Abstract
The human papillomavirus (HPV) E7 oncoprotein exists as a dimer and acts by binding to many cellular factors, preventing or retargeting their function and thereby making the infected cell conducive for viral replication. Dimerization of E7 is attributed primarily to the C-terminal domain, referred to as conserved region 3 (CR3). CR3 is highly structured and is necessary for E7's transformation ability. It is also required for binding of numerous E7 cellular targets. To systematically analyze the molecular mechanisms by which HPV16 E7 CR3 contributes to carcinogenesis, we created a comprehensive panel of mutations in residues predicted to be exposed on the surface of CR3. We analyzed our novel collection of mutants, as well as mutants targeting predicted hydrophobic core residues of the dimer, for the ability to dimerize. The same set of mutants was also assessed functionally for transformation capability in a baby rat kidney cell assay in conjugation with activated ras. We show that some mutants of HPV16 E7 CR3 failed to dimerize yet were still able to transform baby rat kidney cells. Our results identify several novel E7 mutants that abrogate transformation and also indicate that E7 does not need to exist as a stable dimer in order to transform cells.
INTRODUCTION
More than 100 human papillomavirus (HPV) types have been described, and more are presumed to exist (16). HPVs induce papillomas in the cutaneous and mucosal epithelia, where specific HPV types often preferentially infect distinct anatomical sites. HPVs associated with lesions that can progress to carcinogenesis are classified as “high-risk” types, the most common of which is HPV16. In contrast, HPVs associated with benign warts that regress with time are termed “low-risk” viruses (50). Persistent infection by high-risk HPVs is associated with 99.7% of all human cervical cancer cases (15), other genitourinary cancers, and an increasingly growing number of oral cancers (21).
The viral E6 and E7 oncoproteins are consistently expressed in HPV-induced cancers and are necessary to maintain malignant cell growth (3, 6, 29, 42, 43). Repression of their transcription by reexpression of the viral E2 protein induces rapid growth arrest and senescence of cervical cancer cells (22, 23). The HPV E7 oncoprotein binds the product of the retinoblastoma susceptibility locus (pRb) and the related family members p107 and p130 (17). These proteins function as tumor suppressors, maintaining control of the G1/S checkpoint of the cell cycle by binding to the E2F family of transcription factors and by recruiting transcriptional repressor complexes to the E2F-responsive genes (10, 36). Besides pRb, the E7 oncoproteins from high-risk HPV types abrogate the inhibitory activities of the cyclin-dependent kinase inhibitors p21 and p27 by directly binding these factors (19, 48). E7 interacts with the pCAF acetyltransferase, the Mi2β subunit of the NuRD histone deacetylase complex, the S4 component of the 26S proteasome, the HIF-1α and E2F6 transcription factors, and many other important cellular proteins (4, 7, 8, 11, 35).
The N-terminal region of E7 displays sequence similarity to other viral oncoproteins, namely, to a portion of conserved region 1 (CR1) and all of CR2 of adenovirus (Ad) E1A (24, 45). Based on the amino acid sequence similarity between different HPV types, the HPV16 E7 protein can also be separated into three conserved regions, namely, CR1 (amino acids 2 to 15), CR2 (amino acids 16 to 37), and CR3 (amino acids 38 to 98). Binding of many cellular targets of E7 has been mapped to CR3, including a secondary low-affinity pRb binding site (32, 38). This C-terminal zinc binding region of E7 is highly structured and composed of two Cys-X-X-Cys motifs separated by 29 or 30 amino acids, whereas the amino-terminal region (CR1/CR2) is unstructured (34). High-resolution three-dimensional structures of the zinc binding region of E7 have been reported for HPV1 and HPV45 (32, 37). E7 CR3 exhibits a unique β1β2α1β3α2 topology that is not present in any other known zinc binding proteins. The coordinating zinc ions are important for maintaining a folded state of CR3, as removal of zinc causes unfolding of the tertiary structure and formation of large oligomeric complexes (2). E7 CR3 also forms a stable dimer involving the α1 helices of the protomers and the β2 sheet of one protomer and β3 sheet of the other, with dimerization leading to the formation of a hydrophobic core. However, whether E7 exists as a functional dimer in vivo or can function as a protomer has not been established conclusively. Furthermore, it is now known, based on existing structures of E7, that the most highly conserved residues of CR3 are in fact structural components of the protomer core and dimer interface.
The information from structural studies has necessitated a reexamination of much of the existing literature regarding the highly structured CR3 portion of E7. Most of our understanding of this region is based on the analysis of mutants that target the most highly conserved amino acids, which are not surface exposed and are not available for interaction with cellular targets but are likely important structural components. It is thus conceivable that the loss of protein interactions and/or functions observed in studies utilizing these mutants may be attributed to structural effects on E7 rather than to the impairment of specific functions. For instance, this issue has been raised consistently because of mutations in the invariant zinc-coordinating cysteines that decrease protein stability (39, 44, 46) and impair transformation and transactivation functions (18, 34, 39). To explore in detail the functions of CR3, a structure-function analysis requires mutations that preserve the overall structure of this region. In addition, many studies have used short deletion mutants within this region and have reported apparent multiple overlapping interactions with diverse cellular targets, which might simply reflect global disruption of this particular region (25, 27, 28).
Our aim, therefore, was 2-fold: (i) to create a panel of mutations targeting surface-exposed residues within CR3 of E7 and (ii) to establish whether E7 must exist as a dimer for its function. Utilizing the existing structures of HPV1 and HPV45 E7 CR3, we modeled the structure of CR3 of the HPV16 E7 protein. Based on the modeled structure, 24 side chains within CR3 were identified as at least 25% solvent exposed and were chosen as targets for systematic mutagenesis. These residues are likely to be accessible for interaction with cellular proteins and hence may contribute to E7 function. We utilized a substitution strategy in which amino acid residues were replaced with residues of opposite charge, preserving solvent interaction, and thus protein structure, but potentially disrupting specific intermolecular protein interactions. We also targeted the highly conserved hydrophobic core residues for mutagenesis. With this panel of mutants and an analysis of transformation potential, we determined that E7 does not need to exist as a dimer to transform primary baby rat kidney cells (BRK cells), and we identified several new mutants of E7 that abrogate transformation.
MATERIALS AND METHODS
Plasmid construction.
Two methods of site-directed mutagenesis were utilized to construct all mutations targeting the surface-exposed residues, namely, megaprimer (31) and overlap extension (26) methods. A list of primer sequences is available on request. The panel of CR3 mutants was generated with flanking EcoRI and SalI restriction sites and then inserted into the multiple cloning site of the pBAIT (49) yeast expression vector, generating constructs to be used in the yeast two-hybrid assay (LexA DNA-binding-domain fusions). The coding sequence for each mutation in E7 was subsequently subcloned, using EcoRI and SalI, into the same site of a modified pJG4-5+ vector (Clontech) for expression as a fusion with the synthetic B42 transcriptional activation domain. For recombinant protein production and purification, the CR3 region of each E7 construct was cloned into the pGEX4T-1 (Invitrogen) vector, modified to contain the tobacco etch virus (TEV) protease recognition sequence between a glutathione S-transferase (GST) tag and the protein of interest. For transformation assays, E7 mutations were subcloned into the BamHI and XhoI restriction sites of a modified pCMV-Neo-Bam mammalian expression vector. Constructs for the del21-24, L67R, C58G/C91G, and C91G mutations were obtained from K. Munger (Harvard Medical School, Boston, MA), K. H. Vousden (National Cancer Institute at Frederick, Frederick, MD), and D. Galloway (Fred Hutchinson Cancer Research Center, University of Washington) and subcloned as necessary. The pCMV-pRb construct was obtained from F. Dick (University of Western Ontario, London, Ontario, Canada) and subcloned into the pBAIT vector for yeast two-hybrid experiments.
Cell culture and transfection.
Saos2, HEK293, and U2OS cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and penicillin-streptomycin (100 U/ml). For pRb degradation assays, Saos2 cells were seeded into 6-well plates at 3 × 105 cells per well and transfected 24 h later with 2 μg of pRb, with or without 0.2 μg of E7 expression plasmid and with 0.1 μg of green fluorescent protein (GFP) expression plasmid as a control, using FugeneHD (Roche) according to the instructions provided by the manufacturer. The amount of plasmid DNA was balanced with empty pCMV vector where necessary. The amounts of pRb, GFP, and actin were assessed by Western blotting at 48 h posttransfection. For cycloheximide treatment experiments, HEK293 cells were seeded into 6-well plates at a density of 3.5 × 105 cells per well 24 h before transfection. For each mutant, the cells were transfected with 2 μg of E7 pCMV-Neo-Bam expression plasmid along with 0.1 μg of GFP expression plasmid, using the calcium phosphate method (33). At 24 h posttransfection, the cells were treated with 0.5 μg/μl cycloheximide for 0, 15, 30, 60, and 120 min. For immunofluorescence experiments, U2OS cells were seeded on coverslips in 6-well dishes at 1.2 × 105 cells per well. The cells were transfected with 2 μg of E7 expression plasmid by using calcium phosphate (33).
Yeast two-hybrid assay.
Yeast two-hybrid analysis was performed in strain L-40 [MATα his3Δ200 trip1-90 leu2-3,112 ade2 lys2-801am LYS2:(lexAop) 4-HIS3 URA3:(lexAop)8-lacZ Gal4] (provided by D. Mangroo, University of Guelph). Standard yeast culture medium was prepared using previously described methods (1). Yeast transformation was carried out using a modified lithium acetate procedure (20). Transformed yeast cells were plated on selective medium plates and grown at 30°C for 2 to 4 days.
The method for assaying β-galactosidase activity in yeast has been described previously (1). Briefly, transformed yeast colonies were grown in selection medium overnight at 30°C, and a 1.5-ml aliquot of culture was used for the assay. The cells were pelleted and resuspended in 1 ml of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol, pH 7.0), and the optical density at 600 nm (OD600) was determined. To 300 μl of resuspended culture, the following was added: 700 μl Z buffer, 40 μl of 0.1% SDS, and 20 μl of chloroform. The samples were mixed and incubated at 30°C for 15 min. Two hundred microliters of the substrate ortho-nitrophenyl-β-d-galactopyranoside (ONPG; Bioshop) (4 mg/ml) was added to each sample and mixed. The reaction mixtures were incubated at 30°C and, when necessary, terminated by addition of 400 μl of 1 M Na2CO3. The samples were centrifuged for 10 min to remove cellular debris before measuring the absorbance at 420 nm. The β-galactosidase activity was calculated in Miller units by utilizing the following formula: OD420/(OD600 × 0.3 ml [culture volume] × reaction time [min]). For the reported dimerization values, the numbers were corrected for any observed autoactivation for each individual mutant.
For assessing binding between pRb and E7, pRb was fused to the LexA DNA binding domain (bait), and the described E7 constructs were used as prey.
Dimerization in vitro and GST pulldown assays.
Recombinant GST-tagged wild-type E7 CR3 and mutants were expressed in and purified from the BL21 strain of Escherichia coli per protocols provided by the affinity resin manufacturer (Amersham). To obtain untagged E7 CR3, purified recombinant protein was incubated with ∼0.2 μg/ml TEV protease overnight at 4°C. The purified protein concentration was determined using the Bio-Rad assay and was verified by running 5 μg of purified protein in a gel and staining it with Coomassie blue (see Fig. 3b) prior to using the samples in the assay. The ability of E7 CR3 to dimerize was determined by analyzing the capacity of E7 CR3 to interact with GST-E7 CR3. GST pulldown assays were set up at protein concentrations well below the reported dimerization dissociation constant (Kd) of 1.1 μM (14). Briefly, 600 nM E7 CR3 was incubated at room temperature for 1 h in a total volume of 500 μl of buffer (20 mM phosphate, pH 7.0, 200 mM NaCl, 1% Tween 20, 3 μM N-ethylmaleimide); 300 nM GST-E7 CR3 was then added and incubated for an additional 10 min, following by the addition of 20 μl of 50% glutathione Sepharose. GST pulldown assays were carried out for 1 h at 4°C, and the samples were then washed three times with wash buffer (20 mM phosphate, pH 7.0, 200 mM NaCl, 1% Tween 20) and examined by Western blotting for untagged E7 CR3.
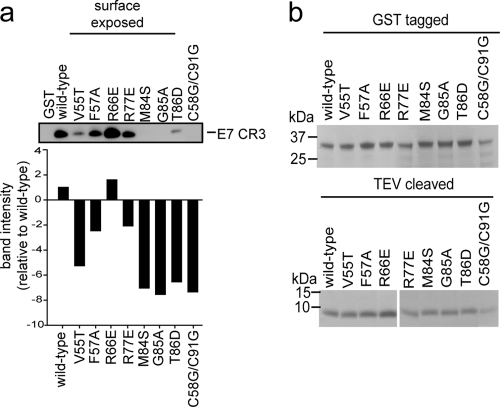
Fig. 3.
Dimerization of E7 CR3 in vitro. Predicted surface-exposed mutants identified in the yeast two-hybrid assay as impaired for dimerization (>55% decrease in activity) were tested for the ability to form dimers in vitro. Both E7 CR3 fused to GST and that cleaved with TEV protease to obtain E7 CR3 alone were purified. A 300 nM concentration of GST-tagged wild-type or mutant E7 CR3 was mixed with 600 nM E7 CR3. The ability of the wild type or mutant to dimerize was assessed by determining the amount of recovered E7 CR3 in a GST pulldown assay. The proteins were resolved by SDS-PAGE, and the amount of E7 CR3 was determined by Western blotting. The bar graph shows band intensities for the representative Western blot, as determined by densitometry. (b) Coomassie blue-stained gels of purified GST-E7 CR3 (top) and TEV-cleaved E7 CR3 (bottom), used as a loading control for the in vitro dimerization assay.
Antibodies.
The following antibodies were used: rabbit anti-LexA (Upstate) at a 1:500 dilution, rat anti-hemagglutinin (anti-HA) (Roche) at 1:2,000, and rabbit anti-glucose-6-phosphate dehydrogenase (anti-G6PD), used as a loading control for yeast samples, at 1:80,000 (Sigma). For detection of E7 CR3 in in vitro dimerization assays, the membranes were blotted with CR3-specific donkey antibody (C20; Santa Cruz) at 1:200. For detection of pRb in the degradation assay, mouse anti-pRb (G3-245; BD Pharmingen) was used at 1:500. GFP was detected using mouse anti-GFP (Living Colors; Clontech) at 1:2,000, and E7 was detected with mouse anti-E7 (8C9; Invitrogen) at 1:200. Horseradish peroxidase (HRP)-conjugated donkey anti-goat secondary antibody was used at 1:10,000 (Santa Cruz). HRP-conjugated goat anti-rabbit (Jackson Laboratories), goat anti-rat (Pierce), and rabbit anti-mouse (Jackson Laboratories) secondary antibodies were used at 1:100,000, 1:20,000, and 1:100,000, respectively.
Western blotting.
To determine expression levels of each E7 mutant in yeast cells, yeast colonies were isolated from selection plates and inoculated into 5 ml of selective liquid medium. Cultures were grown at 30°C overnight; the cell pellet was resuspended in 250 μl of lysis buffer (25 mM Tris, pH 7.5, 125 mM NaCl, 2.5 mM EDTA, 1% Triton X-100) and an equal volume of acid-washed glass beads (Sigma). Samples were mixed vigorously for five cycles of 3 min of vortexing and 3 min on ice. Cellular debris was removed by centrifugation, and 15 μg of protein extract was resolved in NuPAGE Novex Bis-Tris 4 to 12% polyacrylamide gels (Invitrogen). The resolved protein samples were transferred to a polyvinylidene fluoride (PVDF) membrane (Amersham), the samples containing LexA DNA fusions were blotted with rabbit anti-LexA, and the B42 fusions were blotted with rat anti-HA. Mammalian cells were lysed in either NP-40 lysis buffer (50 mM Tris, pH 7.8, 150 mM NaCl, 0.1% NP-40) or directly in 2× SDS sample buffer (100 mM Tris-HCl [pH 6.8], 200 mM dithiothreitol [DTT], 4% SDS, 20% glycerol, 0.2% bromophenol blue), and the cell extracts were separated by SDS-polyacrylamide gel electrophoresis (PAGE) or in NuPAGE Novex Bis-Tris 4 to 12% polyacrylamide gels and blotted onto 0.45-μm-pore-size nitrocellulose membranes (Schleicher and Schuell) or PVDF membranes. Typically, the membranes were blocked at room temperature for 1 h in 5% milk in Tris-buffered saline with 0.1% Tween 20 (TBS-T). The blots were incubated with the appropriate primary antibodies diluted in TBS-T. The incubation time was 1 h at room temperature or overnight at 4°C. After extensive washing, the blots were developed with enhanced chemiluminescence (ECL) or ECL Plus reagent (GE Healthcare) according to the manufacturer's instructions. Protein band intensities were quantitated where possible, using the ImageJ quantification program.
E7 transformation assay.
The ability of wild-type and mutated E7 constructs to transform baby rat kidney cells in cooperation with activated EJ-ras was assessed as described previously (33). Due to the large number of mutants created in this study, transformation experiments were carried out in small groups, together with respective wild-type controls. Each mutant was tested independently at least three times and sometimes up to five times.
Immunofluorescence and microscopy.
Cells were fixed with 3.7% paraformaldehyde in phosphate-buffered saline (PBS) and were permeabilized with 0.1% Triton X-100 in PBS. Primary antibody (anti-E7; 8C9) was incubated for 2 to 3 h at 37°C, washed extensively in PBS, and incubated for 1 h at 37°C with a secondary anti-mouse antibody conjugated to rhodamine (Molecular Probes). Samples were washed several times with PBS and water and were mounted with Vectashield mounting medium (Vector Laboratories) on glass slides. Slides were analyzed with a Leica DMLB fluorescence microscope with a Leica photo camera. The data were collected with a ×40 objective lens.
RESULTS
HPV16 E7 CR3 model and identification of surface-exposed residues.
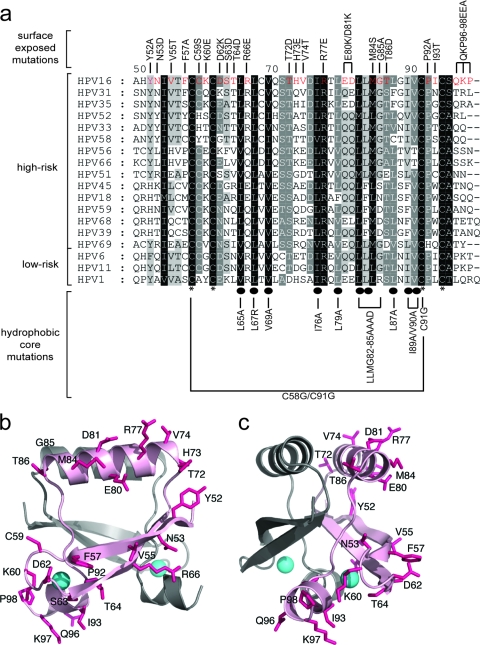
The dimeric CR3 structures for HPV45 (Protein Data Bank [PDB] accession no. 2F8B) and HPV1A (PDB accession no. 2B9D) (32, 37) have been determined, and they are very similar despite representing a high-risk and a low-risk HPV type, respectively. Indeed, these structures are superimposable to a backbone root mean square (RMS) deviation of 1.61 Å. The CR3 structure for HPV16 E7 has not been determined. Utilizing the existing structures of HPV45 and HPV1A E7 CR3 (32, 37), we generated a structural model of CR3 for the HPV16 E7 protein (Fig. 1 b and c). The HPV45 structure served as a template for construction of our HPV16 model, based on the observation that both HPV16 and HPV45 belong to the supergroup A of HPVs and are commonly associated with human cancers. Additionally, the sequences of HPV16 and HPV45 share ∼43% identity and 73% similarity, whereas HPV16 and HPV1A share ∼37% identity and ∼67% similarity. A homology method was chosen to generate the model, as it can produce high-quality structural predictions when the target (HPV16) and the template (HPV45) are closely related (5).
Fig. 1.
(a) Multiple sequence alignment of CR3 regions from representative supergroup A HPVs. Perfectly conserved residues are highlighted in black, with less conserved residues shaded in progressively lighter shades of gray, corresponding to the degree of conservation. The numbering scheme corresponds to HPV16 E7 CR3, starting with residue 49. Residues that are involved in zinc coordination are indicated with asterisks at the bottom of the alignment. Highly conserved hydrophobic core residues which are predicted to stabilize the dimer are labeled with ovals. The panel of mutations created for the purposes of the described studies is labeled at the top and bottom of the alignment. (b and c) Ribbon diagrams of the HPV16 model showing the positions of substitutions introduced into the protein. The model was produced using the PDB coordinates of HPV45 (2F8B) and a sequence alignment between the two proteins. The figures show the dimeric HPV16 model (a) and a view across a portion of the dimerization interface (b). The surface-exposed side chains where substitutions were made are shown as sticks (pink), and the bound zinc atoms are indicated as spheres (cyan). The two protomers are shaded differently, and substitutions are shown on only one protomer for clarity.
The CR3 dimer of HPV45 was analyzed by the web server program VADAR (http://redpoll.pharmacy.ualberta.ca/vadar/), which compiles more than 15 different algorithms for analyzing and assessing peptide and protein structures from atomic coordinate data (47). The HPV45 E7 dimer was analyzed to determine the accessible surface area of the protein and the chi 1 angles. Appropriate residues in HPV45 were then swapped to match those present in HPV16. Often, the quality of a homology model is complicated by the presence of alignment gaps, indicating that a structural region may be present in the target but not in the template, or vice versa. This was not an issue in this case, as the lack of gaps in the alignment of CR3s from the HPV45 and HPV16 E7 proteins made direct substitution of residues possible. The structure was refined by manually adjusting the chi 1 angles in the HPV16 model to superimpose with those in HPV45, followed by 500 steps of energy minimization to relieve poor van der Waals interactions. Atomic overlaps and unnatural strains in the structure were removed, and strong hydrogen bonds were reinforced, while weak ones were broken. As expected from the high level of sequence similarity, the predicted HPV16 E7 CR3 structure superimposes with that of HPV45 E7, with an RMS deviation of 0.34 Å.
The CR3 region of E7 has a specific folded tertiary structure, and therefore various surfaces of the folded conformation of this protein region are available to mediate specific interactions with target cellular proteins. To identify likely residues that may participate in binding of cellular targets, we first identified all amino acid residues whose side chains were at least 25% surface exposed. This analysis resulted in identification of 24 residues within CR3 of HPV16 E7, and from these, we chose to construct 19 point mutations, one double mutation (E80K/D81K), and one mutation targeting the last three amino acids (QKP96-98EEA) of the protein (Fig. 1). The overall aim of the substitution scheme was to target residues in a manner that would help to retain the folded state of the CR3 protomer and dimer and preserve solvent interactions. Given the structure of CR3, mutating polar or charged amino acids by the conventional strategy of alanine-scanning mutagenesis would likely disrupt interaction of these residues with solvent and could lead to precipitation or aggregation. Consequently, we chose to devise an alternative mutational strategy; in general, basic or acidic amino acids were replaced with residues of opposite charge, with the polarity and size of the side chain group being maintained. In the case of proline, glycine, phenylalanine, and tyrosine, there was no clear substitution choice, and these amino acids were replaced with alanine.
In addition to targeting surface-exposed residues, our analysis also included substitutions in the most highly conserved residues within CR3, some of which have been characterized extensively in the literature (11, 12, 30). Based on HPV45 E7 structural studies, the side chains of I73, L75, V77, L84, L87, L90, F91, L95, F97, and V98 contribute to formation of a small hydrophobic core in each protomer (37). The dimer interface is stabilized by contacts of residues V77, E78, T86, L87, L90, L95, and F97; hence, the dimer interface is stabilized by some of the hydrophobic core residues, including V77, L87, L90, L95, and F97. The corresponding hydrophobic core residues in HPV16 CR3 include L65, L67, V69, I76, L79, L82, L83, L87, I89, and V90. As in HPV45, these were also predicted from our model to form the hydrophobic core, and some may contribute to stabilization of the dimer. Alanine mutations targeting these highly conserved hydrophobic residues were therefore perfect candidates to establish whether dimerization of E7 is necessary for function.
Dimerization properties of E7 CR3 mutants in yeast two-hybrid analyses.
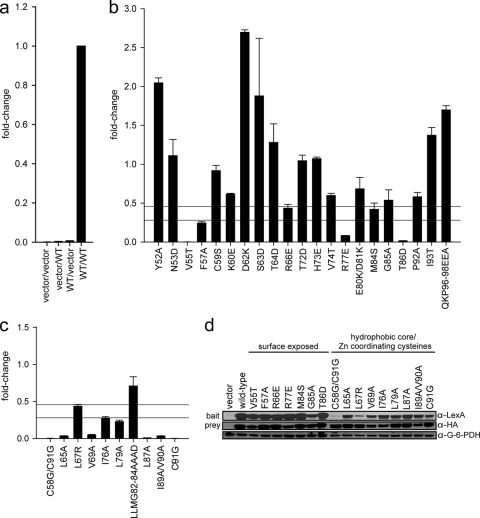
It is possible that despite our mutational strategy, some of the mutations in E7 CR3 may affect the ability of E7 to properly fold and/or dimerize. Therefore, the entire panel of surface-exposed mutants as well as the E7 mutants that targeted the hydrophobic core residues were tested for the ability to dimerize. The yeast two-hybrid test has been utilized previously to demonstrate that E7 can form dimers in vivo (13, 51). HPV16 E7 has been reported to possess an intrinsic ability to activate transcription (51) and interacts with transcriptional regulators such as TATA-binding protein (TBP) (41), which could confound this analysis. However, fusion of wild-type E7 with the LexA DNA binding domain activated transcription of a LexA-responsive reporter gene very poorly in the absence of an interacting prey (Fig. 2 a). Similarly, expression of wild-type E7 fused to the B42 activation domain as prey without an interacting bait had no effect on reporter gene activity. This was also confirmed for each of the mutants in CR3 of E7 (data not shown). Coexpression of wild-type E7 as both bait and prey resulted in a dramatically higher reporter gene activity, indicating dimerization (Fig. 2a). Based on these results, intrinsic transcriptional activation activity by E7 does not contribute significantly to the ability of the E7 homodimer to initiate transcription in the yeast two-hybrid assay. Each mutant was tested for dimerization when coexpressed as a LexA or B42 fusion (Fig. 2b and c). Dimerization of each mutant was calculated relative to wild-type E7 dimerization, with values corrected for the intrinsic transactivation activity of each mutant E7 protein. Mutants with transcriptional activity of <45% of wild-type E7 activity were considered impaired for dimerization. From these data, it is apparent that the majority of mutants with mutations predicted by our model as likely to influence dimerization were unable to dimerize (Fig. 2c). In particular, all but one E7 mutation targeting the hydrophobic core residues and zinc-coordinating cysteines resulted in <45% activity in the yeast two-hybrid assay relative to the wild-type protein (Fig. 2c). This analysis also identified mutations in surface-exposed residues that appeared to have decreased dimerization potential; these included the V55T, F57A, R66E, R77E, M84S, T86D, and possibly G85A mutations (Fig. 2b).
Fig. 2.
E7 dimerization in vivo. (a) Controls for the yeast two-hybrid assay. L-40 yeast cells were cotransformed with a LexA DNA binding domain (bait) and a B42 activation domain (prey). The combination of vectors (bait/prey) used is indicated below each data bar, with “vector” representing an empty vector and “WT” indicating a vector expressing a wild-type HPV16 E7 fusion. Indicated constructs were expressed in L-40 yeast cells containing an integrated LexA-responsive β-galactosidase reporter. Cell extracts were prepared and assayed for β-galactosidase activity. (b) Dimerization of mutants with mutations targeting predicted surface-exposed residues as determined by the yeast two-hybrid assay. HPV16 E7 mutants were expressed as fusions to the LexA DNA binding domain and the B42 activation domain in L-40 yeast cells, and β-galactosidase activity was assessed. The upper horizontal line is set at 45% of wild-type activity, corresponding to a predicted 5-fold increase in Kd; the lower horizontal line corresponds to a predicted 10-fold increase in Kd and is set at 28% of wild-type activity. We considered any mutation with <45% of wild-type activity to be impaired for dimerization. The same criterion also applies to panel c. (c) Dimerization of mutants with mutations targeting predicted hydrophobic core residues and zinc-coordinating cysteines as assessed by the yeast two-hybrid assay. (d) Expression levels of LexA and B42 fusion proteins of dimerization-impaired E7 mutants. Yeast extracts cotransformed with either wild-type E7 or the E7 mutant as both LexA (bait) and B42 (prey) fusions were tested for protein expression by Western blotting using anti-LexA and anti-HA antibodies. An anti-G6PD blot served as a loading control.
Alterations in the metal-binding cysteines have repeatedly been reported to interfere with E7 protein stability, and misfolded proteins are generally degraded quickly in the cell (13, 39, 46). To determine whether the loss of transcriptional activity in the yeast two-hybrid assay was truly due to a defect in the ability to dimerize or simply caused by a lack of sufficient protein expression, Western blots were conducted on yeast extracts for mutants putatively impaired in dimerization (Fig. 2d). Most of these mutants were expressed at levels comparable to the wild-type level, although slightly decreased expression was observed for some of the bait or prey fusions (e.g., for the V55T, I76A, G85A, R77E, I89A/V90A, and C91G mutants). Decreased expression of bait fusions with the C58G/C91G and L67R mutants was observed consistently, whereas the prey counterparts were expressed as well as the wild-type protein. The underlying reasons for this are unclear, although significant differences in the sizes of the bait and prey fusions were most likely a contributing factor, in conjunction with the structural differences between the DNA binding or activation domain and the particular mutation.
Dimerization of E7 mutants in vitro.
As an alternative method to assess dimerization, we tested the ability of E7 CR3 to dimerize in vitro by using the purified recombinant protein. We focused on the surface-exposed E7 mutants that appeared to be impaired for dimerization by the yeast two-hybrid assays. We also included the G85A mutant in this analysis, as this mutant was shown to have 54% ± 13% of wild-type activity in the yeast two-hybrid assay and could fall into the category of proteins impaired for dimerization. We utilized a simple GST pulldown assay where one E7 protomer was expressed and purified as a GST-tagged fusion and the other was protease cleaved to release CR3. The ability of the two proteins to interact with each other was monitored by examining the ability of resin-immobilized GST-CR3 to pull down soluble recombinant CR3 as determined via Western blotting (Fig. 3 a). As expected, a mutant in which the Zn-coordinating cysteine residues (C58G/C91G) were targeted failed to dimerize in vitro. Of the seven surface-exposed E7 mutants with decreased dimerization based on the yeast two-hybrid results, six also showed decreased binding in vitro (V55T, F57A, R77E, M84S, G85A, and T86D mutants). One mutant, the R66E mutant, seemed to be slightly better at binding than the wild-type protein in this system.
Transformation potential of E7 mutants.
HPV16 E7 can transform rodent cells in cooperation with activated ras or extend the life span of primary human cells in cooperation with the viral E6 oncogene (24, 40). These activities require CR1 and CR2. Although not as well characterized, CR3 also plays a role, as certain mutations in CR3 can lead to defective immortalization/transformation activities (25, 30, 34, 39). We tested this panel of E7 CR3 mutants for the ability to transform primary BRK cells in cooperation with activated ras. Most mutations targeting surface-exposed residues had no effect on transformation. However, the E7 M84S and QKP96-98EEA mutants had consistently decreased transformation potentials (Table 1). Interestingly, a number of mutants of E7 demonstrated increased transformation potential, including the V55T, T64D, R66E, and P92A mutants. For the mutants we originally predicted to be part of the hydrophobic core, we identified two different phenotypes. The L67R, V69A, and L79A mutants showed decreased transformation, whereas the L65A, I76A, LLMG82-85AAAD, L87A, and I89A/V90A mutants all behaved like the wild type (Table 2). Both mutations in E7 targeting cysteines (C58G/C91G and C91G) that coordinate zinc gave a decreased transformation potential. This is in agreement with previous studies (30, 34) and can be expected based on the important structural roles of these residues.
Table 1.
Ability of E7 mutants with mutations in predicted surface-exposed residues to transform primary BRK cells in cooperation with activated ras, with a summary of their dimerization potential
| Mutant | Mean (±SD) no. of colonies |
Transformation phenotype | Mean dimerization level (±SD) in vivoa | Dimerization level in vitroa,b | |
|---|---|---|---|---|---|
| Mutant | Wild type | ||||
| Y52A | 9.33 ± 3.85 | 12.00 ± 3.00 | Transforms | 2.05 ± 0.06 | ND |
| N53D | 22.60 ± 17.20 | 26.33 ± 10.33 | Transforms | 1.11 ± 0.21 | ND |
| V55T | 20.00 ± 9.16 | 13.00 ± 3.46 | Increased ability | 0.00 ± 0.00 | − |
| F57A | 13.33 ± 8.08 | 14.00 ± 1.73 | Transforms | 0.24 ± 0.02 | + |
| C59S | 14.20 ± 6.68 | 12.00 ± 2.50 | Transforms | 0.92 ± 0.07 | ND |
| K60E | 19.75 ± 9.32 | 16.25 ± 3.40 | Transforms | 0.62 ± 0.01 | ND |
| D62K | 19.00 ± 9.84 | 13.20 ± 3.83 | Transforms | 2.70 ± 0.03 | ND |
| S63D | 15.25 ± 10.50 | 15.00 ± 4.69 | Transforms | 1.88 ± 0.74 | ND |
| T64D | 20.60 ± 8.32 | 12.00 ± 3.00 | Increased ability | 1.28 ± 0.24 | ND |
| R66E | 22.33 ± 6.80 | 12.00 ± 3.00 | Increased ability | 0.43 ± 0.05 | +++ |
| T72D | 9.66 ± 3.51 | 11.66 ± 3.51 | Transforms | 1.05 ± 0.07 | ND |
| H73E | 14.50 ± 7.77 | 15.00 ± 0.00 | Transforms | 1.07 ± 0.02 | ND |
| V74T | 11.00 ± 1.00 | 12.00 ± 3.00 | Transforms | 0.60 ± 0.03 | ND |
| R77E | 15.00 ± 4.24 | 13.50 ± 2.12 | Transforms | 0.08 ± 0.00 | + |
| E80K/D81K | 11.33 ± 4.93 | 11.00 ± 3.46 | Transforms | 0.68 ± 0.15 | ND |
| M84S | 7.66 ± 5.50 | 12.00 ± 3.00 | Decreased ability | 0.42 ± 0.08 | − |
| G85A | 6.00 ± 4.08 | 10.25 ± 4.27 | Transforms | 0.54 ± 0.13 | − |
| T86D | 10.00 ± 4.24 | 13.62 ± 4.46 | Transforms | 0.02 ± 0.00 | − |
| P92A | 17.00 ± 6.08 | 11.00 ± 3.46 | Increased ability | 0.58 ± 0.06 | ND |
| I93T | 11.50 ± 5.00 | 12.75 ± 2.87 | Transforms | 1.37 ± 0.10 | ND |
| QKP96-98EEA | 5.25 ± 2.98 | 14.50 ± 4.12 | Decreased ability | 1.70 ± 0.05 | ND |
| del21-24 | 2.66 ± 2.31 | 13.00 ± 3.46 | Does not transform | ND | ND |
| C58G/C91G | 4.46 ± 1.52 | 12.00 ± 3.00 | Decreased ability | 0.00 ± 0.00 | − |
| ras only | 1.50 ± 1.73 | 12.00 ± 3.00 | Does not transform | ND | ND |
ND, not determined.
−, little or no dimerization; +, intermediate dimerization; +++, similar to wild type.
Table 2.
Transformation properties of E7 mutants with mutations in predicted hydrophobic core residues and in the zinc-coordinating cysteines, with a summary of dimerization potential in vivo
| Mutant | Mean (±SD) no. of colonies |
Transformation phenotype | Mean dimerization level (±SD) in vivo | |
|---|---|---|---|---|
| Mutant | Wild type | |||
| C58G/C91G | 4.46 ± 1.52 | 12.00 ± 3.00 | Decreased ability | 0.00 ± 0.00 |
| L65A | 12.00 ± 9.93 | 12.75 ± 2.87 | Transforms | 0.03 ± 0.00 |
| L67R | 8.00 ± 3.36 | 16.25 ± 3.40 | Decreased ability | 0.44 ± 0.02 |
| V69A | 7.33 ± 3.51 | 14.16 ± 5.29 | Decreased ability | 0.05 ± 0.01 |
| I76A | 13.00 ± 6.97 | 12.00 ± 3.46 | Transforms | 0.28 ± 0.02 |
| L79A | 4.00 ± 3.60 | 12.00 ± 3.00 | Decreased ability | 0.23 ± 0.02 |
| LLMG82-85AAAD | 7.50 ± 7.45 | 11.25 ± 2.87 | Transforms | 0.71 ± 0.12 |
| L87A | 11.00 ± 7.87 | 10.25 ± 4.27 | Transforms | 0.01 ± 0.00 |
| I89A/V90A | 10.50 ± 6.19 | 10.25 ± 4.27 | Transforms | 0.03 ± 0.01 |
| C91G | 5.25 ± 4.27 | 13.75 ± 4.27 | Decreased ability | 0.00 ± 0.00 |
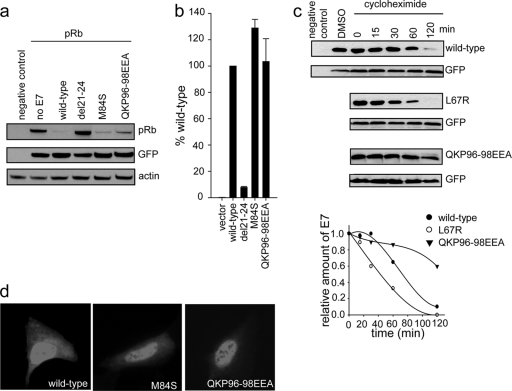
To determine if the two novel mutants identified in the transformation assay as having a decreased ability to transform cells (M84S and QKP96-98EEA mutants) were still able to target pRb, we tested their potential to degrade and bind the retinoblastoma protein (Fig. 4 a and b). Utilizing the pRb null cell line Saos2, we demonstrated that both the M84S and QKP96-98EEA mutants were able to efficiently target pRb for degradation (Fig. 4a). Additionally, employing a yeast two-hybrid approach where pRb was expressed as bait and the E7 wild type or mutant was expressed as prey, we showed that both the M84S and QKP96-98EEA mutants were able to bind pRb comparably to the wild type. Furthermore, these two mutants did not show any changes in subcellular localization (Fig. 4d) as determined by immunofluorescence.
Fig. 4.
Further characterization of a select panel of mutants. (a) E7 mutants with decreased transformation potential. The M84S and QKP96-98EEA mutants, as well as the CR2 del21-24 deletion mutant, which targets the LXCXE motif, were assessed for the ability to target pRb for degradation. Saos2 cells were transfected with a pRb expression plasmid and vectors expressing the indicated E7 mutant; pRb levels were determined by Western blotting at 48 h posttransfection. (b) The ability of E7 mutants to bind pRb was also assessed in a yeast two-hybrid assay. pRb was expressed as bait, with the indicated E7 mutant as prey. β-Galactosidase activity was measured and reported relative to that of wild-type E7. (c) The L67R mutant, with a mutation in the hydrophobic core of E7 which leads to decreased dimerization, and the QKP96-98EEA mutant, which dimerizes but fails to transform cells, were transfected into HEK293 cells together with a GFP expression plasmid. At 24 h posttransfection, cells were treated with cycloheximide for the indicated times and the levels of E7 determined by Western blotting. (d) Cellular localization of M84S and QKP96-98EEA mutants was assessed by immunofluorescence in U2OS cells.
To further investigate whether the introduced mutations could cause instability in the protein, HEK293 cells were transfected with wild-type E7 or L67R or QKP96-98EEA mutant expression plasmids. The cells were treated with cycloheximide for 0 to 120 min at 24 h posttransfection, and the amount of E7 was assessed by Western blotting. The L67R mutant, which targets the hydrophobic core and fails to dimerize, and perhaps causes structural defects in the protein, showed a decreased half-life compared to the wild type (Fig. 4c). In contrast, the QKP96-98EEA mutant appeared to be more stable than wild-type E7. Hence, instability of the QKP96-98EEA mutant was unlikely to contribute to the decreased transformation potential that we observed.
DISCUSSION
Previous studies on the E7 proteins of HPV16 and HPV18 have indicated that the C terminus is required to maintain a stable and functional structure of the protein (9, 13, 34, 51). CR3 contains two Cys-X-X-Cys motifs necessary for zinc binding and subsequent folding of the protomer into a unique viral β1β2α1β3α2 topology. Although the Zn-binding Cys-X-X-Cys motifs do not directly mediate the interprotomer contacts, folding of the individual protomers is necessary to create the dimer interface surface. Specifically, dimerization is mediated by interactions between hydrophobic residues of the α1 helices of the protomers and the formation of an intermolecular two-stranded antiparallel β-sheet between the β2 strand of one protomer and the β3 strand of the other. This forms a continuous hydrophobic core stabilizing the dimeric form of the E7 protein. Point mutations in either of the metal-binding motifs have consistently been shown to significantly reduce the ability of E7 to transform a variety of rodent cells (34, 46) and to immortalize primary human keratinocytes (30). It has not been established definitively if these defects are caused by perturbations in protomer structure that affect binding to specific factors or arise through a loss of dimerization. Indeed, it is not yet known if E7 must exist as a dimer for its transforming activity.
We therefore sought to resolve this issue. Our approach was to utilize the available structural information on E7 proteins (32, 37) and to model the structure of HPV16 E7 CR3. This would allow the identification of residues contributing to the formation of the hydrophobic core, and therefore to dimer stabilization, and also those surface residues which are solvent exposed and available for interaction with E7 target proteins (Fig. 1). This analysis resulted in construction of 21 novel mutations in the putative surface-exposed residues and 8 mutations targeting the highly conserved residues that likely form the hydrophobic core. Some of the latter residues have previously been targeted for mutagenesis (12, 30). As negative controls, we also mutated the Zn-coordinating Cys-X-X-Cys motifs. In total, we created 28 novel mutants and studied their properties in conjunction with several other previously described mutants, such as the L67R mutant. This new panel of E7 CR3 mutants has the potential for many other future applications in this field of study beyond the dimerization properties we assessed.
Early work that addressed the potential of HPV16 E7 to dimerize in vivo utilized the yeast two-hybrid system (13, 51). Employing our extensive panel of mutants and the yeast two-hybrid assay, we tested our bait and prey samples in five combinations: empty vector-mutant E7, mutant E7-empty vector (to determine any possible autoactivation by E7), wild-type E7–mutant E7, mutant E7–wild-type E7, and mutant E7-mutant E7. Although relatively insignificant, any observed autoactivation of the reporter by E7 was subtracted from the reported values (Fig. 2). Yeast two-hybrid data obtained when the mutant E7-mutant E7 combination was tested (Fig. 2) were considered the most biologically relevant.
E7 dimerizes with a reported Kd of 1.1 μM (14). Based on an expression level equivalent to the Kd, a mutation leading to a 5-fold increase in Kd would result in an ∼45% change in activity in our system with respect to wild-type E7; an increase in Kd of 10-fold would result in ∼28% of wild-type activity. Using a 5-fold increase in Kd as a guide, we considered any E7 CR3 mutant with <45% of the wild-type E7 activity to be impaired for dimerization. Using this criterion, all but one of the mutants with mutations predicted to be in the hydrophobic core of the protomer were impaired in the ability to dimerize (Fig. 2c).
Based on the criterion of <45% of wild-type E7 activity, we identified six mutants with mutations in putatively surface-exposed residues with a reduced ability to dimerize, with the possibility that the G85A mutant also falls into this category (Fig. 2b). The apparent inability of these mutant E7 proteins to dimerize could also result from decreased expression, and this could have been the case for several of these proteins (e.g., the V55T, G85A, and R77E mutants). Interestingly, two of the mutants with reduced dimerization had mutations affecting residues G85 and T86, which map close to the junction between the two protomers. These residues may be involved in mediating intersubunit contacts, suggesting that despite their being solvent exposed, a mutation at any of these residues may interfere with stable dimer formation. Additionally, the relatively large positively charged side chain of arginine at position 77 was targeted for mutagenesis in a previous dimerization study and was replaced with a single hydrogen in glycine (13). In that report, the R77G mutant was able to interact strongly with wild-type E7, and it also retained transforming and transcriptional activities in a previous mutagenic analysis conducted in mammalian cells (39). In this study, R77 was replaced with the oppositely charged amino acid glutamate, which should retain solvent interaction. This more dramatic alteration greatly reduced dimerization in vivo. However, this may have been related to decreased expression, as it exhibited only a slight reduction in vitro (Fig. 3). However, this residue is highly conserved across all HPV E7 proteins examined, suggesting that it could play an important nonstructural role (Fig. 1).
As for the remaining surface-exposed mutations in mutants impaired for dimerization (V55T, F57A, R66E, and M84S mutants) (Fig. 2d), these residues are not predicted to lie close to the dimer interface, and a role in stabilizing the dimer is not a likely reason for the lack of interaction between the two protomers. These mutations in E7 may therefore cause other structural defects in the protein. When tested for dimerization in vitro using recombinant protein, all mutants except the R66E mutant had reduced binding (Fig. 3). This provides additional evidence that these mutations in E7 perturb folding of the protomer, which leads to a reduction in the ability to dimerize. It is not known why the in vitro and in vivo assays differed with respect to the R66E mutant. Further biochemical analysis of this mutant is necessary to establish if the introduced change in the protein leads to structural alterations and perhaps aggregation or higher-order oligomerization as opposed to dimerization.
We assessed the ability of wild-type E7 or our novel collection of mutants to transform primary BRK cells in cooperation with ras. Mutations targeting Cys-X-X-Cys motifs have been shown in similar assays to cause impairment in transformation (30, 34), and these mutations served as controls for the assay. Additionally, we also included the del21-24 mutant as another control for these experiments. The del21-24 mutation disrupts the primary pRb binding site in CR2 of E7. It has been studied extensively in the literature and used previously in similar assays (30, 34, 39). Table 1 summarizes the transformation results for the entire panel of surface-exposed mutants, many of which behave like the wild type in this assay. Some have increased transformation potential (V55T mutant, T64D mutant, etc.), indicating that these mutants may perhaps be better at targeting certain cellular proteins/pathways necessary for transformation. Additionally, two mutations in E7, M84S and QKP96-98EEA, were identified as causing decreased transformation ability. These two mutants retained the capacity to both bind and degrade pRb (Fig. 4a and b), and subcellular localization also remained comparable to that of wild-type E7 (Fig. 4d). Additionally, as illustrated by cycloheximide treatment experiments, the QKP96-98EEA mutation did not contribute to any destabilizing effects on the protein, as mutations that target the hydrophobic core might (Fig. 4c). Together, this evidence suggests that the M84S and QKP96-98EEA mutations disrupt interaction with other important cellular targets necessary for transformation, whose identities remain to be elucidated.
Furthermore, Table 2 summarizes transformation results for the hydrophobic core mutants. As expected, the C58G/C91G and C91G mutants had a decreased ability to transform cells. The L67R, V69A, and L79A mutants also had decreased transforming potentials. In contrast, a number of mutants with mutations in this region still transformed cells as effectively as wild-type E7, including the L65A, I76A, LLMG82-85AAAD, L87A, and I89A/V90A mutants. Importantly, our aim was to establish whether E7 must exist as a dimer to function in oncogenic transformation. Correlating transformation ability with dimerization (Table 1 and 2) established two important phenotypes: those mutants that fail to dimerize and have decreased transformation potential (M84S, C58G/C91G, C91G, and L79A mutants) and, notably, numerous mutants that fail to dimerize yet still transform cells as well as wild-type E7 (F57A, R77E, G85A, T86D, I76A, L87A, and I89A/V90A mutants). The second class of mutants clearly illustrates that E7 does not need to exist as a dimer in order to transform BRK cells. This conclusion is also reinforced by the V55T mutation, which results in a protein that is impaired for dimerization yet exhibits an increased potential to transform. Our results also raise the intriguing possibility that additional, as yet unidentified cellular targets of E7 remain to be identified that contribute to E7's transforming activity through interaction with the CR3 region.
In summary, our data strongly suggest that dimerization of E7 is not required for oncogenic transformation of primary rodent cells. In addition, our systematic and rational construction of a new collection of CR3 mutants by use of structural information should provide the field with useful reagents for tackling future questions regarding protein interaction surfaces in CR3 and the functional consequences of abrogating these interactions.
ACKNOWLEDGMENTS
This work was supported by a grant from the Canadian Institute of Health Research, awarded to J.S.M., and by a grant from the Canadian Cancer Society (018414) to G.S.S. L.B. gratefully acknowledges research support from the Associazione Italiana per la Ricerca sul Cancro. B.T. was supported by a Canadian Institute of Health Research doctoral award and a CIHR Michael Smith foreign study supplement.
We thank J. Omichinski for helpful discussions.
Footnotes
Published ahead of print on 20 July 2011.
REFERENCES
- 1. Adams A., Gottschling D. E., Kaiser C. A., Stearns T. 1997. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 2. Alonso L. G., et al. 2004. The HPV16 E7 viral oncoprotein self-assembles into defined spherical oligomers. Biochemistry 43:3310–3317 [DOI] [PubMed] [Google Scholar]
- 3. Androphy E. J., Hubbert N. L., Schiller J. T., Lowy D. R. 1987. Identification of the HPV-16 E6 protein from transformed mouse cells and human cervical carcinoma cell lines. EMBO J. 6:989–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Avvakumov N., Torchia J., Mymryk J. S. 2003. Interaction of the HPV E7 proteins with the pCAF acetyltransferase. Oncogene 22:3833–3841 [DOI] [PubMed] [Google Scholar]
- 5. Baker D., Sali A. 2001. Protein structure prediction and structural genomics. Science 294:93–96 [DOI] [PubMed] [Google Scholar]
- 6. Banks L., et al. 1987. Identification of human papillomavirus type 18 E6 polypeptide in cells derived from human cervical carcinomas. J. Gen. Virol. 68:1351–1359 [DOI] [PubMed] [Google Scholar]
- 7. Berezutskaya E., Bagchi S. 1997. The human papillomavirus E7 oncoprotein functionally interacts with the S4 subunit of the 26S proteasome. J. Biol. Chem. 272:30135–30140 [DOI] [PubMed] [Google Scholar]
- 8. Bodily J. M., Mehta K. P., Laimins L. A. 2011. Human papillomavirus E7 enhances hypoxia-inducible factor 1-mediated transcription by inhibiting binding of histone deacetylases. Cancer Res. 71:1187–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Braspenning J., et al. 1998. The CXXC Zn binding motifs of the human papillomavirus type 16 E7 oncoprotein are not required for its in vitro transforming activity in rodent cells. Oncogene 16:1085–1089 [DOI] [PubMed] [Google Scholar]
- 10. Brehm A., et al. 1998. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391:597–601 [DOI] [PubMed] [Google Scholar]
- 11. Brehm A., et al. 1999. The E7 oncoprotein associates with Mi2 and histone deacetylase activity to promote cell growth. EMBO J. 18:2449–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chesters P. M., Vousden K. H., Edmonds C., McCance D. J. 1990. Analysis of human papillomavirus type 16 open reading frame E7 immortalizing function in rat embryo fibroblast cells. J. Gen. Virol. 71:449–453 [DOI] [PubMed] [Google Scholar]
- 13. Clemens K. E., Brent R., Gyuris J., Munger K. 1995. Dimerization of the human papillomavirus E7 oncoprotein in vivo. Virology 214:289–293 [DOI] [PubMed] [Google Scholar]
- 14. Clements A., Johnston K., Mazzarelli J. M., Ricciardi R. P., Marmorstein R. 2000. Oligomerization properties of the viral oncoproteins adenovirus E1A and human papillomavirus E7 and their complexes with the retinoblastoma protein. Biochemistry 39:16033–16045 [DOI] [PubMed] [Google Scholar]
- 15. Dawar M., Deeks S., Dobson S. 2007. Human papillomavirus vaccines launch a new era in cervical cancer prevention. CMAJ 177:456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. de Villiers E. M., Fauquet C., Broker T. R., Bernard H. U., zur Hausen H. 2004. Classification of papillomaviruses. Virology 324:17–27 [DOI] [PubMed] [Google Scholar]
- 17. Dyson N., Guida P., Munger K., Harlow E. 1992. Homologous sequences in adenovirus E1A and human papillomavirus E7 proteins mediate interaction with the same set of cellular proteins. J. Virol. 66:6893–6902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Edmonds C., Vousden K. H. 1989. A point mutational analysis of human papillomavirus type 16 E7 protein. J. Virol. 63:2650–2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Funk J. O., et al. 1997. Inhibition of CDK activity and PCNA-dependent DNA replication by p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes Dev. 11:2090–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gietz R. D., Schiestl R. H., Willems A. R., Woods R. A. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355–360 [DOI] [PubMed] [Google Scholar]
- 21. Gillison M. L., et al. 2000. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J. Natl. Cancer Inst. 92:709–720 [DOI] [PubMed] [Google Scholar]
- 22. Goodwin E. C., DiMaio D. 2000. Repression of human papillomavirus oncogenes in HeLa cervical carcinoma cells causes the orderly reactivation of dormant tumor suppressor pathways. Proc. Natl. Acad. Sci. U. S. A. 97:12513–12518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goodwin E. C., et al. 2000. Rapid induction of senescence in human cervical carcinoma cells. Proc. Natl. Acad. Sci. U. S. A. 97:10978–10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Halbert C. L., Demers G. W., Galloway D. A. 1991. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J. Virol. 65:473–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Helt A. M., Galloway D. A. 2001. Destabilization of the retinoblastoma tumor suppressor by human papillomavirus type 16 E7 is not sufficient to overcome cell cycle arrest in human keratinocytes. J. Virol. 75:6737–6747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. 1989. Site-directed mutagenesis by overlap extension using the PCR. Gene 77:51–59 [DOI] [PubMed] [Google Scholar]
- 27. Huh K., et al. 2007. Human papillomavirus type 16 E7 oncoprotein associates with the cullin 2 ubiquitin ligase complex, which contributes to degradation of the retinoblastoma tumor suppressor. J. Virol. 81:9737–9747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huh K. W., et al. 2005. Association of the human papillomavirus type 16 E7 oncoprotein with the 600-kDa retinoblastoma protein-associated factor, p600. Proc. Natl. Acad. Sci. U. S. A. 102:11492–11497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jeon S., Allen-Hoffmann B. L., Lambert P. F. 1995. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J. Virol. 69:2989–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jewers R. J., Hildebrandt P., Ludlow J. W., Kell B., McCance D. J. 1992. Regions of human papillomavirus type 16 E7 oncoprotein required for immortalization of human keratinocytes. J. Virol. 66:1329–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kammann M., Laufs J., Schell J., Gronenborn B. 1989. Rapid insertional mutagenesis of DNA by PCR. Nucleic Acids Res. 17:5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu X., Clements A., Zhao K., Marmorstein R. 2006. Structure of the human papillomavirus E7 oncoprotein and its mechanism for inactivation of the retinoblastoma tumor suppressor. J. Biol. Chem. 281:578–586 [DOI] [PubMed] [Google Scholar]
- 33. Massimi P., Banks L. 2005. Transformation assays for HPV oncoproteins. Methods Mol. Med. 119:381–395 [DOI] [PubMed] [Google Scholar]
- 34. McIntyre M. C., Frattini M. G., Grossman S. R., Laimins L. A. 1993. Human papillomavirus type 18 E7 protein requires intact Cys-X-X-Cys motifs for zinc binding, dimerization, and transformation but not for Rb binding. J. Virol. 67:3142–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McLaughlin-Drubin M. E., Huh K. W., Munger K. 2008. Human papillomavirus type 16 E7 oncoprotein associates with E2F6. J. Virol. 82:8695–8705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morris E. J., Dyson N. J. 2001. Retinoblastoma protein partners. Adv. Cancer Res. 82:1–54 [DOI] [PubMed] [Google Scholar]
- 37. Ohlenschlager O., et al. 2006. Solution structure of the partially folded high-risk human papilloma virus 45 oncoprotein E7. Oncogene 25:5953–5959 [DOI] [PubMed] [Google Scholar]
- 38. Patrick D. R., Oliff A., Heimbrook D. C. 1994. Identification of a novel retinoblastoma gene product binding site on human papillomavirus type 16 E7 protein. J. Biol. Chem. 269:6842–6850 [PubMed] [Google Scholar]
- 39. Phelps W. C., Munger K., Yee C. L., Barnes J. A., Howley P. M. 1992. Structure-function analysis of the human papillomavirus type 16 E7 oncoprotein. J. Virol. 66:2418–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Phelps W. C., Yee C. L., Munger K., Howley P. M. 1988. The human papillomavirus type 16 E7 gene encodes transactivation and transformation functions similar to those of adenovirus E1A. Cell 53:539–547 [DOI] [PubMed] [Google Scholar]
- 41. Phillips A. C., Vousden K. H. 1997. Analysis of the interaction between human papillomavirus type 16 E7 and the TATA-binding protein, TBP. J. Gen. Virol. 78:905–909 [DOI] [PubMed] [Google Scholar]
- 42. Schneider-Gadicke A., Schwarz E. 1987. Transcription of human papillomavirus type-18 DNA in human cervical carcinoma cell lines. Haematol. Blood Transfus. 31:380–381 [DOI] [PubMed] [Google Scholar]
- 43. Smotkin D., Wettstein F. O. 1987. The major human papillomavirus protein in cervical cancers is a cytoplasmic phosphoprotein. J. Virol. 61:1686–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Storey A., Almond N., Osborn K., Crawford L. 1990. Mutations of the human papillomavirus type 16 E7 gene that affect transformation, transactivation and phosphorylation by the E7 protein. J. Gen. Virol. 71:965–970 [DOI] [PubMed] [Google Scholar]
- 45. Vousden K. H., Jat P. S. 1989. Functional similarity between HPV16E7, SV40 large T and adenovirus E1a proteins. Oncogene 4:153–158 [PubMed] [Google Scholar]
- 46. Watanabe S., Kanda T., Sato H., Furuno A., Yoshiike K. 1990. Mutational analysis of human papillomavirus type 16 E7 functions. J. Virol. 64:207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Willard L., et al. 2003. VADAR: a web server for quantitative evaluation of protein structure quality. Nucleic Acids Res. 31:3316–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zerfass-Thome K., et al. 1996. Inactivation of the cdk inhibitor p27KIP1 by the human papillomavirus type 16 E7 oncoprotein. Oncogene 13:2323–2330 [PubMed] [Google Scholar]
- 49. Zhang Z., Smith M. M., Mymryk J. S. 2001. Interaction of the E1A oncoprotein with Yak1p, a novel regulator of yeast pseudohyphal differentiation, and related mammalian kinases. Mol. Biol. Cell 12:699–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. zur Hausen H. 1996. Papillomavirus infections—a major cause of human cancers. Biochim. Biophys. Acta 1288:F55–F78 [DOI] [PubMed] [Google Scholar]
- 51. Zwerschke W., Joswig S., Jansen-Durr P. 1996. Identification of domains required for transcriptional activation and protein dimerization in the human papillomavirus type-16 E7 protein. Oncogene 12:213–220 [PubMed] [Google Scholar]