Abstract
There is considerable variability in host susceptibility to human immunodeficiency virus type 1 (HIV-1) infection, but the host genetic determinants of that variability are not well understood. In addition to serving as a block for cross-species retroviral infection, TRIM5 was recently shown to play a central role in limiting primate immunodeficiency virus replication. We hypothesized that TRIM5 may also contribute to susceptibility to mucosal acquisition of simian immunodeficiency virus (SIV) in rhesus monkeys. We explored this hypothesis by establishing 3 cohorts of Indian-origin rhesus monkeys with different TRIM5 genotypes: homozygous restrictive, heterozygous permissive, and homozygous permissive. We then evaluated the effect of TRIM5 genotype on the penile transmission of SIVsmE660. We observed a significant effect of TRIM5 genotype on mucosal SIVsmE660 acquisition in that no SIV transmission occurred in monkeys with only restrictive TRIM5 alleles. In contrast, systemic SIV infections were initiated after preputial pocket exposures in monkeys that had at least one permissive TRIM5 allele. These data demonstrate that host genetic factors can play a critical role in restricting mucosal transmission of a primate immunodeficiency virus. In addition, we used our understanding of TRIM5 to establish a novel nonhuman primate penile transmission model for AIDS mucosal pathogenesis and vaccine research.
INTRODUCTION
There is considerable variability in host susceptibility to primate immunodeficiency virus infections, and the determinants of that variability are not well understood. Efforts have been made to identify host genetic factors that might contribute to the resistance and containment of human immunodeficiency virus type 1 (HIV-1), with the rationale that an understanding of the determinants of AIDS virus transmission and virus replication may guide the design of novel intervention strategies to prevent HIV-1 infection. Although studies have provided evidence that particular major histocompatibility complex (MHC) alleles (6, 24, 30) and chemokine receptor polymorphisms influence the control of HIV-1 in infected individuals, chemokine receptor 5 (CCR5) genetic variants remain the only host genetic factors that have been shown to impact susceptibility to HIV-1 acquisition (4, 23). However, CCR5 polymorphisms account for only a small fraction of exposed, HIV-1-uninfected individuals.
Simian immunodeficiency virus (SIV)-infected rhesus monkeys have provided an important model for studying AIDS immunopathogenesis and evaluating the efficacy of HIV-1 vaccine and microbicide candidates. Infection studies can be initiated in monkeys with specific genetic characteristics, using defined routes of infection and well-characterized virus challenge stocks. The variability in susceptibility to SIV infection and replication in rhesus monkeys exposed to virus by mucosal routes presents an opportunity to elucidate host genetic determinants of AIDS virus restriction that are manifested at mucosal surfaces.
The cytoplasmic tripartite motif protein 5α (TRIM5α) restricts the cross-species transmission of primate immunodeficiency viruses (13, 15, 31, 37, 40). In addition, our group and others have recently shown that allelic variation of TRIM5 modulates the in vitro infectivity and level of in vivo viral replication of SIV strains in susceptible host species (18, 21, 22, 33). Viral inhibition by TRIM5α is mediated by an interaction between the B30.2/SPRY domain and virion Gag capsids that are released into the cytoplasm of the target cell after viral attachment and entry. Two TRIM5 alleles are expressed codominantly in Indian-origin rhesus monkeys. If one or both of those alleles are from the group of alleles 6 to 11, we have previously demonstrated that SIV replicates at a higher level after infection (both peak and set-point viral loads) than if both are from the group of alleles 1 to 5 (21, 22, 33). Accordingly, common variants of TRIM5 in Indian-origin rhesus monkeys can be grouped into three genotypes: homozygous restrictive, heterozygous permissive, and homozygous permissive (21, 22).
We sought to determine whether TRIM5 plays a role in the variable host susceptibility to SIV mucosal infections in rhesus monkeys. We developed a penile challenge model in order to elucidate the role of TRIM5 in mucosal transmission of SIV. We hypothesized that restrictive TRIM5 gene products may mediate an intrinsic host genetic resistance to mucosal SIV infection. Here we describe a host genetic determinant of susceptibility to AIDS virus acquisition in a biologically relevant mucosal transmission model.
MATERIALS AND METHODS
Monkeys and virus.
Outbred, adult male Indian-origin rhesus monkeys (3 to 10 years old) were housed at an AAALAC-accredited institution (Bioqual, Rockville, MD). All procedures were performed with full protocol approvals from the Institutional Animal Care and Use Committee of the Vaccine Research Center, NIAID, NIH, and in accordance with the Guide for the Care and Use of Laboratory Animals (27a). Animals were exposed by trained technicians and veterinary staff to undiluted SIVsmE660 challenge stock, a cell-free uncloned pathogenic stock that was expanded on human peripheral blood mononuclear cells (PBMCs). The env genotypic diversity of this challenge stock is approximately 98% and is well described (17). This virus stock has a concentration of 3 × 108 SIV RNA copies/ml and a 50% tissue culture infectious dose (TCID50) on TZM-bl cells of 1 × 104/ml. We exposed 18 monkeys (3 groups; n = 6/group) to SIVsmE660. All animal studies were approved by the Vaccine Research Center and Harvard Medical School Animal Care and Use Committees.
Preputial pocket inoculation.
All monkeys were anesthetized with 10 mg ketamine/kg of body weight and 0.5 mg/kg xylazine, both administered intramuscularly. After anesthesia, 0.5 ml of undiluted SIV was placed atraumatically in the prepuce pouch by using a syringe without a needle. The foreskin was held opposed for 30 min following exposure to prevent loss of the inoculum and to maximize surface contact. After the 30-min exposure, any excess SIV inoculum was gently wiped away and the animals were returned to their cages to recover from anesthesia. A maximum of 12 weekly penile exposures were carried out using SIVsmE660.
Monkey genotyping.
Genomic DNAs were isolated from lymphocytes from rhesus monkeys by use of a QIAmp DNA kit (Qiagen) and were sequenced for MHC class I alleles (A*01, A*02, B*17, and B*08) and TRIM5 exons as previously described (21, 22).
Cellular immune responses.
Flow cytometric assessments of T lymphocyte subsets utilized the following monoclonal antibodies (MAbs): anti-CD3–Alexa Fluor 700 (SP34.2; BD Biosciences), anti-CD4–peridinin chlorophyll protein (PerCP)–Cy5.5 (L200; BD Biosciences), anti-CD8–allophycocyanin (APC)–H7 (SK1; BD Biosciences), anti-CD28–phycoerythrin (PE)–Cy7 (CD28.2; eBiosciences), anti-CD95–APC (DX2; BD Biosciences), anti-CCR5–PE (3A9; BD Biosciences), and anti-CCR7–fluorescein isothiocyanate (FITC) (150503; R&D Systems).
Viral RNA (vRNA) quantification.
To monitor the infection status of these animals prospectively, whole-blood samples were obtained from each exposed animal every week. Plasma SIV RNA levels were determined using a modified 2-step quantitative reverse transcription-PCR (qRT-PCR) process as previously described (39). Experimental samples were run in parallel with an SIV gag RNA standard on an Applied Biosystems StepOne real-time PCR system. The turnaround time for plasma viral load measurement was 5 days. Monkeys that had a positive viral load were not exposed to further virus exposures. The lower limit of detection using this assay was 500 SIV RNA copies/ml.
Viral RNA extraction and cDNA synthesis.
Viral RNAs were extracted using a QIAamp viral RNA minikit (Qiagen). RNA was eluted and immediately subjected to cDNA synthesis. Reverse transcription of RNA to single-stranded cDNA was performed using SuperScript III reverse transcriptase according to the manufacturer's recommendations (Invitrogen). In brief, each cDNA reaction mixture included 1× RT buffer, a 0.5 mM concentration of each deoxynucleoside triphosphate, 5 mM dithiothreitol, 2 U/ml RNaseOUT (RNase inhibitor), 10 U/ml of SuperScript III reverse transcriptase, and 0.25 mM antisense primer SIVsm/macEnvR1 (5′-TGTAATAAATCCCTTCCAGTCCCCCC-3′). The mixture was incubated at 50°C for 60 min, followed by an increase in temperature to 55°C for an additional 60 min. The reaction mix was then heat inactivated at 70°C for 15 min and treated with 2 U of RNase H at 37°C for 20 min. The newly synthesized cDNA was used immediately or frozen at −80°C.
Single-genome amplification (SGA).
cDNA was serially diluted and distributed among wells of replicate 96-well plates to identify a dilution where PCR-positive wells constituted <30% of the total number of reaction wells, as previously described (16). At this dilution, most positive wells would contain amplicons derived from a single cDNA molecule. This was confirmed for every positive well by direct sequencing of the amplicon and inspection of the sequence for mixed bases, which would be indicative of priming from more than one original template or the introduction of PCR error in early cycles. Any sequence with mixed bases was excluded from further analysis. PCR amplification was performed in the presence of 1× high-fidelity polymerase in a 20-μl reaction mix (Invitrogen). First-round PCR primers included the sense primer SIVsm/macEnvF1 (5′-CCTCCCCCTCCAGGACTAGC-3′) and the antisense primer SIVsm/macEnvR1 (5′-TGTAATAAATCCCTTCCAGTCCCCCC-3′). PCR was performed in MicroAmp 96-well reaction plates (Applied Biosystems) with the following PCR parameters: 1 cycle of 94°C for 2 min; 35 cycles of a denaturing step of 94°C for 15 s, an annealing step of 54°C for 30 s, and an extension step of 68°C for 4 min; and a final extension step of 68°C for 10 min. Next, 2 μl of the first-round PCR product was added to a second-round PCR mix. For second-round PCR, the sense primer SIVsmTatF2 (5′-TATGATAGACATGGAGACACCCTTGAAGGAGC-3′) and the antisense primer SIVsmEnvR2 (5′-ATGAGACATGTCTATTGCCAATTTGTA-3′) were used. The second-round PCR was performed under the same conditions used for first-round PCR, but with a total of 45 cycles. env amplicons were inspected on precast 1% agarose 96-well E-gels (Invitrogen).
Env sequencing and analysis.
Both DNA strands of env amplicons were directly sequenced using partially overlapping fragments. Individual sequence fragments for each amplicon were assembled and edited using the Sequencher 10 program (Gene Codes, Ann Arbor, MI). Inspection of individual chromatograms allowed for the confirmation of amplicons derived from single versus multiple templates. The absence of mixed bases at each nucleotide position throughout env was taken as evidence of SGA from a single vRNA/cDNA template. This quality control measure allowed us to exclude from the analysis amplicons that resulted from PCR-generated in vitro recombination events or Taq polymerase errors and to obtain individual SIV env sequences that proportionately represented those circulating in vivo. All alignments were codon aligned using GeneCutter (http://www.hiv.lanl.gov/content/sequence/GENE_CUTTER/cutter.html) and then reviewed manually to maintain intact reading frames. Maximum likelihood trees of the nucleotide sequences from all monkeys with the SIVsmE660 inoculum were constructed using PhyML (version 2.4.4) and the GTR+gamma+I substitution model.
Transmitted/founder (T/F) virus identification.
The aligned sequences from the earliest time point for each monkey were assessed for a Poisson distribution of intersequence distances with Poisson Fitter (8). In Poisson analysis, failure to reject the null hypothesis indicates a homogeneous, single-founder infection and no selection (16). Because APOBECG3 hypermutation can influence the Poisson distribution of sequence distances, the test was repeated with exclusion of sequences or sites that exhibited effects of hypermutation. The tests were reviewed in aggregate to evaluate which animals exhibited homogeneous infection, and a P value of <0.01 was used to reject the null hypothesis. In monkeys that were not infected with homogeneous founder virus, we further analyzed the env sequences by visualization of phylogenetic trees paired with highlighter polymorphism plots to determine the clades of common ancestry (17).
Gag sequencing.
Viral RNAs were isolated from plasmas of SIV-infected rhesus monkeys and reverse transcribed into cDNA. Primers gag-F (5′-AGCACCATCTAGTGGCAGAGGA-3′) and gag-R (5′-GAAATGGCTCTTTTGGCCCTT-3′) were used to generate bulk PCR products for each infected animal. PCR products were then subjected to T7/SP6 dideoxy sequencing.
Statistical analyses.
We used GraphPad Prism, version 5, for statistical comparisons. The comparisons between the 3 groups of TRIM5 genotypes utilized an unordered, 3 × 2 Fisher's exact test and the log rank test. P values of <0.05 were considered significant.
Nucleotide sequence accession numbers.
All env sequences from ramp-up and peak viremia were deposited in GenBank under accession numbers HM045021 to HM045284.
RESULTS
Sequence alignment of retroviral capsid regions in SIV gag.
In this study, we chose to use the uncloned quasispecies SIVsmE660 as the challenge virus. Given that SIVsmE660 has become increasingly important in AIDS pathogenesis and vaccine challenge studies (14, 20, 28, 32, 36), we felt that it was important to understand the genetic contribution of this virus to mucosal acquisition and pathogenesis. We have previously shown that TRIM5 alleles in Indian-origin rhesus monkeys have a significant effect on the control of SIVmac239 and SIVmac251 replication in lymphocytes both in vitro and in vivo after intravenous infection (21, 22, 33). In addition, we have also shown that TRIM5 alleles affect SIVsmE543 replication in vitro (22). To assess whether TRIM5 would be predicted to have a similar effect on SIVsmE660 replication, we characterized the gag region of our challenge stocks of SIVmac251 and SIVsmE660 quasispecies by using single-genome amplification. Based on a study by Kirmaier et al. (18), we aligned the postulated TRIM5 binding determinants in SIVsmE660 Gag with Gag protein sequences from SIVmac251, SIVmac239, and SIVsmE543. We observed that the capsid binding regions in all quasispecies in our stock of SIVsmE660 were similar to SIVsmE543 (Fig. 1). The serine at position 97 at the base of the helix 5 loop in the capsid was identical in SIVmac251 and SIVmac239, whereas the equivalent residue in SIVsm543 and SIVsmE660 was an arginine. The two glutamine residues (QQ) at positions 89 and 90 in the 4-5 loop of the capsid, which may affect TRIM5-mediated restriction (12, 34), were identical in SIVmac251 and SIVmac239 but disparate from the LPA residues in SIVsmE543 and the majority quasispecies in the SIVsmE660 stock. Based on these data, we concluded that our stock of SIVsmE660 should exhibit sensitivity to TRIM5 restriction similar to that observed with SIVsm543 in vitro.
Fig. 1.
Partial amino acid sequence alignment of SIV Gag. SIVmac251 and SIVsmE660 sequences were obtained by single-genome amplification of viral challenge stocks that were expanded in human PBMCs stimulated with concanavalin A and interleukin-2, while Gag sequences of SIVmac239 and SIVsmE543 were obtained from the Los Alamos HIV Sequence Database. Residues 89, 90, and 97 (SIVmac239 numbering) in the SIV capsid that are postulated to mediate host TRIM5 restriction are highlighted.
Rhesus monkeys expressing only restrictive TRIM5 alleles resist mucosal infection with SIVsmE660.
Having previously shown that the expression of restrictive or permissive TRIM5 alleles impacts the level of SIV replication in rhesus monkeys after infection initiated by intravenous inoculation, we sought to evaluate the contributions of particular TRIM5 alleles to the susceptibility to initiation of mucosal infections. We determined the TRIM5 genotypes of a cohort of adult male rhesus monkeys and divided them into 3 groups: in one group, only TRIM5 alleles 1 to 5 were present (homozygous restrictive); in the second group, one copy of alleles 1 to 5 and one copy of alleles 6 to 11 were present (heterozygous permissive); and in the third group, only alleles 6 to 11 were present (homozygous permissive).
The gross anatomy of the male genitalia of rhesus monkeys is comparable to that of humans. The skin covering the glans and penis in rhesus monkeys is referred to as the prepuce and is similar to the human foreskin. The histologic architecture and populations of potential HIV/SIV target cells in the foreskins of the two species have been shown to be similar (5, 7, 25, 26, 29). For this study, we developed a technique of atraumatically exposing the rhesus monkey prepuce and penile mucosa to SIV. After anesthesia, the monkeys were placed in a supine position and a prepuce pouch was created by pulling up the foreskin by use of blunt forceps. A total of 0.5 ml of undiluted SIV was then placed atraumatically in the prepuce pouch by using a syringe without a needle. The prepuce pouch containing the virus inoculum was held opposed for 30 min. While this technique was designed to maximize exposure of the preputial (foreskin) mucosa to SIV, the virus stock also came into contact with the glans, shaft, and urethral os of the monkeys.
We exposed the penile mucosae of 18 adult male rhesus monkeys (n = 6/group) to SIVsmE660 weekly for 12 weeks. Plasma SIV RNA levels were determined prospectively using quantitative real-time PCR. Strikingly, rhesus monkeys that were homozygous for restrictive TRIM5 alleles resisted infection from repeated preputial SIV exposures (Fig. 2 A). In contrast, infections were initiated in 5 of 6 monkeys in the heterozygous permissive group (Fig. 2B) and 5 of 6 monkeys in the homozygous permissive group (Fig. 2C). The numbers of monkeys in the 3 groups who were infected after 12 preputial SIV exposures were significantly different using unordered Fisher's exact test on a 3 × 2 table (P = 0.005). The numbers of exposures to infection in the 3 groups were also significantly different (P = 0.017; log rank test). It should be noted that most of the difference was between the TRIM5 homozygous restrictive group and the other 2 TRIM5 permissive groups (Fig. 3). There were not enough monkeys that were homozygous for TRIM5 alleles 6 to 11 to differentiate between the effects of having 1 and 2 permissive alleles on mucosal acquisition of infection. These data demonstrate that permissive TRIM5 genes are associated with increased susceptibility to SIV infection via the penis.
Fig. 2.
Systemic infections following repeated penile exposures of rhesus monkeys to SIVsmE660. Rhesus monkeys (n = 6/group) were exposed to SIVsmE660 in a 0.5-ml volume via the penile mucosa for 12 weeks. The first exposure was on week 0, and the last exposure was on week 11. Monkeys were monitored for systemic infection weekly by assessing plasma SIV gag RNA levels by qRT-PCR. The lower limit of detection using qRT-PCR was 500 SIV RNA copies/ml. SIV RNA levels are shown for the first 20 weeks following exposures in TRIM5 homozygous restrictive (A), heterozygous permissive (B), and homozygous permissive (C) monkeys. Viral RNA levels are shown as log10 copies of plasma viral RNA/ml of plasma at each time point. Monkeys that developed a positive plasma virus RNA level were not given further virus exposures. The numbers of monkeys that became infected in each group were statistically different by a 3 × 2 Fisher's exact test (P = 0.005).
Fig. 3.
Effect of TRIM5 genotype on number of exposures before penile infection with SIVsmE660 in rhesus monkeys. The effects of restrictive and permissive TRIM5 alleles on the number of exposures before infection are shown by Kaplan-Meier curves. There were 3 groups of monkeys (n = 6/group): the first group had only restrictive TRIM5 alleles (red), the second group had one permissive allele (blue), and the third group had only permissive alleles (black). The statistical comparison of the numbers of exposures before infection between the three groups of monkeys with different TRIM5 genotypes was made using the log rank test (P = 0.017).
Clinical status following SIV penile exposure.
After infection, there was a 1- to 2-log difference in peak viral replication in both TRIM5 homozygous and heterozygous permissive groups. In addition, we observed that some monkeys controlled SIVsmE660 to undetectable levels after peak viremia, a phenomenon that has been well documented previously for monkeys infected with this strain of SIV (10, 19). The variability in SIVsmE660 replication at peak and set-point viremia in monkeys infected via penile exposures is similar to that in monkeys infected via intrarectal (19) and intravenous (10) inoculations. There were not enough monkeys in the groups to determine whether the expression of particular TRIM5 alleles was associated with variability in viremia.
We then assessed the clinical consequences of infection in the preputially exposed rhesus monkeys by evaluating the percentages of CD4+ T cells and CD28+ CD95+ CCR5− CCR7+ central memory (CM) CD4+ T cells in peripheral blood mononuclear cells of all monkeys during the first 20 weeks (see gating strategy in Fig. S1 in the supplemental material). The percent CD4+ T cells declined in the TRIM5 heterozygous and homozygous permissive monkeys (Fig. 4 A), as determined by a paired Wilcoxon signed rank test (P = 0.04 and 0.02, respectively). However, there was no significant change in the percent CM CD4+ T cells within each group (Fig. 4B). In addition, there was a statistically significant difference in the change in percent CD4+ T cells between the 3 groups of monkeys, as determined by the Kruskal-Wallis test (P = 0.01) (Fig. 4C).
Fig. 4.
Clinical immune status after SIV penile exposures. The percentages of CD4+ T cells (A) and CCR7+ CCR5− CD28+ CD95+ central memory CD4+ T cells (B) were monitored in all 3 cohorts of rhesus monkeys. Asterisks indicate monkeys that resisted mucosal infection in the TRIM5 heterozygous and homozygous permissive groups. (A) There was a decline in the percentage of CD4+ T cells in monkeys in the TRIM5 heterozygous (P = 0.04) and homozygous (P = 0.02) permissive groups. The two-tailed Wilcoxon signed-rank test was used to compare the changes in percentage of CD4+ T cells after 20 weeks. (B) There was no significant decline in the percentage of central memory CD4+ T cells within all three groups. (C) Comparison of differences in percentages of CD4+ T cells after infection between the three groups. Bars representing the median values are shown for each group. The Kruskal-Wallis test was used to evaluate the differences in the percentages of CD4+ T cells between groups (P = 0.01).
All monkeys that were infected after penile exposures to SIV developed systemic antibody responses as detected by Western blotting (data not shown). There was no evidence of SIV-specific antibodies in the 6 uninfected monkeys in the TRIM5 homozygous restrictive cohort, in monkey T8023 in the TRIM5 heterozygous permissive cohort, and in monkey DBFX in the TRIM5 homozygous permissive cohort. There were not enough monkeys in the groups to determine whether the expression of particular TRIM5 alleles was associated with particular immune sequelae of infection.
Penile infection with SIVsmE660 results in limited numbers of transmitted/founder viruses.
Recent studies have shown that approximately 80% of HIV-1 infections after heterosexual exposures are established by a single virus (1, 16). The transmission of a few variants in early infection from a complex viral quasispecies is an important feature of mucosal HIV-1 infection in humans. Therefore, we analyzed the number of T/F SIV variants that established productive clinical infection after penile exposure in these cohorts of monkeys to determine whether TRIM5 alleles impacted the number of SIV variants that were transmitted across the penile mucosa.
The full-length gp160 env sequences from the challenge stock of virus used in this study have been described previously (17). T/F viruses and their progenies were identified by SGA of env from plasma SIV RNA at the times of ramp-up and peak viremia. Direct amplicon sequencing was performed, and a phylogenetic analysis of these sequences was done within the context of a model of random virus evolution (8). A total of 555 full-length gp160 env sequences (range of 14 to 44, with a median of 30, at ramp-up viremia and range of 23 to 48, with a median of 30, at peak viremia) were generated from the 10 productively infected monkeys. Virus diversification conformed to a model of random evolution from a single T/F virus in 7 of 10 infected monkeys.
Analysis of transmitted lineages defined for each infected animal compared to the lineages found in the inoculum revealed no preferential transmission of particular SIVsmE660 env variants, since the T/F env lineages were distributed widely throughout the challenge virus quasispecies (see Fig. S2 in the supplemental material). Among the 5 heterozygous permissive monkeys (Fig. 5 and Table 1), SIV infection was initiated with 1 T/F virus in 2 monkeys (7F8, 9E9). Monkey 4236 was infected with a minimum of 2 T/F viruses (see Fig. S3 in the supplemental material). While we were not able to determine a clear minimum number of T/F variants in monkey AV48 due to infection complicated by viral recombination (see Fig. S4 in the supplemental material), we estimated that this monkey was infected with a minimum of 4 viral variants by analyzing the sequence polymorphisms shared among clades (16). The complexity of infection in AV33X also indicates that the infection was initiated by multiple T/F viruses, since the pattern of diversity did not conform to a Poisson distribtion (Table 1). Even though monkeys AV48 and 4230 were productively infected with more than one T/F virus, the numbers of viral variants identified to have initiated infections in these 2 monkeys were still within the range of the numbers of T/F viruses found in typical acute HIV-1 infections initiated via mucosal surfaces. Of the 5 homozygous permissive monkeys, all were found to have homogeneous infections (Fig. 6 and Table 1). In addition, we also analyzed gag sequences in infected monkeys by bulk amplification during acute infection. The gag genes during ramp-up viremia in all 10 infected TRIM5 permissive monkeys were representative of the gag genes in the SIVsmE660 challenge stock (Fig. 7). These findings demonstrate that although permissive TRIM5 alleles allowed infection at mucosal sites, a mucosal bottleneck was present in the male genital tract of the infected monkeys that limited the number of T/F viruses despite the relatively high-dose SIV exposure.
Fig. 5.
Phylogenetic analyses of T/F viruses in TRIM5 heterozygous permissive monkeys. A maximum likelihood tree of env sequences at ramp-up (open squares) and peak (closed squares) viremia in TRIM5 heterozygous permissive monkeys and of SIVsmE660 inoculum env sequences (black circles) is shown. env sequences from each monkey are displayed as similarly colored symbols. Asterisks indicate monkeys that were infected with homogeneous T/F viruses. Bootstrap values are shown for nodes with at least 50% support. The long branch lengths relative to the T/F virus in some of the monkeys (e.g., AV33X and the single long branch in 9E9) were entirely due to APOBEC-mediated G-to-A hypermutation. This is commonly observed in acute HIV infection and adjusted for in the Poisson modeling of viral diversity.
Table 1.
Characteristics of Indian-origin rhesus monkeys exposed to SIV via the penis
| TRIM5α genotype | Monkey ID | TRIM5 allelesa | Infection status | No. of infected monkeys in group/no. of exposed monkeys (%) | No. of lineagesb | Homogeneous by Poissonc |
|---|---|---|---|---|---|---|
| Homozygous restrictive | AZ43 | 3, 3 | No | 0/6 (0) | NDd | ND |
| FB2B | 1, 1 | No | ND | ND | ||
| T8056 | 1, 4 | No | ND | ND | ||
| AZ40 | 4, 4 | No | ND | ND | ||
| AZ63 | 1, 3 | No | ND | ND | ||
| AZ88 | 2, 2 | No | ND | ND | ||
| Heterozygous permissive | 4236 | 2, 11 | Yes | 5/6 (83) | 2 | No |
| AV33X | 1, 6 | Yes | 1 | No | ||
| T8023 | 2, 8 | No | ND | ND | ||
| AV48 | 2, 7 | Yes | 4–6 | No | ||
| 7F8 | 1, 8 | Yes | 1 | Yes | ||
| 9E9 | 1, 7 | Yes | 1 | Yes | ||
| Homozygous permissive | DBFX | 6, 6 | No | 5/6 (83) | ND | ND |
| DBJL | 6, 7 | Yes | 1 | Yes | ||
| DBME | 7, 9 | Yes | 1 | Yes | ||
| DBGJ | 11, 11 | Yes | 1 | Yes | ||
| DBMT | 7, 11 | Yes | 1 | Yes | ||
| 8E9 | 7, 7 | Yes | 1 | Yes |
Diploid TRIM5 alleles were sequenced and numbered as described previously (22).
Minimum number of transmitted forms indicated by distinct lineages in the phylogenetic tree.
Homogeneous infection indicates that mutations fit a Poisson model of random accumulation in early infection. As observed for early HIV-1 infections, APOBEC mutations were enriched in some animals and excluded for analysis in all but two monkeys (7F8 and DBMT). AV33 appeared as a single lineage in the tree, but a violation of the Poisson model indicates that it may have been infected with multiple highly similar strains.
ND, not determined since the animal was not infected.
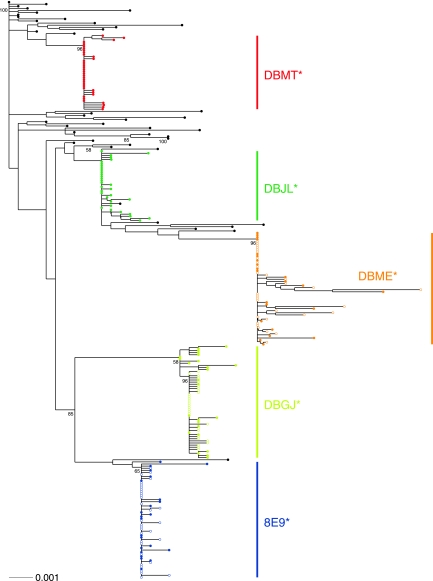
Fig. 6.
Phylogenetic analyses of T/F viruses in TRIM5 homozygous permissive monkeys. A maximum likelihood tree of env sequences at ramp-up (open circles) and peak (closed circles) viremia in TRIM5 homozygous permissive monkeys and of SIVsmE660 inoculum env sequences (black circles) is shown. env sequences from each monkey are displayed as similarly colored symbols. Asterisks indicate monkeys that were infected with homogeneous T/F viruses. Bootstrap values are shown for nodes with at least 50% support. The long branch lengths observed for some of the animals, such as DBME, resulted from APOBEC-mediated hypermutation.
Fig. 7.
Partial amino acid sequence alignment of SIVsmE660 Gag sequences in acutely infected monkeys. Gag sequences were obtained by bulk sequencing of plasmas of 10 infected monkeys at ramp-up viremia (RU) and were aligned with the Gag sequence from the SIVsmE660 challenge stock. Residues 89, 90, and 97 (SIVmac239 numbering) in the SIV capsid that are postulated to mediate host TRIM5 restriction are highlighted.
DISCUSSION
The results of the present study provide direct evidence that polymorphisms in TRIM5 contribute to the variable resistance of Indian-origin rhesus monkeys to mucosal SIV infection in vivo. Recent studies have shown that SIV infections initiated by high-dose intravenous inoculation of rhesus monkeys bearing restrictive TRIM5 alleles result in the attenuation of replication rather than an outright block of infection (18, 21, 22). In the present study, the demonstration that TRIM5 can potently restrict infection following mucosal exposures to SIV in settings that more resemble the conditions of sexual transmission suggests that the viral restriction conferred by TRIM5 alleles may be inversely proportional to the number of virus variants to which the monkeys are exposed. It is likely that a large multiplicity of infection of SIV can override TRIM5 restriction. Thus, the effect of TRIM5 on viral restriction at mucosal surfaces may be more profound than that of TRIM5 restriction in intravenous infections due to the inefficiency of viral transmission at the genital tract. Furthermore, our retrospective analysis of a cohort of rhesus monkeys that was challenged with SIVsmE660 intrarectally showed that there was also a significant association between the expression of two restrictive TRIM5 alleles and protection against SIV acquisition via the rectal mucosa (19). Therefore, the effect of TRIM5 alleles on the susceptibility to mucosal transmission of SIV is likely generalizable to all mucosal compartments in Indian-origin rhesus monkeys.
The variability in set-point viremia in rhesus monkeys in the same cohort suggests that TRIM5 is not the only host genetic factor that influences SIVsmE660 replication in Indian-origin rhesus monkeys. To rule out the possibility that specific MHC class I alleles contributed to the lower levels of viral replication, we evaluated all 18 monkeys in the present study for the presence of the Mamu-A*01, -B*08, and -B*17 alleles. These MHC class I alleles have been implicated in the control of SIV replication in Indian-origin rhesus monkeys. Interestingly, the TRIM5 homozygous permissive monkey (DBFX) that resisted infection after 12 mucosal SIV exposures expressed the Mamu-B*17 allele, an allele that has been associated with low levels of viral replication. This observation raises the possibility that MHC alleles and host adaptive immune responses may have played a role in aborting systemic infection following mucosal exposure to virus. However, since no mucosal specimens were obtained and no other monkeys in this study expressed the Mamu-A*01, -B*08, or -B*17 allele, it is not possible to draw any conclusions regarding the role of MHC class I alleles in the mucosal acquisition of SIV. It is also likely that additional host restriction elements other than TRIM5 also contribute to the control of SIV. These as yet unidentified host restriction factors may have also contributed to the ability of monkeys DBFX and T8023 to resist mucosal infection.
Thus far, only a modest association has been described between TRIM5 alleles and the susceptibility to HIV-1 acquisition and AIDS pathogenesis in humans. The lack of a clear association is likely due to limited polymorphisms in human TRIM5 and retroviral adaptations in the human population (9, 35, 38). However, the present results suggest that genetic factors play a significant role in restricting the mucosal acquisition of HIV-1. Therefore, efforts to illuminate potential genetic determinants that may contribute to host resistance to HIV-1 infection in various cohorts of highly exposed, HIV-1-uninfected individuals should continue.
Although the majority of HIV-1 transmissions in men worldwide occur across the penile mucosa, the cellular events involved in viral acquisition at the portal of entry are not well defined. Despite multiple epidemiologic trials that have demonstrated a reduction in the rate of HIV-1 acquisition in heterosexual men conferred by circumcision (2, 3, 11), the biologic basis for this protective effect remains unclear. An understanding of the early events associated with HIV-1 acquisition in the male genital tract has been hampered by the lack of a robust, physiologically relevant animal penile transmission model. The reason for the absence of this transmission model is that it has been technically difficult to reliably initiate infections via penile exposures in nonhuman primates (NHPs) (27). By exploiting our recent understanding of the role of TRIM5 in the mucosal acquisition of SIV, we have made significant progress toward establishing a novel and robust NHP penile transmission model for AIDS mucosal pathogenesis and vaccine research. Our results show that selecting Indian-origin rhesus monkeys with particular TRIM5 alleles can eliminate host restriction elements to create a more favorable setting for SIV penile transmission in rhesus monkeys. Furthermore, we have shown that this transmission model recapitulates key virologic and immunologic features of mucosal HIV-1 infection, thus lending credibility to the use of this model for HIV-1 mucosal pathogenesis, vaccine, and microbicide research. The SIV-rhesus monkey penile infection model should yield relevant insights into the biology of viral entry and target cells for infection in penile mucosal transmission of HIV/SIV.
The present findings have implications for the design and interpretation of HIV-1 pathogenesis and vaccine efficacy studies. The effect of TRIM5 in restricting viral acquisition should be considered specifically in the selection of rhesus monkeys for vaccine and pathogenesis studies in which SIV mucosal acquisition is an endpoint. TRIM5 genotyping should eliminate some of the variability in mucosal infection that can confound the results of mucosal pathogenesis and vaccine studies. In addition, a penile SIV challenge model will also provide a physiologically relevant challenge route for preclinical testing of candidate HIV-1 vaccines and microbicides. TRIM5 genotyping of rhesus monkeys combined with the use of this physiologically relevant route of penile virus challenge as part of a standardized screening algorithm may be more useful for identifying intervention strategies for blocking HIV-1 acquisition in men than the preclinical animal challenge models that are currently available.
Supplementary Material
ACKNOWLEDGMENTS
We thank Joshua Owuor and Katherine Denison for processing of monkey blood. Saran Bao prepared the inocula for preputial exposures, and Alyse Zajac helped with the protocol approvals at the Vaccine Research Center. Anthony Cook provided veterinary oversight, and we thank the animal care staff at BioQual. We are grateful to Sarah Schaake and Kathryn Furr for carrying out the flow cytometric analyses. We thank Rebecca Gelman for helpful discussions on statistics. We also thank everyone at the Immunology Quality Assurance Center at the Duke Human Vaccine Institute for carrying out viral load monitoring.
This work was supported by NIAID grants K08 AI069995 and R21 AI093199 (W.W.Y.), the intramural research program of the Vaccine Research Center, NIAID, NIH, Center for HIV/AIDS Vaccine Immunology (grant U19 AI067854), Harvard Clinical and Translational Science Center grant UL1 RR 025758 (W.W.Y.), a Shore Fellowship from Harvard Medical School (W.W.Y.), the Pfizer Young Investigator Award in Vaccine Development from the Infectious Diseases Society of America (W.W.Y.), and Harvard University Center for AIDS Research grant P30AI060354 (W.W.Y.).
The authors declare no competing financial interests.
W.W.Y., S.S.R., and N.L.L. designed the experiments. W.W.Y., S.S.R., J.Z., C.L., J.P.T., A.D., L.S., A.P.B., L.M.B., and J.B.W. performed the research. W.W.Y., P.T.H., S.-Y.L., L.M.B., B.T.K., G.J.N., J.R.M., and N.L.L. analyzed the data. W.W.Y. and N.L.L. wrote the paper.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 20 July 2011.
REFERENCES
- 1. Abrahams M. R., et al. 2009. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-Poisson distribution of transmitted variants. J. Virol. 83:3556–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Auvert B., et al. 2005. Randomized, controlled intervention trial of male circumcision for reduction of HIV infection risk: the ANRS 1265 Trial. PLoS Med. 2:e298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bailey R. C., et al. 2007. Male circumcision for HIV prevention in young men in Kisumu, Kenya: a randomised controlled trial. Lancet 369:643–656 [DOI] [PubMed] [Google Scholar]
- 4. Dean M., et al. 1996. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science 273:1856–1862 [DOI] [PubMed] [Google Scholar]
- 5. Dinh M. H., McRaven M. D., Kelley Z., Penugonda S., Hope T. J. 2010. Keratinization of the adult male foreskin and implications for male circumcision. AIDS 24:899–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fellay J., et al. 2007. A whole-genome association study of major determinants for host control of HIV-1. Science 317:944–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fischetti L., Barry S. M., Hope T. J., Shattock R. J. 2009. HIV-1 infection of human penile explant tissue and protection by candidate microbicides. AIDS 23:319–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Giorgi E. E., et al. 2010. Estimating time since infection in early homogeneous HIV-1 samples using a Poisson model. BMC Bioinformatics 11:532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goldschmidt V., et al. 2006. Role of common human TRIM5alpha variants in HIV-1 disease progression. Retrovirology 3:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Goldstein S., Brown C. R., Dehghani H., Lifson J. D., Hirsch V. M. 2000. Intrinsic susceptibility of rhesus macaque peripheral CD4(+) T cells to simian immunodeficiency virus in vitro is predictive of in vivo viral replication. J. Virol. 74:9388–9395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gray R. H., et al. 2007. Male circumcision for HIV prevention in men in Rakai, Uganda: a randomised trial. Lancet 369:657–666 [DOI] [PubMed] [Google Scholar]
- 12. Hatziioannou T., Cowan S., Von Schwedler U. K., Sundquist W. I., Bieniasz P. D. 2004. Species-specific tropism determinants in the human immunodeficiency virus type 1 capsid. J. Virol. 78:6005–6012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hatziioannou T., Perez-Caballero D., Yang A., Cowan S., Bieniasz P. D. 2004. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5alpha. Proc. Natl. Acad. Sci. U. S. A. 101:10774–10779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johnston R. E., et al. 2005. Vaccination of macaques with SIV immunogens delivered by Venezuelan equine encephalitis virus replicon particle vectors followed by a mucosal challenge with SIVsmE660. Vaccine 23:4969–4979 [DOI] [PubMed] [Google Scholar]
- 15. Keckesova Z., Ylinen L. M., Towers G. J. 2004. The human and African green monkey TRIM5alpha genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. U. S. A. 101:10780–10785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Keele B. F., et al. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105:7552–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Keele B. F., et al. 2009. Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J. Exp. Med. 206:1117–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kirmaier A., et al. 2010. TRIM5 suppresses cross-species transmission of a primate immunodeficiency virus and selects for emergence of resistant variants in the new species. PLoS Biol. 8:e1000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Letvin N. L., et al. 2011. Immune and genetic correlates of vaccine protection against mucosal infection by SIV in monkeys. Sci. Transl. Med. 3:81ra36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lifson J. D., et al. 2004. Evaluation of the safety, immunogenicity, and protective efficacy of whole inactivated simian immunodeficiency virus (SIV) vaccines with conformationally and functionally intact envelope glycoproteins. AIDS Res. Hum. Retroviruses 20:772–787 [DOI] [PubMed] [Google Scholar]
- 21. Lim S. Y., et al. 2010. Contributions of Mamu-A*01 status and TRIM5 allele expression, but not CCL3L copy number variation, to the control of SIVmac251 replication in Indian-origin rhesus monkeys. PLoS Genet. 6:e1000997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lim S. Y., et al. 2010. TRIM5alpha modulates immunodeficiency virus control in rhesus monkeys. PLoS Pathog. 6:e1000738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu R., et al. 1996. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86:367–377 [DOI] [PubMed] [Google Scholar]
- 24. Martin M. P., et al. 2002. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat. Genet. 31:429–434 [DOI] [PubMed] [Google Scholar]
- 25. McCoombe S. G., Short R. V. 2006. Potential HIV-1 target cells in the human penis. AIDS 20:1491–1495 [DOI] [PubMed] [Google Scholar]
- 26. Miller C. J. 1998. Localization of simian immunodeficiency virus-infected cells in the genital tract of male and female rhesus macaques. J. Reprod. Immunol. 41:331–339 [DOI] [PubMed] [Google Scholar]
- 27. Miller C. J., et al. 1990. Effect of virus dose and nonoxynol-9 on the genital transmission of SIV in rhesus macaques. J. Med. Primatol. 19:401–409 [PubMed] [Google Scholar]
- 27a. National Research Council 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, DC [Google Scholar]
- 28. Ourmanov I., et al. 2009. Improved survival in rhesus macaques immunized with modified vaccinia virus Ankara recombinants expressing simian immunodeficiency virus envelope correlates with reduction in memory CD4+ T-cell loss and higher titers of neutralizing antibody. J. Virol. 83:5388–5400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Patterson B. K., et al. 2002. Susceptibility to human immunodeficiency virus-1 infection of human foreskin and cervical tissue grown in explant culture. Am. J. Pathol. 161:867–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pereyra F., et al. 2010. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330:1551–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Perron M. J., et al. 2004. TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. U. S. A. 101:11827–11832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reynolds M. R., et al. 2008. Macaques vaccinated with live-attenuated SIV control replication of heterologous virus. J. Exp. Med. 205:2537–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rogers T. F., et al. 2010. Variability in a dominant block to SIV early reverse transcription in rhesus monkey cells predicts in vivo viral replication and time to death. Virol. J. 7:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sakuma R., Mael A. A., Ikeda Y. 2007. Alpha interferon enhances TRIM5alpha-mediated antiviral activities in human and rhesus monkey cells. J. Virol. 81:10201–10206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Speelmon E. C., et al. 2006. Genetic association of the antiviral restriction factor TRIM5alpha with human immunodeficiency virus type 1 infection. J. Virol. 80:2463–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Staprans S. I., et al. 2004. Enhanced SIV replication and accelerated progression to AIDS in macaques primed to mount a CD4 T cell response to the SIV envelope protein. Proc. Natl. Acad. Sci. U. S. A. 101:13026–13031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stremlau M., et al. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848–853 [DOI] [PubMed] [Google Scholar]
- 38. van Manen D., et al. 2008. The effect of Trim5 polymorphisms on the clinical course of HIV-1 infection. PLoS Pathog. 4:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Whitney J. B., et al. 2009. Monitoring HIV vaccine trial participants for primary infection: studies in the SIV/macaque model. AIDS 23:1453–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yap M. W., Nisole S., Lynch C., Stoye J. P. 2004. Trim5alpha protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. U. S. A. 101:10786–10791 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.