Abstract
Susceptibility to respiratory syncytial virus (RSV) infection in mice is genetically determined. While RSV causes little pathology in C57BL/6 mice, pulmonary inflammation and weight loss occur in BALB/c mice. Using major histocompatibility complex (MHC)-congenic mice, we observed that the H-2d allele can partially transfer disease susceptibility to C57BL/6 mice. This was not explained by altered viral elimination or differences in the magnitude of the overall virus-specific cytotoxic T lymphocyte (CTL) response. However, H-2d mice showed a more focused response, with 70% of virus-specific CTL representing Vβ8.2+ CTL directed against the immunodominant epitope M2-1 82, while in H-2b mice only 20% of antiviral CTL were Vβ9+ CTL specific for the immunodominant epitope M187. The immunodominant H-2d-restricted CTL lysed target cells less efficiently than the immunodominant H-2b CTL, probably contributing to prolonged CTL stimulation and cytokine-mediated immunopathology. Accordingly, reduction of dominance of the M2-1 82-specific CTL population by introduction of an M187 response in the F1 generation of a C57BL/6N × C57BL/6-H-2d mating (C57BL/6-H-2dxb mice) attenuated disease. Moreover, disease in H-2d mice was less pronounced after infection with an RSV mutant failing to activate M2-1 82-specific CTL or after depletion of Vβ8.2+ cells. These data illustrate how the MHC-determined diversity and functional avidity of CTL responses contribute to disease susceptibility after viral infection.
INTRODUCTION
Respiratory syncytial virus (RSV) causes significant morbidity and mortality in infants, immunocompromised adults, and the elderly (4, 17). The nature and severity of disease vary widely between infected individuals. Viral, host, and environmental factors probably all contribute to this variable disease expression, but the relative roles of these factors in human RSV disease remain difficult to evaluate. A series of recent genetic association studies have suggested that polymorphisms in a range of genes encoding cytokines, chemokines, surfactant proteins, or toll-like receptors influence the disease phenotype in humans (1, 8, 19, 21, 23, 35, 45, 52, 57). However, it remains unclear how and to what extent these factors contribute to disease pathogenesis.
Inbred mouse strains have also been used to identify genetic factors that influence susceptibility to RSV infection. In a study of 20 strains of inbred mice, permissiveness to viral replication has been found to differ by up to a factor of 100 between certain strains (44). Disease susceptibility was not analyzed in that study. A second study compared virus titers and RSV-induced weight loss in 8 strains of mice and reported low virus replication and no disease in “resistant” C57BL/6 mice, whereas viral titers and disease were more pronounced in “susceptible” AKR, 129P3, or BALB/c mice (51). The difference in viral replication and weight loss between BALB/c and C57BL/6 mice was confirmed in two further studies (7, 49), although airway obstruction and airway hyperresponsiveness were similar (7). Furthermore, in a model of vaccine-induced eosinophilia during RSV infection, BALB/c mice were more prone to develop a pathological eosinophilic response than C57BL/6 mice and this was partly linked to the H-2 locus (20). Higher viral replication could be a reasonable explanation for the different outcomes of infection in the two strains (33) but may not be the only relevant factor.
Both in humans and in mice, cytotoxic T lymphocytes (CTL) have been identified as important mediators of virus control and disease. Infants with congenital T cell deficiencies cannot eliminate RSV (11, 12, 18), and depletion of T cells leads to persistent infection in BALB/c mice (14). Adoptive transfer of RSV-specific T cells can eliminate RSV from infected mice but also aggravates disease (5). In immunodeficient human infants persistently infected with RSV who are undergoing bone marrow transplantation, virus control has been observed in parallel to donor T cell reconstitution, and this was associated with significant deterioration of lung disease (11). Both CD4+ and CD8+ T cells can eliminate virus and cause immunopathology independently, but CD8+ T cells appear to be more effective (14). The composition and quality of the CTL response to viruses is determined by the major histocompatibility complex (MHC) haplotype. An important contribution of the MHC to susceptibility to viral infections has been documented in murine herpesvirus hominis type 1 (HVH-1) (34) and lymphocytic choriomeningitis virus (LCMV) infection (31) and has been linked to the impact of the MHC on T cell repertoire and T cell avidity (34). For RSV infection, a recent study documented that the MHC influences the extent of pulmonary CTL infiltrates and disease susceptibility in adult mice reinfected after neonatal priming (54). However, it remained unclear how the MHC governs disease-inducing CTL responses and how these observations in a reinfection model relate to primary infection.
In this study, we addressed the question of to what extent and how the MHC-determined CD8+ T cell response contributes to the different outcomes after primary RSV infection. For this, we not only compared C57BL/6 to BALB/c mice but analyzed three C57BL/6 mouse strains differing only in their MHC haplotype: C57BL/6 (H-2b), C57BL/6-H-2d, and the F1 generation of a C57BL/6N × C57BL/6-H-2d mating (C57BL/6-H-2dxb). This allowed studying the impact of the MHC independent of strain-specific background genes. Our results show that the MHC has no impact on virus elimination but influences weight loss and pulmonary inflammation. These manifestations of RSV-induced disease are determined by a highly focused CTL response exhibiting a low functional avidity (28, 50) and therefore contributing to prolonged cytokine-mediated immunopathology.
MATERIALS AND METHODS
Mice.
Specific-pathogen-free BALB/c, C57BL/6N, and C57BL/6-H-2d (B6.C-H-2d/bByJ) mice were obtained from Charles River (Sulzfeld, Germany) or Jackson Laboratories (Bar Harbor, ME) and used at 6 to 12 weeks of age. C57BL/6-H-2dxb mice are the F1 generation of a C57BL/6N × C57BL/6-H-2d mating. Mice were kept in an individual ventilated cage (IVC) unit (BioZone, Kent, United Kingdom). All animal experiments were performed in accordance with guidelines of the local animal care commission (accreditation no. 35/9185.81/G-08/49).
Virus infection and in vivo T cell depletion.
Human RSV (RSV) A2 and RSV 8A (55) were grown on HEp-2 cells and stored at −80°C until use. Mice were anesthetized intraperitoneally (i.p.) with ketamine and xylazine and inoculated intranasally (i.n.) with 1 × 106 PFU of RSV. RSV titers from lung homogenates were determined as described previously (5). To deplete T cells bearing T cell receptors (TCRs) with the Vβ8.2 (F23.2 [26]) chain, mice were injected i.p. with 100 μg of F23.2 in 200 μl phosphate-buffered saline (PBS) 1 day prior to and 2 and 5 days after infection (24). Efficiency of depletion was determined by flow cytometry.
Flow cytometry.
Cells were isolated either by performing a bronchoalveolar lavage (BAL) as described previously (40) or from lung parenchyma as described previously (56). Cells were stained with the following antibodies: CD3-allophycocyanin (clone 145-2C11), CD8-phycoerythrin (PE) Cy5 (clone 53-6.7), CD4-PE (clone RM4-5), CD25-fluorescein isothiocyanate (FITC) (clone 7D4), and Vβ chain FITC (all from BD Bioscience, Heidelberg, Germany), and with MHC-Kd M2-1 82-90 tetramers (provided by the National Institutes of Health tetramer facility, Emory University, Atlanta, GA). The frequency of gamma interferon (IFN-γ)-producing T cells was determined by restimulating 1 × 105 BAL fluid cells for 3 h with the respective peptide (RSV M2-1 82-90 [M2-1 82] or M187-196 [M187], synthesized by PolyPeptide Group, Strasbourg, France) at a concentration of 0.1 μg/ml or with decreasing peptide concentrations in a total volume of 150 μl/well in the presence of monensin (Golgistop; BD). Cells were surface stained with anti-CD3 and anti-CD8 antibodies and then stained intracellularly with an anti-IFN-γ antibody (clone XMG1.2; BD) using the Cytofix/Cytoperm kit according to the manufacturer's instructions (BD). In some experiments, 1 × 105 BAL fluid cells were restimulated with 4 × 104 RAW309CR.1 (TIB-69; ATCC), a murine macrophage-like cell line expressing both H-2d and H-2b MHC class I molecules, which had been infected 1 day previously with RSV at a multiplicity of infection (MOI) of 5. To determine the frequency of Th17 cells, 1 × 105 BAL fluid cells were restimulated in the presence of monensin with phorbol myristate acetate (PMA)-ionomycin for 3 h. After harvesting, the cells were surface stained with anti-CD3 and anti-CD4 antibodies, followed by intracellular staining using anti-interleukin 17A (IL-17A) (clone TC11-18H10; BD) antibody. For regulatory T cell (Treg) staining, BAL fluid cells were surface stained without restimulation, followed by intracellular staining for FoxP3 (clone MF23; BD). Cells were analyzed on a FACSsort cytometer using the Cellquest Pro v4.02 software program.
Cytometric bead array.
BAL was performed with 0.8 ml of phosphate-buffered saline. Fifty microliters of supernatants of BAL fluid or of lung homogenates were analyzed for secreted cytokines (IL-6, IL-10, monocyte chemoattractant protein 1 [MCP-1], IFN-γ, tumor necrosis factor alpha [TNF-α], and IL-12p70) using a CBA mouse inflammation kit (BD) according to the manufacturer's instructions. The data were analyzed by using the CBA software program (BD).
Cytotoxicity assays.
Ex vivo CTL assays were performed under “Mini-Killer” conditions (41). P815 cells (H-2d; TIB-64; ATCC) or RAW309CR.1 cells (H-2dxb) were used as target cells. They were labeled with decreasing peptide concentrations from 2 × 10−5 to 2 × 10−13 M and 51Cr for 2 h. BAL fluid cells were then incubated with target cells for 5 h in a total volume of 100 μl Iscove's modified Dulbecco's medium (IMDM). After this incubation period, 50 μl of the supernatant was removed and analyzed for 51Cr release in a γ counter. Spontaneous 51Cr release was below 20% in all experiments.
Statistics.
Data were analyzed using Student's t test in the case of a normal distribution of raw data and equality of standard deviations. When standard deviations differed significantly, the Welch t test was used. When comparing more than two experimental groups, a one-way analysis of variance (ANOVA) with posttest was performed. Tests were performed with the GraphPad InStat software program, version 3.06. Differences were considered significant at a P value less than 0.05.
RESULTS
The MHC haplotype is an important determinant of disease susceptibility following RSV infection.
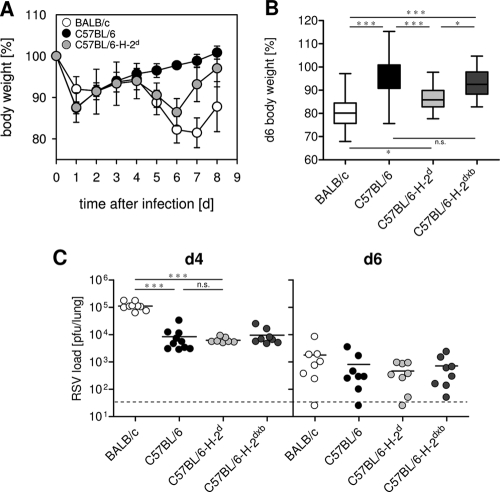
After infection with 106 PFU of RSV, BALB/c mice develop an inflammatory pneumonia that is clinically associated with signs of illness, such as weight loss, ruffled fur, and inactivity (16). These signs of illness are not observed after RSV infection of C57BL/6 mice (51). To analyze to what extent these differences are determined by the MHC locus, we performed experiments with MHC congenic mice. BALB/c (H-2d) mice, C57BL/6 (H-2b) mice, MHC congenic C57BL/6-H-2d mice, and an F1 backcross (H-2dxb) of these mice to C57BL/6 mice were infected with 106 PFU RSV and weighed daily. As expected, C57BL/6 mice did not show any signs of illness, while BALB/c mice significantly lost weight from day 5 to day 7 after infection (Fig. 1A and B). Weight loss in C57BL/6-H-2d mice was comparable to that in BALB/c mice, but C57BL/6-H-2d mice recovered earlier (Fig. 1A), while C57BL/6-H-2dxb did not lose weight after RSV infection (Fig. 1B). These results show that disease susceptibility can in part be transferred with the H-2d MHC allele and that the protective effect of the H-2b allele is dominant.
Fig. 1.
Weight loss after RSV infection is more pronounced in mice carrying the H-2d MHC haplotype, but disease is independent of peak virus titers and elimination. Mice were infected intranasally (i.n.) with 1 × 106 PFU RSV. (A and B) Weight was monitored for 8 days. (A) The data show mean values and SD from mice in one experiment representative of 6 independent experiments. (B) Each box represents the weight on day six after infection in 12 independent experiments for BALB/c and C57BL/6 mice (n = 44). In six of those experiments, C57BL/6-H-2d and C57BL/6-H-2dxb mice were included (n = 18). *, P < 0.05; ***, P < 0.001; n.s. (not significant), P > 0.05. (C) Pulmonary virus load was determined at days four and six after infection. Data were pooled from two independent experiments with 3 to 4 mice per group and time point. The dashed line indicates the detection limit. ***, P < 0.001; n.s. (not significant), P > 0.05.
RSV-induced disease is not determined by peak virus titers or virus elimination kinetics.
RSV replicates to higher titers in lungs of BALB/c mice than in those of C57BL/6 mice (44, 51). To determine whether the disease observed in C57BL/6-H-2d mice is due to differences in peak virus titers or to a different kinetic of viral elimination, we compared lung RSV titers in the four mouse strains at day 4 (d4) and d6 after infection. As expected, at the peak of viral replication, BALB/c mice had 10-fold-higher pulmonary virus titers than C57BL/6 mice, while the virus load in lungs of C57BL/6-H-2d and C57BL/6-H-2dxb mice was similar to that for C57BL/6 mice (Fig. 1C). At d6 after infection, all groups had similar virus titers (Fig. 1C), and at d7, virus was eliminated below the detection limit in all strains (data not shown). The poor correlation between signs of illness and viral titers suggests that different kinetics of virus growth and elimination are not responsible for the different infection outcomes in C57BL/6 and MHC-congenic C57BL/6-H-2d mice.
Introduction of the H-2d MHC results in increased cytokine production in the lungs of C57BL/6 mice.
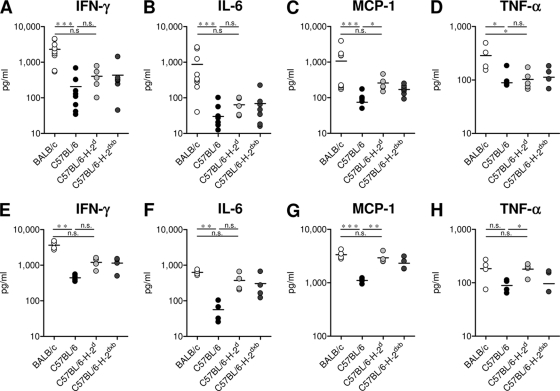
To determine whether these differences in weight loss were also reflected by the extent of inflammation in the infected lungs, we analyzed a panel of inflammatory cytokines in supernatants of BAL fluid on d7 (Fig. 2A to D) and of lung homogenates on d6 after infection (Fig. 2E to H). BALB/c mice showed significantly higher levels of IFN-γ, MCP-1, IL-6, and TNF-α than C57BL/6 mice (Fig. 2A to H), while no differences were found in the levels of IL-10 and IL-12p40 (data not shown). C57BL/6 mice carrying the H-2d haplotype had significantly higher levels of MCP-1 and TNF-α than C57BL/6 mice with the H-2b haplotype, although the pulmonary inflammatory response was less pronounced than that in BALB/c mice (Fig. 2A to H). These data show that the MHC H-2d locus influences the pulmonary cytokine levels in response to infection, mainly MCP-1 and TNF-α. However, the extent of weight loss was not fully reflected by the investigated cytokines, indicating that other factors also contribute to this manifestation of immunopathology. Thus, C57BL/6-H-2dxb mice had levels of cytokines similar to those for C57BL/6-H-2d mice but showed less weight loss.
Fig. 2.
Different cytokine patterns in supernatants of BALF and of lung homogenates of BALB/c, C57BL/6, C57BL/6-H-2d, and C57BL/6-H-2dxb mice. Mice were infected with 1 × 106 PFU RSV, and the indicated cytokines, IFN-γ (A and E), IL-6 (B and F), MCP-1 (C and G), and TNF-α (D and H), were determined in supernatants of BAL fluid on d7 (A to D) or of lung homogenates on d6 (E to H) following infection by cytometric bead array. The graphs show results of two independent experiments (n = 5 to 9). *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s. (not significant), P > 0.05.
The pulmonary CTL response is of similar magnitude in MHC-congenic mice.
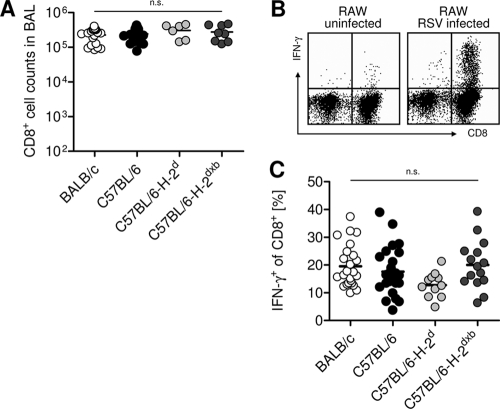
Differences in MHC result in a different extent and composition of T cell responses. Since T cells and in particular CD8+ cytotoxic T cells play a major role in RSV-induced pathology in BALB/c mice (14), we compared the CTL responses to RSV infection in the different mouse strains. Seven days after RSV infection, cells recruited to the airways were eluted via bronchoalveolar lavage (BAL) and restimulated with RSV-infected RAW309Cr.1 cells expressing both MHC haplotypes, H-2d and H-2b. The percentage of IFN-γ-producing cells among CD8+ T cells was determined by intracellular cytokine staining. The absolute numbers of CTL eluted by BAL (Fig. 3A) and the total numbers of CTL in lung parenchyma (data not shown) were similar in all groups. Moreover, the fraction of virus-specific CTL among total BAL fluid CTL was similar in all 4 experimental groups (Fig. 3B and C). Thus, disease susceptibility was not determined by the amount of virus-specific CTL recruited to the lung.
Fig. 3.
Total antiviral CTL responses are comparable in all groups. Mice were infected i.n. with 1 × 106 PFU RSV. Seven days later, BAL fluid CTL were analyzed for total numbers of CD8 T cells and IFN-γ production after restimulation with RSV-infected RAW macrophages. (A) Absolute numbers of CD8 T cells in BAL fluid were determined by a combination of microscopic cell counts and flow cytometry. (B) Representative fluorescence-activated cell sorter (FACS) plots gated on CD3+ T cells are shown. (C) Frequency of IFN-γ-producing CD8 T cells following restimulation with RAW cells. Pooled data from 6 independent experiments with 3 to 5 mice/group for BALB/c and C57BL/6 mice are shown. In two (A) or three (C) of those experiments, C57BL/6-H-2d and C57BL/6-H-2dxb mice were included. n.s. (not significant), P > 0.05.
Since pulmonary inflammatory responses may also be influenced by regulatory or inflammatory helper T cell responses, we determined the number and percentage of FoxP3-expressing and IL-17-expressing CD4+ T cells in the BAL fluid of BALB/c and C57BL/6 mice. On d7 after RSV infection, both strains showed similar numbers and percentages of BAL fluid CD4+ T cells expressing IL-17 or FoxP3 (data not shown).
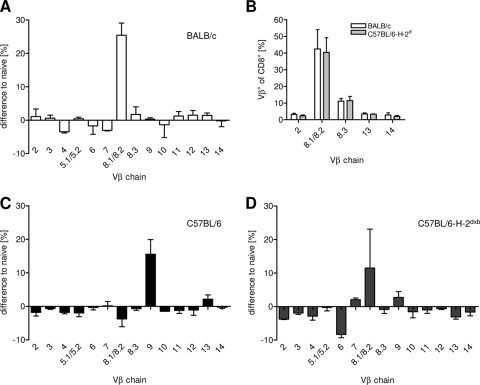
Vβ skewing of pulmonary CTL is more pronounced in BALB/c and C57BL/6-H-2d mice than in C57BL/6 mice.
Although the MHC differences did not lead to a difference in the overall number of pulmonary virus-specific CTL, their composition and quality could be influenced by the MHC. To address this issue, we analyzed Vβ chain usage of pulmonary CTL and compared it to the Vβ usage of CTL obtained from the spleens of naive mice. In these experiments, we initially used an antibody recognizing Vβ8.1/8.2. We later obtained a Vβ8.2-specific hybridoma (F23.2 [26]) and showed that more than 90% of the cells recognized by the Vβ8.1/8.2 antibody expressed Vβ8.2 (see also Fig. 6C). Seven days after RSV infection, there was a significant increase in Vβ8.1/8.2-expressing CTL in the BAL fluid of BALB/c and C57BL/6-H-2d mice, while there were few changes in the frequency of CTL using other chains (Fig. 4A and B). In C57BL/6 mice, we observed a relative increase in the dominant Vβ9-expressing population, but this was less pronounced (Fig. 4C). In C57BL/6-H-2dxb mice, the expansion of the Vβ8.1/8.2-expressing population was attenuated, and there was only a slight increase in the Vβ9-expressing population (Fig. 4D). Overall, the response was highly oligoclonal in the H-2d strains, while this was less pronounced in the strains carrying H-2b alleles.
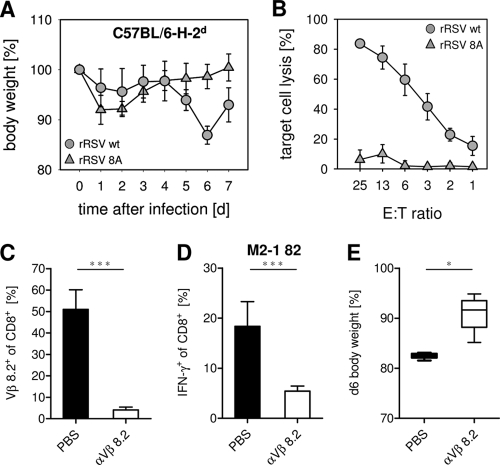
Fig. 6.
Reduced weight loss after infection of C57BL/6-H-2d mice with an RSV strain carrying a mutation in the immunodominant epitope or after depletion of Vβ8.2+ CTL in BALB/c mice. (A) C57BL/6-H-2d mice were infected i.n. with 1 × 106 PFU of recombinant RSV (rRSV) wild type (wt) or rRSV 8A. Weight was monitored for 7 days. Data show mean values and SD for 6 mice per group, obtained in 2 independent experiments. (B) Specific lysis of target cells labeled with M2-1 82 peptide is shown for BAL fluid CTL of mice infected with rRSV 8A or rRSV wt. (C to E) Mice were treated with the hybridoma supernatant (αVβ8.2 [F23.2] d-1, d2, and d5) i.p. and infected i.n. with 1 × 106 PFU of RSV. (C) Efficacy of depletion was monitored by flow cytometry of BAL fluid CTL at d7 after infection. (D) After in vitro restimulation with M2-1 82, the percentage of IFN-γ+ CD8+ T cells is shown. (E) Percent body weight is shown for depleted and PBS-treated mice (d6). Results shown are from one of two independent experiments with 4 mice per group. *, P < 0.05; ***, P < 0.001.
Fig. 4.
TCR repertoire of BALB/c, C57BL/6-H-2d, C57BL/6, and C57BL/6-H-2dxb mice after RSV infection. Mice were infected i.n. with 1 × 106 PFU RSV. Seven days later, BAL fluid cells were stained with antibodies against CD3, CD8, and the indicated Vβ chains and analyzed by flow cytometry. The mean percentages of the indicated Vβ+ CD8+ T cells from BAL fluid of RSV-infected BALB/c (A), C57BL/6 (C), or C57BL/6-H-2dxb (D) mice (n = 4), normalized by subtraction of the mean values from spleens of naive control mice, are shown. (B) Vβ usage among BAL fluid CTL from BALB/c and C57BL/6-H-2d mice is shown for 5 selected Vβ chains. The experiments were repeated with similar results.
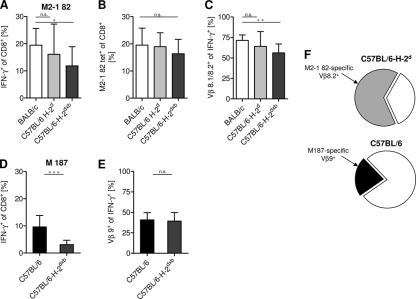
The epitope-specific pulmonary CTL response is more focused in H-2d mice than in H-2b mice.
To further investigate the T cell repertoire in the different mouse strains, we analyzed defined virus-derived CTL responses. After RSV infection of BALB/c mice, the majority of the RSV-specific CTL response is directed against the immunodominant CTL epitope M2-1 82-90 (M2-1 82), presented by H-2Kd (29, 30), while in C57BL/6 mice, the immunodominant epitope is M187-195 (M187), presented by H-2Db (48). On d7 after RSV infection, in BALB/c and C57BL/6-H-2d mice, about 20% of the pulmonary CTL produced IFN-γ in response to M2-1 82 stimulation (Fig. 5A). This high frequency of M2-1 82-specific CTL, representing the majority (∼80%) of the virus-specific CTL (38, 39, 55), was confirmed by tetramer staining (Fig. 5B). In contrast, only about 10% of total BAL fluid CTL, representing about 50% of the virus-specific CTL of C57BL/6 mice (32), were specific for the immunodominant epitope M187 (Fig. 5D). Analysis of the Vβ usage of these immunodominant populations revealed that in BALB/c and C57BL/6-H-2d mice, about 75% of the M2-1 82-specific CTL carried the dominant Vβ8.2 chain (Fig. 5C) (3, 55). In contrast, only about 40% of the M187-specific CTL of C57BL/6 mice carried the dominant Vβ9 chain (Fig. 5E) (3). These results revealed significant differences in the composition of the virus-specific CTL repertoire, with a highly focused response in H-2d mice (about 70% of virus-specific CTL represented by a single Vβ-expressing dominant population; Fig. 5F) and a broader, less focused response (about 20% represented by a single Vβ-expressing population) in H-2b mice (Fig. 5F). While we were preparing the manuscript, similar results regarding the Vβ usage of epitope-specific CTL (using the different IMGT [international ImMunoGeneTics information system] nomenclature) in BALB/c and C57BL/6 mice were published by Graham and colleagues (3). Of note, the coexpression of the H-2b allele, which protected from RSV-induced weight loss, led to a significant reduction in the dominant H-2d-restricted CTL population and to a more diverse CTL response (Fig. 5A to E).
Fig. 5.
CTL response against the immunodominant peptide epitopes of BALB/c and C57BL/6 mice. Seven days after intranasal inoculation with 1 × 106 PFU RSV, epitope-specific BAL fluid CTL were quantified by flow cytometry. (A and D) Intracellular IFN-γ production was measured following restimulation with the indicated peptides: H-2Kd epitope M2-1 82-90 (A) or H-2Db epitope M187-195 (D). (B) M2-1 82-specific CTL were quantified by tetramer staining. (C and E) Percentages of Vβ8.1/8.2+ cells among the M2-1 82-specific IFN-γ+ cells (C) or percentages of Vβ9+ cells among the M187-specific IFN-γ+ cells (E). (A to E) Pooled data from 3 to 5 independent experiments are shown, with 3 to 5 mice per group. (F) Estimated contribution of the dominant CTL population to the overall RSV-specific response. *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s. (not significant), P > 0.05.
Vβ8.2+ M2-1 82-specific CTL response is responsible for RSV-induced disease in H-2d mice.
Collectively, the obtained results indicated that the focused M2-1 82-specific CTL response in H-2d mice could be a significant determinant of RSV-induced disease. This would predict that failure to activate this T cell population or depletion of Vβ8.2+ cells should attenuate the disease. To test this hypothesis, we infected C57BL/6-H-2d mice either with RSV or with RSV 8A, carrying a loss-of-recognition point mutation in the immunodominant M2-1 82 epitope (55). Indeed, in mice infected with the mutant virus, weight loss was significantly reduced (Fig. 6A) in the absence of the M2-1 82-specific CTL population (Fig. 6B). In a second set of experiments, Vβ8.2+ cells were depleted via injecting an anti-Vβ8.2 depleting antibody 1 day before and 2 and 5 days after RSV infection of BALB/c mice. This treatment depleted about 90% of Vβ8.2+ CD8+ T cells (Fig. 6C) and about 70% of M2-1 82-specific CTL (Fig. 6D). Figure 6E shows that this partial depletion significantly attenuated weight loss after RSV infection of BALB/c mice.
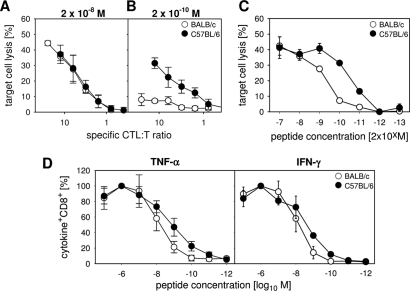
H-2b-restricted M187-specific CTL show higher functional avidity than H-2d-restricted M2-1 82-specific CTL.
To compare functional avidities of the two immunodominant CTL populations, we first analyzed their abilities to lyse peptide-loaded target cells. For this, we labeled 51Cr-loaded RAWdxb cells with decreasing concentrations of both peptides (M2-1 82 and M187) and incubated them with BAL fluid cells obtained 7 days after RSV infection. BAL fluid cell numbers in the assay were adapted to contain the same numbers of M187-specific CTL (for C57BL/6 mice) and M2-1 82-specific CTL (for BALB/c mice), as determined by IFN-γ analysis. While the specific lysis by RSV-specific CTL was comparable at higher peptide concentrations (Fig. 7A and C), M187-specific CTL from C57BL/6 mice were able to lyse their target cells more efficiently at lower peptide concentrations than M2-1 82-specific CTL from BALB/c mice (Fig. 7B and C). Similar results were obtained by analysis of cytokine production after restimulation with decreasing peptide concentrations. While there was a good IFN-γ and TNF-α response to saturating peptide concentrations (10−6 M) in both strains (defined as 100%), the dose-response curves showed higher proportions of TNF-α- and IFN-γ-producing CTL for C57BL/6 mice than for BALB/c mice at more limiting peptide concentrations (Fig. 7D). These data indicate that the dominant M2-1 82-specific CTL population in H-2d mice has a lower functional avidity for its cognate antigen/MHC complex than the M187-specific CTL population in H-2b mice. Therefore, a less efficient killing of antigen-presenting cells (APC) may contribute to prolonged stimulation of CTL, resulting in enhanced secretion of pathogenic cytokines.
Fig. 7.
The highly focused M2-1 82-specific CTL population has a lower avidity than the M187-specific CTL population. The specific lysis of target cells loaded with a peptide concentration of 10−8 M (A) or 10−10 M (B) or loaded with peptides in decreasing concentrations (C) is shown for BAL fluid CTL of BALB/c and C57BL/6 mice 7 days following infection with RSV. (D) Dose-response curves of M2-182- and M187-specific CD8 T cells stimulated with decreasing peptide concentrations are shown for TNF-α+ (left panel) or IFN-γ+ (right panel) CTL. Values obtained after stimulation with a peptide concentration of 10−6 M represented the maximum response and were set as 100% response. Mean values and SD for 5 mice per group are shown.
DISCUSSION
This study shows that the MHC locus can contribute to disease severity after a respiratory viral infection by determining the diversity and functional avidity of the antiviral CTL response. Limited diversity and dominance of a CTL population that is not very efficient in target cell lysis may lead to increased cytokine production and subsequent immune-mediated disease. In contrast, a more diverse CTL response mediating more effective elimination of target cells attenuates the disease. Of note, in a viral infection that is eliminated independent of perforin, these differences in disease severity can occur despite similar kinetics of viral elimination.
Starting from the observation that RSV-induced weight loss and the pulmonary inflammatory response are more pronounced in BALB/c (H-2d) mice than in C57BL/6 (H-2b) mice, we analyzed the contribution of the MHC-determined CTL response to differences in disease susceptibility using MHC-congenic mice. In contrast to previous studies using an F1 hybrid mouse model (47, 49), we used MHC-congenic mice on the same genetic background (C57BL/6) which differed only in their MHC expression. This allowed us to study the impact of the MHC independent of strain-specific background genes. We found that the disease observed in BALB/c mice following RSV infection was in part transferred with the H-2d allele to mice on the C57BL/6 genetic background. In contrast, C57BL/6 mice carrying both MHC haplotypes, H-2d and H-2b, were found to be less susceptible to disease, indicating that the H-2b haplotype is dominant and confers resistance to RSV-induced disease. It should be stated that an increased disease susceptibility remained in BALB/c mice compared to C57BL/6-H-2d mice, indicating a relevant role of additional genetic factors carried by these different mouse strains.
The key parameters determined by the MHC are the composition and efficacy of CD4+ and CD8+ T cell responses, both of which can contribute to disease pathogenesis. Of note, regulatory helper T cells were recently shown to limit immunopathology following RSV infection (13, 46). Moreover, a critical role for IL-17 in disease development after influenza infection was observed (9). In our experimental model, there were no differences in the overall number of CD4+ helper T cells or in the number of regulatory T cells or Th17 cells recruited to the lung after RSV infection of H-2b mice versus results for H-2d mice. As expected from previous observations (14, 15), disease was clearly determined by CD8+ T cells and was dependent on the presence of the immunodominant population of H-2d-restricted M2-1 82-specific CTL. Thus, only infection with wild-type virus but not with a mutant virus carrying a single amino acid substitution in the dominant epitope (RSV 8A), which failed to induce this particular CTL population, caused disease in C57BL/6-H-2d mice. This was similar to our previous observations with BALB/c mice (55). In addition, depletion of a large fraction of this immunodominant CTL population by anti-Vβ8.2 treatment also significantly reduced the pathology.
How do MHC-determined CTL responses contribute to disease in our model? A recent study showed that increased recruitment of CD8+ T cells to the lung contributes to MHC-dependent increased disease susceptibility in mice reinfected with RSV after neonatal priming (54). Increased CTL recruitment was not the key factor in our primary infection model, where the MHC did not affect the extent of the overall pulmonary CTL infiltration or the extent of the virus-specific CTL response. We identified two other relevant factors. First, the MHC determined the diversity of the virus-specific CTL response. In H-2d mice, about 70% of virus-specific CTL represented a particular Vβ-expressing population directed against a single epitope, while these were only 20% of CTL in H-2b mice. Of note, C57BL/6 mice expressing both H-2b and H-2d alleles did not develop disease. In that situation, reduction of the relative dominance of the M2-1 82-specific population in favor of an additional M187-specific CTL population provided an antiviral CTL repertoire that was not associated with disease. These results extend previous observations by Ruckwardt et al. In elegant experiments using different viral epitope mutant RSV isolates, they showed with C57BL/6/BALB/c F1 mice that the extent of dominance of the M2-1 82 population over the M187 population was related to disease (47). Similar to results of our experiments, a very narrow M2-1 82-specific response was associated with more-severe disease, while broadening the response by M187-reactive CTL was beneficial to the host.
The second factor was the quality of the M2-1 82-specific CTL response. These H-2d-restricted CTL had a lower functional avidity than M187-specific CTL mediating the immunodominant response in H-2b mice, and on a per-cell basis, their efficacy in mediating target cell lysis was significantly lower. A link between reduced CTL cytotoxicity and enhanced cytokine-mediated disease has been observed in several other experimental models. For example, in perforin-deficient mice, impaired target cell lysis does not significantly delay clearance of RSV infection but leads to an increase in inflammatory cytokines, in particular IFN-γ, associated with enhanced disease manifestations (2). Increased cytokine-mediated immunopathology as a consequence of impaired target cell lysis is also the key pathogenetic principle in a human genetic disorder of lymphocyte cytotoxicity called familial hemophagocytic lymphohistiocytosis (FHL) (10). In that disease, which can be reproduced by LCMV infection of perforin-deficient mice (25), defective lysis of APC leads to a prolonged stimulation of virus-specific CTL with subsequent IFN-γ-mediated disease (25, 42). We have recently shown that quite subtle differences in CTL cytotoxicity that can be detected only under the low peptide concentrations also used in this study are highly relevant and can be a deciding factor regarding recovery versus lethal disease in the FHL model (B. Jessen, A. Maul-Pavicic, H. Ufheil, T. Vraetz, A. Enders, K. Lehmberg, A. Längler, U. Gross-Wieltsch, A. Bay, Z. Kaya, Y. T. Bryceson, E. Koscielniak, S. Badawy, G. Davies, M. Hufnagel, A. Schmitt-Gräff, P. Aichele, U. zur Stadt, K. Schwarz, and S. Ehl, submitted for publication).
In the context of these examples, we would like to argue that after infection of H-2d mice, the MHC-determined generation of the large immunodominant M2-1 82-specific CTL population is a disadvantage to the host because it is of low functional avidity. This does not impair virus elimination but may lead to delayed elimination of APC and prolonged stimulation of antiviral CTL, which contribute to cytokine-mediated immunopathology. We thus provide an additional aspect illustrating how T cell diversity and functional avidity determined by the MHC can contribute to increased susceptibility to viral infections. While previous data obtained in a mouse model of herpesvirus infection showed that these factors can determine control of virus replication and survival after viral infection (34), our results indicate that they can be equally important in the determination of infection-induced immunopathology.
In humans, MHC-disease associations are much more difficult to prove. While in mice defined single homozygous MHC alleles are present on a constant non-MHC background, codominant expression of 6 MHC class I alleles in the human outbred population introduces significant complexity. Moreover, individual MHC-restricted CTL responses may be disease causing or protective, even after infection with the same virus, depending on viral parameters (36). This may explain why association studies between particular HLA class I antigens and severe RSV bronchiolitis in infants failed to reveal obvious correlations (22). Nevertheless, MHC-disease correlations have been observed in several human infectious diseases. For example, the major genetic determinants of HIV-1 control affect HLA class I peptide presentation (43), and certain MHC alleles have been shown to delay onset of disease in HIV-1-infected patients (6, 27, 53). Furthermore, clearance of hepatitis C virus infection has been associated with certain MHC class I alleles (37).
Apart from improving our understanding of MHC-disease associations in viral infections, our findings also have implications for vaccine design. If antiviral responses are dominated by ineffective CTL, this may have consequences for viral clearance and for infection-associated immunopathology. If the vaccine is designed to elicit a broad and diverse T cell response, the risk of inducing one dominant response of limited quality is significantly smaller.
ACKNOWLEDGMENTS
This study was supported by the German Ministry for Education and Research (BMBF 01 EO 0803) and the German Research Foundation (SFB620, TPA4).
We acknowledge the excellent technical assistance of Nadja Goos. We are indebted to Peter L. Collins for providing the RSV reverse genetics system for generation of the RSV 8A mutant.
Footnotes
Published ahead of print on 27 July 2011.
REFERENCES
- 1. Amanatidou V., Apostolakis S., Spandidos D. A. 2009. Genetic diversity of the host and severe respiratory syncytial virus-induced lower respiratory tract infection. Pediatr. Infect. Dis. J. 28:135–410 [DOI] [PubMed] [Google Scholar]
- 2. Aung S., Rutigliano J. A., Graham B. S. 2001. Alternative mechanisms of respiratory syncytial virus clearance in perforin knockout mice lead to enhanced disease. J. Virol. 75:9918–9924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Billam P., et al. 2011. T cell receptor clonotype influences epitope hierarchy in the CD8+ T cell response to respiratory syncytial virus infection. J. Biol. Chem. 286:4829–4841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brandt C. D., et al. 1973. Epidemiology of respiratory syncytial virus infection in Washington, D.C. 3. Composite analysis of eleven consecutive yearly epidemics. Am. J. Epidemiol. 98:355–364 [DOI] [PubMed] [Google Scholar]
- 5. Cannon M. J., Openshaw P. J., Askonas B. A. 1988. Cytotoxic T cells clear virus but augment lung pathology in mice infected with respiratory syncytial virus. J. Exp. Med. 168:1163–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carrington M., et al. 1999. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 283:1748–1752 [DOI] [PubMed] [Google Scholar]
- 7. Chavez-Bueno S., Mejias A., Jafri H. S., Ramilo O. 2005. Respiratory syncytial virus: old challenges and new approaches. Pediatr. Ann. 34:62–68 [DOI] [PubMed] [Google Scholar]
- 8. Choi E. H., Lee H. J., Yoo T., Chanock S. J. 2002. A common haplotype of interleukin-4 gene IL4 is associated with severe respiratory syncytial virus disease in Korean children. J. Infect. Dis. 186:1207–1211 [DOI] [PubMed] [Google Scholar]
- 9. Crowe C. R., et al. 2009. Critical role of IL-17RA in immunopathology of influenza infection. J. Immunol. 183:5301–5310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Saint Basile G., Menasche G., Fischer A. 2010. Molecular mechanisms of biogenesis and exocytosis of cytotoxic granules. Nat. Rev. Immunol. 10:568–579 [DOI] [PubMed] [Google Scholar]
- 11. El Saleeby C. M., Suzich J., Conley M. E., DeVincenzo J. P. 2004. Quantitative effects of palivizumab and donor-derived T cells on chronic respiratory syncytial virus infection, lung disease, and fusion glycoprotein amino acid sequences in a patient before and after bone marrow transplantation. Clin. Infect. Dis. 39:e17–e20 [DOI] [PubMed] [Google Scholar]
- 12. Fishaut M., Tubergen D., McIntosh K. 1980. Cellular response to respiratory viruses with particular reference to children with disorders of cell-mediated immunity. J. Pediatr. 96:179–186 [DOI] [PubMed] [Google Scholar]
- 13. Fulton R. B., Meyerholz D. K., Varga S. M. 2010. Foxp3+ CD4 regulatory T cells limit pulmonary immunopathology by modulating the CD8 T cell response during respiratory syncytial virus infection. J. Immunol. 185:2382–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Graham B. S., Bunton L. A., Wright P. F., Karzon D. T. 1991. Role of T lymphocyte subsets in the pathogenesis of primary infection and rechallenge with respiratory syncytial virus in mice. J. Clin. Invest. 88:1026–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Graham B. S., Johnson T. R., Peebles R. S. 2000. Immune-mediated disease pathogenesis in respiratory syncytial virus infection. Immunopharmacology 48:237–247 [DOI] [PubMed] [Google Scholar]
- 16. Graham B. S., Perkins M. D., Wright P. F., Karzon D. T. 1988. Primary respiratory syncytial virus infection in mice. J. Med. Virol. 26:153–162 [DOI] [PubMed] [Google Scholar]
- 17. Hall C. B. 1999. Respiratory syncytial virus: a continuing culprit and conundrum. J. Pediatr. 135:2–7 [PubMed] [Google Scholar]
- 18. Hall C. B., et al. 1986. Respiratory syncytial viral infection in children with compromised immune function. N. Engl. J. Med. 315:77–81 [DOI] [PubMed] [Google Scholar]
- 19. Hull J., Thomson A., Kwiatkowski D. 2000. Association of respiratory syncytial virus bronchiolitis with the interleukin 8 gene region in UK families. Thorax 55:1023–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hussell T., et al. 1998. Host genetic determinants of vaccine-induced eosinophilia during respiratory syncytial virus infection. J. Immunol. 161:6215–6222 [PubMed] [Google Scholar]
- 21. Inoue Y., et al. 2007. CD14-550 C/T, which is related to the serum level of soluble CD14, is associated with the development of respiratory syncytial virus bronchiolitis in the Japanese population. J. Infect. Dis. 195:1618–1624 [DOI] [PubMed] [Google Scholar]
- 22. Isaacs D., Taylor C. J., Ting A., McMichael A. J. 1989. HLA class I antigens in severe RSV bronchiolitis. Tissue Antigens 34:210–212 [DOI] [PubMed] [Google Scholar]
- 23. Janssen R., et al. 2007. Genetic susceptibility to respiratory syncytial virus bronchiolitis is predominantly associated with innate immune genes. J. Infect. Dis. 196:826–834 [DOI] [PubMed] [Google Scholar]
- 24. Johnson T. R., Varga S. M., Braciale T. J., Graham B. S. 2004. Vbeta14(+) T cells mediate the vaccine-enhanced disease induced by immunization with respiratory syncytial virus (RSV) G glycoprotein but not with formalin-inactivated RSV. J. Virol. 78:8753–8760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jordan M. B., Hildeman D., Kappler J., Marrack P. 2004. An animal model of hemophagocytic lymphohistiocytosis (HLH): CD8+ T cells and interferon gamma are essential for the disorder. Blood 104:735–743 [DOI] [PubMed] [Google Scholar]
- 26. Kappler J. W., Staerz U., White J., Marrack P. C. 1988. Self-tolerance eliminates T cells specific for Mls-modified products of the major histocompatibility complex. Nature 332:35–40 [DOI] [PubMed] [Google Scholar]
- 27. Kaslow R. A., et al. 1996. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2:405–411 [DOI] [PubMed] [Google Scholar]
- 28. Kuball J., et al. 2009. Increasing functional avidity of TCR-redirected T cells by removing defined N-glycosylation sites in the TCR constant domain. J. Exp. Med. 206:463–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kulkarni A. B., et al. 1995. Cytotoxic T cells specific for a single peptide on the M2 protein of respiratory syncytial virus are the sole mediators of resistance induced by immunization with M2 encoded by a recombinant vaccinia virus. J. Virol. 69:1261–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kulkarni A. B., Connors M., Firestone C. Y., Morse III H. C., Murphy B. R. 1993. The cytolytic activity of pulmonary CD8+ lymphocytes, induced by infection with a vaccinia virus recombinant expressing the M2 protein of respiratory syncytial virus (RSV), correlates with resistance to RSV infection in mice. J. Virol. 67:1044–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leist T., Althage A., Haenseler E., Hengartner H., Zinkernagel R. M. 1989. Major histocompatibility complex-linked susceptibility or resistance to disease caused by a noncytopathic virus varies with the disease parameter evaluated. J. Exp. Med. 170:269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lukens M. V., et al. 2006. Characterization of the CD8+ T cell responses directed against respiratory syncytial virus during primary and secondary infection in C57BL/6 mice. Virology 352:157–168 [DOI] [PubMed] [Google Scholar]
- 33. Mejias A., et al. 2004. Anti-respiratory syncytial virus (RSV) neutralizing antibody decreases lung inflammation, airway obstruction, and airway hyperresponsiveness in a murine RSV model. Antimicrob. Agents Chemother. 48:1811–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Messaoudi I., Guevara Patino J. A., Dyall R., LeMaoult J., Nikolich-Zugich J. 2002. Direct link between MHC polymorphism, T cell avidity, and diversity in immune defense. Science 298:1797–1800 [DOI] [PubMed] [Google Scholar]
- 35. Miyairi I., DeVincenzo J. P. 2008. Human genetic factors and respiratory syncytial virus disease severity. Clin. Microbiol. Rev. 21:686–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moskophidis D., et al. 1995. Role of virus and host variables in virus persistence or immunopathological disease caused by a non-cytolytic virus. J. Gen. Virol. 76(Pt. 2):381–391 [DOI] [PubMed] [Google Scholar]
- 37. Neumann-Haefelin C., et al. 2006. Dominant influence of an HLA-B27 restricted CD8+ T cell response in mediating HCV clearance and evolution. Hepatology 43:563–572 [DOI] [PubMed] [Google Scholar]
- 38. Nicholas J. A., Rubino K. L., Levely M. E., Adams E. G., Collins P. L. 1990. Cytolytic T-lymphocyte responses to respiratory syncytial virus: effector cell phenotype and target proteins. J. Virol. 64:4232–4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Openshaw P. J., Anderson K., Wertz G. W., Askonas B. A. 1990. The 22,000-kilodalton protein of respiratory syncytial virus is a major target for Kd-restricted cytotoxic T lymphocytes from mice primed by infection. J. Virol. 64:1683–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ostler T., Hussell T., Surh C. D., Openshaw P., Ehl S. 2001. Long-term persistence and reactivation of T cell memory in the lung of mice infected with respiratory syncytial virus. Eur. J. Immunol. 31:2574–2582 [DOI] [PubMed] [Google Scholar]
- 41. Ostler T., et al. 2001. An improved protocol for measuring cytotoxic T cell activity in anatomic compartments with low cell numbers. J. Immunol. Methods 257:155–161 [DOI] [PubMed] [Google Scholar]
- 42. Pachlopnik Schmid J., et al. 2009. Neutralization of IFNgamma defeats haemophagocytosis in LCMV-infected perforin- and Rab27a-deficient mice. EMBO Mol. Med. 1:112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pereyra F., et al. 2010. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330:1551–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Prince G. A., Horswood R. L., Berndt J., Suffin S. C., Chanock R. M. 1979. Respiratory syncytial virus infection in inbred mice. Infect. Immun. 26:764–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Puthothu B., Krueger M., Forster J., Heinzmann A. 2006. Association between severe respiratory syncytial virus infection and IL13/IL4 haplotypes. J. Infect. Dis. 193:438–441 [DOI] [PubMed] [Google Scholar]
- 46. Ruckwardt T. J., Bonaparte K. L., Nason M. C., Graham B. S. 2009. Regulatory T cells promote early influx of CD8+ T cells in the lungs of respiratory syncytial virus-infected mice and diminish immunodominance disparities. J. Virol. 83:3019–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ruckwardt T. J., et al. 2010. Responses against a subdominant CD8+ T cell epitope protect against immunopathology caused by a dominant epitope. J. Immunol. 185:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rutigliano J. A., Rock M. T., Johnson A. K., Crowe J. E., Jr., Graham B. S. 2005. Identification of an H-2D(b)-restricted CD8+ cytotoxic T lymphocyte epitope in the matrix protein of respiratory syncytial virus. Virology 337:335–343 [DOI] [PubMed] [Google Scholar]
- 49. Rutigliano J. A., Ruckwardt T. J., Martin J. E., Graham B. S. 2007. Relative dominance of epitope-specific CD8+ T cell responses in an F1 hybrid mouse model of respiratory syncytial virus infection. Virology 362:314–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Slifka M. K., Whitton J. L. 2001. Functional avidity maturation of CD8(+) T cells without selection of higher affinity TCR. Nat. Immunol. 2:711–717 [DOI] [PubMed] [Google Scholar]
- 51. Stark J. M., et al. 2002. Genetic susceptibility to respiratory syncytial virus infection in inbred mice. J. Med. Virol. 67:92–100 [DOI] [PubMed] [Google Scholar]
- 52. Tal G., et al. 2004. Association between common Toll-like receptor 4 mutations and severe respiratory syncytial virus disease. J. Infect. Dis. 189:2057–2063 [DOI] [PubMed] [Google Scholar]
- 53. Trachtenberg E., et al. 2003. Advantage of rare HLA supertype in HIV disease progression. Nat. Med. 9:928–935 [DOI] [PubMed] [Google Scholar]
- 54. Tregoning J. S., et al. 2010. Genetic susceptibility to the delayed sequelae of neonatal respiratory syncytial virus infection is MHC dependent. J. Immunol. 185:5384–5391 [DOI] [PubMed] [Google Scholar]
- 55. Vallbracht S., et al. 2007. Influence of a single viral epitope on T cell response and disease after infection of mice with respiratory syncytial virus. J. Immunol. 179:8264–8273 [DOI] [PubMed] [Google Scholar]
- 56. Vallbracht S., Unsold H., Ehl S. 2006. Functional impairment of cytotoxic T cells in the lung airways following respiratory virus infections. Eur. J. Immunol. 36:1434–1442 [DOI] [PubMed] [Google Scholar]
- 57. Zeng R., Li C., Li N., Wei L., Cui Y. 2011. The role of cytokines and chemokines in severe respiratory syncytial virus infection and subsequent asthma. Cytokine 53:1–7 [DOI] [PubMed] [Google Scholar]