImageJ software v1.40 (NIH, Bethesda, MD). The relative sizes normalized to GUS are shown. Error bars represent the standard deviations. (D) Cell-to-cell movement analysis of PVX-GFPΔp25 in leaves expressing p25 mutants (none [–], p25T117A, or p25A104V) and a silencing suppressor (GUS as a control, p19, pvmTGBp1, or pvmCRP). Cells expressing GFP were visualized using fluorescence microscopy and photographs were taken at 5 dpi. Scale bars, 50 μm.
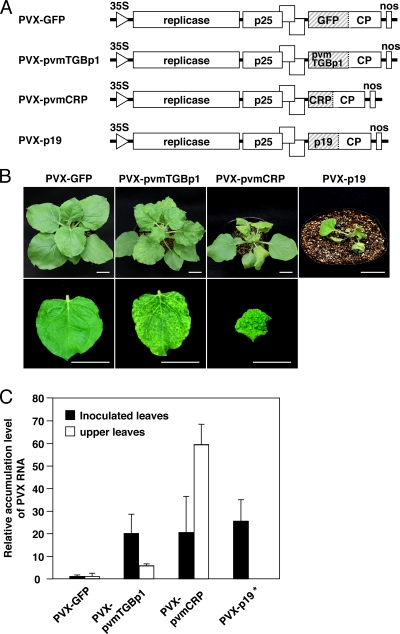
Fig. 4.
Effects of pvmTGBp1 and pvmCRP on PVX pathogenicity. (A) Schematic representation of recombinant PVX variants. The GFP, pvmTGBp1, pvmCRP, or TBSV p19 sequence is inserted downstream from the coat protein promoter in an infectious PVX cDNA clone. (B) Symptoms of the PVX-GFP, PVX-pvmTGBp1, PVX-pvmCRP, or PVX p19-infected plants. Photographs of whole plants (upper panels) and of systemically infected leaves (lower panels) were taken at 15 dpi. Scale bars, 3 cm. (C) Quantitative real-time PCR (qRT-PCR) analysis of PVX coat protein (CP) RNA accumulation in each recombinant virus-inoculated plant. Total RNA was extracted from inoculated leaves at 5 dpi (▪) or upper leaves at 16 dpi (□). The accumulation levels relative to PVX-GFP were calculated via the second derivative method using ubiquitin transcripts as the internal standard. The error bars represent the standard deviations of three replicates. Note that the upper leaves of the plant inoculated with PVX-p19 are not shown due to necrosis of the entire plant.
Preparation and inoculation of protoplasts.
Tobacco protoplasts were prepared from BY-2 suspension cell cultures and inoculated with 10 μg of each PVX recombinant cDNA by electroporation, as described previously (54). Electroporated protoplasts were incubated in the dark from 0 to 96 h at 28°C, after which they were harvested by low-speed centrifugation (1,000 × g for 5 min) for RNA extraction.
Immunoblot analysis.
Immunoblot analysis was performed as described previously (42). Total protein extracted from leaf tissue expressing the GFP-tagged pvmCRP variants was separated on an sodium dodecyl sulfate (SDS)-polyacrylamide gel, blotted onto a polyvinylidene difluoride membrane, and detected by an anti-GFP antibody. Equal volumes of total protein were separated on an SDS-polyacrylamide gel and stained with Coomassie brilliant blue as a loading control.
RNA isolation and analysis.
RNA isolation and Northern blot analysis of GFP mRNA and siRNAs were performed as described previously (42). Total RNA was extracted from infiltrated or infected leaves or inoculated protoplasts. For quantitative real-time PCR (qRT-PCR) analysis, total RNA was treated with RNase-free recombinant DNase I (TaKaRa, Shiga, Japan). A 0.5-μg aliquot was used for cDNA synthesis with a high-capacity cDNA RT kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. qRT-PCR was performed with the primers PVXCP159F (5′-CGC AAC AAA TGA GGA CCT CAG CAA T-3′) and PVXCP239R (5′-GCA GCC TGT GCC ATA GTG TCT GTG-3′). Ubiquitin expression in N. benthamiana was assayed as an internal standard using the primers UBI26F (5′-CGG CAT GCT TAA CAC ATG CA-3′) and UBI136R (5′-AGC CGT TTC CAG CTG TTG TTC-3′). SYBR Premix ExTaq II (TaKaRa) and a thermal cycler dice real-time system (TaKaRa) were used for analysis. The PCR amplification efficiency and relative gene expression were calculated by setting the threshold with the second derivative maximum, and the fold changes were calculated using thermal cycler dice real-time system single software (version 1.00).
GFP fluorescence analysis.
For coexpression assays, the GFP fluorescence of infiltrated leaves and whole plants was observed under long-wavelength UV light (B-100 Black-Ray long-wave UV lamp; UV Products, Upland, CA) and photographed using an EOS Kiss digital camera (Canon, Tokyo, Japan) with a yellow filter. For the viral movement assay, GFP fluorescence was observed using a fluorescence microscope (MZ16F; Leica Microsystems, Wetzler, Germany) and a confocal laser scanning microscope (TCS SP5; Leica Microsystems). All images were edited using Adobe Photoshop version 7.0 software (Adobe Systems, San Jose, CA).
RESULTS
CRP of PVM suppresses local RNA silencing, but TGBp1 does not.
PVM encodes two candidate RNA silencing suppressor proteins in its genome: TGBp1, a homolog of a potexvirus protein that functions as an RNA silencing suppressor (42, 49), and CRP, the homologs of which prevent RNA silencing in various plant RNA viruses (1, 13, 16, 53, 57). To investigate whether these two proteins encoded by PVM function as RNA silencing suppressors, we performed an Agrobacterium-mediated transient-coexpression assay (21). In this assay, suppression of transgene-induced RNA silencing in patches infiltrated with A. tumefaciens was examined using GFP as a marker. A plasmid expressing GFP and a plasmid expressing either TGBp1 or CRP (hereafter referred to as pvmTGBp1 and pvmCRP, respectively) were cointroduced into N. benthamiana leaves by agroinfiltration. As negative and positive controls, either GUS or the well-characterized RNA silencing suppressor p19 from TBSV (50), respectively, was coexpressed with GFP.
GFP fluorescence was observed in each agroinfiltrated patch under UV light at 4 days postinfiltration (dpi). The patches expressing GFP and GUS showed decreased green fluorescence at 4 dpi as a consequence of RNA silencing activation (Fig. 1B, left upper patch of each panel) (21). In contrast, patches expressing p19, together with GFP, showed bright green fluorescence at 4 dpi (Fig. 1B, left lower patch of each panel), indicating that RNA silencing was suppressed. In the patch expressing pvmTGBp1, GFP fluorescence decreased to a level similar to that of the negative control at 4 dpi (Fig. 1B, right patch of left panel). Conversely, when pvmCRP was coexpressed with GFP, strong fluorescence was maintained at 4 dpi (Fig. 1B, right patch of right panel). Consistent with these observations, Northern blot analysis revealed that the accumulation levels of GFP mRNA were very low in patches expressing GUS and pvmTGBp1 (Fig. 1C). In contrast, much higher accumulation levels of GFP mRNA were observed in patches expressing p19 and pvmCRP (Fig. 1C).
To test whether these differences in the accumulation levels of GFP mRNA accompanied alterations in siRNA accumulation, GFP-specific siRNA accumulation was analyzed. Inversely correlated with the levels of GFP mRNA, the levels of GFP-specific siRNAs were significantly lower in patches expressing p19 and pvmCRP than in those expressing GUS and pvmTGBp1 (Fig. 1D). Collectively, these data indicate that pvmCRP inhibits local RNA silencing induced in agroinfiltrated areas, but pvmTGBp1 does not function as a suppressor of local silencing.
Two functional motifs in CRP are required for local RNA silencing suppression.
Carlavirus CRPs contain a basic motif consisting of highly basic residues and a putative zinc-finger motif characterized by a series of cysteine residues in the central region of the protein (Fig. 2A) (18, 25). These two motifs are characteristic of eukaryotic transcription factors and are suggested to be involved in nucleic acid binding properties (18, 31). To examine whether these motifs play a role in the suppression of local RNA silencing, we engineered two pvmCRP mutants with alanines replacing either the arginine residues of the basic motif (CRPmBM) or cysteine residues of the zinc finger motif (CRPmZF) (Fig. 2A). First, we confirmed the accumulation of these mutant proteins. Total protein was extracted from the patches introduced with the tagged-pvmCRP variants and p19 (to eliminate the influence of silencing triggered against the pvmCRP transgenes) and the accumulation levels of the wild-type pvmCRP and pvmCRP mutants were examined by immunoblot analysis. As shown in Fig. 2B, CRPmBM accumulated at a rather high level, and CRPmZF at a similar level, compared to wild-type pvmCRP, demonstrating that the mutations did not lead to an instability of the protein. We then performed the Agrobacterium-mediated transient coexpression assay with CRPmBM and CRPmZF to determine whether these mutants have RNA silencing suppressor activity. At 4 dpi, GFP fluorescence almost completely disappeared in the patch expressing CRPmBM with GFP (Fig. 2C, left panel). In the patch expressing CRPmZF, GFP fluorescence was slightly stronger than in patches coexpressing GUS and GFP but significantly lower than in patches expressing wild-type pvmCRP (Fig. 2C, right panel). These observations were confirmed by Northern blot analysis of GFP mRNA (Fig. 2D). In addition, the levels of GFP-specific siRNAs coincided with the suppression levels of GFP fluorescence and GFP mRNA (Fig. 2E). Therefore, these results demonstrate that both the basic motif and the zinc finger motif are involved in the suppressor activity of pvmCRP.
Both TGBp1 and CRP suppress the spread of silencing.
In plants, after the establishment of local RNA silencing, a mobile silencing signal spreads between cells and then enters the phloem to induce the systemic silencing of homologous sequences in upper leaves (35, 48, 51), and some viral proteins are known to prevent the spread of silencing (19, 49, 52). Therefore, we investigated whether TGBp1 and CRP of PVM inhibit the spread of silencing.
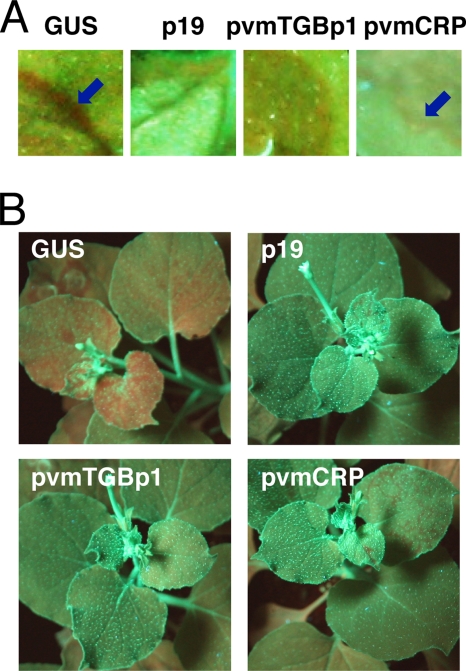
When the GFP and GUS genes were introduced into the GFP-expressing N. benthamiana line 16c plants (7) by agroinfiltration, the short-range movement of GFP silencing was observed as a red border at the edge of the infiltrated patch (Fig. 3A), and the GFP fluorescence disappeared along the veins of upper noninfiltrated leaves at 13 dpi, indicating induction of systemic silencing (Fig. 3B). The induction of silencing in upper leaves was observed in 81.8% of the plants (18 of 22 infiltrated plants) (Table 1). In contrast, no red border was observed at the edge of the patch infiltrated with the GFP and p19 genes (Fig. 3A) and only 7.1% (two of 28) of the plants showed GFP silencing in their upper leaves (Fig. 3B; Table 1), indicating that p19 blocked both the short-range and systemic spread of silencing. Similar to p19, pvmTGBp1-introduced plants showed no obvious red border, although local RNA silencing was induced and the GFP fluorescence was reduced (Fig. 3A). The efficiency of systemic silencing induction was also significantly lower (35.7%) than in GUS-introduced plants (Fig. 3B; Table 1). In pvmCRP-introduced plants, a rim of GFP silenced cells was faintly visible around the infiltrated patch, and the red vein phenotype was partially observed in upper leaves; however, the extent of the border and the efficiency of systemic silencing induction were significantly lower (28.6%) than in GUS-introduced plants (Fig. 3B; Table 1). Collectively, these results demonstrate that both TGBp1 and CRP encoded by PVM are able to inhibit the spread of silencing.
Fig. 3.
Suppressor activity of pvmTGBp1 and pvmCRP on the spread of RNA silencing. (A and B) GFP fluorescence images of infiltrated (A) or systemic (B) leaves of N. benthamiana line 16c infiltrated with Agrobacterium mixtures containing a vector expressing GFP and either GUS, p19, pvmTGBp1, or pvmCRP are shown. Photographs were taken under UV light at 13 dpi. The short-range movement of GFP silencing is represented by the red ring on the border of the patch (blue arrow). Note that the pvmTGBp1-introduced patch shows the disappearance of GFP fluorescence because of local RNA silencing induction, which is unrelated to the red border. Also, the pvmCRP-introduced patch shows GFP fluorescence due to suppression of local RNA silencing, whereas the red border is faintly visible.
Table 1.
Efficiency of systemic silencing induction in the N. benthamiana line 16c plants shown in Fig. 3
| Treatment | No. of plants infiltrated | No. of plants systemically silenced | Efficiency of systemic silencing induction (%) |
|---|---|---|---|
| GFP + GUS | 22 | 18 | 81.8 |
| GFP + p19 | 28 | 2 | 7.1 |
| GFP + pvmTGBp1 | 28 | 10 | 35.7 |
| GFP + pvmCRP | 28 | 8 | 28.6 |
TGBp1 and CRP of PVM enhance the pathogenicity of PVX.
Numerous studies have shown that viral silencing suppressors increase the severity of viral symptoms when they are expressed from a heterologous viral expression vector (7, 11). To examine the effect on viral pathogenicity, we constructed potato virus X (PVX) recombinant viruses expressing either pvmTGBp1 (PVX-pvmTGBp1) or pvmCRP (PVX-pvmCRP) and recombinants expressing GFP or p19 as controls (Fig. 4A). When the infectious PVX-GFP cDNA clone was mechanically inoculated into N. benthamiana, the plants showed faint chlorosis in the upper leaves at 12 dpi, which evolved into a mild mosaic (Fig. 4B). In contrast, PVX-pvmTGBp1 caused visible systemic symptoms more rapidly, consisting of a clear mosaic in the upper leaves at 7 dpi, followed by strongly exacerbated symptoms with vein yellowing (Fig. 4B). The plants inoculated with PVX-pvmCRP showed severe deformities, consisting of leaf crinkle, rugose, curl, mosaic, and petiole twisting in the upper younger leaves at 7 dpi, and then developed necrotic spots and whole-plant stunting by 20 dpi (Fig. 4B; data not shown). PVX-p19 induced necrosis in petioles, veins, and mesophyll tissue of the upper leaves at 5 dpi and eventually resulted in the death of the entire plant (Fig. 4B).
We then examined accumulation of these recombinant viruses by qRT-PCR. In plants inoculated with PVX-pvmTGBp1, PVX-pvmCRP, or PVX-p19, marked increases in viral RNA accumulation were observed compared to PVX-GFP in both inoculated and upper leaves (Fig. 4C). The RNA accumulation levels of these suppressor-expressing recombinant viruses were about 20- to 25-fold higher than that of PVX-GFP in inoculated leaves. Furthermore, the accumulation level increased ∼60-fold in leaves systemically infected with PVX-pvmCRP, whereas inoculation with PVX-pvmTGBp1 resulted in a smaller but substantial increase (∼5-fold) of viral RNA in the upper leaves compared to PVX-GFP. Taken together, these results demonstrate that the TGBp1 and CRP of PVM enhance virulence of the heterologous virus PVX, which is associated with enhanced virus accumulation in both the inoculated and systemically infected leaves.
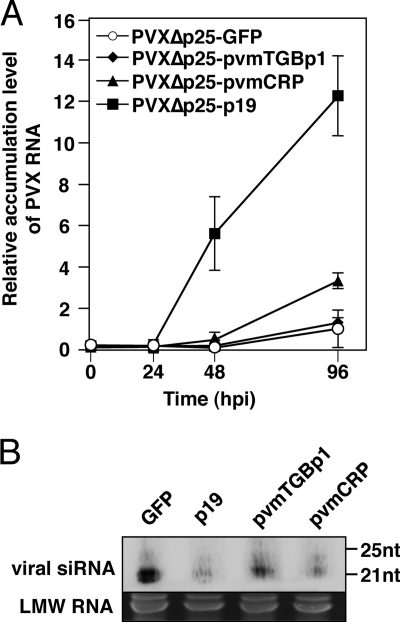
CRP increases viral accumulation in protoplasts.
We have shown that pvmTGBp1 and pvmCRP enhance virus accumulation even in inoculated leaves of N. benthamiana plants. These findings suggest that these suppressor proteins act either at the cellular level by enhancing viral replication or viral RNA stability or play a role in the cell-to-cell movement of the virus. To further elucidate the role of these proteins in the viral infection cycle, we first investigated their effect on virus accumulation in protoplasts. For an exact evaluation of the activity of each protein, we used the PVX mutant that lacks its own suppressor, TGBp1 (p25). When the control virus PVXΔp25-GFP was inoculated into N. tabacum BY-2 protoplasts, accumulation of viral RNA was readily detected by 96 h postinoculation (hpi), suggesting the occurrence of viral replication (Fig. 5A). In protoplasts inoculated with PVXΔp25-pvmTGBp1, no significant difference was observed in viral RNA accumulation relative to the PVXΔp25-GFP control during the experimental period. Conversely, PVXΔp25-pvmCRP accumulated at significantly higher levels (∼3-fold higher) than PVXΔp25-GFP at 96 hpi. The levels of PVXΔp25-p19 RNA increased more rapidly, such that the level of PVX-p19 RNA was higher than those of PVXΔp25-GFP or PVXΔp25-pvmTGBp1 as early as 48 hpi and reached ∼12-fold higher at 96 hpi. Inversely correlated with the levels of viral RNA, the levels of virus-specific siRNAs were lower in protoplasts inoculated with PVXΔp25-p19 and PVXΔp25-pvmCRP than in those inoculated with PVXΔp25-GFP and PVXΔp25-pvmTGBp1 (Fig. 5B). Therefore, these data indicate that CRP of PVM increases the accumulation of viral RNA in its replication step by inhibiting RNA silencing, whereas TGBp1 does not affect viral accumulation at the protoplast level.
Fig. 5.
Effects of pvmTGBp1 and pvmCRP on viral replication. (A) PVXΔp25-GFP, PVXΔp25-pvmTGBp1, PVXΔp25-pvmCRP, or PVXΔp25-p19 was inoculated into N. tabacum BY-2 protoplasts, and qRT-PCR analysis of PVX coat protein (CP) RNA was performed with total RNA extracted from the protoplasts at 0, 24, 48, and 96 hpi. The accumulation levels relative to PVX-GFP at 96 hpi were calculated as described in Fig. 4C. The error bars represent the standard deviations of two independent experiments. (B) Northern blot analysis of viral siRNAs extracted from the protoplasts at 96 hpi as shown in panel A. Ethidium bromide-stained low-molecular-weight RNA (LMW RNA) is shown below each panel as a loading control. Molecular size markers are indicated on the right.
TGBp1 and CRP of PVM promote viral cell-to-cell movement by inhibiting RNA silencing.
Next, we investigated the effects of pvmTGBp1 and pvmCRP on viral cell-to-cell movement. To this end, a GFP-tagged virus (PVX-GFP, Fig. 6A) was cointroduced into N. benthamiana with GUS (negative control), p19, pvmTGBp1, or pvmCRP, and viral spread in the inoculated leaves was monitored with GFP fluorescence imaging. Note that in order to avoid RDR6-dependent transgene silencing triggered by agroinfiltration, PVX-GFP was inoculated into the leaves mechanically. As shown in Fig. 6B, green fluorescent spots were observed in all leaves cointroduced with PVX-GFP and each expression construct at 6 dpi, indicating viral propagation and subsequent movement from cell to cell. However, fluorescence microscopy revealed that the sizes of the fluorescent spots differed depending on the construct cointroduced. The mean size of 20 randomly selected spots in leaves expressing p19, pvmTGBp1, or pvmCRP was more than 3-fold greater than that of leaves expressing GUS, suggesting that these suppressor proteins promote the cell-to-cell movement of PVX-GFP (Fig. 6C).
The promotion of movement of PVX-GFP by p19, pvmTGBp1, and pvmCRP can be interpreted as supporting the idea that silencing suppression has a role in viral cell-to-cell movement, as shown in previous studies (5, 37). To test this, we developed a viral movement complementation assay (VMCA) using mutant variants of p25 encoded by PVX (5). p25 functions as both an MP and an RNA silencing suppressor (46), and the p25T117A mutant, which contains a single amino acid substitution (Tyr117 to Ala; Fig. 6A), lost the ability to suppress RNA silencing and to complement the movement of p25-deficient PVX. However, it can complement the movement of p25-deficient PVX if a silencing suppressor such as TBSV p19 or PVY HC-Pro is additionally expressed, indicating that this mutant lacks only the suppressor activity required for cell-to-cell movement (5). Thus, a protein with a suppressor ability required for movement can complement the movement of p25-deficient PVX-GFP when it is expressed together with p25T117A. We performed a VMCA utilizing p25T117A for p19, pvmTGBp1, and pvmCRP. When GFP-tagged and TGBp1-deficient PVX (PVXΔp25-GFP, Fig. 6A) were cointroduced with p25T117A and GUS into N. benthamiana leaves by agroinfiltration, the virus was restricted to a single epidermal cell. However, expression of p19 instead of GUS resulted in the spread of PVXΔp25-GFP (Fig. 6D, top panels), as described previously (5). In leaves expressing p25T117A and pvmTGBp1, PVXΔp25-GFP spread to multiple cells, although the GFP fluorescence in each cell was slightly lower than that of p19-expressing leaves (Fig. 6D, top panels). In leaves expressing p25T117A and pvmCRP, the virus also moved efficiently, as shown by the cluster of bright fluorescent cells (Fig. 6D, top panels). In a control experiment, we used another p25 mutant, p25A104V (substitution of Ala104 with Val; Fig. 6A), which is defective in viral movement but retains the ability to suppress RNA silencing. As described previously (5), PVXΔp25-GFP did not spread in leaves expressing p25A104V and p19, similar to the negative control GUS (Fig. 6D, middle panels). PVXΔp25-GFP was also restricted to a single epidermal cell in leaves expressing either pvmTGBp1 or pvmCRP with p25A104V (Fig. 6D, middle panels). These results show that not just any suppressor can complement the inability of p25A104V to move PVX from cell to cell. As an additional control experiment, PVXΔp25-GFP was cointroduced with GUS (negative control), p19, pvmTGBp1, or pvmCRP (i.e., VMCA without any p25 mutant). PVXΔp25-GFP was restricted to a single epidermal cell in all tested leaves even over a 7-day period (Fig. 6D, bottom panels). We confirmed that the cell-to-cell movement of PVXΔp25-GFP was restored by complementary expression of p25 at 4 dpi (data not shown). Therefore, these data demonstrate that p19, pvmCRP, or even pvmTGBp1 (which is homologous to p25) cannot complement the entire function of p25. In conclusion, these results suggest that in addition to p19, both pvmTGBp1 and pvmCRP assist viral cell-to-cell movement by inhibiting RNA silencing.
DISCUSSION
In this study, we analyzed RNA silencing suppressor encoded by PVM and found that PVM employs a “dual strategy” to block host antiviral silencing. In assays using a GFP reporter gene, although pvmTGBp1 showed suppressor activity only on the spread of silencing, pvmCRP suppressed both local and the spread of silencing. Furthermore, by analyzing the effect of two suppressors on viral infection, we demonstrated that pvmTGBp1 only facilitated viral movement, whereas pvmCRP increased the accumulation of viral RNA at the single-cell level and facilitated viral movement by suppressing RNA silencing. These results demonstrate that PVM prevents host antiviral silencing at two steps during infection by utilizing two mechanistically distinct suppressors. At the same time, our results suggest that suppressors of local and the spread of silencing encoded by plant viruses may have roles to assist viral replication and movement, respectively, by suppressing host antiviral silencing. Furthermore, this report is the first of the RNA silencing suppressor in the genus Carlavirus.
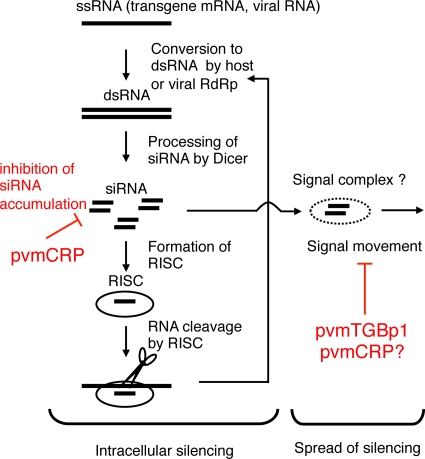
RNA silencing plays a key role in the plant defense response against foreign nucleic acids such as viruses and transgenes (8, 12, 45). As depicted in Fig. 7, the presence of viruses or transgenes in host cells evokes the RNA silencing machinery and leads to the cleavage of the cognate RNA guided by siRNAs. Some siRNAs generated through the silencing pathway in the initial cells spread to neighboring cells and systemically over long distances to mediate RNA silencing (14, 36). This RNA silencing movement probably protects surrounding naive cells against the invading nucleic acids (15, 34, 39).
Fig. 7.
Model of RNA silencing directed against exogenous RNA species and its suppression by viral suppressor proteins. pvmCRP suppresses intracellular silencing and inhibits the accumulation of siRNA. These suppressors also interfere with the spread of silencing, either due to the inhibition of signal siRNA accumulation at the single cell level or due to interference with the signal movement/recipient step in addition to intracellular silencing. pvmTGBp1 prevents the spread of silencing by specifically targeting the signal movement/recipient step.
In the present study, we found that pvmCRP inhibited local RNA silencing induced in the agroinfiltrated area (Fig. 1) and also increased the accumulation of viral RNA at the single-cell level (Fig. 5A). These results show that a suppressor of local RNA silencing can inhibit RNA silencing at the single-cell level (i.e., intracellular silencing) and thus reveal that pvmCRP is a suppressor of intracellular silencing (Fig. 7). They also suggest that pvmCRP is effective primarily at the viral replication step because it prevents viral replication intermediates or their progeny from being degraded by RNA silencing. Because site-directed mutagenesis of the two nucleic acid binding motifs (basic motif, zinc-finger motif) compromised its suppressor activity without affecting its accumulation (Fig. 2), pvmCRP might suppress intracellular silencing by binding to dsRNAs, which is a common mechanism of viral suppressors (26, 33). pvmCRP also inhibited the spread of GFP silencing (Fig. 3) and, correspondingly, promoted viral movement (Fig. 6). One possible explanation for this is that the suppression of intracellular silencing inhibits signal siRNA accumulation and thereby prevents silencing from spreading (Fig. 7), which means a lack of antiviral response in distant tissues. In fact, the level of GFP-specific siRNAs and viral siRNAs in patches and protoplasts expressing pvmCRP was significantly reduced (Fig. 1D, 2D, and 5B). An alternative explanation is that pvmCRP independently targets the two distinct RNA silencing pathways, intracellular silencing and the spread of silencing (Fig. 7), although the effect on the latter is rather weak (Fig. 3A).
In contrast to pvmCRP, pvmTGBp1 did not inhibit local RNA silencing in the GFP transient coexpression assay (Fig. 1). Nevertheless, pvmTGBp1 did suppress the spread of GFP silencing (Fig. 3), which means that pvmTGBp1 specifically targets a step related to the spread of silencing, such as formation of the signal complex, signal movement, or signal reception (Fig. 7). Related to this, although pvmTGBp1 did not affect viral accumulation at the protoplast level (Fig. 5A), it promoted cell-to-cell movement of PVX-GFP when expressed in trans (Fig. 6B and C). pvmTGBp1 did not complement the entire function of p25, but instead only facilitated the movement of PVX-GFPΔp25 when expressed together with p25T117A (Fig. 6D), indicating that pvmTGBp1 most likely promotes viral movement by inhibiting the spread of RNA silencing. Since pvmTGBp1 blocked the short-range movement of GFP silencing (Fig. 3A) and its effect on systemic silencing was rather modest (an ∼2-fold decrease of plants exhibiting systemic silencing; Table 1), pvmTGBp1 might act as a silencing suppressor specifically on the cell-to-cell silencing movement step. This also concurs with the observations that the increase in viral RNA of PVX-pvmTGBp1 in inoculated leaves was as strong as those of PVX-pvmCRP and PVX-p19 but that the effect of pvmTGBp1 was much more modest in systemic leaves (Fig. 4C). Further studies are required to elucidate the mechanism of spread-specific silencing suppression by pvmTGBp1 and its correlation with viral movement.
From the present study, we conclude that PVM establishes a successful infection by inhibiting antiviral silencing at both the replication and the movement steps using two distinct suppressors. Inhibiting antiviral silencing at multiple infection steps with distinct suppressors may be a general strategy used by diverse plant viruses, although there is still limited knowledge of this. Currently, multiple suppressors that have different functions have been reported only in members of the Closteroviridae family. For instance, CP, p20, and p23 of citrus tristeza virus (CTV; genus Closterovirus) have been shown in reporter-based assays to be silencing suppressors that inhibit distinct steps of the silencing pathway (29). These suppressors encoded by closteroviruses could facilitate different viral infection steps, as shown in the present study. Moreover, it is possible that many potential silencing suppressors encoded by plant viruses may escape identification as silencing suppressors. Although the classical GFP transient-coexpression assay is convenient and has been widely used to identify silencing suppressors, this assay is unable to identify spread-specific silencing suppressors such as pvmTGBp1. Accordingly, a VMCA such as the p25 mutant assay performed in the present study or the turnip crinkle virus (TCV) vector-based assay (which utilizes recombinant TCV expressing GFP instead of CP [37]) will help to identify such suppressors.
Considering that the onset of RNA silencing in initially infected cells lags behind viral replication, the major antiviral impact of RNA silencing might require the spread of silencing. In fact, “recovery” plants initially infected with viruses subsequently develop new leaves completely lacking disease symptoms and viral accumulation. Thus, inhibiting the spread of host antiviral silencing is likely to be essential for successful viral infection. MP (p50) of apple chlorotic leaf spot virus (ACLSV) functions as a suppressor of systemic silencing, but ACLSV does not encode any protein that can suppress intracellular silencing (52). In addition, the suppressor activity of p25 is dispensable for PVX replication, but is essential for its cell-to-cell movement (5, 46, 47). In the present study, we demonstrated that PVM possesses as many as two silencing suppressors that can promote viral cell-to-cell movement (Fig. 6). This finding further supports the importance of inhibiting the spread of silencing.
Some members of the family Flexiviridae encode CRPs in their genome (genera Carlavirus, Allexivirus, Mandarivirus, Vitivirus, and some members of Torichovirus). Previous reports showed that CRP of chrysanthemum virus B (CVB; genus Carlavirus) binds to nucleic acids but has no suppressor activity (30, 31). These observations can be explained by a previous study showing that suppressor activity differs even among homologous proteins encoded by members of the same genus (42). It is possible that although the original host of CVB is species in the Asteraceae, PVM originally infected Solanaceae plants, so pvmCRP might show its suppressor activity in N. benthamiana (Solanaceae), but CRP of CVB might not. In addition to pvmCRP, CRP of grape virus A (genus Vitivirus) has been shown to suppress local RNA silencing. Furthermore, in genus members outside the Flexiviridae family, CRP functions as a silencing suppressor. These data suggest that the suppressor activity of CRP is conserved across a wide variety of plant viruses.
The genera Carlavirus and Potexvirus both belong to the family Flexiviridae, and the members of these genera share quite similar genomic organization, except that carlavirus has an extra gene encoding CRP at the 3′ proximal end of its genome. Phylogenetic analysis of the flexiviral replicases suggests that carlavirus and potexvirus diverged from a common ancestor that possessed (at least) a replicase, triple gene block proteins (TGBps), and CP, whereas CRP is considered to have been acquired independently by carlavirus (32). The fact that CRPs encoded by different groups of plant RNA viruses share no significant sequence similarity and that they are related to different families of nucleic acid-binding proteins originating from plants, animal viruses, or humans also supports this idea (25). Our data show that pvmCRP suppresses both intracellular silencing and the spread of silencing, but pvmTGBp1 suppresses only the spread of silencing. Notably, TGBp1 encoded by potexviruses is able to suppress both intracellular silencing and silencing spread (5, 49). Thus, it is speculated that the ancestor virus of potexvirus and carlavirus possessed a TGBp1 with the ability to suppress both intracellular silencing and silencing spread such as potexvirus TGBp1. However, because of the acquisition of a CRP with strong suppressor activity, carlavirus TGBp1 gradually lost part of its suppression ability, resulting in the loss of its ability to suppress intracellular silencing. Alternatively, TGBp1 of the common ancestor did not possess the ability to suppress intracellular silencing, and later the potexvirus TGBp1 might have acquired it instead of CRP. Notably, viruses in which the CRP functions as a silencing suppressor have not been reported to possess another suppressor protein. Although barley stripe mosaic virus (genus Hordeivirus) and peanut clump virus (genus Pecluvirus) have both TGBp1 and CRP (similar to carlavirus), only CRP has been shown to suppress RNA silencing (13, 53). It is possible that the acquisition of CRP causes the loss of all or part (against intracellular silencing) of the silencing suppression ability.
Plant viruses have evolved diverse strategies to counteract host antiviral silencing. Our study showed that PVM inhibits silencing at different steps of viral infection using multiple suppressors. In contrast, potexvirus utilizes a single suppressor in multiple infection steps. Moreover, some viruses (such as ACLSV) only encode a suppressor that specifically inhibits the spread of RNA silencing. The strategy for silencing suppression could depend on a trade-off between factors such as genome size, replication efficiency, and the manner of movement. Further analysis of silencing suppressors and other viral factors will be required to uncover the sophisticated survival strategies of plant viruses, organisms with limited genetic material.
ACKNOWLEDGMENTS
We thank D. C. Baulcombe (University of Cambridge, Cambridge, United Kingdom) for providing the cDNA clone of PVX, the plasmid pBin19, and the Nicotiana benthamiana line 16c.
This study was supported by a grant-in-aid from the Japan Society for the Promotion of Science (grant 21-5744) and a grant from the Program for Promotion of Basic Research Activities for Innovative Bioscience.
Footnotes
Published ahead of print on 13 July 2011.
REFERENCES
- 1. Alvarado V., Scholthof H. B. 2009. Plant responses against invasive nucleic acids: RNA silencing and its suppression by plant viral pathogens. Semin. Cell Dev. Biol. 20:1032–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Azevedo J., et al. 2010. Argonaute quenching and global changes in Dicer homeostasis caused by a pathogen-encoded GW repeat protein. Genes Dev. 24:904–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baulcombe D. 2004. RNA silencing in plants. Nature 431:356–363 [DOI] [PubMed] [Google Scholar]
- 4. Baumberger N., Tsai C. H., Lie M., Havecker E., Baulcombe D. C. 2007. The polerovirus silencing suppressor P0 targets Argonaute proteins for degradation. Curr. Biol. 17:1609–1614 [DOI] [PubMed] [Google Scholar]
- 5. Bayne E. H., Rakitina D. V., Morozov S. Y., Baulcombe D. C. 2005. Cell-to-cell movement of potato potexvirus X is dependent on suppression of RNA silencing. Plant J. 44:471–482 [DOI] [PubMed] [Google Scholar]
- 6. Bortolamiol D., Pazhouhandeh M., Marrocco K., Genschik P., Ziegler-Graff V. 2007. The polerovirus F box protein P0 targets Argonaute1 to suppress RNA silencing. Curr. Biol. 17:1615–1621 [DOI] [PubMed] [Google Scholar]
- 7. Brigneti G., et al. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17:6739–6746 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8. Brodersen P., Voinnet O. 2006. The diversity of RNA silencing pathways in plants. Trends Genet. 22:268–280 [DOI] [PubMed] [Google Scholar]
- 9. Cronin S., Verchot J., Haldeman-Cahill R., Schaad M. C., Carrington J. C. 1995. Long-distance movement factor: a transport function of the potyvirus helper component proteinase. Plant Cell 7:549–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cruz S. S., et al. 1996. Assembly and movement of a plant virus carrying a green fluorescent protein overcoat. Proc. Natl. Acad. Sci. U. S. A. 93:6286–6290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Csorba T., Pantaleo V., Burgyan J. 2009. RNA silencing: an antiviral mechanism. Adv. Virus Res. 75:35–71 [DOI] [PubMed] [Google Scholar]
- 12. Ding S. W., Voinnet O. 2007. Antiviral immunity directed by small RNAs. Cell 130:413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dunoyer P., et al. 2002. Identification, subcellular localization and some properties of a cysteine-rich suppressor of gene silencing encoded by peanut clump virus. Plant J. 29:555–567 [DOI] [PubMed] [Google Scholar]
- 14. Dunoyer P., et al. 2010. Small RNA duplexes function as mobile silencing signals between plant cells. Science 328:912–916 [DOI] [PubMed] [Google Scholar]
- 15. Fagard M., Vaucheret H. 2000. Systemic silencing signal(s). Plant Mol. Biol. 43:285–293 [DOI] [PubMed] [Google Scholar]
- 16. Ghazala W., Waltermann A., Pilot R., Winter S., Varrelmann M. 2008. Functional characterization and subcellular localization of the 16K cysteine-rich suppressor of gene silencing protein of tobacco rattle virus. J. Gen. Virol. 89:1748–1758 [DOI] [PubMed] [Google Scholar]
- 17. Giner A., Lakatos L., Garcia-Chapa M., Lopez-Moya J. J., Burgyan J. 2010. Viral protein inhibits RISC activity by Argonaute binding through conserved WG/GW motifs. PLoS Pathog. 6:e1000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gramstat A., Courtpozanis A., Rohde W. 1990. The 12-kDa protein of potato virus M displays properties of a nucleic acid-binding regulatory protein. FEBS Lett. 276:34–38 [DOI] [PubMed] [Google Scholar]
- 19. Guo H. S., Ding S. W. 2002. A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J. 21:398–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hofmann C., Sambade A., Heinlein M. 2007. Plasmodesmata and intercellular transport of viral RNA. Biochem. Soc. Trans. 35:142–145 [DOI] [PubMed] [Google Scholar]
- 21. Johansen L. K., Carrington J. C. 2001. Silencing on the spot Induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol. 126:930–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kalantidis K., Schumacher H. T., Alexiadis T., Helm J. M. 2008. RNA silencing movement in plants. Biol. Cell 100:13–26 [DOI] [PubMed] [Google Scholar]
- 23. Kasschau K. D., Carrington J. C. 2001. Long-distance movement and replication maintenance functions correlate with silencing suppression activity of potyviral HC-Pro. Virology 285:71–81 [DOI] [PubMed] [Google Scholar]
- 24. Kasschau K. D., Cronin S., Carrington J. C. 1997. Genome amplification and long-distance movement functions associated with the central domain of tobacco etch potyvirus helper component-proteinase. Virology 228:251–262 [DOI] [PubMed] [Google Scholar]
- 25. Koonin E. V., Boyko V. P., Dolja V. V. 1991. Small cysteine-rich proteins of different groups of plant RNA viruses are related to different families of nucleic acid-binding proteins. Virology 181:395–398 [DOI] [PubMed] [Google Scholar]
- 26. Lakatos L., et al. 2006. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 25:2768–2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lakatos L., Szittya G., Silhavy D., Burgyan J. 2004. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 23:876–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li F., Ding S. W. 2006. Virus counterdefense: diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol. 60:503–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lu R., et al. 2004. Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proc. Natl. Acad. Sci. U. S. A. 101:15742–15747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lukhovitskaia N. I., Solov'eva A. G., Koshkina T. E., Zavriev S. K., Morozov S. 2005. Interaction of cysteine-rich protein of Carlavirus with plant defense system. Mol. Biol. (Moscow) 39:896–904 [PubMed] [Google Scholar]
- 31. Lukhovitskaya N. I., et al. 2009. Role of the zinc-finger and basic motifs of chrysanthemum virus B p12 protein in nucleic acid binding, protein localization and induction of a hypersensitive response upon expression from a viral vector. J. Gen. Virol. 90:723–733 [DOI] [PubMed] [Google Scholar]
- 32. Martelli G. P., Adams M. J., Kreuze J. F., Dolja V. V. 2007. Family Flexiviridae: a case study in virion and genome plasticity. Annu. Rev. Phytopathol. 45:73–100 [DOI] [PubMed] [Google Scholar]
- 33. Merai Z., et al. 2006. Double-stranded RNA binding may be a general plant RNA viral strategy to suppress RNA silencing. J. Virol. 80:5747–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mlotshwa S., Pruss G. J., Vance V. 2008. Small RNAs in viral infection and host defense. Trends Plant Sci. 13:375–382 [DOI] [PubMed] [Google Scholar]
- 35. Mlotshwa S., et al. 2002. RNA silencing and the mobile silencing signal. Plant Cell 14(Suppl.):S289–S301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Molnar A., et al. 2010. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328:872–875 [DOI] [PubMed] [Google Scholar]
- 37. Powers J. G., et al. 2008. A versatile assay for the identification of RNA silencing suppressors based on complementation of viral movement. Mol. Plant-Microbe Interact. 21:879–890 [DOI] [PubMed] [Google Scholar]
- 38. Pruss G., Ge X., Shi X. M., Carrington J. C., Bowman Vance V. 1997. Plant viral synergism: the potyviral genome encodes a broad-range pathogenicity enhancer that transactivates replication of heterologous viruses. Plant Cell 9:859–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ratcliff F., Harrison B. D., Baulcombe D. C. 1997. A similarity between viral defense and gene silencing in plants. Science 276:1558–1560 [DOI] [PubMed] [Google Scholar]
- 40. Scholthof H. B., Scholthof K. B., Jackson A. O. 1995. Identification of tomato bushy stunt virus host-specific symptom determinants by expression of individual genes from a potato virus X vector. Plant Cell 7:1157–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Scholthof H. B., Scholthof K. B., Kikkert M., Jackson A. O. 1995. Tomato bushy stunt virus spread is regulated by two nested genes that function in cell-to-cell movement and host-dependent systemic invasion. Virology 213:425–438 [DOI] [PubMed] [Google Scholar]
- 42. Senshu H., et al. 2009. Variability in the level of RNA silencing suppression caused by triple gene block protein 1 (TGBp1) from various potexviruses during infection. J. Gen. Virol. 90:1014–1024 [DOI] [PubMed] [Google Scholar]
- 43. Silhavy D., Burgyan J. 2004. Effects and side-effects of viral RNA silencing suppressors on short RNAs. Trends Plant Sci. 9:76–83 [DOI] [PubMed] [Google Scholar]
- 44. Silhavy D., et al. 2002. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 21:3070–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vaucheret H. 2006. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 20:759–771 [DOI] [PubMed] [Google Scholar]
- 46. Verchot-Lubicz J. 2005. A new cell-to-cell transport model for potexviruses. Mol. Plant-Microbe Interact. 18:283–290 [DOI] [PubMed] [Google Scholar]
- 47. Verchot-Lubicz J., Ye C. M., Bamunusinghe D. 2007. Molecular biology of potexviruses: recent advances. J. Gen. Virol. 88:1643–1655 [DOI] [PubMed] [Google Scholar]
- 48. Voinnet O., Baulcombe D. C. 1997. Systemic signaling in gene silencing. Nature 389:553. [DOI] [PubMed] [Google Scholar]
- 49. Voinnet O., Lederer C., Baulcombe D. C. 2000. A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103:157–167 [DOI] [PubMed] [Google Scholar]
- 50. Voinnet O., Pinto Y. M., Baulcombe D. C. 1999. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. U. S. A. 96:14147–14152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Voinnet O., Vain P., Angell S., Baulcombe D. C. 1998. Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell 95:177–187 [DOI] [PubMed] [Google Scholar]
- 52. Yaegashi H., et al. 2007. Apple chlorotic leaf spot virus 50-kDa movement protein acts as a suppressor of systemic silencing without interfering with local silencing in Nicotiana benthamiana. J. Gen. Virol. 88:316–324 [DOI] [PubMed] [Google Scholar]
- 53. Yelina N. E., Savenkov E. I., Solovyev A. G., Morozov S. Y., Valkonen J. P. 2002. Long-distance movement, virulence, and RNA silencing suppression controlled by a single protein in hordei- and potyviruses: complementary functions between virus families. J. Virol. 76:12981–12991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yoshii A., et al. 2008. NTH201, a novel class II KNOTTED1-like protein, facilitates the cell-to-cell movement of Tobacco mosaic virus in tobacco. Mol. Plant-Microbe Interact. 21:586–596 [DOI] [PubMed] [Google Scholar]
- 55. Zavriev S. K., Kanyuka K. V., Levay K. E. 1991. The genome organization of potato virus M RNA. J. Gen. Virol. 72:9–14 [DOI] [PubMed] [Google Scholar]
- 56. Zhang X., et al. 2006. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 20:3255–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhou Z., et al. 2006. Identification of an RNA-silencing suppressor in the genome of Grapevine virus A. J. Gen. Virol. 87:2387–2395 [DOI] [PubMed] [Google Scholar]