Abstract
The retroviral restriction factor TRIMCyp, derived from the TRIM5 gene, blocks replication at a postentry step. TRIMCyp has so far been found in four species of Asian macaques, Macaca fascicularis, M. mulatta, M. nemestrina, and M. leonina. M. fascicularis is commonly used as a model for AIDS research, but TRIMCyp has not been analyzed in detail in this species. We analyzed the prevalence of TRIMCyp in samples from Indonesia, Indochina, the Philippines, and Mauritius. We found that TRIMCyp is present at a higher frequency in Indonesian than in Indochinese M. fascicularis macaques and is also present in samples from the Philippines. TRIMCyp is absent in Mauritian M. fascicularis macaques. We then analyzed the restriction specificity of TRIMCyp derived from three animals of Indonesian origin. One allele, like the prototypic TRIMCyp alleles described for M. mulatta and M. nemestrina, restricts human immunodeficiency virus type 2 (HIV-2) and feline immunodeficiency virus (FIV) but not HIV-1. The others restrict HIV-1 and FIV but not HIV-2. Mutagenesis studies confirmed that polymorphisms at amino acid residues 369 and 446 in TRIMCyp (or residues 66 and 143 in the cyclophilin A [CypA] domain) confer restriction specificity. Additionally, we identified a polymorphism in the coiled-coil domain that appears to affect TRIMCyp expression or stability. Taken together, these data show that M. fascicularis has the most diverse array of TRIM5 restriction factors described for any primate species to date. These findings are relevant to our understanding of the evolution of retroviral restriction factors and the use of M. fascicularis models in AIDS research.
INTRODUCTION
Primates express several intrinsic restriction factors that can combat retroviral infection (15). One of the best studied is TRIM5α, which binds to the retroviral capsid and restricts replication at a postentry step (31). TRIM5α belongs to the TRIM (tripartite motif) gene family, a large family found throughout vertebrates with numerous members that have been implicated in immune responses (20, 24). TRIM genes contain, in order, RING, B-box, and coiled-coil domains (23). TRIM5α also has a C-terminal B30.2/SPRY domain, which is required for binding to retroviral capsids (27, 32).
In some primates, the retrotransposition of a cyclophilin A (CypA) sequence into the TRIM5 gene allows the expression of the TRIMCyp isoform. Alternative splicing to this insertion results in the replacement of the capsid-binding B30.2/SPRY domain with a CypA domain, which is also capable of binding some retroviral capsids (14). Thus, TRIMCyp, like TRIM5α, has retroviral restriction activity, but its specificity is determined by the CypA domain rather than the B30.2/SPRY domain. Interestingly, TRIMCyp has evolved independently by separate retrotransposition events in two distantly related primate genera, Aotus (owl monkeys; New World monkeys), and Macaca (macaques; Old World monkeys) (2, 11, 18, 25, 36, 37). In macaques, the CypA insertion is linked to a single-nucleotide polymorphism (SNP) at the exon 7 splice acceptor site, which changes the canonical AG splice acceptor sequence to AT and allows the skipping of exons 7 and 8 and splicing to the CypA sequence in the 3′ untranslated region (UTR) of the gene (2, 3).
These genetic changes are thought to be fixed in Macaca nemestrina and M. leonina; all individuals in these species examined so far express TRIMCyp but not TRIM5α (2, 3, 10). In contrast, TRIMCyp is found in approximately 25% of Indian Macaca mulatta animals and has not been found in at least 76 Chinese M. mulatta animals reported so far (17, 37). Both TRIMCyp and TRIM5α are expressed in Macaca fascicularis macaques (2), but the prevalence of the two forms in this species has not been previously examined.
M. fascicularis, known as the cynomolgus, longtail, or crab-eating macaque, occupies a geographic range in Southeast Asia, including parts of Indochina, Indonesia, and the Philippines (30, 34). These animals can be sorted into genetically distinct subpopulations (7, 28). However, all M. fascicularis macaques analyzed in the TRIMCyp literature to date have been of Indonesian origin (2, 39). Given the known geographic variability of TRIMCyp in M. mulatta (37), we hypothesized that similar geographic variations may exist in M. fascicularis TRIMCyp. Additionally, we sought to expand our knowledge of the allelic variability of M. fascicularis TRIMCyp.
Aotus TRIMCyp binds to and restricts the same viruses that bind to parental CypA, including human immunodeficiency virus type 1 (HIV-1) and feline immunodeficiency virus (FIV) (18, 25). However, the prototypical Old World TRIMCyp sequences from M. mulatta and M. nemestrina restrict HIV-2 but not HIV-1, due to two amino acid changes in the CypA domain (2, 21, 36, 37). The M. fascicularis TRIMCyp allele originally described by our laboratory had no detectable restriction activity (2). However, whether this lack of activity was a true reflection of the allelic diversity of M. fascicularis TRIMCyp or was the result of a cloning error cannot be definitively ascertained. Recently, Ylinen et al. described a second allele of M. fascicularis TRIMCyp and showed that its CypA domain can bind to and restrict HIV-1 (39). Therefore, we also sought to examine the functional diversity of TRIMCyp restriction factors among M. fascicularis macaques.
Here, we show that TRIMCyp-linked genotypes are present at significantly different frequencies in different M. fascicularis populations. We show that M. fascicularis has alleles of TRIMCyp that can restrict either HIV-1 or HIV-2 and that both alleles are found in all TRIMCyp-expressing M. fascicularis populations. These findings are likely to have critical implications for the use of M. fascicularis in AIDS-related research.
MATERIALS AND METHODS
Samples.
Peripheral blood from unrelated M. fascicularis animals housed at the Washington National Primate Research Center (WaNPRC) was collected in heparinized Vacutainer tubes. Blood was collected by venipuncture when animals were under sedation (ketamine at 10 to 15 mg/kg of body weight). DNA and RNA were isolated by using a QIAamp DNA blood minikit or a QIAamp RNA blood minikit (Qiagen), respectively, on fresh whole blood.
Blood samples from 16 M. fascicularis animals were obtained from Alpha Genesis, Inc., including 4 each derived from Indochina (2 from Vietnam and 2 from Cambodia), Indonesia, Mauritius, and the Philippines. Five additional Indonesian and 10 additional Indochinese samples were obtained from different breeders to ensure that animals were unrelated. DNAs were extracted by using the MagneSil genomic DNA extraction procedure (Promega, Inc.), and the animal ancestries were confirmed by a 66-SNP assay developed to distinguish macaque ancestry (30). The SNP assay also verified each animal to be unique.
Genomic DNAs from 40 feral Mauritian-origin M. fascicularis macaques housed at the Wisconsin National Primate Research Center (WNPRC) were purified from 2 ml EDTA-treated peripheral blood samples by using a MagNA Pure LC DNA isolation kit (Roche).
All animals were cared for according to the regulations and guidelines of the Institutional Animal Care and Use Committees at their respective institutions.
Cell lines.
293T cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum. CrFK cells were grown in DMEM containing 8% bovine growth serum. CrFK cell lines stably transduced with TRIMCyp and previously produced stable control cell lines containing M. nemestrina TRIMCyp and M. mulatta TRIM5α (2) were grown in DMEM containing 8% bovine growth serum and 0.4 μg/ml puromycin.
PCR and restriction analysis.
For the NsiI restriction assay, genomic DNA was amplified by using Platinum PCR Supermix High Fidelity (Invitrogen) with forward primer MfaT5ex6F (primer 2) (ATC TGA AAC GAA TGC TAG ACA TG) and reverse primer 3TrmNotI (primer 4) (ATC TAG GCG GCC GCT TAA GAG CTT GGT GAG CAC AGA GTC ATG). PCR products were digested by using FastDigest NsiI enzyme (Fermentas) (17).
To test for the CypA insertion, genomic DNA was PCR amplified by using Platinum PCR Supermix High Fidelity (Invitrogen) with forward primer 3UTRF (primer 3) (TGA CTC TGT GCT CAC CAA GCT CTT G) and reverse primer 3UTRRLong (primer 6) (TCA CCC TAC TAT GCA ATA AAA CAT TAG GAC), as described previously by Wilson et al. (37). CypA-containing PCR products were cloned and sequenced (at least two clones per animal).
For reverse transcriptase PCR (RT-PCR), RNA was reverse transcribed and PCR amplified by using a SuperScript III One-Step RT-PCR kit (Invitrogen) with forward primer XhoIHATRIM5 (primer 1) (CTA GAT CTC GAG ATG TAC CCA TAT GAC GTT CCA GAC TAC GCG GCT TCT GGA ATC CTG GTT AAT GTA AAG) and reverse primer CypARMCSNotI (primer 5) (GTA TAT GCG GCC GCT TAT TCG AGT TGT CCA CAG TCA G).
To confirm cDNA sequences, genomic DNA was amplified in two segments due to the length of intron 4. Exons 2 to 4 were amplified by using forward primer XhoITRIM5 (CTA GAT CTC GAG ATG GCT TCT GGA ATC CTG GTT AAT GTA AAG) and reverse primer MfaT5int4R (CAG CAA CTG ATT CAG AAA AGC GGT CC). Exons 5 and 6 were amplified by using forward primer MfaT5int4F (CAC TGT CAC CAT CTC CAT TCT CAG CAC) and reverse primer MfaT5int6R (CCC ATC AGC AGC AGA CAA TAT CCA). Two clones from each animal were sequenced.
Brightness and contrast were adjusted in gel photos by using Adobe Photoshop CS4 software. All adjustments were made across entire images.
Cloning.
PCR products for sequencing were cloned by using a StrataClone Blunt PCR cloning kit (Stratagene). Sequencing was performed by the University of Washington Biochemistry Sequencing Center.
For the stable transduction of cell lines, RT-PCR products were cloned by using a StrataClone PCR cloning kit (Stratagene). The resulting pSC-A plasmids containing hemagglutinin (HA)-tagged TRIMCyp were digested by using XhoI and NotI restriction enzymes (New England BioLabs). The excised TRIMCyp bands were subcloned into the pLPCX retroviral expression vector (Clontech).
TRIMCyp-containing pLPCX plasmids were mutagenized at amino acid positions 178, 369, and 446 (numbering is based on the full-length untagged TRIMCyp sequence of M. fascicularis translated from cDNA) by using a QuikChange II XL site-directed mutagenesis kit (Stratagene).
Sequence analysis.
Sequence analysis was performed by using Geneious 4.8 (6) and MEGA 5 (33). Sequences for the intron 6-exon 8 region of M. mulatta (extracted from chromosome 14 of the rhesus macaque genome [GenBank accession number NC_007871.1]), one M. nemestrina sequence (accession number EU371641.1), and the homologous Homo sapiens sequence (extracted from chromosome 11 of build 37.1 of the human genome [accession number NT_009327.18]) were obtained from GenBank. Sequences from Papio hamadryas, Papio cynocephalus anubis, Macaca sylvanus, M. nigra, M. thibetana, and six WaNPRC M. fascicularis animals were described previously (GenBank accession numbers HM468434 to HM468446) (5). These sequences were aligned with newly derived M. fascicularis sequences. A phylogenetic tree of this region was generated with MEGA 5, using a maximum likelihood algorithm with 1,000 bootstrap replicates. The human sequence was used as an outgroup.
Production of stable TRIMCyp-expressing cell lines.
293T cells were cotransfected with plasmids expressing vesicular stomatitis virus G (VSV-G) envelope protein (pL-VSV-G) and the murine leukemia virus (MLV) Gag-Pol backbone (mGP) and with pLPCX plasmids containing HA-tagged M. fascicularis TRIMCyp using linear polyethylenimine (22). Supernatants were harvested after 48 h. CrFK cells in 6-well plates were transduced with 1 ml of virus-containing supernatant and spinoculated for 1 h at 650 × g. Cells were then allowed to grow to confluence before selection was begun with 6 μg/ml puromycin.
For immunoblotting, cells were lysed in M-PER mammalian protein extraction reagent (Thermo Scientific). HA-tagged proteins were bound with monoclonal antibody HA.11 (Covance) and detected with a goat anti-mouse alkaline phosphatase-conjugated secondary antibody (Sigma-Aldrich).
Pseudotyped virus production.
293T cells were cotransfected with the VSV-G envelope protein and envelope-negative, green fluorescent protein (GFP)-expressing virus backbones (HIV-1 pNL4-3, HIV-2 ROD, simian immunodeficiency virus [SIV] SIVmac239, and FIV) by using linear polyethylenimine (22). In the case of FIV, the packaging plasmid pGinSin was also cotransfected. Virus-containing supernatants were harvested after 48 h and concentrated by using PEG-It (System Biosciences). The amount of reverse transcriptase in viral preparations was quantified by use of a colorimetric reverse transcriptase assay (Roche).
Infectivity assay.
CrFK cells, and CrFK-derived cells expressing TRIMCyp, were seeded and grown overnight in 6-well plates with nonselective medium. The cells were then infected with dilutions of virus, normalized by the RT concentration, in DMEM containing 5 μg/ml Polybrene. After 2 h, the medium was replaced with fresh growth medium, and the cells were grown for an additional 48 h. GFP-expressing cells were enumerated by flow cytometry on a FACSCalibur instrument (Becton Dickinson), and data were analyzed with FlowJo 7.1.3 software (Tree Star, Inc.). Normalized percent infectivity was calculated relative to CrFK cells.
Nucleotide sequence accession numbers.
All sequences were deposited in GenBank under accession numbers HQ834751 to HQ834786 and HQ840753 to HQ840761.
RESULTS
Prevalence of TRIMCyp varies among M. fascicularis populations.
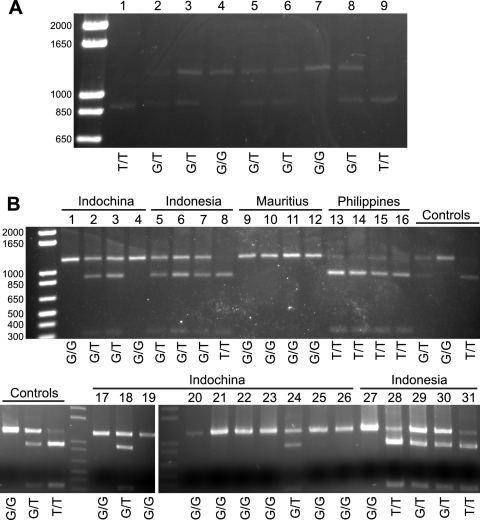
In order to determine the prevalence of TRIMCyp in different M. fascicularis populations, we performed an NsiI restriction assay described previously by Newman et al. (17). This assay takes advantage of a second polymorphism closely linked to the splice-site polymorphism that is required for TRIMCyp expression (Fig. 1). Sequences with this NsiI restriction site have the T allele at the splice site, which is linked to the presence of the CypA insertion and the ability to produce TRIMCyp. Sequences lacking the NsiI site have the G allele at the splice site, lack the CypA insertion, and cannot produce TRIMCyp. Initially, we tested nine animals of Indonesian origin housed at the WaNPRC (Fig. 2A). At least three clones of this region from each animal were also sequenced to ensure the accuracy of the restriction assay (GenBank accession numbers HQ834776 to HQ834786). We then genotyped an additional 9 animals of Indonesian origin as well as 14 animals of Indochinese origin, 4 of Mauritian origin, and 4 from the Philippines (Fig. 2B). All four animals from Mauritius were G/G homozygotes, while all 4 animals from the Philippines were T/T homozygotes. In contrast, the samples from both Indonesia and Indochina were genetically variable. The T allele was significantly more prevalent in Indonesian than in Indochinese animals (P = 0.0008 by Fisher's exact test) (Table 1).
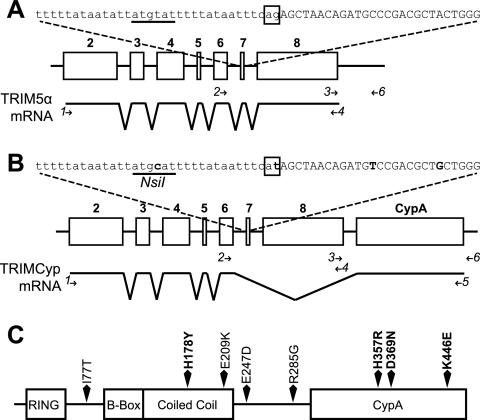
Fig. 1.
Genomic organization and mRNA splicing of TRIM5 alleles. (A) TRIM5α-expressing allele. At the top is the sequence of the intron 6-exon 7 junction, with the intronic sequence in lowercase type and exon 7 in uppercase type. The canonical AG splice acceptor dinucleotide is boxed. The underlined sequence represents the location of the NsiI restriction site in the other allele. Below is a schematic of the TRIM5 gene, with open boxes representing exons 2 to 8, numbered in boldface type. The mRNA splicing pattern is indicated below. (B) TRIMCyp-expressing allele. Sequence changes from the sequence in panel A are in boldface type, and the NsiI site is labeled. Below, the CypA insertion in the TRIMCyp-expressing allele is located as indicated. Minor splice isoforms in both alleles are not depicted. Primers used for analyses described in the subsequent figures and the text are depicted as arrows and numbered in italics. Primers used for genomic analysis are shown above the mRNA diagram, and those used for RT-PCR analysis are shown below. Primer 4 was used for both analyses. (C) Primary structure of TRIMCyp, showing polymorphisms found in M. fascicularis. Residues known to affect protein function are marked in boldface type.
Fig. 2.
Restriction analysis of geographically diverse M. fascicularis macaques. NsiI restriction analysis was used to determine the splice site genotype. The uncut band, which is associated with the G allele and the absence of TRIMCyp, is expected to be approximately 1,194 bp; the cut band associated with the T allele and the presence of TRIMCyp is expected to be approximately 891 bp. The inferred genotype is shown below the gel. (A) Animals of Indonesian origin housed at the WaNPRC. (B) Samples from Indonesia, Indochina, Mauritius, and the Philippines. Samples tested in panel A were used as controls (labeled) to test for the completion of digestion in independent assays. For inferring the genotype, the band density was quantified by using Quantity One software (Bio-Rad) and compared to those of controls processed at the same time.
Table 1.
Frequency of TRIMCyp-linked alleles in M. fascicularis populations
| Origin | No. of animals | No. of animals of genotypea: |
Allele frequency (%) |
|||
|---|---|---|---|---|---|---|
| GG | GT | TT | G | T | ||
| Indonesia | 18 | 3 | 10 | 5 | 44.4 | 55.6 |
| Indochina | 14 | 10 | 4 | 0 | 85.7 | 14.3 |
| Mauritius | 44 | 44 | 0 | 0 | 100 | 0 |
| Philippines | 4 | 0 | 0 | 4 | 0 | 100 |
G or T refers to the single-nucleotide polymorphism (SNP) at the exon 7 splice acceptor site linked to the expression of TRIMCyp (Fig. 1). GG or TT refers to homozygotes, and GT refers to heterozygotes.
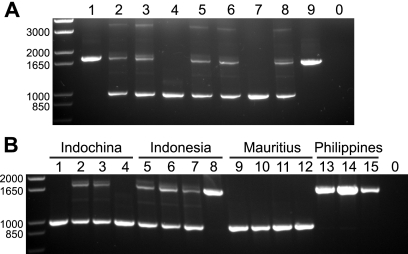
To further verify these findings, we tested a subset of the samples by using a PCR assay designed to detect the presence or absence of the CypA insertion (37). These data confirmed the results of the restriction assay (Fig. 3), lending support to the idea that the CypA insertion and the splice site T allele are invariably linked in macaques.
Fig. 3.
PCR analysis of the CypA insertion. PCR was performed by using primers that spanned the 3′ UTR and the putative CypA insertion. The band containing the insertion is expected to be 1,750 bp; the band without the insertion is expected to be 994 bp. (A) WaNPRC animals. (B) Geographically diverse samples shown in Fig. 2B (top). In both parts, negative-control PCRs with no template are shown in lane 0.
M. fascicularis animals of Mauritian origin have been developed recently as an AIDS model (19, 38). Because this population is genetically relatively homogeneous, we hypothesized that these animals completely lack TRIMCyp, based on its absence in the four samples that we initially tested. To test this hypothesis, we analyzed an additional 40 Mauritian M. fascicularis samples from animals currently in use for AIDS model studies. All were negative for TRIMCyp by both the NsiI restriction assay and PCR.
TRIMCyp-linked sequences form a monophyletic group.
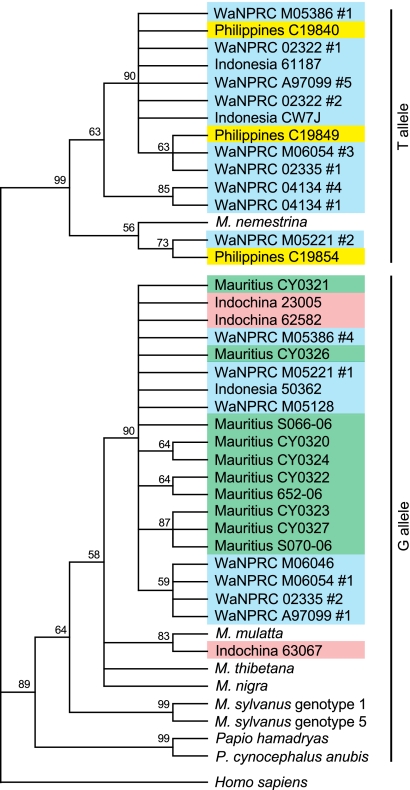
We sequenced clones of the region used in the NsiI restriction assay, comprising introns 6 and 7 and exons 7 and 8 (approximately 1,194 bp) (GenBank accession numbers HQ834757 to HQ834786). We found that these sequences clustered depending on their genotype at the exon 7 splice site (Fig. 4). As previously reported (5), sequences containing the T allele, which are presumably linked to the CypA insertion, formed a monophyletic group, separate from those with the G allele, which are expected to lack the CypA insertion. M. fascicularis sequences grouped together, although interestingly, one Indochinese animal (animal 63067) clustered with the only available M. mulatta sequence. As expected, sequences from Mauritian animals clustered within a single clade, closely related to some sequences from Indonesian and Indochinese animals.
Fig. 4.
Phylogenetic tree of the intron 6-exon 8 region. The intron 6-exon 8 region of the TRIM5 gene was cloned and sequenced. M. fascicularis animals are classified based on geographic origin; other species are as indicated. All WaNPRC M. fascicularis animals are of Indonesian origin. Multiple clones were sequenced from each WaNPRC animal; thus, heterozygotes are present in two locations on the tree. Only one clone was sequenced from each of the other individuals. Sequences from Papio, M. thibetana, M. nigra, M. sylvanus, and six of the WaNPRC M. fascicularis animals were described previously (5). The tree was built using a maximum parsimony algorithm (33), using the human sequence as an outgroup. Similar results were also found by use of maximum likelihood and neighbor-joining algorithms (not shown). Bootstrap values from 1,000 replicates are shown.
Diverse TRIMCyp genotypes in M. fascicularis.
We cloned and sequenced full-length TRIMCyp cDNA from six of the seven TRIMCyp-positive M. fascicularis animals from the WaNPRC analyzed previously (GenBank accession numbers HQ834751 to HQ834756) (Table 2 and Fig. 1C and 2A). We also sequenced the TRIM5 gene in genomic DNA to determine whether the SNPs in our cDNA clones resulted from cloning errors. We found six likely cloning errors. Two of these were in the linker 2 region: M246T in animal M05386 and K272R in animal M06054. The other four cloning errors were in the CypA domain: F310S in animal 02335, F310I and F370L in animal 04134, and N440D in animal M05386. These likely artifacts are not included in Table 2. None of these artifacts per se appeared to affect restriction activity or specificity (see below).
Table 2.
Polymorphisms in M. fascicularis TRIMCyp
| Animal | Residue at amino acida: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 77 | 178 | 209 | 247 | 285 | 310 | 357 | 369 | 446 | |
| M. nemestrinab | I | H | E | E | G | S | H | N | E |
| 04116b | T | H | K | E | R | F | R | D | K |
| MafaTC2c | I | H | E | D | R | F | H | D | K |
| A97099 | I | H | K | E | R | F | H | D | K |
| 02335 | I | H | E | E | R | F | H | D | K |
| M06054 | I | H | E | E | R | F | H | D | K |
| M05221 | I | Y | E | D | G | F | H | D | K |
| M05386 | I | Y | E | D | R | F | H | N | E |
| 04134 | I | H | E | E | G | F | H | N | E |
Residue 77 is in the L1 domain (linker 1 region between the RING and B-box domains), residues 178 and 209 are in the coiled-coil domain, residues 247 and 285 are in the linker 2 domain, and residues 310, 357, 369, and 446 are in the CypA domain. Boldface type indicates a difference from the consensus.
See reference 2.
See reference 39.
Amino acids 369 and 446 were previously described to be important for restriction specificity (21, 39). Two of the WaNPRC animals had asparagine residues at position 369 and glutamic acid at position 446 (N369/E446 [henceforth referred to as NE]), while four animals had aspartic acid and lysine, respectively (D369/K446 [DK]).
To examine these variable TRIMCyp genotypes in geographically diverse populations, we sequenced the CypA insertion region from additional animals: four Indonesian, two Indochinese, and three Filipino animals (GenBank accession numbers HQ840753 to HQ840761) (Fig. 3). The only polymorphisms found in these sequences were at positions 369 and 446; the CypA sequences were otherwise identical to the consensus found for the Indonesian WaNPRC samples. All three animals from the Philippines had the DK genotype (D369/K446). In contrast, one Indochinese animal had DK, and the other had NE (N369/E446). Two Indonesian animals had NE, one had DK, and, interestingly, one had DE (D369/E446). The DE genotype has not previously been described for natural isolates, but an isolated CypA domain of this genotype has been created by mutagenesis and was found to restrict both HIV-1 and HIV-2 (39). Unfortunately, because RNA from that animal is not available, we were not able to clone full-length TRIMCyp cDNA in order to determine the restriction function of this genotype from a natural isolate. However, we did confirm the restriction specificity of DE-containing TRIMCyp proteins by mutagenesis (see Fig. 6).
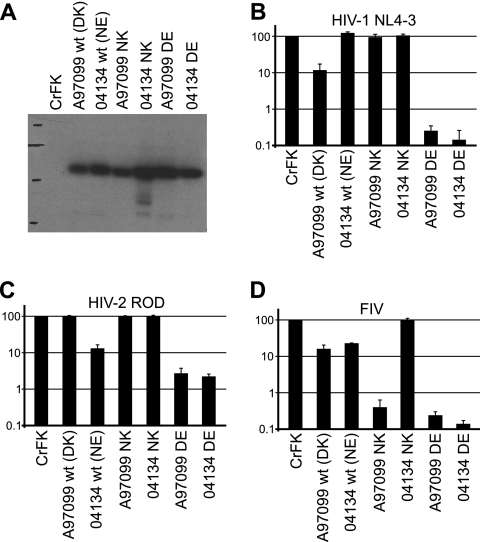
Fig. 6.
Amino acids 369 and 446 determine TRIMCyp restriction activity. (A) Anti-HA immunoblot of control and mutagenized TRIMCyp-expressing CrFK cell lines. (B) Restriction activity against HIV-1 NL4-3. (C) Restriction activity against HIV-2 ROD. (D) Restriction activity against FIV. Genotypes at amino acid positions 369 and 446 are shown, as described in the legend of Fig. 5. Data shown are means ± standard deviations from three independent experiments using one representative dilution of virus (11 ng RT/well for HIV-1 and HIV-2 and 33 ng RT/well for FIV). wt, wild type.
M. fascicularis TRIMCyp alleles have two distinct restriction activities.
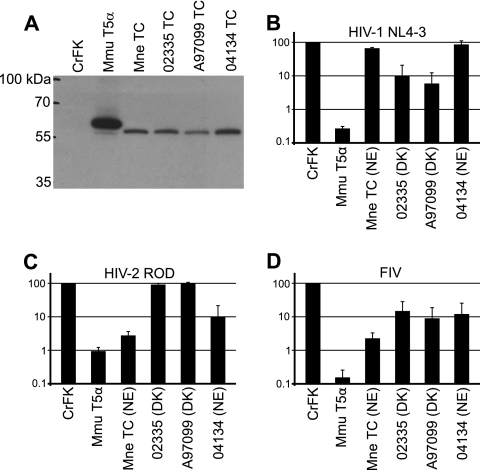
To characterize the restriction activity of the M. fascicularis TRIMCyp variants described above, we expressed HA-tagged versions of three of the WaNPRC M. fascicularis TRIMCyp alleles in TRIM5-negative feline CrFK cells (Fig. 5A). One of these, from animal 04134, had the NE genotype. The other two alleles, from animals 02335 and A97099, had the DK genotype. We then infected these cells with GFP-expressing, VSV-G-pseudotyped HIV-1 NL4-3, HIV-2 ROD, SIVmac, and FIV (Fig. 5). None of the alleles restricted SIVmac (data not shown), and all restricted FIV. The allele from animal 04134 (NE genotype) restricted HIV-2 but not HIV-1, like TRIMCyp isolates with the NE allele previously described for the M. mulatta and M. nemestrina TRIMCyp alleles (2, 21, 36, 37). The other two alleles, those from animals A97099 and 02335 (DK genotype), restricted HIV-1 but not HIV-2, as recently described for an independent M. fascicularis TRIMCyp allele, which also had the DK genotype (39).
Fig. 5.
Expression and antiviral restriction activity of M. fascicularis TRIMCyp. (A) Anti-HA immunoblot of CrFK and CrFK-derived TRIM5α- and TRIMCyp-expressing cell lines. (B) Restriction activity of CrFK and TRIMCyp-expressing cell lines against HIV-1 NL4-3. (C) Restriction activity against HIV-2 ROD. (D) Restriction activity against FIV. Controls are TRIM5α from M. mulatta (Mmu T5α) and TRIMCyp from M. nemestrina (Mne TC). Genotypes at amino acid positions 369 and 446 are shown as NE (N369/E446) or DK (D369/K446). Data shown are means ± standard deviations from three independent experiments using one representative dilution of virus (11 ng RT/well for HIV-1 and HIV-2 and 33 ng RT/well for FIV).
Amino acids 369 and 446 are determinants of TRIMCyp restriction specificity.
To verify and extend previous findings on the restriction determinants in Old World monkey TRIMCyp, we created D369N and K446E mutants in the A97099 background (referred to as A97099 NK and DE, respectively) and similarly created N369D and E446K mutants in the 04134 background (referred to as 04134 DE and NK, respectively). TRIMCyp in either background containing DE residues restricted both HIV-1 and HIV-2, as previously described (39), as well as FIV (Fig. 6). TRIMCyp with the NK residues restricted neither HIV-1 nor HIV-2.
Aside from the differences at positions 369 and 446, there are four additional amino acid differences between TRIMCyp of animal A97099 and that of animal 04134, at positions 209, 285, 310, and 370 (Table 2 [the differences at positions 310 and 370 are probably cloning artifacts that were not found in genomic DNA and are not shown in Table 2]). Interestingly, NK-containing TRIMCyp restricted FIV in the A97099 background but not in the 04134 background (Fig. 6). Therefore, one of the four differences between these two TRIMCyp proteins must have additional critical effects on the restriction of FIV.
Residue 178 affects TRIMCyp protein expression or stability.
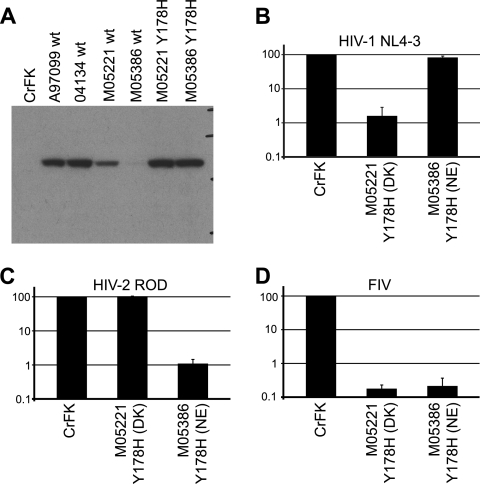
We also attempted to produce cell lines expressing TRIMCyp from animals M05221 (DK) and M05386 (NE). After two independent transductions, we were not able to produce a cell line expressing these alleles at levels comparable to those attained for the other three TRIMCyp alleles analyzed (Fig. 7 and data not shown). Cell lines expressing low levels of these TRIMCyp proteins had no or negligible restriction activity (data not shown). These two alleles share an H178Y polymorphism in the coiled-coil domain of TRIMCyp (Table 2). We therefore mutagenized this site to the consensus histidine residue. Cells transduced with the resulting mutant H178 alleles expressed TRIMCyp stably and at high levels, whereas parallel transductions done with wild-type M05221 and M05386 alleles did not (Fig. 7A). The restriction specificity of both Y178H mutants was as predicted by their genotype at amino acid positions 369 and 446 (Fig. 7).
Fig. 7.
Amino acid 178 affects TRIMCyp protein expression or stability. (A) Anti-HA immunoblot of control and mutagenized TRIMCyp-expressing CrFK cell lines. Cell lines expressing wild-type and H178Y TRIMCyp from animals M05221 and M05386 were transduced and grown in parallel. (B) Restriction activity against HIV-1 NL4-3. (C) Restriction activity against HIV-2 ROD. (D) Restriction activity against FIV. Genotypes at amino acid positions 369 and 446 are shown, as described in the legend of Fig. 5. Data shown are means ± standard deviations from three independent experiments using one representative dilution of virus (11 ng RT/well for HIV-1 and HIV-2 and 33 ng RT/well for FIV). Cell lines expressing wild-type TRIMCyp from animals M05221 and M05386 had no or negligible restriction activity (not shown).
DISCUSSION
Here, we present a detailed description of TRIMCyp in M. fascicularis. We show that M. fascicularis is more diverse than any other species described to date, with respect to the presence or absence of TRIMCyp and to its restriction specificity. We found differences among animals both within and between geographic populations. TRIMCyp was present in three of four populations tested. Within Indonesian and Indochinese populations of M. fascicularis, three active antiretroviral TRIM5 proteins with different specificities coexist, namely, TRIM5α, TRIMCyp DK, and TRIMCyp NE. We also show that one individual had a DE genotype, which has not previously been found in natural isolates. It will be of interest to determine the prevalence of this genotype in natural populations. Although we were not able to create a cDNA clone from that individual, our mutagenesis results as well as the results of others suggest that it likely has an expanded restriction specificity. The identification of the DE allele in naturally occurring M. fascicularis populations, even at a low frequency, suggests that this genotype should be taken into consideration for the design of HIV studies involving M. fascicularis.
Our phylogenetic analysis is consistent with our previous findings in which TRIMCyp-linked sequences (with T at the exon 7 splice acceptor site) clustered together, separately from all other sequences (with G at the splice acceptor site) (5). These findings suggest that TRIMCyp arose early in the evolution of the Asian macaques. The clear differentiation between T-containing and G-containing alleles is likely due to a combination of founder effects and changed selection pressures. At one time in evolutionary history there would presumably have been an intermediate state, containing the CypA insertion in the absence of the splice acceptor polymorphism or vice versa; however, such sequences have not been found in existing populations.
The current work also serves to expand our understanding of the evolution of TRIMCyp in M. fascicularis. The geographic range of Indochinese M. fascicularis animals overlaps with that of Chinese M. mulatta animals, and genetic evidence indicates that the two species have experienced and likely are currently experiencing hybridization and introgression (7, 30, 34). Interestingly, TRIMCyp is apparently absent in Chinese M. mulatta macaques (17, 37). Introgression with TRIMCyp-negative Chinese M. mulatta animals may have decreased the prevalence of TRIMCyp in Indochinese M. fascicularis animals, potentially explaining the lower prevalence of TRIMCyp in Indochinese than in Indonesian M. fascicularis animals. Such introgression likely also explains the fact that the TRIM5 sequence from an Indochinese M. fascicularis animal clustered phylogenetically with an M. mulatta sequence rather than with other M. fascicularis sequences. Because gene flow between the species appears to be unidirectional from M. mulatta to M. fascicularis, it is unlikely that Indochinese M. fascicularis macaques have contributed TRIMCyp to M. mulatta, although this possibility cannot be entirely ruled out (1, 29).
M. fascicularis animals were introduced to Mauritius in historical times, likely from an Indonesian (Sumatran) stock (7, 28, 35). The initial founding population was small, and genetic signs of founder effects are apparent in the population (7, 35, 38). The fact that TRIMCyp is apparently absent in Mauritian individuals but common in the parental Indonesian population underscores the genetic bottleneck involved in the founding of the Mauritius colony. Previous genetic studies have found that sequences from Mauritian individuals tend to cluster apart from those from other M. fascicularis animals (28). Although data from our phylogenetic analysis are consistent with this finding, we were not able to resolve the relationships among TRIMCyp-negative M. fascicularis macaques due to the limited sequence data available. Because of the low genetic diversity in the Mauritian M. fascicularis population, these animals are of particular interest as an AIDS model (4, 9, 19, 38). The finding that these animals uniformly lack TRIMCyp, which could be a confounding factor in infection studies, indicates the potential for additional advantages of working with this model.
Our group previously reported an M. fascicularis TRIMCyp clone (animal 04116, also referred to as MafaTC1) (39) that had no detectable antiviral activity (2). This lack of activity was likely due to an H357R mutation, which prevents binding to the viral capsid (39). However, we were unable to replicate this sequence using archival samples, and no animals that we subsequently tested had this genotype. Since the initial cloning of the 04116 allele was performed by using a low-fidelity polymerase, the H357R mutation may have been an artifact introduced during cloning. Thus, its physiological relevance is uncertain. We have subsequently optimized the procedure to minimize such artifacts and, whenever possible, to confirm the cDNA sequence with genomic DNA sequencing.
We tested three TRIMCyp alleles and three mutated versions for restriction activity. Our mutagenesis analysis confirmed previous reports that residues 369 and 446 are important for restriction specificity (21, 39). Furthermore, because at least one animal sequenced had the DE genotype, our finding supports the potential biological relevance of DE-containing TRIMCyp proteins (39). In contrast, the NK genotype, which has not previously been analyzed and was not found in any of the animals tested, restricted neither HIV-1 nor HIV-2. Interestingly, only one of the two NK-containing proteins tested restricted FIV. This result may provide interesting clues about the determinants required for this restriction activity, although it may result from one of the two introduced mutations in the CypA domain of TRIMCyp from animal 04134.
Our data also suggest that a polymorphism in the coiled-coil region, H178Y, affects the ability to stably express TRIMCyp proteins. It is possible that this polymorphism either decreases protein stability or increases toxicity; we have not examined the mechanisms behind this effect. The amino acid at this position is a histidine in all other Old World TRIMCyp proteins described to date and in M. nemestrina TRIM5η, which is also produced by alternative splicing from the same allele as TRIMCyp. Interestingly, however, the amino acid at this position is a tyrosine in all existing Old World monkey and ape TRIM5α sequences (and is an asparagine in both TRIM5α and TRIMCyp from New World primates). It is therefore tempting to speculate that the histidine allele at amino acid 178 could have been selected in some Old World primates due to improved TRIMCyp expression levels.
TRIM5α alleles have evolved under both positive and balancing selection (16, 26). The balancing selection is likely due to pressure from pathogens restricted by different alleles (16). Although TRIMCyp evolved recently in Old World primates, as it is found only in Asian macaques (5), it is also likely to be under balancing selection. The presence of multiple restriction phenotypes in M. fascicularis TRIMCyp, in addition to TRIM5α, may reflect a history of such balancing forces in this species and its ancestors. Current TRIM5α and TRIMCyp expression patterns have also likely been influenced by evolutionary pressures such as genetic drift and founder effects, particularly in the Mauritius and Philippines populations (7, 28, 35), in addition to possible pressures from past retroviral infections.
Recently, it has become clear that differences in TRIM5 genotypes have dramatic effects on the outcome of SIV infection in M. mulatta (8, 12, 13). It is likely that the TRIM5 and TRIMCyp diversity in M. fascicularis has also played a role in the outcomes of infection during previous studies. It will be of interest to test the restriction of M. fascicularis TRIMCyp alleles against relevant SIV strains and examine historical samples from infected M. fascicularis animals to determine whether the TRIMCyp genotype could have affected pathogenesis. More in-depth knowledge of restriction factors in model species will inform the choice of appropriate models for AIDS research and the proper interpretation of results.
ACKNOWLEDGMENTS
We thank Summer Street for help with sample collection, Lisa Jones-Engel and Alicia Wilbur for helpful discussions, James Ha for help with statistical analysis, and Kendan Jones-Isaac for help with the NsiI restriction assay. For plasmids used in this study, we thank Matthias Dittmar (HIV-2 ROD), David Evans (SIVmac239), Michael Emerman (mGP, pL-VSV-G, and FIV), and the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH (pNL4-3-deltaE-EGFP from Haili Zhang, Yan Zhou, and Robert Siliciano).
This work was supported by NIH P51 RR000166 (to the Washington National Primate Research Center), NIH K08 AI067138 (to G.B.), NIH P51 RR000167 and P40 RR019995 (to the Wisconsin National Primate Research Center), and NIH P51 RR000163 (to the Oregon National Primate Research Center).
Footnotes
Published ahead of print on 27 July 2011.
REFERENCES
- 1. Bonhomme M., Cuartero S., Blancher A., Crouau-Roy B. 2009. Assessing natural introgression in 2 biomedical model species, the rhesus macaque (Macaca mulatta) and the long-tailed macaque (Macaca fascicularis). J. Hered. 100:158–169 doi:10.1093/jhered/esn093 [DOI] [PubMed] [Google Scholar]
- 2. Brennan G., Kozyrev Y., Hu S. L. 2008. TRIMCyp expression in Old World primates Macaca nemestrina and Macaca fascicularis. Proc. Natl. Acad. Sci. U. S. A. 105:3569–3574 doi:10.1073/pnas.0709511105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brennan G., Kozyrev Y., Kodama T., Hu S. L. 2007. Novel TRIM5 isoforms expressed by Macaca nemestrina. J. Virol. 81:12210–12217 doi:10.1128/JVI.02499-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cafaro A., et al. 2010. Impact of viral dose and major histocompatibility complex class IB haplotype on viral outcome in Mauritian cynomolgus monkeys vaccinated with Tat upon challenge with simian/human immunodeficiency virus SHIV89.6P. J. Virol. 84:8953–8958 doi:10.1128/JVI.00377-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dietrich E. A., Jones-Engel L., Hu S. L. 2010. Evolution of the antiretroviral restriction factor TRIMCyp in Old World primates. PLoS One 5:e14019 doi:10.1371/journal.pone.0014019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Drummond A. J., et al. 2009. Geneious 4.8. Biomatters, Auckland, New Zealand [Google Scholar]
- 7. Kanthaswamy S., et al. 2008. Hybridization and stratification of nuclear genetic variation in Macaca mulatta and M. fascicularis. Int. J. Primatol. 29:1295–1311 doi:10.1007/s10764-008-9295-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kirmaier A., et al. 2010. TRIM5 suppresses cross-species transmission of a primate immunodeficiency virus and selects for emergence of resistant variants in the new species. PLoS Biol. 8:e1000462.http://dx.doi.org/10.1371%2Fjournal.pbio.1000462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Krebs K. C., Jin Z., Rudersdorf R., Hughes A. L., O'Connor D. H. 2005. Unusually high frequency MHC class I alleles in Mauritian origin cynomolgus macaques. J. Immunol. 175:5230–5239 [DOI] [PubMed] [Google Scholar]
- 10. Kuang Y., et al. 2009. Genotyping of TRIM5 locus in northern pig-tailed macaques (Macaca leonina), a primate species susceptible to human immunodeficiency virus type 1 infection. Retrovirology 6:58 http://www.retrovirology.com/content/6/1/58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liao C. H., Kuang Y. Q., Liu H. L., Zheng Y. T., Su B. 2007. A novel fusion gene, TRIM5-cyclophilin A in the pig-tailed macaque determines its susceptibility to HIV-1 infection. AIDS 21(Suppl. 8):S19–S26 doi:10.1097/01.aids.0000304692.09143.1b [DOI] [PubMed] [Google Scholar]
- 12. Lim S. Y., et al. 2010. Contributions of Mamu-A*01 status and TRIM5 allele expression, but not CCL3L copy number variation, to the control of SIVmac251 replication in Indian-origin rhesus monkeys. PLoS Genet. 6:e1000997.doi:10.1371/journal.pgen.1000997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lim S. Y., et al. 2010. TRIM5alpha modulates immunodeficiency virus control in rhesus monkeys. PLoS Pathog. 6:e1000738 doi:10.1371/journal.ppat.1000738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lin T. Y., Emerman M. 2006. Cyclophilin A interacts with diverse lentiviral capsids. Retrovirology 3:70 doi:10.1186/1742-4690-3-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Neil S., Bieniasz P. 2009. Human immunodeficiency virus, restriction factors, and interferon. J. Interferon Cytokine Res. 29:569–580 doi:10.1089/jir.2009.0077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Newman R. M., et al. 2006. Balancing selection and the evolution of functional polymorphism in Old World monkey TRIM5α. Proc. Natl. Acad. Sci. U. S. A. 103:19134–19139 doi:10.1073/pnas.0605838103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Newman R. M., et al. 2008. Evolution of a TRIM5-CypA splice isoform in Old World monkeys. PLoS Pathog. 4:e1000003.doi:10.1371/journal.ppat.1000003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nisole S., Lynch C., Stoye J. P., Yap M. W. 2004. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc. Natl. Acad. Sci. U. S. A. 101:13324–13328 doi:10.1073/pnas.0404640101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. O'Connor S. L., et al. 2010MHC heterozygote advantage in simian immunodeficiency virus-infected Mauritian cynomolgus macaques. Sci. Transl. Med. 2:22ra18–22ra18 doi:10.1126/scitranslmed.3000524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ozato K., Shin D. M., Chang T. H., Morse III H. C. 2008. TRIM family proteins and their emerging roles in innate immunity. Nat. Rev. Immunol. 8:849–860 doi:10.1038/nri2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Price A. J., et al. 2009. Active site remodeling switches HIV specificity of antiretroviral TRIMCyp. Nat. Struct. Mol. Biol. 16:1036–1042 doi:10.1038/nsmb.1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reed S. E., Staley E. M., Mayginnes J. P., Pintel D. J., Tullis G. E. 2006. Transfection of mammalian cells using linear polyethylenimine is a simple and effective means of producing recombinant adeno-associated virus vectors. J. Virol. Methods 138:85–98 doi:10.1016/j.jviromet.2006.07.024 [DOI] [PubMed] [Google Scholar]
- 23. Reymond A., et al. 2001. The tripartite motif family identifies cell compartments. EMBO J. 20:2140–2151 doi:10.1093/emboj/20.9.2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sardiello M., Cairo S., Fontanella B., Ballabio A., Meroni G. 2008. Genomic analysis of the TRIM family reveals two groups of genes with distinct evolutionary properties. BMC Evol. Biol. 8:225 doi:10.1186/1471-2148-8-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sayah D. M., Sokolskaja E., Berthoux L., Luban J. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430:569–573 doi:10.1038/nature02777 [DOI] [PubMed] [Google Scholar]
- 26. Sawyer S. L., Wu L. I., Emerman M., Malik H. S. 2005. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. U. S. A. 102:2832–2837 doi:10.1073/pnas.0409853102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sebastian S., Luban J. 2005. TRIM5α selectively binds a restriction-sensitive retroviral capsid. Retrovirology 2:40 doi:10.1186/1742-4690-2-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Smith D. G., McDonough J. W., George D. A. 2007. Mitochondrial DNA variation within and among regional populations of longtail macaques (Macaca fascicularis) in relation to other species of the fascicularis group of macaques. Am. J. Primatol. 69:182–198 doi:10.1002/ajp.20337 [DOI] [PubMed] [Google Scholar]
- 29. Stevison L. S., Kohn M. H. 2009. Divergence population genetic analysis of hybridization between rhesus and cynomolgus macaques. Mol. Ecol. 18:2457–2475 doi:10.1111/j.1365-294X.2009.04212.x [DOI] [PubMed] [Google Scholar]
- 30. Street S. L., Kyes R. C., Grant R., Ferguson B. 2007. Single nucleotide polymorphisms (SNPs) are highly conserved in rhesus (Macaca mulatta) and cynomolgus (Macaca fascicularis) macaques. BMC Genomics 8:480 doi:10.1186/1471-2164-8-480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stremlau M., et al. 2004. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427:848–853 doi:10.1038/nature02343 [DOI] [PubMed] [Google Scholar]
- 32. Stremlau M., et al. 2006. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5α restriction factor. Proc. Natl. Acad. Sci. U. S. A. 103:5514–5519 doi:10.1073/pnas.0509996103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tamura K., et al. 4 May 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. doi:10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tosi A. J., Morales J. C., Melnick D. J. 2003. Paternal, maternal, and biparental molecular markers provide unique windows onto the evolutionary history of macaque monkeys. Evolution 57:1419–1435 [DOI] [PubMed] [Google Scholar]
- 35. Tosi A. J., Coke C. S. 2007. Comparative phylogenetics offer new insights into the biogeographic history of Macaca fascicularis and the origin of the Mauritian macaques. Mol. Phylogenet. Evol. 42:498–504 doi:10.1016/j.ympev.2006.08.002 [DOI] [PubMed] [Google Scholar]
- 36. Virgen C. A., Kratovac Z., Bieniasz P. D., Hatziioannou T. 2008. Independent genesis of chimeric TRIM5-cyclophilin proteins in two primate species. Proc. Natl. Acad. Sci. U. S. A. 105:3563–3568doi:10.1073/pnas.0709258105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wilson S. J., et al. 2008. Independent evolution of an antiviral TRIMCyp in rhesus macaques. Proc. Natl. Acad. Sci. U. S. A. 105:3557–3562 doi:10.1073/pnas.0709003105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wiseman R. W., et al. 2007. Simian immunodeficiency virus SIVmac239 infection of major histocompatibility complex-identical cynomolgus macaques from Mauritius. J. Virol. 81:349–361 doi:10.1128/JVI.01841-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ylinen L. M. J., et al. 2010. Conformational adaptation of Asian macaque TRIMCyp directs lineage specific antiviral activity. PLoS Pathog. 6:e1001062.http://dx.doi.org/10.1371%2Fjournal.ppat.1001062 [DOI] [PMC free article] [PubMed] [Google Scholar]