Abstract
Murine cytomegalovirus (MCMV) Smith strain has been cloned as a bacterial artificial chromosome (BAC) named pSM3fr and used for analysis of virus gene functions in vitro and in vivo. When sequencing the complete BAC genome, we identified a frameshift mutation within the open reading frame (ORF) encoding MCMV chemokine homologue MCK-2. This mutation would result in a truncated MCK-2 protein. When mice were infected with pSM3fr-derived virus, we observed reduced virus production in salivary glands, which could be reverted by repair of the frameshift mutation. When looking for the source of the mutation, we consistently found that virus stocks of cell culture-passaged MCMV Smith strain are mixtures of viruses with or without the MCK-2 mutation. We conclude that the MCK-2 mutation in the pSM3fr BAC is the result of clonal selection during the BAC cloning procedure.
INTRODUCTION
Herpesviruses cloned as bacterial artificial chromosomes (BACs) have opened a new era of analysis of herpesviral gene functions (48). The major advantage to conventional virus mutagenesis by homologous recombination in cell culture is the fast mutagenesis and clonal propagation in bacteria.
When a virus is cloned as a BAC, the growth of reconstituted virus in cell culture and, if possible, spread in animals, are compared to those of the parental wild-type virus. If these parameters are comparable and genome organization is identical, the BAC-cloned virus is commonly defined as wild type-like (2, 4, 44, 47). For murine cytomegalovirus (MCMV) and also murine gammaherpesvirus 68 (MHV-68), whose growth phenotypes can also be tested in animals, it was soon recognized that insertion of a BAC cassette attenuated the virus in vivo. For both viruses, strategies were developed to remove the BAC cassette (1, 47). Thus, the in vivo wild-type growth characteristics could be restored.
The MCMV BAC pSM3fr cloned from MCMV Smith strain is the prototype of BAC-cloned MCMV (30, 47). It has been shown to infect newborn mice like wild-type Smith strain (47). Since then, it has been used by several groups to study MCMV biology in vitro and in vivo. Mutations were introduced in pSM3fr, and the properties of reconstituted mutants were compared to those of the parental virus reconstituted from pSM3fr (named C3X or MW97.01) (6, 15, 19, 32, 35, 49).
Here, we show that after infection of adult BALB/c mice, pSM3fr-derived virus showed a defect in virus production in salivary glands and that this was due to a frameshift mutation in the MCMV MCK-2 open reading frame (ORF). MCK-2 is a CC(β) chemokine homologue which is thought to be involved in the recruitment of leukocyte subsets that serve as vehicles for viral dissemination (17, 27, 39). In the MCMV wild-type strains Smith and K181, the MCK-2 ORF codes for a 280-amino-acid-long protein. In the pSM3fr-derived virus, the MCK-2 frameshift mutation would shorten the protein by 144 C-terminal amino acids. Repair of the frameshift mutation restored high titers in the salivary glands of mice infected with pSM3fr-derived virus. We could show that cell culture-passaged MCMV Smith strain is consistently a mixture of viruses with intact or mutant MCK-2 ORFs and that after passage through salivary glands of BALB/c mice, the MCK-2 mutant genomes are suppressed. Thus, we propose that cloning of the pSM3fr BAC has selected a MCK-2 mutant genome resulting in the observed growth phenotype in salivary glands. We propose to routinely sequence BAC-cloned viruses.
MATERIALS AND METHODS
Cells and viruses.
Primary mouse embryonal fibroblasts (MEF) from BALB/c mice, M2-10B4 cells (CRL-1972; ATCC), and NIH 3T3 cells (CRL-1658; ATCC) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with either 5 or 10% fetal calf serum, 100 units/ml penicillin, and 100 units/ml streptomycin.
Wild-type MCMV Smith and K181 strains have been propagated in MEF or M2-10B4 cells. pSM3fr-derived virus is a BAC-derived virus which was cloned from MCMV Smith strain (47). VpSM3fr-Δm157-ΔM27 is derived from a pSM3fr mutant with deletions of m157 and M27 (38).
Mice and infection.
Female BALB/c mice were either purchased from Elevate Janvier (Le Genest Saint Isle, France) at the age of 5 weeks and housed at the animal facility of the Max von Pettenkofer-Institute (University of Munich) for at least 1 week before use in experiments or were bred at the Department for Histology and Embryology (University of Rijeka, Croatia) and used at the age of 10 to 12 weeks. All animals were kept under specific-pathogen-free (SPF) conditions. The mice were infected intraperitoneally (i.p.) or intravenously (i.v.) with 3 × 105 50% tissue culture infective doses (TCID50) of MCMV in a volume of 500 μl, or the mice were infected in the right hind footpad with 1.5 × 106 TCID50 in a volume of 25 μl. Virus titers in organs were determined as described previously (11). Animal experiments were approved by the responsible office of the State of Bavaria or by the Ethics Committee of the Medical Faculty, University of Rijeka.
454 sequencing of BAC-cloned MCMV.
pSM3fr, pSM3fr-Δm157-ΔM27, and pSM3fr-ΔloxP DNA (38, 47) from Escherichia coli were prepared using the NucleoBond AX kit (Macherey-Nagel, Düren, Germany) following the manufacturer's instructions. DNA was digested with at least three restriction enzymes and run on an 0.8% agarose gel to control integrity. Virus particles were purified on a linear Nycodenz gradient (Axis-shield PoC, AS, Oslo, Norway) as described previously (14). Virion DNA was isolated (13) and tested for integrity and purity by restriction enzyme digestion. Emulsion PCR was performed as described elsewhere (26), and 454 sequencing was carried out on the Roche 454 FLX and titanium platforms.
Sequencing of PCR-amplified viral sequences.
BAC DNA isolated from E. coli as described above or viral DNA isolated from infected cells, organs, or cell culture supernatants using the DNeasy blood and tissue kit (Qiagen) were used as templates to amplify fragments containing the sequence deviations from MCMV Smith strain observed for pSM3fr. The primer pairs used for amplification are listed in Table 1. These fragments then served as templates for subsequent sequencing.
Table 1.
PCR primers used to amplify viral gene fragments
| Gene | Primer direction | Primer sequence |
|---|---|---|
| M12 | Forward | 5′-CCGCGAATAAACAAGTGGAT-3′ |
| Reverse | 5′-AGATTTTCGTCCGTGACT-3′ | |
| M45 | Forward | 5′-GAGACGATGGCGGTGTAGAG-3′ |
| Reverse | 5′-GGGAGACGACGAAGATCAGT-3′ | |
| M49 | Forward | 5′-GTGTGATTGCCCTCCAACTT-3′ |
| Reverse | 5′-TTTCGTCGAACGCTACTGTC-3′ | |
| M58 | Forward | 5′-TCACAATCCTCACGCAAGC-3′ |
| Reverse | 5′-TGCTACGAGACCACAAATCG-3′ | |
| M86 | Forward | 5′-GGGTAGCGGAAGAACTCCAG-3′ |
| Reverse | 5′-CGTGAGCCAGAACATCAACA-3′ | |
| M102 | Forward | 5′-GAGGCACTTTCGCATCAAC-3′ |
| Reverse | 5′-ACGCGCTTGCAGATGTAGTT-3′ | |
| m107 | Forward | 5′-TGTCCGATGCGATCAAGTAA-3′ |
| Reverse | 5′-TGCGGATGTTCAACCTAGTG-3′ | |
| m117.1 | Forward | 5′-GCATGAAAGGCAGAGGTAGC-3′ |
| Reverse | 5′-CGTGCTGCTTAGCGTTTTTA-3′ | |
| M119.1 | Forward | 5′- TCGATCCATAGTCCCGAAAG-3′ |
| Reverse | 5′-AAGTGACGGAACCCACTGAG-3′ | |
| M122 | Forward | 5′-CAGAGGGAGGTGTGAAAGGA-3′ |
| Reverse | 5′-TGAGAGTGAGGCAGATGAGG-3′ | |
| m129/1 | Forward | 5′-GGGGCGCGATAGGGGGTGTC-3′ |
| Reverse | 5′-GAAGTCTAACAATCTCTCGG-3′ | |
| m129/2 | Forward | 5′-TGTGAGATCACCTGGGTCGG-3′ |
| Reverse | 5′-GTATGTCATGGTTTTTAATC-3′ | |
| m131 | Forward | 5′-GCTTAACGTTTCACTGATTC-3′ |
| Reverse | 5′-AGGACACGCAGTCTGGAATC-3′ |
BAC mutagenesis of the MCK-2 locus.
Markerless BAC mutagenesis was performed to introduce changes in the MCK-2 locus as described previously (23, 45). Briefly, an I-SceI-aphAI cassette was amplified from the plasmid pEPKAN-S by PCR using primers m129repair-for (5′-ACAGACACACTCTATCCAGTTTTCACCGTCCTTCACCTCGTAGGGGTACTTGCCCACCG TGAGGATGACGACGATAAGTAGGG-3′) and m129repair-rev (5′-GATGACGGAATCGCCACGCATCACGGTGGGCAAGTACCCCTACGAGG TGAAGGACGGTGAAACAACCAATTAACCAATTCTGATTAG-3′). In the first Red recombination, this PCR fragment was inserted in pSM3fr, resulting in a BAC carrying a kanamycin resistance cassette flanked by an I-SceI restriction site. The kanamycin resistance cassette was then removed from kanamycin-resistant clones by an I-SceI digestion and a subsequent Red recombination, resulting in a markerless insertion of a T·A base pair at position 187786 of the MCMV Smith strain. The resulting BACmid was called pSM3fr-MCK-2fl (fl stands for full-length).
Deletions and insertions were controlled by restriction pattern analysis and subsequent sequencing.
Reconstitution of virus from recombinant BACs and preparation of virus stocks.
BACs were reconstituted to virus by transfection of BAC DNA into MEF using Superfect transfection reagent (Qiagen, Germany) according to the manufacturer's instructions. Transfected cells were propagated until viral plaques appeared, and supernatants from these cultures were used for further propagation.
Virus stocks were prepared by pelleting virus from supernatants of infected M2-10B4 cells (26,000 × g for 3 h), and the pellets were resuspended in VSB buffer (0.05 M Tris-HCl, 0.012 M KCl, and 0.005 M EDTA, adjusted to pH 7.8) and then purified by centrifugation through a 15% sucrose cushion in VSB buffer (53,000 × g).
Virus titers of stocks were determined by a TCID50 assay performed on MEF on 96-well plates.
RESULTS
Sequence divergency of pSM3fr and pSM3fr-derived virus.
When we infected adult BALB/c mice with BAC (pSM3fr)-derived MCMV, only relatively low titers of virus were found in the salivary glands. This was also shown by other groups (10, 12, 15, 21, 32) but was never addressed, although it stood in marked contrast to the high salivary gland titers observed when BALB/c mice were infected with MCMV strains not cloned as BACs (17, 22, 28, 39). To find out whether this phenotype of the pSM3fr-derived virus was due to genomic differences between the MCMV Smith strain and the pSM3fr genome, we sequenced the complete genomes of pSM3fr, two independent pSM3fr-derived mutants (pSM3fr-Δm157-ΔM27 and pSM3fr-ΔloxP) and a virus stock derived from pSM3fr-Δm157-ΔM27. Both pSM3fr-derived mutants had undergone two consecutive targeted deletions of either m157 and M27 or the two lox-P sites in pSM3fr as described before (38). The virus stock has been passaged 6 times on MEF. Table 2 shows deviations from the BAC-cloned genomes to the published sequence of MCMV Smith strain (36). Since the EcoRI digestion fragments O, b, f, and g, comprising the ORFs m151 to m158 in the BAC-cloned genomes were derived from MCMV strain K181 introduced during the BAC cloning procedure (29, 47), this region was excluded from the analysis. Only divergency which was found in at least two of the four sequences and which lead to changes in the amino acid composition of viral proteins was considered. In total, 22 positions of sequence divergency to the published MCMV Smith sequence could be found, affecting 15 ORFs, most of them in accordance with previously published data (3, 5, 9, 16, 25, 41, 46). Many sequence deviations of the pSM3fr sequence reported by us and others (41) presumably are sequence failures of the deposited MCMV Smith strain sequence. When we sequenced PCR-amplified fragments of MCMV Smith strain-derived DNA (ATCC-VR-1399) covering these positions, we found the same deviations from the published Smith strain sequence (Table 2, group B). Overall, the sequencing data show that the pSM3fr sequence and the Smith strain sequence are nearly identical, which once again reveals that MCMV genomes are very stable during in vivo and in vitro passage (8).
Table 2.
Differences between the Smith strain sequencea and pSM3fr sequence
| Group and gene | Position | Mutation | Change(s) in protein | Reference(s) |
|---|---|---|---|---|
| Group A (sequence failures of the deposited Smith sequence identified before) | ||||
| M20 | 20957 | ΔG | Frameshift, ORF extended | 25,41 |
| M29/m29.1 | 36198 | +G | M29 frameshift, ORF truncated; m29.1 frameshift, ORF extended | 3,41 |
| M30 | 37260 | +C | Frameshift, ORF extended | 3,41 |
| M31 | 38803 | +G | Frameshift, ORF extended | 25,41 |
| m143 | 201403 | +G | Frameshift, ORF extended | 9,41 |
| Group B (sequence deviations of pSM3fr that are due to sequence failures of the deposited Smith sequence) | ||||
| M45 | 61918 | +C | Frameshift, fusion of M45 and m45.1 | 5,41 |
| M49 | 74719 | T→C | Ser271 → Gly271 | |
| M58 | 92088 | ΔA | Protein sequence deviation from amino acid 110 to amino acid 220 | 41 |
| 92178 | C→GCG | Protein sequence deviation from amino acid 110 to amino acid 220 | ||
| 92206 | CG→GC | Protein sequence deviation from amino acid 110 to amino acid 220 | ||
| 92226 | G→A | Protein sequence deviation from amino acid 110 to amino acid 220 | ||
| 92230 | G→A | Protein sequence deviation from amino acid 110 to amino acid 220 | ||
| 92358 | ΔG | Protein sequence deviation from amino acid 110 to amino acid 220 | ||
| M86 | 123941 | A→C | Leu1107 → Arg1107 | 41 |
| 123951 | T→G | Asn1104 → His1104 | ||
| m107/m108 | 162374 | T→C | m107 Leu131→ Pro131 m108 Ser133→ Gly133 | 16,41 |
| 162380 | T→C | m107 Leu133 →Ser133 m108 Asn131→ Asp131 | ||
| m117.1/m117 | 170652 | A→C | m117.1 Asp371 →Ala371 m117 Ser87 → Ala87 | |
| Group C (sequence deviations of pSM3fr) | ||||
| M12 | 12052 | G→A | Glu123 → Lys123 | |
| M102 | 146853 | G→A | Asp420 → Asn420 | 41 |
| M122 Ex5 | 179136 | G→A | Ser227 → Phe227 | 41 |
| m129 | 187786 | ΔT | Frameshift, ORF truncated | 41 |
Restoration of the full-length MCK-2 ORF reversed the salivary gland phenotype.
Most interestingly, the major pSM3fr-specific sequence deviation is a deletion of a T·A base pair corresponding to position 187786 of the MCMV Smith strain sequence. This frameshift results in a stop codon in the reading frame of the chemokine homologue MCK-2 formed by the ORFs m131 and m129 (37) and thus, would lead to a truncated MCK-2 protein (Fig. 1). Moreover, infection of mice with viruses having mutations and deletions of MCK-2 have been shown to result in reduced numbers of infected peripheral blood leukocytes (PBLs) and reduced titers in salivary glands, whereas titers in other organs were not affected (17, 39, 40). We hypothesized from this finding that the loss of the C-terminal part of MCK-2 might be the cause for the attenuated salivary gland phenotype of pSM3fr-derived virus.
Fig. 1.
Schematic presentation of the deletion found in the MCK-2 ORF of pSM3fr. The nucleotide sequence of MCK-2 exons and introns is shown. The initiating methionine residues for the m131 and m129 ORFs and the intron sequence are circled and boxed, respectively. The inset indicates the position of the T·A deletion at MCK-2 position 486 and the resulting stop mutation in pSM3fr.
The full-length MCK-2 ORF was restored by inserting a T·A base pair at position 187786 of pSM3fr using a markerless mutagenesis protocol (23, 45). Two clones of pSM3fr-MCK-2fl (full length) (clones 3.3 and 4.1) were reconstituted to virus, and the virus was tested for its in vitro and in vivo growth patterns. pSM3fr- and pSM3fr-MCK-2fl-derived viruses showed comparable growth curves in NIH 3T3 cells infected at a low multiplicity of infection (MOI) (Fig. 2), which was in accordance with comparisons of the in vitro growth curves of MCMV Smith strain and its MCK-2 mutants (17).
Fig. 2.
Comparison of growth of unrepaired and repaired pSM3fr-derived viruses. Multistep growth curves of virus derived from pSM3fr or pSM3fr-MCK-2fl clone 3.3 or 4.1 are shown. NIH 3T3 cells were infected at an MOI of 0.1, and supernatants were harvested daily and titrated. The titers of supernatants are expressed as TCID50 per ml.
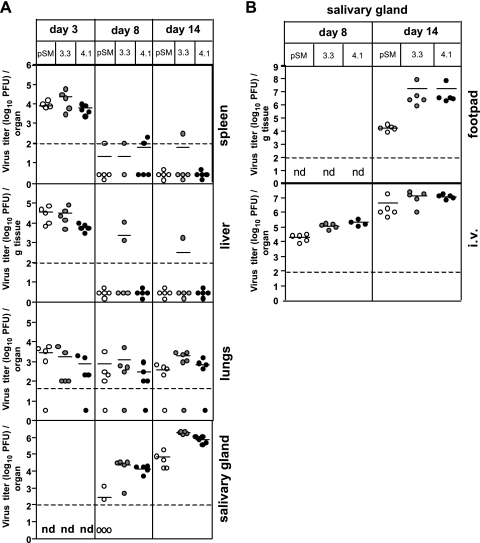
BALB/c mice were infected by the intraperitoneal route, and virus titers were determined in organs 3, 8, and 14 days after infection. The only obvious difference between the pSM3fr-derived virus and the two MCK-2 full-length clones were elevated virus titers in the salivary glands after restoration of MCK-2 (Fig. 3 A). This salivary gland phenotype could also be observed when mice were infected in the periphery via footpads (Fig. 3B). The low levels of salivary gland virus observed for MCK-2 mutants are considered to be the result of a deficiency in dissemination in the absence of an intact MCK-2 (17, 39). This hypothesis was strengthened by our finding that after intravenous infection, the differences between salivary gland titers of viruses with a full-length MCK-2 ORF and viruses with a disrupted MCK-2 ORF were smaller (Fig. 3B). Here, the route of virus administration already ensures dissemination via the bloodstream.
Fig. 3.
Infection of BALB/c mice with virus derived from pSM3fr or pSM3fr-MCK-2fl clone 3.3 or 4.1. (A) Virus titers in the spleen, liver, lungs, and salivary glands of BALB/c mice injected i.p. with 3 × 105 TCID50 of pSM3fr (pSM) (open circles) or pSM3fr-MCK-2fl clone 3.3 (gray circles) or 4.1 (black circles). The organs were harvested on days 3, 8, and 14 postinfection (p.i.). (B) Virus titers in salivary glands of BALB/c mice infected with pSM3fr (pSM) (open circles) or pSM3fr-MCK-2fl clone 3.3 (gray circles) or 4.1 (black circles) via the footpad or i.v. with 1.5 × 106 and 3 × 105 TCID50, respectively. The organs were harvested on day 8 and 14 p.i. Each circle shows the value for an individual mouse, and mean values for the groups of mice are indicated by the short horizontal bars. The broken lines indicate the detection limit. nd, not determined.
Cell culture-passaged MCMV Smith strain is a mixture of genomes carrying intact or mutant MCK-2 ORFs.
Cloning of viruses as BACs involves a recombination step between the viral genome and the BAC cassette in infected cells and a subsequent transfer of the recombinant genomes to E. coli followed by clonal propagation of transformed E. coli. We wondered whether the MCK-2 mutation found in the pSM3fr BAC was introduced into the MCMV genome during the BAC cloning procedure or whether it was the result of clonal selection of a mutant preexisting in the virus inoculum used to infect cells for integration of the BAC cassette. Therefore, we tested whether MCMV Smith virus stocks are heterogeneous with respect to their MCK-2 sequences. We prepared DNA from an MCMV Smith virus stock originating from the time when the pSM3fr BAC was cloned in our laboratory (ATCC-VR-194; Smith strain Munich) and also from an MCMV Smith virus stock recently sold by ATCC (ATCC-VR-1399; Smith strain ATCC). Both virus stocks were derived from cells infected in vitro. We amplified the MCK-2 sequence spanning the locus of the mutation found in pSM3fr by PCR and subsequently sequenced the amplified fragments. We found that with respect to the MCK-2 mutation, both virus stocks were mixtures (Fig. 4 A). The electropherograms of the sequenced PCR products showed differences in the proportions of wild-type and mutant sequences for the selected stocks. In addition, we tested several cell culture-derived Smith strain virus stocks from other laboratories and found comparable mixtures for the MCK-2 mutation as depicted in Fig. 4A (data not shown). When we infected BALB/c mice with Smith strain ATCC and isolated salivary gland virus, the proportion of the nonmutated MCK-2 sequence quickly increased in passage 1, and already after two passages, the electropherograms no longer showed the mutated sequence (Fig. 4B). When we recultured salivary gland virus from salivary passage 1/mouse 1 for two passages in MEF, the mutation reappeared (Fig. 4C). When we compared salivary gland titers of mice infected with a Smith strain virus stock showing an MCK-2 mixture phenotype like Smith strain Munich with pSM3fr-derived virus, we observed only slightly higher salivary gland titers for the Smith strain (Fig. 4D). This was in accordance with the dominance of the MCK-2 mutation in this virus mixture, and it strongly supported our hypothesis that the salivary gland phenotype of pSM3fr-derived virus can be completely attributed to the MCK-2 mutation.
Fig. 4.
Cell culture-passaged MCMV Smith strain exhibits mixtures of MCK-2 wild-type and mutant genomes. (A) The locus of the MCK-2 gene spanning the pSM3fr base pair deletion was amplified from two MCMV Smith virus stocks by PCR, and the PCR products were sequenced. The electropherograms indicate that with respect to nucleotide position 486 of MCK-2, the stocks are mixtures. For each stock, the more abundant sequence is depicted above the electropherogram and the less abundant sequence is indicated below the electropherogram. The 5′→3′ orientation respective to the MCK-2 ORF and nucleotide position 486 (Fig. 1) are indicated. The sequence resulting in a full-length MCK-2 ORF contains a T enclosed in a red box. The sequencing direction is indicated by the white open arrow. (B) Suppression of the mutant population by passage of cell culture-derived MCMV Smith strain in salivary glands. Three BALB/c mice (animals 1, 2, and 3) were injected i.p. with 2 × 105 TCID50 cell culture-passaged Smith strain, and the salivary glands were harvested on day 14 p.i. Then, salivary gland-derived virus was used for another round of infection (animals 1′, 2′, and 3′). Viral DNA from all salivary glands was amplified as described above for panel A and sequenced. The electropherograms of the mutant locus are shown. (C) Salivary gland-derived virus from mouse 1 was used to infect MEF cells and passaged twice. Viral DNA was amplified from supernatant virus and sequenced. The electropherogram of the mutant locus is shown. (D) BALB/c mice were infected as described in the legend to Fig. 3A with cell culture-passaged Smith strain (small white squares), pSM3fr-derived virus (pSM) (open circles), or pSM3fr-MCK-2fl clone 3.3 (small gray circles). The salivary glands were harvested on days 8 and 14 p.i. Each symbol shows the value for an individual mouse, and mean values for the groups of mice are indicated by the short horizontal bars. The broken lines indicate the detection limit.
In addition, we sequenced BAC-derived clonal MCMV to exclude the possibility that the MCK-2 gene locus is unstable in E. coli or during cell culture propagation. To this end, we isolated DNA from several pSM3fr-derived mutants and virus progeny derived from these mutants and amplified the relevant MCK-2 sequence by PCR. For all BAC mutants that had undergone several rounds of BAC mutagenesis in E. coli and all virus progeny of these mutants that had been passaged either in cell culture (at least 3 passages) or were derived from the salivary glands of infected mice, the MCK-2 sequences were absolutely stable, preserving either the mutation (pSM3fr-derived DNA or virus) or the repaired MCK-2 sequence (DNA or virus derived from pSM3fr-MCK-2fl clones) (data not shown).
When we sequenced cell culture-derived Smith strain, sequence mixtures were detected for only the MCK-2 gene and for none of the other sequences amplified (primers listed in Table 1) (data not shown). To find out whether the appearance of mixtures of MCK-2 sequences after cell culture passage is due to a specific property of genome position 187786 or whether it reflects a property of the MCK-2 gene, we sequenced the complete MCK-2 ORF, including the intron of cell culture-derived Smith strain, and compared it to salivary gland-passaged Smith strain and cell culture-derived BAC-cloned virus. For the cell culture-passaged Smith strain, but not for salivary gland-passaged and pSM3fr-derived virus, we could detect not only the described subpopulations having or lacking a T·A base pair at position 486 of MCK-2 but also another subpopulation carrying a 9-bp deletion at the 3′ end of the intron, which would remove the splice junction and thus also interrupt the MCK-2 ORF. Interestingly, cell culture-passaged MCMV strain K181 also showed subpopulations carrying either frameshifts or a base pair exchange, all of which would result in truncated MCK-2 proteins (data not shown). Yet, a frameshift mutation at position 486 was not found for strain K181 (data not shown). Thus, the appearance of different MCK-2 truncating mutations strongly suggests that a property of MCK-2 results in selection or counterselection of genomes carrying MCK-2-inactivating mutations in cell culture or in salivary glands, respectively.
DISCUSSION
Cloning of herpesvirus genomes as bacterial artificial chromosomes (BACs) has introduced a new and fast technique of manipulating viral genomes in the field of herpesvirus research. It allows us to mutate or to modify viral genomes in E. coli, to reconstitute virus from these mutated genomes, and to analyze the virus progeny in vitro and in vivo. One major advantage of BAC cloning to classical methods based on recombination of viral genomes with plasmids in cells and subsequent selection of recombinant viruses was the avoidance of selection steps to separate mutant from wild-type viruses. Instead, during the mutagenesis procedure, truly clonal genome progeny are created in E. coli.
Here, we sequenced the complete genomes of the Smith strain-derived BAC pSM3fr, two mutants of this BAC and virus progeny derived from one of the mutants, and compared the sequences to the sequence published for MCMV Smith strain (36). Most of the sequence deviations we observed have been described recently for a pSM3fr-derived spontaneous M112/113 mutant (41). We could now show that the majority of these deviations are presumably sequence failures of the published sequence of MCMV Smith strain. Only four of the observed differences could not be found in Smith strain-derived DNA (Table 2, group C). Thus, the sequence of the pSM3fr-BAC is nearly identical to the sequence of the Smith strain.
The most striking difference between the Smith strain sequence and the pSM3fr BAC was a mutation in the MCK-2 ORF that would truncate MCK-2. Sequencing of the MCK-2 ORF of the MCMV Smith strain and MCMV K181 strain had shown undisrupted ORFs coding for 280-amino-acid-long, highly conserved proteins (17, 27, 36, 43). Thus, we posed the following two questions: is the MCK-2 mutation accountable for the low salivary gland titers observed after infection of BALB/c mice with pSM3fr-derived virus and how was this mutation introduced into pSM3fr?
Mutations disrupting the MCK-2 ORF are well characterized, and one hallmark has been low salivary gland titers in mice infected with these mutants (17, 39). This was also observed when pSM3fr-derived viruses were characterized in vivo (10, 12, 15, 21, 32) and stood in marked contrast to high salivary gland titers obtained after infection with MCMV strains not cloned as BACs (17, 22, 28, 39). We could restore the phenotype of high virus titers in salivary glands by repairing the pSM3fr mutation and thus provide evidence that pSM3fr-derived virus shows an attenuated growth phenotype in salivary glands due to a mutation in MCK-2. In accordance with earlier publications which characterized MCK-2 deletion mutants and showed that, independent of the route of infection, virus attenuation was restricted to salivary glands (17, 39), pSM3fr-derived attenuation was also restricted to salivary glands when we analyzed the intraperitoneal route of infection.
Usually, publications on in vivo growth phenotypes do not compare MCMV wild-type strain properties with growth properties of BAC-cloned viruses, and thus, the growth discrepancy between wild-type virus and pSM3fr-derived virus was never revealed. Additionally, as we could show here, cell culture-passaged Smith strain usually is a mixture containing genomes with intact and mutant MCK-2 ORFs, and depending on the proportion of mutant genomes, salivary gland virus titers will probably not differ from titers achieved by pSM3fr-derived virus. Interestingly, we observed that in vivo passage in salivary glands suppressed detectable populations of mutant MCK-2 sequences, which quickly reappeared when virus was recultured in cells. This fast switch reminds us of earlier publications showing a loss of virulence of MCMV Smith strain after cell culture passage and an increase in virulence for salivary gland-derived virus (24, 34). Whether Smith strain virulence, as defined for example by 50% lethal dose (LD50) values for survival of infected newborn mice, and MCK-2 mutations correlate will have to be determined in the future.
The MCK-2 locus of MCMV Smith strain has been sequenced before from PCR-amplified genomic fragments, but neither mixed populations nor mutations interrupting the MCK-2 ORF have been described (43). One reason might be that plaque purified virus was used instead of unpurified supernatant virus, and thus, the virus progeny of a single “MCK-2 genotype” were selected. It is striking that the MCK-2 sequence populations which we observed for cell culture-passaged Smith or K181 strain all consisted of mutations interrupting the MCK-2 ORF. Our data imply that salivary gland passage selects nonmutated virus, which could be explained by the reduced titers of MCK-2 mutants in salivary glands. Yet, our data also imply that cell culture passage selects the mutant phenotype, although wild-type and mutant viruses replicate equally well in cell culture (Fig. 2). The fast switch in the relative proportions of mutant and wild-type sequences suggests that the mutations are preexisting in the virus preparations and become visible or disappear depending on whether the virus is replicating in cell culture or in mice. It is noteworthy that mixed sequences only become apparent in electropherograms when at least 10% of the sequences are aberrant. In contrast to the wild-type MCMV strains, passage of BAC-cloned viruses, either in cell culture or in mice, neither reversed the MCK-2 mutation nor introduced new mutations (data not shown). Thus, the pSM3fr-MCK-2fl-derived viruses might be an alternative to salivary gland-derived virus for infection of mice.
BAC cloning is usually done using virus isolates. Yet, cytomegalovirus (CMV) isolates for example are consistently found to be mixtures of different genotypes (7, 18, 20, 31, 42, 43). Due to clonal propagation steps in E. coli, BAC cloning of viral genomes will automatically result in selection of a single genotype from a mixture, which might have a different phenotype than the genotypically heterogeneous parental virus isolate. We therefore propose to sequence each BAC-cloned genome to determine its specific genotype, a practice which has been followed in the last years for many BAC-cloned human CMVs (33, 42, 44).
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft (AD131/3-1) and the Studienstiftung des deutschen Volkes (S. Jordan). S. Jonjic was supported by EU FP7 REGPOT CAPRI2010.
We thank H. Adler for critically reading the manuscript.
Footnotes
Published ahead of print on 3 August 2011.
REFERENCES
- 1. Adler H., Messerle M., Koszinowski U. H. 2001. Virus reconstituted from infectious bacterial artificial chromosome (BAC)-cloned murine gammaherpesvirus 68 acquires wild-type properties in vivo only after excision of BAC vector sequences. J. Virol. 75:5692–5696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Adler H., Messerle M., Wagner M., Koszinowski U. H. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 74:6964–6974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ahasan M. M., Sweet C. 2007. Murine cytomegalovirus open reading frame m29.1 augments virus replication both in vitro and in vivo. J. Gen. Virol. 88:2941–2951 [DOI] [PubMed] [Google Scholar]
- 4. Borst E.-M., Hahn G., Koszinowski U. H., Messerle M. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320–8329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brune W., Menard C., Heesemann J., Koszinowski U. H. 2001. A ribonucleotide reductase homolog of cytomegalovirus and endothelial cell tropism. Science 291:303–305 [DOI] [PubMed] [Google Scholar]
- 6. Bubic I., et al. 2004. Gain of virulence caused by loss of a gene in murine cytomegalovirus. J. Virol. 78:7536–7544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chandler S. H., Handsfield H. H., McDougall J. K. 1987. Isolation of multiple strains of cytomegalovirus from women attending a clinic for sexually transmitted disease. J. Infect. Dis. 155:655–660 [DOI] [PubMed] [Google Scholar]
- 8. Cheng T. P., Valentine M. C., Gao J., Pingel J. T., Yokoyama W. M. 2010. Stability of murine cytomegalovirus genome after in vitro and in vivo passage. J. Virol. 84:2623–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Child S. J., Hanson L. K., Brown C. E., Janzen D. M., Geballe A. P. 2006. Double-stranded RNA binding by a heterodimeric complex of murine cytomegalovirus m142 and m143 proteins. J. Virol. 80:10173–10180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cicin-Sain L., Brune W., Bubic I., Jonjic S., Koszinowski U. H. 2003. Vaccination of mice with bacteria carrying a cloned herpesvirus genome reconstituted in vivo. J. Virol. 77:8249–8255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cicin-Sain L., Podlech J., Messerle M., Reddehase M. J., Koszinowski U. H. 2005. Frequent coinfection of cells explains functional in vivo complementation between cytomegalovirus variants in the multiply infected host. J. Virol. 79:9492–9502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cicin-Sain L., et al. 2008. Dominant-negative FADD rescues the in vivo fitness of a cytomegalovirus lacking an antiapoptotic viral gene. J. Virol. 82:2056–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deryckere F., Burgert H. G. 1997. Rapid method for preparing adenovirus DNA. Biotechniques 22:868–870 [DOI] [PubMed] [Google Scholar]
- 14. Dohner K., Radtke K., Schmidt S., Sodeik B. 2006. Eclipse phase of herpes simplex virus type 1 infection: efficient dynein-mediated capsid transport without the small capsid protein VP26. J. Virol. 80:8211–8224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dolken L., et al. 2010. Cytomegalovirus microRNAs facilitate persistent virus infection in salivary glands. PLoS Pathog. 6:e1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dolken L., et al. 2007. Mouse cytomegalovirus microRNAs dominate the cellular small RNA profile during lytic infection and show features of posttranscriptional regulation. J. Virol. 81:13771–13782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fleming P., et al. 1999. The murine cytomegalovirus chemokine homolog, m131/129, is a determinant of viral pathogenicity. J. Virol. 73:6800–6809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gerna G., et al. 2003. Rescue of human cytomegalovirus strain AD169 tropism for both leukocytes and human endothelial cells. J. Gen. Virol. 84:1431–1436 [DOI] [PubMed] [Google Scholar]
- 19. Ghazal P., Messerle M., Osborn K., Angulo A. 2003. An essential role of the enhancer for murine cytomegalovirus in vivo growth and pathogenesis. J. Virol. 77:3217–3228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grazia Revello M., et al. 2001. In vitro selection of human cytomegalovirus variants unable to transfer virus and virus products from infected cells to polymorphonuclear leukocytes and to grow in endothelial cells. J. Gen. Virol. 82:1429–1438 [DOI] [PubMed] [Google Scholar]
- 21. Gustems M., Busche A., Messerle M., Ghazal P., Angulo A. 2008. In vivo competence of murine cytomegalovirus under the control of the human cytomegalovirus major immediate-early enhancer in the establishment of latency and reactivation. J. Virol. 82:10302–10307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Henson D., Smith R. D., Gehrke J., Neapolitan C. 1967. Effect of cortisone on nonfatal mouse cytomegalovirus infection. Am. J. Pathol. 51:1001–1011 [PMC free article] [PubMed] [Google Scholar]
- 23. Jiang X. J., et al. 2008. UL74 of human cytomegalovirus contributes to virus release by promoting secondary envelopment of virions. J. Virol. 82:2802–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jordan M. C., Takagi J. L. 1983. Virulence characteristics of murine cytomegalovirus in cell and organ cultures. Infect. Immun. 41:841–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kattenhorn L. M., et al. 2004. Identification of proteins associated with murine cytomegalovirus virions. J. Virol. 78:11187–11197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Krause J., et al. 2010. A complete mtDNA genome of an early modern human from Kostenki, Russia. Curr. Biol. 20:231–236 [DOI] [PubMed] [Google Scholar]
- 27. MacDonald M. R., Burney M. W., Resnick S. B., Virgin IV H. W. 1999. Spliced mRNA encoding the murine cytomegalovirus chemokine homolog predicts a beta chemokine of novel structure. J. Virol. 73:3682–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Manning W. C., Stoddart C. A., Lagenaur L. A., Abenes G. B., Mocarski E. S. 1992. Cytomegalovirus determinant of replication in salivary glands. J. Virol. 66:3794–3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mercer J. A., Marks J. R., Spector D. H. 1983. Molecular cloning and restriction endonuclease mapping of the murine cytomegalovirus genome (Smith strain). Virology 129:94–106 [DOI] [PubMed] [Google Scholar]
- 30. Messerle M., Crnkovic I., Hammerschmidt W., Ziegler H., Koszinowski U. H. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. U. S. A. 94:14759–14763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Meyer-Konig U., Ebert K., Schrage B., Pollak S., Hufert F. T. 1998. Simultaneous infection of healthy people with multiple human cytomegalovirus strains. Lancet 352:1280–1281 [DOI] [PubMed] [Google Scholar]
- 32. Mohr C. A., et al. 2010. A spread-deficient cytomegalovirus for assessment of first-target cells in vaccination. J. Virol. 84:7730–7742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Murphy E., et al. 2003. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc. Natl. Acad. Sci. U. S. A. 100:14976–14981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Osborn J. E., Walker D. L. 1971. Virulence and attenuation of murine cytomegalovirus. Infect. Immun. 3:228–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pinto A. K., Munks M. W., Koszinowski U. H., Hill A. B. 2006. Coordinated function of murine cytomegalovirus genes completely inhibits CTL lysis. J. Immunol. 177:3225–3234 [DOI] [PubMed] [Google Scholar]
- 36. Rawlinson W. D., Farrell H. E., Barrell B. G. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833–8849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reddehase M. J., Podlech J., Grzimek N. K. 2002. Mouse models of cytomegalovirus latency: overview. J. Clin. Virol. 25(Suppl. 2):S23–S36 [DOI] [PubMed] [Google Scholar]
- 38. Sacher T., et al. 2008. The major virus-producing cell type during murine cytomegalovirus infection, the hepatocyte, is not the source of virus dissemination in the host. Cell Host Microbe 3:263–272 [DOI] [PubMed] [Google Scholar]
- 39. Saederup N., Aguirre S. A., Sparer T. E., Bouley D. M., Mocarski E. S. 2001. Murine cytomegalovirus CC chemokine homolog MCK-2 (m131-129) is a determinant of dissemination that increases inflammation at initial sites of infection. J. Virol. 75:9966–9976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Saederup N., Lin Y. C., Dairaghi D. J., Schall T. J., Mocarski E. S. 1999. Cytomegalovirus-encoded beta chemokine promotes monocyte-associated viremia in the host. Proc. Natl. Acad. Sci. U. S. A. 96:10881–10886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schumacher U., Handke W., Jurak I., Brune W. 2010. Mutations in the M112/M113-coding region facilitate murine cytomegalovirus replication in human cells. J. Virol. 84:7994–8006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sinzger C., et al. 2008. Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J. Gen. Virol. 89:359–368 [DOI] [PubMed] [Google Scholar]
- 43. Smith L. M., Shellam G. R., Redwood A. J. 2006. Genes of murine cytomegalovirus exist as a number of distinct genotypes. Virology 352:450–465 [DOI] [PubMed] [Google Scholar]
- 44. Stanton R. J., et al. 2010. Reconstruction of the complete human cytomegalovirus genome in a BAC reveals RL13 to be a potent inhibitor of replication. J. Clin. Invest. 120:3191–3208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tischer B. K., von Einem J., Kaufer B., Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197 [DOI] [PubMed] [Google Scholar]
- 46. Valchanova R. S., Picard-Maureau M., Budt M., Brune W. 2006. Murine cytomegalovirus m142 and m143 are both required to block protein kinase R-mediated shutdown of protein synthesis. J. Virol. 80:10181–10190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wagner M., Jonjic S., Koszinowski U. H., Messerle M. 1999. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 73:7056–7060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wagner M., Ruzsics Z., Koszinowski U. H. 2002. Herpesvirus genetics has come of age. Trends Microbiol. 10:318–324 [DOI] [PubMed] [Google Scholar]
- 49. Zimmermann A., et al. 2005. A cytomegaloviral protein reveals a dual role for STAT2 in IFN-gamma signaling and antiviral responses. J. Exp. Med. 201:1543–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]