Abstract
Japanese encephalitis virus (JEV), a mosquito-borne zoonotic pathogen, is one of the major causes of viral encephalitis worldwide. Previous phylogenetic studies based on the envelope protein indicated that there are four genotypes, and surveillance data suggest that genotype I is gradually replacing genotype III as the dominant strain. Here we report an evolutionary analysis based on 98 full-length genome sequences of JEV, including 67 new samples isolated from humans, pigs, mosquitoes, midges. and bats in affected areas. To investigate the relationships between the genotypes and the significance of genotype I in recent epidemics, we estimated evolutionary rates, ages of common ancestors, and population demographics. Our results indicate that the genotypes diverged in the order IV, III, II, and I and that the genetic diversity of genotype III has decreased rapidly while that of genotype I has increased gradually, consistent with its emergence as the dominant genotype.
INTRODUCTION
Japanese encephalitis virus (JEV), a member of the genus Flavivirus in the family Flaviviridae, is a major cause of viral encephalitis and is endemic in several regions of Asia and the Pacific (4, 13), causing an estimated 35,000 to 50,000 infections and 10,000 to 15,000 deaths annually (4, 13, 27). Fifty percent of survivors suffer from lingering neurological effects (7, 27, 30). Japanese encephalitis (JE) was first reported in Japan in 1924, and JE cases were subsequently reported in many other Asian countries (4, 6, 7, 13, 22, 27, 30). JE was first reported in Australia in 1995 (8, 9, 31). Thus, JE has become a major cause of mosquito-transmitted viral encephalitis on two continents (15, 16, 25).
JEV, the pathogen of JE, has a genome comprising a positive-sense, single-stranded RNA molecule of approximately 11 kb that is capped at the 5′ end and is not polyadenylated at the 3′ end. It carries a single open reading frame (ORF) encoding a polyprotein that is processed into three structural (C, M, and E) and seven nonstructural (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) proteins, flanked by 5′ and 3′ nontranslated regions (NTRs) (13).
Until the latter part of the 20th century, studies indicated that the predominant genotype was genotype III. Since then, there have been multiple reports of genotype I displacing genotype III in many regions (12, 18, 19, 20, 24, 32, 34, 35), and in many areas genotype I is now recognized as the dominant strain.
As part of a national encephalitis surveillance program, we collected samples from a variety of vectors (mosquitoes and midges), host animals (bats and pigs), and patients with cases of encephalitis in areas where the disease is epidemic, and we isolated viruses from a selection of the JEV-positive samples and sequenced their full genomes. We combined these sequences with other, publicly available full-length genome sequences for a final set of 98 genome sequences. With this set we performed the first detailed evolutionary analysis of JEV based on full-length genome sequences and investigated the epidemiology of genotype I relative to that of genotype III.
MATERIALS AND METHODS
Sample collection and genome sequencing.
As part of a national encephalitis surveillance program, samples were collected from around China. Sixty-seven of these samples were selected for isolation, identification, and full-genome sequencing.
The samples were amplified as described elsewhere (12, 17, 28, 32, 33, 34, 35). Briefly, viral RNA was isolated using the Viral RNA Mini kit (Qiagen, Hilden, Germany). First-strand cDNA was synthesized using the Ready-To-Go kit (Amersham Pharmacia Biotech, Uppsala, Sweden). Whole genomes were amplified using primer sets described previously (32, 33); PCR products were recovered using a gel purification kit (Qiagen, Valencia, CA); and sequences were determined using an ABI Prism 3730 sequence analyzer (Applied Biosystems, Foster City, CA). The SeqMan program in the DNAStar software package was used to splice, edit, and correct sequence fragments. A subset of the complete set of sequences that was representative of the sampled geographical regions, hosts, and vectors was submitted to GenBank (Table 1).
TABLE 1.
JEV isolates analyzed in this study
| Strain | Date | Country | Hosta | GenBank accession no. b |
|---|---|---|---|---|
| 47 | 1950s | China, Heilongjiang | CSF | JF706269* |
| 14178 | 2001 | India | — | EF623987 |
| 57434 | 2005 | India | — | EF623988 |
| 04940-4 | 2002 | India | — | EF623989 |
| B58 | 1989 | China, Yunnan | Bat | FJ185036 |
| Beijing-1 | 1949 | China | Human brain | L48961 |
| BL06-50 | 2006 | China, Guangxi | Culex tritaeniorhynchus | JF706270* |
| BL06-54 | 2006 | China, Guangxi | Culex tritaeniorhynchus | JF706271* |
| CBH | 1954 | China, Fujian | CSF | JN381860* |
| CH-13 | 1957 | China, Sichuan | CSF | JN381870* |
| CH1392 | 1990 | Taiwan | Culex tritaeniorhynchus | AF254452 |
| CTS | 1955 | China, Fujian | CSF | GQ429184 |
| CZX | 1954 | China, Fujian | CSF | JN381865* |
| DH107 | 1989 | China, Yunnan | Aedes lineatopennis | JN381873* |
| DL04-29 | 2004 | China, Yunnan | Culex theileri | JF706272* |
| DL04-45 | 2004 | China, Yunnan | Armigeres subalbatus and Mansonia uniformis | JN381854* |
| Fj02-29 | 2002 | China, Fujian | CSF | JF706273* |
| Fj02-76 | 2002 | China, Fujian | Human blood | JN381867* |
| FJ03-39 | 2003 | China, Fujian | Human blood | JN381859* |
| FJ03-94 | 2003 | China, Fujian | Human blood | JN381858* |
| FU | 1995 | Australia | Human serum | AF217620 |
| G35 | 1954 | China, Fujian | Mosquito pool | GQ429185 |
| GB30 | 1997 | China, Yunnan | Murina aurata brain tissue | FJ185037 |
| GP78 | 1978 | India | Human brain | AF075723 |
| GS07-TS11 | 2007 | China, Gansu | Culex tritaeniorhynchus | JN381843* |
| GSBY0801 | 2008 | China, Gansu | Culex tritaeniorhynchus | JF706274* |
| GSBY0804 | 2008 | China, Gansu | Culex tritaeniorhynchus | JN381844* |
| GSBY0810 | 2008 | China, Gansu | Culex tritaeniorhynchus | JN381840* |
| GSBY0816 | 2008 | China, Gansu | Culex tritaeniorhynchus | JN381842* |
| GSBY0827 | 2008 | China, Gansu | Culex tritaeniorhynchus | JN381845* |
| GSBY0861 | 2008 | China, Gansu | Culex tritaeniorhynchus | JN381833* |
| GSS | 1960s | China, Beijing | CSF | JF706275* |
| GX0519 | 2005 | China, Guanxi | Culex tritaeniorhynchus | JN381835* |
| GX0523/44 | 2005 | China, Guanxi | Culex tritaeniorhynchus | JN381832* |
| GZ04-2 | 2004 | China, Guizhou | Armigeres | JN381857* |
| GZ56 | 2006 | China, GuiZhou | CSF | HM366552 |
| Ha-3 | 1960s | China, Heilongjiang | CSF | JN381872* |
| HB49 | 1990 | China, Yunnan | Rousettus leschenaulti blood | JF706284* |
| HB97 | 1990 | China, Yunnan | Rousettus leschenaulti blood | JF706285* |
| HLJ02-134 | 2002 | China, Heilongjiang | Genus Culicoides | JF706276* |
| HN04-11 | 2004 | China, Henan | Culex | JN381831* |
| HN04-21 | 2004 | China, Henan | Culex | JN381841* |
| HN06129 | 2006 | China, Henan | Armigeres | JF706277* |
| HN0621 | 2006 | China, Henan | Culex | JN381830* |
| HN0626 | 2006 | China, Henan | Culex | JN381837* |
| HVI | 1965 | Taiwan | Mosquito | AF098735 |
| HYZ | 1979 | China, Yunnan | Patient blood | JN381853* |
| Ishikawa | 1994 | Japan | Culex tritaeniorhynchus | AB051292 |
| JaGAr 01 | 1959 | Japan | Culex | AF069076 |
| JaOArS982 | 1982 | Japan | Mosquito | M18370 |
| JaOH0566/Japan/1966/human | 1966 | Japan | Human | AY508813 |
| JEV/sw/Mie/40/2004 | 2004 | Japan | Swine serum | AB241118 |
| JEV/sw/Mie/41/2002 | 2002 | Japan | Swine serum | AB241119 |
| JH04-18 | 2004 | China, Yunnan | Culex whitmorei and Anopheles sinensis | JN381855* |
| JKT6468 | 1981 | Indonesia | Mosquito | AY184212 |
| K87P39 | 1987 | South Korea | Mosquito | AY585242 |
| KV1899 | 1999 | Korea | Swine | AY316157 |
| LFM | 1955 | China, Fujian | Human blood | JN381863* |
| Ling | 1965 | Taiwan | Human brain | L78128 |
| LN02-102 | 2002 | China, Liaoning | Culex modestus | JF706278* |
| LN0716 | 2007 | China, Liaoning | Culex tritaeniorhynchus | JN381849* |
| LYZ | 1957 | China, Fujian | CSF | JN381869* |
| M28 | 1977 | China, Yunnan | Culex pseudovishnui | JF706279* |
| Nakayama | 1935 | Japan | Human brain | EF571853 |
| p3 | 1949 | China, Beijing | Human brain | U47032 |
| RP-2ms | 1985 | Taiwan | Mosquito | AF014160 |
| RP-9 | 1985 | Taiwan | Mosquito | AF014161 |
| SA14 | 1954 | China | Mosquito | U14163 |
| SC04-12 | 2004 | China, Sichuan | Culex | JN381839* |
| SC04-15 | 2004 | China, Sichuan | Culex tritaeniorhynchus | JN381838* |
| SD0810 | 2008 | China, Shandong | Culex tritaeniorhynchus | JF706286* |
| SH03-103 | 2003 | China, Shanghai | Culex tritaeniorhynchus | JN381847* |
| SH03-105 | 2003 | China, Shanghai | Culex tritaeniorhynchus | JN381846* |
| SH04-10 | 2004 | China, Shanghai | Culex tritaeniorhynchus | JN381856* |
| SH04-5 | 2004 | China, Shanghai | Culex tritaeniorhynchus | JN381866* |
| SH17 M-07 | 2007 | China | — | EU429297 |
| SH-3 | 1987 | China, Shanghai | CSF | JN381864* |
| SH-53 | 2001 | China, Shanghai | Culex tritaeniorhynchus | JN381850* |
| SH-80 | 2001 | China, Shanghai | Culex tritaeniorhynchus | JN381848* |
| T1P1 | 1997 | Taiwan | Armigeres subalbatus | AF254453 |
| TLA | 1971 | China, Liaoning | CSF | JN381868* |
| Vellore P20778 | 1958 | India | Human brain | AF080251 |
| XJ69 | 2007 | China | Culex pipiens pallens | EU880214 |
| XJP613 | 2007 | China | Culex tritaeniorhynchus | EU693899 |
| XZ0938 | 2009 | China, Xizang | Culex tritaeniorhynchus | HQ652538* |
| YLG | 1955 | China, Fujian | CSF | JF706280* |
| YN | 1954 | China, Yunnan | CSF | JN381871* |
| YN05124 | 2005 | China, Yunnan | Culex tritaeniorhynchus | JF706281* |
| YN05155 | 2005 | China, Yunnan | Culex tritaeniorhynchus | JN381852* |
| YN0623 | 2006 | China, Yunnan | Culex tritaeniorhynchus | JN381836* |
| YN0911 | 2009 | China, Yunnan | Culex tritaeniorhynchus | JF706267* |
| YN0967 | 2009 | China, Yunnan | Culex tritaeniorhynchus | JF706268* |
| YN79-Bao83 | 1979 | China, Yunnan | Culex tritaeniorhynchus | JN381851* |
| YN82-BN8219 | 1982 | China, Yunnan | Mosquito | JN381834* |
| YN83-Meng83-54 | 1983 | China, Yunnan | Lasiohelea taiwana (Shiraki) | JF706282* |
| YN98-A151 | 2003 | China, Yunnan | Mosquitoes | JN381861* |
| ZMT | 1955 | China, Fujian | CSF | JF706283* |
| ZSZ | 1955 | China, Fujian | CSF | JN381862* |
—, information not available.
Asterisks indicate strains newly sequenced in this study.
JEV genome sequence data set.
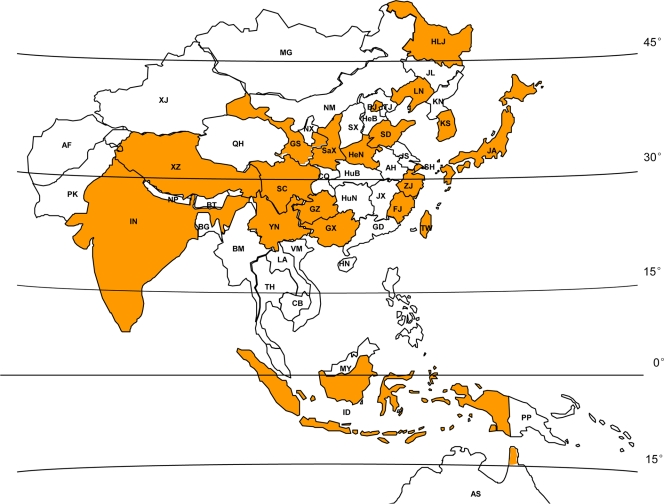
Additional full-length genome sequences were downloaded from GenBank, combined with the new samples, and aligned using ClustalW, version 2.0 (29). Vaccine or derivative strains were excluded, and sequences sharing more than 98% similarity were removed from the data set by analyzing the alignment with the T-COFFEE software package (21), to leave a final data set containing 98 sequences. The complete sequence set contained samples isolated from a variety of hosts: mosquitoes and other insects (n = 55), with Culex tritaeniorhynchus as the major species (n = 30), midges (n = 3), bats (n = 4), a pig (n = 1), and humans (n = 28). The isolation dates ranged from 1935 to 2009, and samples were collected from the entire region in which JEV cases have been identified (latitude 15°S to latitude 45°N) (Fig. 1).
FIG. 1.
Worldwide distribution of identified JEV cases. The provinces in China and the other countries from which JEV was isolated and used in this study are shaded. AF, Afghanistan; PK, Pakistan; IN, India; NP, Nepal; BT, Bhutan; BG, Bangladesh; BM, Burma; TH, Thailand; LA, Laos; VM, Vietnam; CB, Cambodia; MY, Malaysia; ID, Indonesia; PP, Papua New Guinea; AS, Australia; KN, North Korea; KS, South Korea; JA, Japan. Chinese provinces: HLJ, Heilongjiang Province; JL, Jilin Province; LN, Liaoning Province; NM, Neimenggu; XJ, Xinjiang; BJ, Beijing, TJ, Tianjin; HeB, Hebei Province; SX, Shanxi Province; SaX, Shaanxi Province; GS, Gansu Province; QH, Qinghai Province; NX, Ningxia; SD, Shandong Province; SH, Shanghai; JS, Jiangsu Province; AH, Anhui Province; HeN, Henan Province; XZ, Xizang; ZJ, Zhejiang Province; JX, Jiangxi Province; HuB, Hubei Province; CQ, Chongqing; SC, Sichuan Province; HuN, Hunan Province; GZ, Guizhou Province; YN, Yunnan Province; FJ, Fujian Province; GD, Guangdong Province; GX, Guangxi; HN, Hainan; TW, Taiwan; MG, Mongolia;.
Bayesian Markov chain Monte Carlo (MCMC) analysis of JEV.
The GTR+I+G substitution model was selected by MrModelTest (23), and the rate of nucleotide substitution, model and rate of population growth, and age of the most recent common ancestor (TMRCA) were estimated using the BEAST software package (2). Rates were estimated for the relaxed clock model, and the chain length was 1,000,000,000 generations with 10% burn-in. The demographics of genotypes I and III were compared by generating their respective Bayesian skyline plots with an uncorrelated log-normal relaxed molecular clock. There were insufficient data to allow the analysis of genotypes II and IV.
Construction of the JEV E gene data set.
To obtain a simple estimate of the relative abundances of genotype I and III strains over time, we downloaded all JEV E gene sequences in GenBank as of January 2011. Derivative and genetically modified JEV strains were excluded, and only sequences with background information describing the place and time of isolation were retained, for a final data set of 537 elements.
Nucleotide sequence accession numbers.
The virus sequences determined in this study have been deposited in GenBank under accession numbers JF706267 to JF706286 and JN381830 to JN381873.
RESULTS
Phylogenetic analysis.
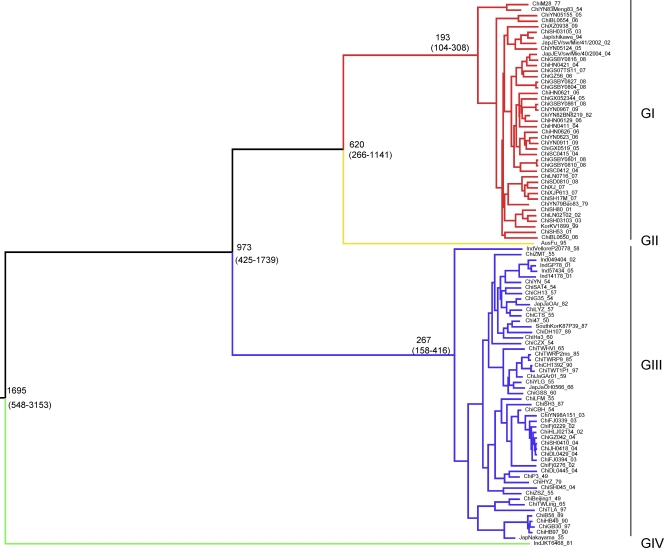
The maximum clade credibility (MCC) tree for the whole genomes of JEV is shown in Fig. 2. The tree contains four distinct clades corresponding to genotypes IV, III, II, and I. The most recent common ancestor for all genotypes is estimated to have occurred 1,695 years ago (95% highest posterior density [HPD], −548 to −3,153 years). The branching of the lineages occurred in the following order: genotype IV, genotype III at −973 years (95% HPD, −425 to −1,739 years), genotype II at −620 years (95% HPD, −266 to −1,141 years), and genotype I at −193 years (95% HPD, −104 to −308 years). The width of most of the 95% HPD intervals is due to the fact that for genotypes II and IV, only a single full-length sequence is available.
FIG. 2.
Maximum clade credibility tree for 98 whole-genome sequences of JEV. Consistent with previous studies, the tree identifies four distinct lineages: genotype I (GI) (red), genotype II (yellow), genotype III (blue), and genotype IV (green). Estimated TMRCAs of these lineages (with their 95% HPD values in parentheses) are shown.
The JEV strains isolated from mosquitoes, midges, and JE patients were distributed throughout the evolutionary branches. Furthermore, no host adaptation was detected, and no specific branches were associated with isolation time, sample category, or geographical distribution, suggesting the absence of geographical or species barriers.
Rate of evolutionary change in the JEV genome.
Based on the Bayesian MCMC approach and assuming an uncorrelated log-normal molecular clock, the mean nucleotide substitution rate for the entire sequence set was estimated as 1.11 × 10−4 substitution per site per year (95% HPD, 6.04 × 10−5 to 1.69 × 10−4). For genotypes I and III, the estimated rates were 5.82 × 10−5 (95% HPD, 3.42 × 10−9 to 1.57 × 10−4) and 7.91 × 10−5 (95% HPD, 4.28 × 10−5 to 1.18 × 10−4) substitution per site per year, respectively.
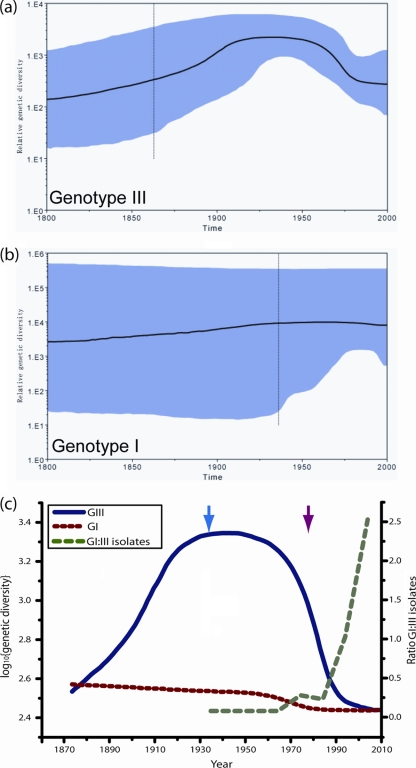
Skyline plot and genetic diversity.
The skyline plots for genotypes III and I are shown in Fig. 3a and b, respectively. For genotype III, there is an increase in genetic diversity for the first half of the plot, followed by a subsequent order of magnitude decrease. For the genotype I plot, the diversity appears to have remained relatively constant over the entire period. However, one notable feature of the plot for genotype I is the broad 95% HPD values; this uncertainty is also reflected in the 95% HPD intervals of the estimated average substitution rate for genotype I, given in the preceding section. This uncertainty is a consequence of the genome sequence set; genotype I has become prevalent only in recent years (the earliest isolate is from 1977), so a degree of extrapolation is involved in estimating quantities, and this is reflected in both of these 95% HPD intervals. Conversely, since genotype III was the dominant genotype in the latter half of the 20th century, and the earliest sampled sequence dates to 1935, the corresponding estimates are more robust. However, the uncertainty in these estimates can be seen to increase prior to this date.
FIG. 3.
(a and b) Bayesian skyline plots for genotype III (a) and genotype I (b). Highlighted areas correspond to 95% HPD intervals. The broad 95% HPD for genotype I is a consequence of the sequence set used in the estimation. (c) Medians of the skyline plots for both genotypes drawn on the same scale. Dotted red line, genotype I; solid blue line, genotype III; dashed green line, ratio of the number of genotype I isolates to the number of genotype III isolates deposited in GenBank by year. The blue arrow marks the earliest isolate of genotype III; the red arrow marks the earliest isolate of genotype I. The order of magnitude drop in the estimated genetic diversity of genotype I is matched by a corresponding order of magnitude increase in the ratio of genotype I to genotype III isolates. See the text for full details.
Figure 3c shows a plot of the median genetic diversity values of both genotypes on the same scale and the ratio of dated genotype I to genotype III isolates submitted to GenBank over time. The genotypes show a marked difference in their variation in genetic diversity over time. Since the first genotype III sample was isolated in 1935 (Fig. 3c, blue arrow), the diversity of this genotype has decreased by an order of magnitude, while the diversity of genotype I (first isolated in 1977 [Fig. 3c, red arrow]) has remained almost constant. Figure 3c also shows a plot of the ratio of genotype I to genotype III E gene isolates based on the E gene data set described in Materials and Methods. Although this is a simple estimate of the relevant abundances of the two genotypes, the features of the plot are consistent with the estimates of genetic diversity for both genotypes. In particular, the most rapid decrease in the genetic diversity of genotype III precedes the most rapid increase in the number of genotype I samples collected.
DISCUSSION
The earliest observations of JEV were recorded in the late 19th century in Japan, and subsequent reports trace its gradual spread into neighboring regions in Southeast Asia (4, 16, 31). Compared to those of other viruses, the JEV genome is highly conserved, and previous phylogenetic studies have been based on the more variable prM or E gene sequences. Many of these studies have focused on the classification of isolates by genotype and have highlighted the gradual displacement of genotype III by genotype I (14, 19, 22, 32, 35). The earliest comprehensive attempt to investigate the origin of JEV found that the virus probably originated from Indonesia/Malaysia (26). With the development of more-sophisticated phylogenetic analysis techniques (2), some recent studies have reinvestigated the relationships between the different genotypes in an effort to understand the origin of JEV. These studies also found that tropical Southeast Asia plays an important role in the introduction of new strains and indicated that birds and windblown mosquitoes may be responsible for importing these new strains from mainland China into Taiwan and Japan (10, 18). All of these studies were based on prM and E gene sequences. In this work we report the first detailed phylogenetic analysis of the epidemiology of JEV based on full-length genome sequences. Unlike previous investigators, in addition to examining the origin of the virus, we also considered the roles of genotypes I and III in the spread of the virus and the displacement of genotype III strains by genotype I.
Our results lead to an estimate that the most recent common ancestor of JEV occurred 1,695 years ago, with the divergence of the four JEV genotypes occurring in the following order: genotype IV (1,695 years ago), genotype III (973 years ago), genotype II (620 years ago), and genotype I (193 years ago). Our prediction that genotype I is the youngest genotype is supported by growing evidence that genotype I is replacing genotype III in several regions. Acute encephalitis syndrome (AES) induced by genotype I JEV has been reported in Japan (11), India (5), Yunnan Province in southwest China (35, 36), and Gansu (35) and Shanxi (34) Provinces in northwest China, covering a region spanned by latitude 24°37′ to 42°57′N and longitude 92°13′ to 111°15′E in China. Vietnam and Thailand (20) first reported the isolation of genotype I JEV in the 1980s. In Japan, all samples isolated after 1994 belonged to genotype I (14), and in Thailand, all samples isolated from insects and pigs after 2000 belonged to genotype I (20). National JEV surveillance data for China revealed that from 2001 to 2005, genotype I samples accounted for 71% of all isolates (32). The region in which genotype I samples have been isolated now spans latitudes 10°N (Thailand) to 35°N (Gansu, China).
Our work also provides insight into the relationship between genotype I and genotype III, the two major JEV genotypes in Asia. Our results indicate that genotype I, as the youngest genotype, began to replace genotype III to become the dominant genotype approximately 20 years ago. Genotype I appeared to be a minor strain until the early 1970s, at which point it seemed to reach some critical size and expanded rapidly through the region, accompanied by a rapid drop in the number of genotype III isolates and a corresponding drop in the genetic diversity of genotype III. It is also interesting that by the time the first genotype III isolate was collected, the genetic diversity of this genotype had already reached a plateau (Fig. 3c); i.e., although surveillance data suggest that genotype III was dominant during this period, it seems it already had a limited selective advantage.
Japanese encephalitis is a vaccine-preventable disease, and vaccines demonstrate cross-protection between genotypes (1, 3). Nevertheless, the currently available live attenuated and inactivated vaccines were derived when genotype III was dominant and cases of genotype I JE were relatively rare. Since genotype III has now been displaced, and given a recent report of isolation of a JEV genotype I strain from the cerebrospinal fluid (CSF) of a vaccinated JE patient in Yunnan Province (36), it would be prudent to monitor the protective effect of current vaccines closely.
ACKNOWLEDGMENTS
This work was supported by grants from The Ministry of Science and Technology, China (2003BA712A08-01, 2008ZX10004-008, 2009ZX10004-202, and 2011CB504702), a Development Grant of the State Key Laboratory for Infectious Disease Prevention and Control (2008SKLID105), The Japan Health Science Foundation, the National Natural Science Foundation of China (31070145), and the China CDC—U.S. CDC Cooperative Agreement (U19-GH000004).
Footnotes
Published ahead of print on 22 June 2011.
REFERENCES
- 1. Beasley D. W., Lewthwaite P., Solomon T. 2008. Current use and development of vaccines for Japanese encephalitis. Expert. Opin. Biol. Ther. 8:95–106 [DOI] [PubMed] [Google Scholar]
- 2. Drummond A. J., Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Duggan S. T., Plosker G. L. 2009. Japanese encephalitis vaccine (inactivated, adsorbed) [IXIARO]. Drugs 69:115–122 [DOI] [PubMed] [Google Scholar]
- 4. Erlanger T. E., Weiss S., Keiser J., Utzinger J., Wiedenmayer K. 2009. Past, present, and future of Japanese encephalitis. Emerg. Infect. Dis. 15:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fulmali P. V., et al. 2011. Introduction of Japanese encephalitis virus genotype I, India. Emerg. Infect. Dis. 17:319–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gao X., Nasci R., Liang G. 2010. The neglected arboviral infections in Mainland China. PLoS Negl. Trop. Dis. 4:e624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ghosh D., Basu A. 2009. Japanese encephalitis—a pathological and clinical perspective. PLoS Negl. Trop. Dis. 3:e437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hanna J. N., et al. 1996. An outbreak of Japanese encephalitis in the Torres Strait, Australia, 1995. Med. J. Aust. 165:256–260 [DOI] [PubMed] [Google Scholar]
- 9. Hanna J. N., et al. 1999. Japanese encephalitis in north Queensland, Australia, 1998. Med. J. Aust. 170:533–536 [DOI] [PubMed] [Google Scholar]
- 10. Huang J. H., et al. 2010. Molecular epidemiology of Japanese encephalitis virus, Taiwan. Emerg. Infect. Dis. 16:876–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kuwayama M., et al. 2005. Japanese encephalitis virus in meningitis patients, Japan. Emerg. Infect. Dis. 11:471–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li Y. X., et al. 2011. Japanese encephalitis, Tibet, China. Emerg. Infect. Dis. 17:934–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lindenbach B. D., Thiel H. J., Rice C. M. 2007. Flaviviridae: the viruses and their replication, p. 1101–1152 In Knipe D. M., et al. (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 14. Ma S. P., et al. 2003. A major genotype of Japanese encephalitis virus currently circulating in Japan. Am. J. Trop. Med. Hyg. 69:151–154 [PubMed] [Google Scholar]
- 15. Mackenzie J. S., Gubler D. J., Petersen L. R. 2004. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat. Med. 10:S98–S109 [DOI] [PubMed] [Google Scholar]
- 16. Mackenzie J. S., Williams D. T., Smith D. W. 2007. Japanese encephalitis virus: the geographic distribution, incidence, and spread of a virus with a propensity to emerge in new areas, p. 201–268 In Tabor E. (ed.), Emerging viruses in human populations. Elsevier B.V., Amsterdam, Netherlands [Google Scholar]
- 17. Mo Z. J., et al. 2010. Isolation of genotype I Japanese encephalitis virus in Beiliu, Guangxi. Dis. Surveill. 25:115–119 (In Chinese.) [Google Scholar]
- 18. Nabeshima T., et al. 2009. Evidence of frequent introductions of Japanese encephalitis virus from south-east Asia and continental east Asia to Japan. J. Gen. Virol. 90:827–832 [DOI] [PubMed] [Google Scholar]
- 19. Nga P. T., et al. 2004. Shift in Japanese encephalitis virus (JEV) genotype circulating in northern Vietnam: implications for frequent introductions of JEV from Southeast Asia to East Asia. J. Gen. Virol. 85:1625–1631 [DOI] [PubMed] [Google Scholar]
- 20. Nitatpattana N., et al. 2008. Change in Japanese encephalitis virus distribution, Thailand. Emerg. Infect. Dis. 14:1762–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Notredame C., Higgins D. G., Heringa J. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205–217 [DOI] [PubMed] [Google Scholar]
- 22. Parida M., et al. 2006. Japanese encephalitis outbreak, India, 2005. Emerg. Infect. Dis. 12:1427–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Posada D. 2003. Using MODELTEST and PAUP* to select a model of nucleotide substitution. Curr. Protoc. Bioinformatics 2003:Chapter 6, Unit 6.5. [DOI] [PubMed] [Google Scholar]
- 24. Saito M., Taira K., Itokazu K., Mori N. 2007. Recent change of the antigenicity and genotype of Japanese encephalitis viruses distributed on Okinawa Island, Japan. Am. J. Trop. Med. Hyg. 77:737–746 [PubMed] [Google Scholar]
- 25. Solomon T. 2006. Control of Japanese encephalitis—within our grasp? N. Engl. J. Med. 355:869–871 [DOI] [PubMed] [Google Scholar]
- 26. Solomon T., et al. 2003. Origin and evolution of Japanese encephalitis virus in southeast Asia. J. Virol. 77:3091–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Solomon T., et al. 2000. Japanese encephalitis. J. Neurol. Neurosurg. Psychiatry 68:405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sun X., et al. 2009. Distribution of arboviruses and mosquitoes in northwestern Yunnan Province, China. Vector Borne Zoonotic Dis. 9:623–630 [DOI] [PubMed] [Google Scholar]
- 29. Thompson J. D., Gibson T. J., Higgins D. G. 2002. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics 2002:Chapter 2, Unit 2.3. [DOI] [PubMed] [Google Scholar]
- 30. United Nations 2005. The United Nations urbanization prospects: the 2005 revision. POP/DB/WUP/Rev.2005/1/F1. United Nations, New York, NY.
- 31. van den Hurk A. F., Ritchie S. A., Mackenzie J. S. 2009. Ecology and geographical expansion of Japanese encephalitis virus. Annu. Rev. Entomol. 54:17–35 [DOI] [PubMed] [Google Scholar]
- 32. Wang H. Y., et al. 2007. Molecular epidemiological analysis of Japanese encephalitis virus in China. J. Gen. Virol. 88:885–894 [DOI] [PubMed] [Google Scholar]
- 33. Wang J. L., et al. 2009. Japanese encephalitis viruses from bats in Yunnan, China. Emerg. Infect. Dis. 15:939–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang L. H., et al. 2007. Japanese encephalitis Yuncheng, China, 2006. Emerg. Infect. Dis. 13:1123–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang L., et al. 2010. Identification and isolation of Genotype-I Japanese encephalitis virus from encephalitis patients. Virol. J. 7:345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang J. S., et al. 2011. Isolation and genetic characteristics of human genotype 1 Japanese encephalitis virus, China, 2009. PLoS One 6:e16418. [DOI] [PMC free article] [PubMed] [Google Scholar]