Abstract
Eukaryotic translation initiation factor 2B (eIF2B) is a heteropentameric guanine nucleotide exchange factor that converts protein synthesis initiation factor 2 (eIF2) from a GDP-bound form to the active eIF2-GTP complex. Cellular stress can repress translation initiation by activating kinases capable of phosphorylating the alpha subunit of eIF2 (eIF2α), which sequesters eIF2B to prevent exchange activity. Previously, we demonstrated that tumor cells are sensitive to viral replication, possibly due to the occurrence of defects in eIF2B that overcome the inhibitory effects of eIF2α phosphorylation. To extend this analysis, we have investigated the importance of eIF2Bα function and report that this subunit can functionally substitute for its counterpart, GCN3, in yeast. In addition, a variant of mammalian eIF2Bα harboring a point mutation (T41A) was able overcome translational inhibition invoked by amino acid depravation, which activates Saccharomyces cerevisiae GCN2 to phosphorylate the yeast eIF2α homolog SUI2. Significantly, we also demonstrate that the loss of eIF2Bα, or the expression of the T41A variant in mammalian cells, is sufficient to neutralize the consequences of eIF2α phosphorylation and render normal cells susceptible to virus infection. Our data emphasize the importance of eIF2Bα in mediating the eIF2 kinase translation-inhibitory activity and may provide insight into the complex nature of viral oncolysis.
INTRODUCTION
The initiation of translation requires the coordinate actions of several protein factors to complete a multiphase process that culminates in the formation of an 80S ribosome on a nascent mRNA molecule (30). The first of these steps is the formation of a stable ternary complex composed of one charged initiator Met-tRNAi molecule, GTP, and eukaryotic initiation factor 2 (eIF2), comprised of its α, β, and γ subunits. Configuration of the ternary complex is required to bring the Met-tRNAi molecule into contact with the 40S ribosomal subunit, which subsequently recognizes mRNA molecules through the eIF4F cap-binding complex (35). These events lead to the recognition of the AUG start codon of the mRNA and the binding of the 60S ribosomal subunit to the 40S subunit (26). At the end of initiation and upon the creation of the 80S ribosome, the accompanying protein factors are released from the ribosome-mRNA molecule to be recycled for future use. In addition to initiation factor release, the GTP bound to eIF2 is hydrolyzed by the action of eIF5, such that the resultant eIF2 is liberated as an inactive binary complex bound to GDP (31, 36). In order for subsequent rounds of initiation to occur, it is necessary to generate new active eIF2-GTP from the existing eIF2-GDP, a reaction catalyzed by the heteropentameric guanine nucleotide exchange factor (GEF) eIF2B (41).
eIF2B provides a key regulatory mechanism for cells to decrease protein synthesis rates during periods of stress. Selected cellular stress, such as the accumulation of viral RNA species following infection or misfolded proteins in the endoplasmic reticulum (ER), causes the phosphorylation of eIF2 at the serine 51 residue on the alpha subunit (42). The phosphorylated alpha subunit of eIF2 (eIF2α) still associates with eIF2B, although there is a decrease in the overall GEF exchange activity that leads to a decline in ternary complex formation (33). While the actual mechanism of inhibition remains speculative, it is possible that when eIF2α is phosphorylated, it binds with a higher affinity to eIF2B than to nonphosphorylated eIF2α. This stringent binding is thought to hinder conformational changes in eIF2B, thereby preventing GEF activity. Alternatively, eIF2B simply may not be able to dissociate from eIF2α when the latter is phosphorylated (21, 44). Because the cell contains much larger quantities of eIF2 than eIF2B, it appears that even low levels of eIF2α phosphorylation can lead to reduced levels of protein synthesis (21). Despite its functional resemblance to other nucleotide exchange factors, the structure of eIF2B appears more intricate. Whereas members of the Ras family of GTPases are typically small monomers, eIF2B is quite large, with its 5 proteins that are distinguished by their roles in maintaining either catalytic or regulatory function (Fig. 1a). The catalytic activity of eIF2B is contained primarily in its ε subunit, which cooperates with the other subunits to exhibit full GEF activity (10). In Saccharomyces cerevisiae, a complex consisting of eIF2Bγ and eIF2Bε is sufficient for full catalytic activity, whereas in mammals all of the subunits except eIF2Bα are required (9). eIF2Bα is a 30-kDa protein that couples with two other proteins, eIF2Bβ and -δ, all of which share moderate sequence homology and form the regulatory subcomplex of eIF2B (46). The regulatory subunits are responsible for the initial binding to eIF2; once eIF2B binds eIF2, it is thought that the eIF2B proteins undergo a conformational change that brings the catalytic subcomplex into closer proximity with eIF2 such that it exerts guanine nucleotide exchange (27).
Fig. 1.
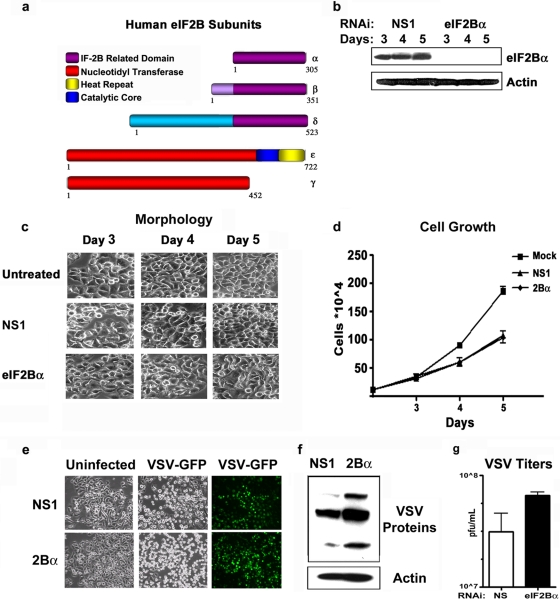
Loss of the alpha subunit of eIF2B does not induce immediate cell death. (a) Schematic of human eIF2B subunits showing conserved regions of the regulatory and catalytic subcomplexes. (b) Immunoblot analysis of HeLa cell lysates, in triplicate, following treatment with nonspecific (NS1) or eIF2Bα-specific RNAi on days 3, 4, and 5 posttransfection. (c) Light microscopy depicting the morphology of HeLa cells following RNAi treatment for 3, 4, or 5 days with NS1 and eIF2Bα-specific RNAi or of untreated HeLa cells. (d) Growth rates of HeLa cells either untreated, treated with NS1 RNAi, or treated with eIF2Bα-specific RNAi. (e) HeLa cells transfected with NS1 or eIF2Bα-specific RNAi for 72 h were infected with VSV-GFP at an MOI of 0.1 for 24 h. Light (left and middle) and fluorescent (right) microscopy images were taken to demonstrate cytopathic effects (CPE) and viral GFP expression. (f) Viral protein synthesis was determined by immunoblot analysis of lysates using murine anti-VSV or anti-actin antibody. (g) Quantitation of viral plaque assays performed on tissue culture medium from panel e (error bars indicate standard deviations [SD] of data from duplicate samples from 3 independent experiments).
A number of kinases are now known to recognize cellular stress and phosphorylate eIF2α to repress protein synthesis rates (42). For example, the interferon (IFN)-inducible double-stranded RNA (dsRNA)-dependent kinase (PKR) interacts with viral dsRNA species following infection and autophosphorylates, yielding an active kinase dimer, which is then able to phosphorylate eIF2α. Other eIF2 kinases include the PKR-endoplasmic reticulum (ER)-related kinase (PERK), which responds to the accretion of misfolded proteins within the ER; GCN2, which responds to amino acid deprivation; and the hemin-regulated inhibitor kinase (HRI), which is activated following heme deprivation in reticulocytes (13, 16, 25, 43). In the case of GCN2- and PERK-mediated eIF2α phosphorylation, the reduction of global protein synthesis is coupled to the upregulation of stress-induced genes, such as ATF4 in mammalian cells and GCN4 in yeast. These mRNAs are translated by a regulated reinitiation mechanism involving the altered translation of upstream open reading frames (uORFs) in the 5′ leaders of each mRNA (12, 14, 45).
Previous reports using yeast and insect cell models indicated that the smallest subunit of eIF2B, eIF2Bα, is necessary for the recognition of phosphorylated eIF2α to inhibit protein synthesis. However, it is not clear whether eIF2Bα is essential for GEF activity under nonstressful, normal cellular conditions in mammalian cells (17, 28, 44). Additionally, reports in yeast have shown that certain point mutations in the yeast eIF2Bα subunit GCN3 are able to overcome the growth-inhibitory effects of PKR overexpression but fail to induce GCN4 expression in the presence of constitutive GCN2 activity or under starvation conditions. Interestingly, such point mutations can exhibit a stronger phenotype than the deletion of the GCN3 gene (28).
Other studies relating to mammalian eIF2B have involved attempts to understand mechanisms of virus-mediated oncolysis. For example, tumor cells appear to be remarkably susceptible to virally induced lysis compared to normal cells. These observations have led to an interest in the utilization of viruses such as vesicular stomatitis virus (VSV) as an oncolytic agent for the treatment of malignant disease. Previous studies indicated that while PKR commonly retains the ability to phosphorylate eIF2α in tumor cells following virus infection, GEF exchange rates remain surprisingly high (2). Subsequent analyses of susceptible tumor cells indicated possible defects in eIF2B function, which may neutralize eIF2α phosphorylation. Specifically, an excess expression of the catalytic eIF2Bε subunit was found in some sensitive tumor cells (2).
To extend these studies, we sought to determine the importance of the alpha subunit with respect to eIF2B function, including its role in the regulation of translation rates following virus infection. Here, using an RNA interference (RNAi) approach, we show that eIF2Bα is dispensable for normal growth and translation in cell lines and demonstrate an important requirement for the alpha subunit of eIF2B in translational regulatory innate immune responses to VSV infection in mammalian cells. Our studies indicate that normal cells depleted of eIF2Bα were significantly more resistant to the inhibition of translation following the phosphorylation of eIF2α by kinases such as PKR and, as a consequence, exhibited greatly increased susceptibility to VSV infection. Furthermore, we show that reconstitution with eIF2Bα variants harboring single missense mutations can also neutralize the eIF2α translation-inhibitory pathway and similarly render cells predisposed to virus-mediated lysis. These data further imply that a dysregulation of eIF2B may be sufficient to make cells more prone to viral infection and could contribute to the susceptibilities of different tumor cells to viral oncolysis.
MATERIALS AND METHODS
Plasmids.
eIF2Bα and eIF2Bβ cDNA clones were purchased from the ATCC and cloned into pCMV-tag2B (Stratagene) using the EcoRI/XhoI sites. Site-directed mutagenesis was performed on eIF2Bα by using a Stratagene PCR site-directed mutagenesis kit to yield point mutations using the following primers: AGGGTCTGAGGGCCAATCTCACCAGTG and CACTGGTGAGATTCGCCCTCAGACCCT for T41A, TGGCGGGGAGCTCCTCCTTCCGCTTCA and TGAAGCGGAGGAGGAGCTCCCCGCCA for F72L, AGTTTGTCCGGTCTCTTCCCACTAAAC and GTTTAGTGGAAAGAGCCGGACAAACT for F239L, and CTGACACCCCGAGCAGTCAGCGATG and CATCGCTGACTGCTCGGGGTGTCAG for S294R. Point mutations and wild-type eIF2Bα were subcloned into yeast vector pEMBLyex4 using the BamHI and HindIII sites. For baculovirus experiments, constructs were subcloned into the pFASTBAC vector using the NotI site upstream of the ATG in pCMV-tag2B and the XhoI site. The eIF2Bγ (Open Biosystems) construct was inserted between the HindIII and ApaI sites in pCMV-tag2B and then subcloned into pFASTBAC using the NotI and KpnI sites. The eIF2Bδ (Invitrogen) construct was inserted between the EcoRV and HindIII sites in pCMV-tag2B and subcloned into pFASTBAC using the NotI site and the HindIII site. The eIF2Bε (ATCC) construct was inserted between the EcoRV and HindIII sites in pCMV-tag2B and subcloned into pFASTBAC using the NotI site and the KpnI site.
Immunoblotting.
Polyclonal antiserum to VSV was obtained by immunizing BALB/c mice with wild-type VSV. Rabbit polyclonal antibodies specific to eIF2Bα (DDKELIEYFKSQMKEDPD) and eIF2Bβ (FIESFVTLKGGGPRS) were generated to peptides through Invitrogen Antibody Services. The following other commercially available antibodies were obtained: anti-murine PKR (mPKR), rabbit polyclonal anti-eIF2α, and monoclonal anti-β-actin (SantaCruz); anti-phospho-eIF2α (Biosource International); and monoclonal anti-M2-FLAG (Sigma). Nitrocellulose was developed by using Super-Signal ECL (Pierce).
Cells and transfections.
HeLa cells (ATCC) and primary murine embryonic fibroblasts (MEFs) (kind gift of Wen-Chen Yeh) were propagated in Dulbecco's modified Eagle's medium (DMEM) (VWR) supplemented with 10% fetal bovine serum (VWR), 5% penicillin-streptomycin (VWR), and 1 ml Normocin (Invivogen). Transfections were carried out by using Lipofectamine/Plus or Lipofectamine 2000 (Invitrogen) transfection reagent in Opti-mem (Invitrogen-Gibco) according to the manufacturer's protocol. Cell growth curves were determined by the quantitation of live cells using trypan blue exclusion assays.
Measurement of protein synthesis by 35S labeling.
DNA-transfected and/or small interfering RNA (siRNA)-treated HeLa cells were pulsed with 35S-labeled methionine and cysteine (0.2 mCi/ml) for 20 min. Cells were washed and immediately lysed in cold radioimmunoprecipitation assay (RIPA) buffer following virus infection or the indicated treatments. The incorporation of label into total cellular proteins was measured by a standard trichloroacetic acid (TCA) precipitation. Washed and dried protein pellets were resuspended in 2× SDS loading dye. Two microliters of lysate was spotted onto Whatman filters and dried, and the incorporated radioactivity was measured with a scintillation counter, or the lysate was run on SDS-PAGE gels. 35S counts were corrected for the total protein concentration as determined by a Bradford assay using a commercial kit (Pierce). Chemical treatments were as follows: poly(IC) (Amersham) was transfected at 2 μg/ml after being complexed in Opti-mem (Invitrogen-Gibco) using Lipofectamine 2000 (Invitrogen), and tunicamycin and thapsigargin (Sigma-Aldrich) were used at concentrations of 2 μg/ml and 400 nM/ml, respectively.
Virus infections.
siRNA-treated HeLa or MEF cells were grown to 70% confluence for 72 h following the siRNA knockdown of the indicated genes. After washing with 1× phosphate-buffered saline (PBS), cells were infected with either VSV-GFP (Indian strain), encephalomyocarditis virus (EMCV) (ATCC), or influenza virus using the indicated multiplicities of infection (MOIs) for all infections. Cells were incubated with virus for 1 h at 37°C in DMEM, with rocking every 10 min. After the removal of the medium and two washes with 1× PBS, complete medium was added to cells. Supernatant samples, protein lysates, and green fluorescent protein (GFP) fluorescence (for VSV-GFP infections) were detected at the indicated time points, usually 24 h postinfection. For beta interferon protection, cells were incubated 24 h prior to infection with 100 U/ml beta interferon (catalog number I-9032; Sigma).
Viral titers.
For the determination of viral titers, BHK cells were employed for VSV-GFP and EMCV. Cells were plated to 90% confluence and incubated with 4 dilutions of the collected supernatants. After 1 h of incubation with virus, 1.0% low-melting-point agarose in complete medium was added to the cells. After plaque formation (approximately 18 h for VSV and EMCV), cells and plaques were fixed and visualized with 0.1% crystal violet and 30% methanol. Viral titers are expressed in PFU. Determinations of influenza virus titers were performed similarly, with the exception that Vero cells were plated in place of BHK cells, and plaque formation was observed at 48 h.
RNAi.
HeLa cells (100,000 cells per well) grown in 6-well plates were transfected by using 100 pmol siRNA and 6 μl of Oligofectamine (Invitrogen, Carlsbad, CA) for the formation of DNA-liposome complexes, according to the manufacturer's recommendations, in Opti-mem (Invitrogen-Gibco). Mouse embryonic fibroblasts were transfected by using an Amaxa Nucleofector apparatus (program A-023) and MEF Nucleofector kit 1 according to the manufacturer's recommendations (Amaxa Biosystems). Cells were replated in 6-well plates. Assays were typically performed 72 h after siRNA treatment, when gene knockdown was found to be maximal, and cells were 75% to 80% confluent. The following chemically synthesized 21-nucleotide sense and antisense RNA oligonucleotides were obtained from Dharmacon: nonsilencing (NS) D-001810-01; murine eIF2Bα-silencing J-052907-05, J-052907-06, and J-052907-07; murine PKR-silencing J-040807-06; and human eIF2Bα-silencing M-020180-00, D-020180-01, and D-020180-02.
Yeast strains and genetic methods.
Standard methods were used to transform the previously described isogenic yeast strains H1515 (GCN2 GCN3), GP3153 (GCN2 gcn3Δ::LEU2), and Y117 (gcn2Δ gcn3Δ::LEU2) (28) with the indicated plasmids. The growth of transformants on selective medium was monitored by growing liquid cell cultures to an A600 of 0.3, from which 5-fold serial dilutions were made. Three microliters of each dilution was then spotted onto the appropriate medium and incubated at 30°C. SD (synthetic dextrose) medium contains 2% glucose, and S-Gal medium (Sigma) contains 4% galactose and 2% raffinose as carbon sources. 3-Aminotriazole (3AT; Fluka) was added at a 50 mM final concentration where indicated.
Plasmids.
The following yeast plasmids expressing yeast eIF2Bα (yeIF2Bα) (GCN3) were described previously: p2304 (2μm URA3 GCN3) (46), pWY03 (2μm URA3 GCN3-T41A), and pWY115 (2μm URA3 GCN3-S293R) (28). Plasmids to express N-terminally FLAG-tagged human eIF2Bα (heIF2Bα) in yeast were constructed by subcloning the wild-type and mutated heIF2Bα ORFs into a yeast expression plasmid with galactose-inducible promoter pGAL-GCN2-FH so that the GCN2 ORF and the His6 tag were replaced with the appropriate heIF2Bα ORF using the SmaI and HindIII enzymes. The resulting plasmids were pAV1852 (2μm URA3 pGAL-heIF2Bα), pAV1853 (2μm URA3 pGAL-heIF2Bα-T41A), pAV1854 (2μm URA3 pGAL-heIF2Bα-F72L), pAV1855(2μm URA3 pGAL-heIF2Bα-F239L), and pAV1856 (2μm URA3 pGAL-heIF2Bα-S294R). Insert DNA sequences were confirmed. For an unknown reason, heIF2Bα-S294R mutant protein expression was not detected with either FLAG or heIF2Bα antiserum, and in all yeast phenotypic tests, this construct behaved as the vector control (data not shown). pGAL-FH-GCN2 is a derivative of pEMBLyex4 (2μm URA3 pGAL). The latter was used as a vector control throughout (34).
Yeast protein analysis.
Whole-yeast-cell protein extraction and immunoblotting were performed as previously described (34). For Immunopreciptiations with anti-FLAG antisera, 1 mg of total cell extracts in buffer (20 mM Tris-HCl [pH 7.5], 100 mM NaCl, 2 mM MgCl2, 0.1% Triton X-100, 5 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride [PMSF], 0.7 μg/ml pepstatin, 1 μg/ml leupeptin,1 μg/ml aprotinin, 1× complete-EDTA protease inhibitors [Roche]), prepared using glass beads and a Fastprep machine, was bound to 40 μl FLAG-M2 EZ-View resin (Sigma) for 2 h at 4°C. The pellets were washed extensively in the same buffer, and proteins were eluted into sample buffer.
RESULTS
Loss of the alpha subunit of eIF2B neutralizes sensitivity to thapsigargin.
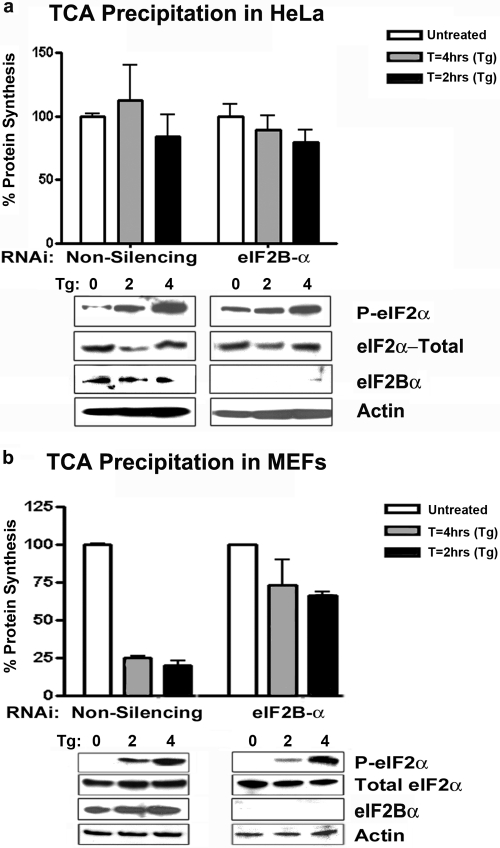
In yeast, eIF2Bα has been shown to be the only nonessential subunit of the eIF2B complex (11). However, it is not clear whether eIF2Bα is similarly dispensable in mammalian cells. To start to evaluate whether eIF2Bα is essential for normal cell growth and viability in mammalian cells, we used siRNA to repress eIF2Bα gene expression in HeLa cells. RNAi oligonucleotides specific to eIF2Bα and nonspecific (NS1) RNAi oligonucleotides were used for these studies, and knockdown was confirmed by immunoblotting using a novel peptide-derived antibody generated to the eIF2Bα subunit (Fig. 1a and b). These experiments were repeated with two additional RNAi oligonucleotides specific to eIF2Bα and a second nonsilencing RNAi (data not shown). Following up to 5 days of silencing, eIF2Bα-depleted cells did not exhibit any detectable changes in cell morphology or replication compared to nonspecific-RNAi-treated cells (Fig. 1c and d). These data indicate that eIF2Bα may not be critically required to maintain cell viability, at least over a short period. These results contrast with data from our previous report on the eIF2Bε subunit, where we observed a precipitous decrease in cell viability at approximately 4 days post-RNAi silencing (2). Because the eIF2-eIF2B checkpoint has been shown to play a vital role in controlling VSV replication, we next sought to determine whether the depletion of eIF2Bα would increase HeLa cell susceptibility to VSV infection. Following 72 h of RNAi transfection, NS1 and eIF2Bα RNAi-treated cells were infected with VSV-GFP for 18 h at an MOI of 0.1. Cells were examined for increases in GFP fluorescence, viral protein expression, and viral replication. eIF2Bα-depleted cells showed very slight increases in GFP and viral protein expression levels compared to the NS1-treated cells; however, there was no significant difference in viral replication (Fig. 1e to g). Again, these observations contrast with earlier studies on eIF2Bε where RNAi silencing of the eIF2B catalytic subunit yielded viral titers 100-fold lower than those of nonspecific-RNAi-treated HeLa cells, presumably because eIF2Bε is a critical component of GTP exchange. However, that report also indicated that HeLa cells are already extremely sensitive to VSV infection and have high GEF rates even in the presence of phosphorylated eIF2α, implying that the regulatory subunits in these cells may not function properly (2). These observations argue for a defect at the level of eIF2α-eIF2B regulation in HeLa cells and potentially may explain why these cells exhibit such a high degree of susceptibility to VSV infection. To evaluate this further, we treated NS1 or eIF2Bα RNAi-transfected HeLa cells with thapsigargin, an activator of the eIF2α-specific kinase PERK, for 2 and 4 h, respectively (13). Following thapsigargin treatment, HeLa cells were labeled with [35S]methionine, and protein synthesis rates were examined by scintillation analysis. These results indicated that despite the efficient phosphorylation of eIF2α, neither the NS1 control-treated nor the eIF2Bα-depleted HeLa cells showed a significant reduction in protein synthesis rates following exposure to thapsigargin (Fig. 2a). These data agree with previously reported observations indicating a potential defect in eIF2B activity in HeLa cells as well as other types of transformed cells (2). Because this pathway appears to be largely unresponsive in HeLa cells, we repeated the same experiment with primary mouse embryonic fibroblasts (MEFs) using an RNAi duplex specific to the murine eIF2Bα gene. First, we confirmed that RNAi efficiently inhibited eIF2Bα in normal MEFs (C57BL/6), to greater than 90% over 3 days (Fig. 2b). Subsequently, we observed that thapsigargin treatment resulted in a 70 to 80% decrease in protein synthesis in MEFs treated with control RNAi, as demonstrated by a reduction in 35S incorporation (Fig. 2b). In contrast, we found that the RNAi depletion of eIF2Bα in primary MEFs resulted in a dramatic neutralization of sensitivity to eIF2α phosphorylation (Fig. 2b). For primary MEFs we observed an approximately 20 to 30% reduction in translation rates in the eIF2Bα RNAi-treated cells. Immunoblot analysis confirmed that eIF2α was phosphorylated to comparable levels after thapsigargin treatment in both the NS1 and eIF2Bα RNAi-treated MEFs. These data indicate that the loss of eIF2Bα in mammalian cells results in a loss of sensitivity to the phosphorylation of eIF2α as a consequence of thapsigargin treatment and PERK activity.
Fig. 2.
eIF2Bα does not cause a reduction in protein synthesis rates but is required for sensitivity to eIF2 phosphorylation. (a) HeLa cells were transfected with RNAi (either NS1 or eIF2Bα specific), treated with thapsigargin (Tg) (400 nM) for the indicated times, and labeled with [35S]methionine during the last 20 min of incubation. Cell lysates were electrophoresed on a 12% SDS-PAGE gel, subjected to Western blotting, and probed with antibodies for the indicated proteins. Lysates were also precipitated by using trichloroacetic acid (TCA) to remove free probe. Twenty-microgram samples were blotted onto Whatman circles, and 35S counts were determined by scintillation. (Error bars indicate SD, where n = 3.) (b) Primary murine embryonic fibroblasts were transfected with RNAi (either NS1 or eIF2Bα specific), treated with thapsigargin (400 nM) for the indicated times, and labeled with [35S]methionine during the last 20 min of incubation. Cell lysates were subjected to Western blot analysis as described above for data in panel A. (Error bars indicate SD, where n = 3.)
Loss of eIF2Bα renders primary MEFs susceptible to VSV.
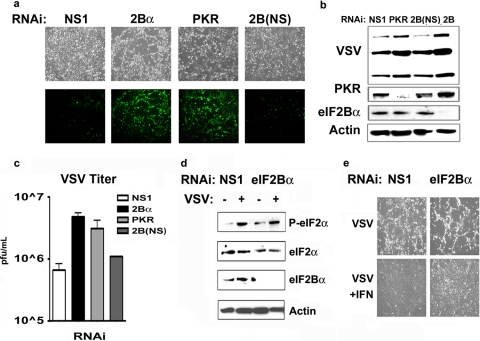
Because our results shown in Fig. 2 indicated that eIF2Bα is necessary for the inhibition of translation following thapsigargin treatment, we predicted that the depletion of eIF2Bα would influence VSV replication in primary cells, since viral dsRNA intermediates are known to activate PKR, which causes the phosphorylation of eIF2α. Once eIF2α is phosphorylated, it represses translation initiation by binding eIF2B in a manner that prevents eIF2B GEF activity, as described above. To evaluate this possibility, we treated normal MEFs with RNAi specific to eIF2Bα or PKR as a positive control (the loss of PKR has been shown to render MEFs sensitive to VSV infection) (3). Two negative controls were also adopted: a nonspecific RNAi and an RNAi duplex generated for the human eIF2Bα gene that contained a 2-bp mismatch with its murine counterpart. Immunoblot analysis confirmed the specific knockdown of eIF2Bα or PKR (Fig. 3b). After 3 days of RNAi treatment, MEFs were infected with VSV expressing a GFP construct (VSV-GFP) (MOI of 1 for 24 h). Following infection, our results indicated that primary MEFs treated with RNAi specific to eIF2Bα allowed almost a full log increase in viral replication compared to NS1 and eIF2Bα scrambled control RNAi-treated cells (Fig. 3a to c). Cells transfected with RNAi specific to PKR were also susceptible to VSV infection compared to negative controls, as expected (Fig. 3a to c). The increases in viral replication occurred despite the observed equivalent levels of phosphorylated eIF2α in both the NS1 and eIF2Bα cells (Fig. 3d). The data indicate that the loss of eIF2Bα in normal MEFs renders them more sensitive to VSV infection, possibly due to a loss of sensitivity to the PKR-mediated phosphorylation of eIF2α. Therefore, the loss of a single subunit of eIF2B in normal cells can dramatically affect the outcome of viral replication, similarly to the loss of PKR.
Fig. 3.
Loss of eIF2Bα results in increased susceptibility to VSV. (a) Light and fluorescence microscopy of primary MEFs transfected with NS1, 2Bα, PKR, or 2Bα-NS (RNAi generated to eIF2Bα that does not result in knockdown) and infected with VSV-GFP at an MOI of 1 for 24 h. Pictures illustrate both CPE, indicated by cell rounding and a loss of adherence, and viral GFP expression, indicated by the fluorescent images. (b) Immunoblot showing viral protein expression of MEFs from panel a. The blot was also probed with antibodies generated to PKR, eIF2Bα, and actin to demonstrate RNAi knockdown and equivalent loading. (c) Quantitation of a viral plaque assay performed with supernatants from panel a. (Error bars represent SD of data from duplicate samples from 3 independent experiments.) (d) Western blot analysis of lysates from MEFs treated with NS1 or eIF2Bα-specific RNAi following infection with VSV-GFP at an MOI of 1 for 24 h, showing efficient phosphorylation of eIF2α. (e) MEFs transfected with NS1 or eIF2Bα-specific RNAi were pretreated with 100 U/ml beta interferon for 24 h prior to infection with VSV-GFP at an MOI of 1. Pictures were taken at 24 h postinfection.
Previous reports indicated that defects in interferon (IFN) signaling can also render cells susceptible to virus replication (3). To evaluate whether the loss of eIF2Bα influences IFN signaling, we transfected MEFs with control RNAi or RNAi specific to eIF2Bα and after 2 days pretreated them with murine IFN-β. After 24 h, MEFs were infected with VSV-GFP (MOI = 1). This study indicated that while the loss of eIF2Bα rendered cells more susceptible to VSV than controls, IFN was able to overcome the deficit of eIF2Bα and prevent virus replication (Fig. 3e). Thus, the loss of eIF2Bα does not affect IFN-mediated signaling or antiviral activity, at least against VSV infection. Nevertheless, these data suggest that eIF2Bα is a key component in an effective early host defense against virus infection, probably prior to the induction of IFN-mediated antiviral gene expression. We next sought to determine whether eIF2Bα depletion rendered MEFs sensitive to a broad range of viruses or if these results are specific to VSV. To address this question, we treated MEFs with control or eIF2Bα-specific RNAi and then infected these cells with either EMCV (MOI = 0.1) or influenza virus (MOI = 1). In contrast to our previous results using VSV, we show that the loss of eIF2Bα does not result in observable increases in viral replication of either EMCV or influenza virus in MEFs (see Fig. S1 in the supplemental material). These results indicate that the increased susceptibility of MEFs to VSV replication following eIF2Bα silencing may be virus specific or that EMCV or influenza virus may have devised methods to avoid the activation of this pathway (16).
eIF2Bα variants can neutralize sensitivity to eIF2α phosphorylation in yeast.
Previously, Hannig and Hinnebusch found that the loss of the yeast eIF2Bα gene (GCN3) did not reduce growth rates in response to amino acid deprivation (11). Moreover, it was found that a variety of specific missense mutations in any of three subunits of yeast eIF2B (α-GCN3, β-GCD7, and δ-GCD2) could strongly neutralize the sensitivity of eIF2B to phosphorylated yeast eIF2α (28). All alleles reduced the sensitivity of eIF2B to the presence of phosphorylated eIF2α and failed to derepress the translation of GCN4. Subsequent biochemical analyses of a subset of these alleles revealed that they reduced the affinity of the mutant eIF2B complex for phosphorylated eIF2α and allowed phosphorylated eIF2 to be a substrate of eIF2B (21, 28, 40).
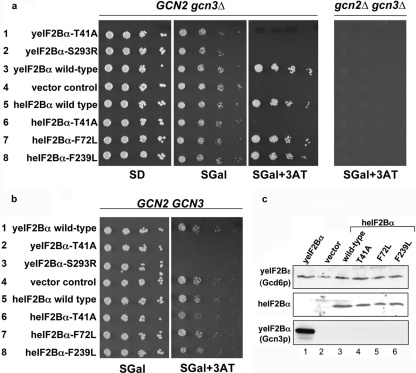
To determine whether mutations within human eIF2Bα (heIF2Bα) would have similar effects on the eIF2B function in yeast, a cDNA encoding human eIF2Bα (EIF2B1, sharing approximately 36% homology with its yeast counterpart) was cloned with an N-terminal FLAG tag into a yeast expression plasmid under the control of a galactose-inducible promoter (28). This plasmid was transformed into yeast cells (gcn3Δ) lacking the yeast eIF2Bα gene. As reported previously, gcn3Δ yeast cells grow normally on nonstarvation medium but fail to induce GCN4 translation upon amino acid starvation, which activates GCN2 and phosphorylates yeast eIF2 (11). Therefore, gcn3Δ yeast cells cannot grow in the presence of 3-aminotriazole (3AT), which inhibits histidine biosynthesis (Fig. 4a, row 4). Human eIF2Bα conferred resistance to 3AT that was dependent on the presence of the protein kinase Gcn2p (Fig. 4a, row 5). This showed that human eIF2Bα can replace the function of yeast eIF2Bα, similar to results previously described for rat and Drosophila melanogaster eIF2Bα (19, 44). In terms of the point mutants, we found that two alleles behaved identically to wild-type heIF2Bα (F72L and F239L), while one behaved similarly to the corresponding yeast mutations (T41A) and conferred sensitivity to 3AT when grown on galactose-containing medium (Fig. 4a, rows 6 to 8). Immunoblotting with anti-heIF2Bα serum revealed that wild-type heIF2Bα and all mutants were expressed to equal amounts (Fig. 4c). These data indicate that select point mutations occurring in the alpha subunit of eIF2B can neutralize the activity of eIF2α phosphorylation in yeast.
Fig. 4.
Expression of human eIF2Bα variants in yeast. (a) Growth of serially diluted (5-fold) yeast strains (derived from a GCN2 gcn3Δ or a gcn2Δ gcn3Δ strain) bearing human eIF2Bα or yeast GCN3 missense mutations on minimal synthetic dextrose medium (SD), galactose-raffinose medium (SGAL), and SGAL supplemented with 3-aminotriazole (3AT). (b) Growth of serially diluted yeast strains (derived from a GCN2 GCN3 strain) bearing human eIF2Bα or yeast GCN3 missense mutations on galactose-raffinose medium (SGAL) and SGAL supplemented with 3AT. (c) Immunoblot analysis of protein extracts to show expression of the human eIF2B yeast constructs grown on SGAL in panel A. Membranes were probed for FLAG expression.
Point mutations in eIF2Bα result in differences in eIF2B subunit associations.
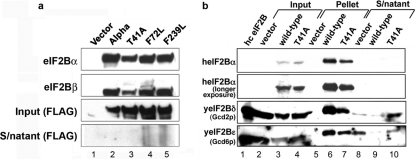
To determine whether the 3AT-sensitive (3ATs) phenotype of heIF2Bα-T41A was caused by this protein forming a complex with the yeast eIF2B subunits, further experiments were done. First, a genetic experiment was undertaken to determine the dominance of the 3ATs phenotype. The overexpression of the yeast 3ATs alleles T41A and S293R conferred the 3ATs phenotype even in the presence of chromosomally encoded yeIF2Bα (GCN3) (Fig. 4b, rows 2 and 3). Similarly, heIF2Bα-T41A weakened the growth on 3AT plates, indicating that the expression of this human point mutant can substitute for the homologous yeast mutant and produce a similar, albeit slightly weaker, phenotype (Fig. 4b, row 6). Second, to demonstrate the expression and stable association of our point mutants within the eIF2B complex, we overexpressed each of the FLAG-tagged point mutants, as well as the empty vector or wild-type eIF2Bα gene, in 293T cells and used FLAG-conjugated beads to immunoprecipitate eIF2B complexes. We then examined the precipitates by immunoblot analysis for the presence of eIF2Bα as well as another eIF2B subunit, eIF2Bβ, using antibodies generated in our laboratory. Our results show that eIF2Bβ can efficiently coprecipitate with eIF2Bα, indicating that the eIF2Bα variants were complexing with eIF2B (Fig. 5a). We next examined the ability of wild-type human eIF2Bα and the mutant alleles to associate with a yeast eIF2B complex. Protein complexes were immunoprecipitated with anti-FLAG antibody beads, and the coassociation of yeast eIF2B subunits was assessed by immunoblotting using anti-yeast eIF2B antibody. The results showed that both wild-type heIF2Bα and the T41A mutant can associate with yeast eIF2B subunits but not as completely as with heIF2B (Fig. 5b). Taken together, these data provide evidence that the human eIF2Bα-T41A mutant subunit can compete with endogenous yeIF2Bα, enter the yeIF2B complex, and reduce the ability of yeast cells to respond to eIF2α phosphorylation. This experiment provides evidence of the equivalence of the function of the eIF2Bα T41 residue in sensitizing yeast eIF2B and human eIF2B to the phosphorylation status of eIF2.
Fig. 5.
Point mutations in eIF2Bα. (a) Immunoblot of 293T cell lysates from cells transfected with FLAG-tagged wild-type or mutant eIF2Bα constructs, which were immunoprecipitated using anti-FLAG agarose beads to pull down eIF2B complexes. Blots were probed for eIF2Bα and eIF2Bβ to show associations. For input lanes, 20 μl of lysate supernatant (S/natant) was probed by Western blotting for FLAG expression to ensure equal protein concentrations of the starting material. The remaining supernatant following pulldown with anti-FLAG agarose was also subjected to Western blotting and probed for FLAG to ensure the complete removal of FLAG constructs by coimmunoprecipitation. (b) Extracts of yeast cells carrying human FLAG-tagged wild-type eIF2B or eIF2B variants were immunoprecipitated by using anti-FLAG agarose and examined by immunoblotting using yeast eIF2 antibodies.
Point mutations in eIF2Bα do not fully reconstitute proper translational control in response to VSV infection.
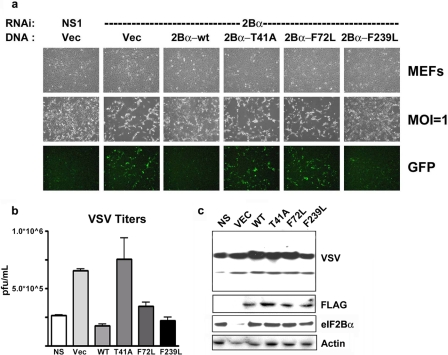
Results from our studies in yeast indicated that the expression of eIF2Bα variants, such as the T41A mutation, may neutralize the effects of eIF2α phosphorylation in mammalian cells, similar to the complete loss of the eIF2Bα gene using RNAi approaches. Possibly, this could render cells more sensitive to VSV infection. To evaluate this possibility, primary MEFs were treated with RNAi to ablate the expression of the endogenous eIF2Bα protein. After 3 days of treatment, the MEFs were transiently transfected with plasmids encoding FLAG-tagged wild-type eIF2Bα or variants of eIF2Bα to reconstitute eIF2Bα protein expression. The RNAi knockdown of eIF2Bα and the expression of FLAG-tagged transfected wild-type eIF2Bα or eIF2Bα variants was confirmed by immunoblot analysis (Fig. 6c). Similar to previous results observed following the ablation of eIF2Bα by RNAi, the vector-control-transfected cells depleted of eIF2Bα exhibited increased VSV-GFP protein expression and viral replication levels, while in contrast, the expression of wild-type eIF2Bα appeared to restore antiviral translational control (Fig. 6). Conversely, the transfection of the eIF2Bα mutants resulted in variable levels of sensitivity to VSV infection. Most notably, the expression of the T41A mutant yielded viral titers comparable to those of vector control transfectants lacking eIF2Bα, indicating that this variant was insensitive to eIF2α phosphorylation in MEFs, similar to its observed effects in yeast (Fig. 6b). Thus, eIF2Bα with select point mutations can overcome PKR-mediated antiviral activity, similar to the loss of eIF2Bα expression in mammalian cells, as in yeast. Collectively, our data indicate that eIF2Bα is a key regulator of translation at the point of eIF2α phosphorylation and that the loss or select mutation of this gene renders stress-induced kinases ineffective at inhibiting protein synthesis.
Fig. 6.
Point mutations in eIF2Bα may function similarly to the loss of eIF2Bα. (a) MEFs were cotransfected with either NS or murine-specific eIF2Bα RNAi and empty vector, human wild-type eIF2Bα (WT), or human eIF2Bα point mutants. At 72 h posttransfection cells were infected with VSV-GFP at an MOI of 1 for 24 h and examined by light and fluorescence microscopy. Cells were examined for both CPE and GFP fluorescence. (b) Quantitation of viral plaque assays performed with supernatants from panel a. (Error bars indicate SD of data from duplicate samples from two independent experiments.) (c) Western blot analysis of viral protein synthesis and overexpression of FLAG-tagged constructs performed with lysates from panel a. Blots were probed by using antiserum to VSV, FLAG, and actin.
DISCUSSION
Vesicular stomatitis virus is currently being explored for its potential use as a therapeutic treatment for cancer; therefore, an understanding of the mechanisms by which VSV is able to infect and kill tumor cells is an important objective for furthering the clinical exploitation of this virus. VSV is a cytoplasmic virus that relies entirely upon host translational machinery to synthesize its own proteins, and as such, it is susceptible to host defense countermeasures that regulate these pathways (4, 37). The PKR-eIF2-eIF2B pathway serves as a key checkpoint to prevent protein synthesis following virus infection. Accordingly, defects that interrupt this association, such as the loss of PKR, have been shown to increase cellular susceptibility to virus infection (3). In this report, we emphasize the importance of ternary complex regulation in host defense through the RNAi depletion of the eIF2B subunit eIF2Bα, which is sufficient to cause an increase in VSV replication and cell death in normal mammalian cells. These data accentuate a definitive requirement for eIF2Bα in the regulation of eIF2B activity by phosphorylated eIF2α in mammalian cells. Our data also suggest that there is not a strong requirement for eIF2Bα to maintain normal rates of protein synthesis or cell viability in mammalian cells, at least using RNAi, which suppresses eIF2Bα expression over a 4-day period. However, it remains plausible that complete knockdown has not been entirely obtained (we routinely achieved greater than 95% suppression) and that residual amounts of eIF2Bα are sufficient to maintain eIF2B activity under normal conditions. Nevertheless, our observations agree with data from previous studies of eIF2Bα in both yeast and insect cells, where the loss of this subunit did not affect cell growth or normal GEF activity (8, 18, 28). Additionally, it was reported previously that eIF2Bα is required for the derepression of GCN4 in yeast following amino acid starvation, an event that is dependent upon eIF2α phosphorylation (1, 6). In contrast, a further study indicated eIF2B complexes lacking the alpha subunit do not maintain full exchange activity when examined in vitro. These studies went on to show that reconstitution with rat eIF2Bα does restore some eIF2B function, and those authors concluded that this implies that the loss of eIF2B in eIF2BΔα complexes is attributable directly to the absence of this subunit (44). Possibly, the discrepancies in such observations are a reflection of differences in the assays. For example, while GEF in vitro assays are a measure of eIF2B activity using purified protein extracts, it is quite possible that they do not entirely reflect in vivo situations where issues of protein folding or other unknown regulatory mechanisms may be important. We also cannot rule out that there might be another gene substituting for eIF2Bα in our RNAi-treated cells, although this is unlikely given that cells lacking the RNAi-targeted eIF2Bα are extremely sensitive to virus infection. RNAi analysis with MEFs indicated that cells lacking eIF2Bα have similar rates of [35S]methionine incorporation compared to the basal incorporation rates in NS RNAi-treated cells. This implies that the loss of eIF2Bα does not result in a significant decrease in the translation rates, at least under nonstressful steady-state conditions. This assertion is further supported by the fact that cell morphology and growth remain normal after RNAi depletion.
It is of interest that defects in eIF2B have been associated with the human disorder vanishing white matter with leukoencephalopathy (VWM) (38). Mutational analysis of patients with this neuropathological disorder revealed the presence of point mutations in eIF2B that have since been genetically linked to the disease (24). To date, all five eIF2B subunits have been linked to VWM; however, less than 2% of the known mutations identified have been reported to occur in eIF2Bα (32). In the case of VWM mutations, patient samples show slightly lower eIF2B exchange rates, but these rates are accompanied by higher levels of ATF4 expression and unfolded-protein responses, indicating the presence of low levels of constitutive phospho-eIF2α sensitivity. Contrary to those findings, our analysis showed that eIF2Bα point mutations and RNAi depletion of eIF2Bα did not lead to higher levels of ATF4 induction (data not shown). These studies highlight the fact that point mutations in different subunits of eIF2Bα may result in contrasting phenotypes. It is of interest that the eIF2B mutations specifically impair glial cells and appear to spare other cell types. For example, two studies concerning eIF2B and VWM further showed that decreased eIF2B activity does not reduce the protein synthesis, defective translation regulation, or cell growth of patient-derived lymphoblasts and fibroblasts (15, 39). Future studies are needed to clarify these issues and resolve the discrepancies observed among the various assay systems. Nevertheless, our study confirms that eIF2Bα is not strongly required to maintain cellular translation rates but is required to confer eIF2B sensitivity to eIF2α phosphorylation in mammalian cells.
Previous studies have also implicated deficiencies in eIF2B regulation and function in malignant cells by demonstrating that tumor cells lack sensitivity to eIF2α phosphorylation. Data indicated the presence of variations in the expression levels of eIF2B subunits in different tumor cell types, and the cells exhibited high GEF exchange rates in comparison to those of their normal counterparts (2). Levels of eIF2Bε were noted to be much higher in HeLa cells than in normal murine fibroblasts (data not shown). In this study, we demonstrate that the loss of eIF2Bα has little effect on the VSV susceptibility of HeLa cells, presumably because either this checkpoint is already defective or another pathway is overriding its effects on these cells. Regardless, it appears that eIF2-eIF2B regulation is largely nonresponsive in HeLa cells, and this seems to be a common characteristic among many tumor cells (2). Furthermore, while we demonstrate here that the loss of eIF2Bα with a T41A mutation can deregulate eIF2α phosphorylation, it is highly plausible that mutations in the other eIF2B subunits may function similarly. This event remains to be seen for mammalian cells; however, missense mutations in yeast eIF2Bβ and eIF2Bδ (GCD7 and GCD2) have been shown to produce a phenotype similar to that of eIF2Bα (GCN3) mutations (28).
An important question raised by studies such as those mentioned here remains whether or not translational dysregulation is a cause or a result of the process of transformation. To date, the majority of known oncogenes come from the transcription factor family, yet there are some examples of translation factors and their regulators acting as oncogenes. Mutant forms of PKR and eIF2α have been shown to transform cells, but the overexpression of eIF2Bα point mutants was not able to transform NIH 3T3 cells, nor was the overexpression of eIF2Bε (2, 7, 20, 29; data not shown). Additionally, sequence analysis did not reveal the natural occurrence of point mutations in eIF2Bα in HeLa cells (data not shown). Another translation factor, eIF4E, has also been shown to transform immortalized cell lines by simple overexpression and to cooperate with both v-myc and E1A to enhance transformation (5, 22, 23). Further studies of the role of translational regulation in tumorigenesis are needed to determine how this important cellular function is altered as cells transform. Nevertheless, it is highly likely that the oncolytic abilities of VSV are, at least in part, related to the loss of the regulation of protein synthesis in these cells. Additional studies will now have to address how we can take advantage of faulty translational machinery in cancer to develop the next generation of oncolytic therapeutics.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sharon Rannels and Lydia Kutzler for their contributions to the work presented in the manuscript.
This work was supported by the following grants: RO1CA095924-05 and RO1CA086431-09 to Glen N. Barber and NIH DK13499 to Scot R. Kimball.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 27 July 2011.
REFERENCES
- 1. Abastado J. P., Miller P. F., Hinnebusch A. G. 1991. A quantitative model for translational control of the GCN4 gene of Saccharomyces cerevisiae. New Biol. 3:511–524 [PubMed] [Google Scholar]
- 2. Balachandran S., Barber G. N. 2004. Defective translational control facilitates vesicular stomatitis virus oncolysis. Cancer Cell 5:51–65 [DOI] [PubMed] [Google Scholar]
- 3. Balachandran S., et al. 2000. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity 13:129–141 [DOI] [PubMed] [Google Scholar]
- 4. Barber G. N. 2005. VSV-tumor selective replication and protein translation. Oncogene 24:7710–7719 [DOI] [PubMed] [Google Scholar]
- 5. De Benedetti A., Rhoads R. E. 1990. Overexpression of eukaryotic protein synthesis initiation factor 4E in HeLa cells results in aberrant growth and morphology. Proc. Natl. Acad. Sci. U. S. A. 87:8212–8216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dever T. E., et al. 1992. Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68:585–596 [DOI] [PubMed] [Google Scholar]
- 7. Donze O., Jagus R., Koromilas A. E., Hershey J. W., Sonenberg N. 1995. Abrogation of translation initiation factor eIF-2 phosphorylation causes malignant transformation of NIH 3T3 cells. EMBO J. 14:3828–3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fabian J. R., Kimball S. R., Heinzinger N. K., Jefferson L. S. 1997. Subunit assembly and guanine nucleotide exchange activity of eukaryotic initiation factor-2B expressed in Sf9 cells. J. Biol. Chem. 272:12359–12365 [DOI] [PubMed] [Google Scholar]
- 9. Fabian J. R., Kimball S. R., Jefferson L. S. 1998. Reconstitution and purification of eukaryotic initiation factor 2B (eIF2B) expressed in Sf21 insect cells. Protein Expr. Purif. 13:16–22 [DOI] [PubMed] [Google Scholar]
- 10. Gomez E., Mohammad S. S., Pavitt G. D. 2002. Characterization of the minimal catalytic domain within eIF2B: the guanine-nucleotide exchange factor for translation initiation. EMBO J. 21:5292–5301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hannig E. M., Hinnebusch A. G. 1988. Molecular analysis of GCN3, a translational activator of GCN4: evidence for posttranslational control of GCN3 regulatory function. Mol. Cell. Biol. 8:4808–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harding H. P., et al. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6:1099–1108 [DOI] [PubMed] [Google Scholar]
- 13. Harding H. P., Zhang Y., Ron D. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397:271–274 [DOI] [PubMed] [Google Scholar]
- 14. Hinnebusch A. G. 1993. Gene-specific translational control of the yeast GCN4 gene by phosphorylation of eukaryotic initiation factor 2. Mol. Microbiol. 10:215–223 [DOI] [PubMed] [Google Scholar]
- 15. Kantor L., et al. 2005. Heightened stress response in primary fibroblasts expressing mutant eIF2B genes from CACH/VWM leukodystrophy patients. Hum. Genet. 118:99–106 [DOI] [PubMed] [Google Scholar]
- 16. Katze M. G. 1995. Regulation of the interferon-induced PKR: can viruses cope? Trends Microbiol. 3:75–78 [DOI] [PubMed] [Google Scholar]
- 17. Kimball S. R., Fabian J. R., Pavitt G. D., Hinnebusch A. G., Jefferson L. S. 1998. Regulation of guanine nucleotide exchange through phosphorylation of eukaryotic initiation factor eIF2alpha. Role of the alpha- and delta-subunits of eIF2b. J. Biol. Chem. 273:12841–12845 [DOI] [PubMed] [Google Scholar]
- 18. Kimball S. R., Heinzinger N. K., Horetsky R. L., Jefferson L. S. 1998. Identification of interprotein interactions between the subunits of eukaryotic initiation factors eIF2 and eIF2B. J. Biol. Chem. 273:3039–3044 [DOI] [PubMed] [Google Scholar]
- 19. Kimball S. R., Horetsky R. L., Jagus R., Jefferson L. S. 1998. Expression and purification of the alpha-subunit of eukaryotic initiation factor eIF2: use as a kinase substrate. Protein Expr. Purif. 12:415–419 [DOI] [PubMed] [Google Scholar]
- 20. Koromilas A. E., Roy S., Barber G. N., Katze M. G., Sonenberg N. 1992. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science 257:1685–1689 [DOI] [PubMed] [Google Scholar]
- 21. Krishnamoorthy T., Pavitt G. D., Zhang F., Dever T. E., Hinnebusch A. G. 2001. Tight binding of the phosphorylated alpha subunit of initiation factor 2 (eIF2alpha) to the regulatory subunits of guanine nucleotide exchange factor eIF2B is required for inhibition of translation initiation. Mol. Cell. Biol. 21:5018–5030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lazaris-Karatzas A., Montine K. S., Sonenberg N. 1990. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature 345:544–547 [DOI] [PubMed] [Google Scholar]
- 23. Lazaris-Karatzas A., Sonenberg N. 1992. The mRNA 5′ cap-binding protein, eIF-4E, cooperates with v-myc or E1A in the transformation of primary rodent fibroblasts. Mol. Cell. Biol. 12:1234–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leegwater P. A., et al. 2001. Subunits of the translation initiation factor eIF2B are mutant in leukoencephalopathy with vanishing white matter. Nat. Genet. 29:383–388 [DOI] [PubMed] [Google Scholar]
- 25. Lu L., Han A. P., Chen J. J. 2001. Translation initiation control by heme-regulated eukaryotic initiation factor 2alpha kinase in erythroid cells under cytoplasmic stresses. Mol. Cell. Biol. 21:7971–7980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pain V. M. 1996. Initiation of protein synthesis in eukaryotic cells. Eur. J. Biochem. 236:747–771 [DOI] [PubMed] [Google Scholar]
- 27. Pavitt G. D., Ramaiah K. V., Kimball S. R., Hinnebusch A. G. 1998. eIF2 independently binds two distinct eIF2B subcomplexes that catalyze and regulate guanine-nucleotide exchange. Genes Dev. 12:514–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pavitt G. D., Yang W., Hinnebusch A. G. 1997. Homologous segments in three subunits of the guanine nucleotide exchange factor eIF2B mediate translational regulation by phosphorylation of eIF2. Mol. Cell. Biol. 17:1298–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Perkins D. J., Barber G. N. 2004. Defects in translational regulation mediated by the alpha subunit of eukaryotic initiation factor 2 inhibit antiviral activity and facilitate the malignant transformation of human fibroblasts. Mol. Cell. Biol. 24:2025–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pestova T. V., Kolupaeva V. G. 2002. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 16:2906–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pestova T. V., et al. 2001. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. U. S. A. 98:7029–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pronk J. C., van Kollenburg B., Scheper G. C., S. van der Knaap M. 2006. Vanishing white matter disease: a review with focus on its genetics. Ment. Retard. Dev. Disabil. Res. Rev. 12:123–128 [DOI] [PubMed] [Google Scholar]
- 33. Ramaiah K. V., Davies M. V., Chen J. J., Kaufman R. J. 1994. Expression of mutant eukaryotic initiation factor 2 alpha subunit (eIF-2 alpha) reduces inhibition of guanine nucleotide exchange activity of eIF-2B mediated by eIF-2 alpha phosphorylation. Mol. Cell. Biol. 14:4546–4553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Richardson J. P., Mohammad S. S., Pavitt G. D. 2004. Mutations causing childhood ataxia with central nervous system hypomyelination reduce eukaryotic initiation factor 2B complex formation and activity. Mol. Cell. Biol. 24:2352–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Richter J. D., Sonenberg N. 2005. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433:477–480 [DOI] [PubMed] [Google Scholar]
- 36. Singh C. R., et al. 2006. An eIF5/eIF2 complex antagonizes guanine nucleotide exchange by eIF2B during translation initiation. EMBO J. 25:4537–4546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sonenberg N., Dever T. E. 2003. Eukaryotic translation initiation factors and regulators. Curr. Opin. Struct. Biol. 13:56–63 [DOI] [PubMed] [Google Scholar]
- 38. van der Knaap M. S., Pronk J. C., Scheper G. C. 2006. Vanishing white matter disease. Lancet Neurol. 5:413–423 [DOI] [PubMed] [Google Scholar]
- 39. van Kollenburg B., et al. 2006. Regulation of protein synthesis in lymphoblasts from vanishing white matter patients. Neurobiol. Dis. 21:496–504 [DOI] [PubMed] [Google Scholar]
- 40. Vazquez de Aldana C. R., Hinnebusch A. G. 1994. Mutations in the GCD7 subunit of yeast guanine nucleotide exchange factor eIF-2B overcome the inhibitory effects of phosphorylated eIF-2 on translation initiation. Mol. Cell. Biol. 14:3208–3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Webb B. L., Proud C. G. 1997. Eukaryotic initiation factor 2B (eIF2B). Int. J. Biochem. Cell Biol. 29:1127–1131 [DOI] [PubMed] [Google Scholar]
- 42. Wek R. C., Jiang H. Y., Anthony T. G. 2006. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 34:7–11 [DOI] [PubMed] [Google Scholar]
- 43. Wek S. A., Zhu S., Wek R. C. 1995. The histidyl-tRNA synthetase-related sequence in the eIF-2 alpha protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol. Cell. Biol. 15:4497–4506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Williams D. D., Price N. T., Loughlin A. J., Proud C. G. 2001. Characterization of the mammalian initiation factor eIF2B complex as a GDP dissociation stimulator protein. J. Biol. Chem. 276:24697–24703 [DOI] [PubMed] [Google Scholar]
- 45. Yang R., Wek S. A., Wek R. C. 2000. Glucose limitation induces GCN4 translation by activation of Gcn2 protein kinase. Mol. Cell. Biol. 20:2706–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang W., Hinnebusch A. G. 1996. Identification of a regulatory subcomplex in the guanine nucleotide exchange factor eIF2B that mediates inhibition by phosphorylated eIF2. Mol. Cell. Biol. 16:6603–6616 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.