Abstract
Human immunodeficiency virus type 1 (HIV-1) Vpu enhances the release of viral particles from infected cells by targeting BST-2/tetherin, a cellular protein inhibiting virus release. The widely used HIV-1NL4-3 Vpu functionally inactivates human BST-2 but not murine or monkey BST-2, leading to the notion that Vpu antagonism is species specific. Here we investigated the properties of the CXCR4-tropic simian-human immunodeficiency virus DH12 (SHIVDH12) and the CCR5-tropic SHIVAD8, each of which carries vpu genes derived from different primary HIV-1 isolates. We found that virion release from infected rhesus peripheral blood mononuclear cells was enhanced to various degrees by the Vpu present in both SHIVs. Transfer of the SHIVDH12 Vpu transmembrane domain to the HIV-1NL4-3 Vpu conferred antagonizing activity against macaque BST-2. Inactivation of the SHIVDH12 and SHIVAD8 vpu genes impaired virus replication in 6 of 8 inoculated rhesus macaques, resulting in lower plasma viral RNA loads, slower losses of CD4+ T cells, and delayed disease progression. The expanded host range of the SHIVDH12 Vpu was not due to adaptation during passage in macaques but was an intrinsic property of the parental HIV-1DH12 Vpu protein. These results demonstrate that the species-specific inhibition of BST-2 by HIV-1NL4-3 Vpu is not characteristic of all HIV-1 Vpu proteins; some HIV-1 isolates encode a Vpu with a broader host range.
INTRODUCTION
The human immunodeficiency virus type 1 (HIV-1) Vpu protein enhances virus release from virus-infected cells (54, 55). This effect has been reported to be cell type dependent (44) and species specific (10, 14, 15, 24, 32, 41, 45, 58, 61, 62). Recently, BST-2 (also known as CD317, HM1.24, or tetherin) was identified to be the Vpu-sensitive cellular factor responsible for restricting HIV-1 virion release (36, 57). In this regard, Vpu and BST-2 are both integral membrane proteins, albeit with different membrane topologies. BST-2 has a short N-terminal cytoplasmic domain, with the bulk of the protein comprising the C-terminal ectodomain, while Vpu has virtually no ectodomain and essentially consists of an N-terminal transmembrane (TM) domain and a C-terminal cytoplasmic domain. It is generally accepted that the BST-2 TM domain is critical for interference by Vpu (9, 11, 15, 27, 32, 33, 40, 41, 62) and is consistent with our previous observation of the importance of the Vpu TM domain for regulating particle release (46). Indeed, the physical interaction of Vpu and human BST-2 and the critical importance of the BST-2 TM domain for this interaction were demonstrated in several coimmunoprecipitation studies (9, 10, 15, 23, 24, 32, 41) and bimolecular fluorescence complementation analyses (27). The inability of HIV-1 Vpu to target macaque BST-2 has been attributed to sequence differences in the TM domains of human and monkey BST-2 proteins (45, 62). However, virtually all of the published work assessing the interaction of HIV-1 Vpu with BST-2 has used the HIV-1NL4-3 molecular clone (1), a prototypical HIV-1 strain used in laboratories worldwide. These studies have led to the conclusion that Vpu function is species specific (viz., it is directed against human BST-2) (14, 24, 32, 58, 61).
More recently, simian immunodeficiency virus (SIV) Nef and the Env glycoprotein of some SIV and HIV-2 isolates were found to have Vpu-like activity capable of antagonizing macaque and human BST-2s, respectively (16, 17, 24, 28, 45, 61, 63). The current study was inspired by a recent comprehensive evaluation of Vpu proteins from multiple strains of HIV-1 that included members of three different viral groups (HIV-1 groups M, N, and O) and suggested that certain HIV-1 Vpu proteins may have a broader anti-BST-2 activity than NL4-3 Vpu (45). Our study was further motivated by the observation that during the serial passaging of CXCR4 (X4)-tropic simian-human immunodeficiency virus DH12 (SHIVDH12), the pathogenic virus which emerged (SHIVDH12R) (20) and its molecularly cloned derivative SHIVDH12-CL7 (43) each carried a vpu gene that differed genetically from the homologue present in HIV-1NL4-3. This led us to investigate whether the Vpu encoded by SHIVDH12-CL7, despite its presumed inability to target macaque BST-2 and despite encoding a SIVmac239 Nef protein with presumed Vpu-like function, might contribute to the augmented pathogenic potential observed in inoculated rhesus macaques. Surprisingly, we found that the SHIVDH12-CL7 Vpu protein was able to functionally inactivate rhesus BST-2 in ex vivo assays while retaining its activity against human BST-2. We also observed that the expanded host range of SHIVDH12-CL7 Vpu was not the result of serial passaging in monkeys but was an intrinsic property of the Vpu protein encoded by the parental HIVDH12 isolate. The capacity of a second vpu gene, derived from CCR5 (R5)-tropic HIV-1Ada (13, 56) and also present in the recently described R5-tropic SHIVAD8 (38), to antagonize macaque BST-2 was similarly evaluated. Compared to SHIVDH12-CL7 Vpu, the SHIVAD8 Vpu was less potent in abrogating monkey BST-2 in the same in vitro assays. Nonetheless, a majority of monkeys inoculated with Vpu-defective X4- or R5-tropic SHIVs generated lower set-point viremia, exhibited better maintenance of CD4+ T lymphocyte levels, and experienced delayed disease onset compared to SHIVs carrying wild-type vpu genes. Taken together, these results suggest that HIV-1 Vpu proteins are functionally different and that the inability of the prototypic HIV-1NL4-3 Vpu to inhibit macaque BST-2 may not be a typical property of Vpu proteins encoded by primary HIV-1 isolates.
MATERIALS AND METHODS
Plasmids.
The construction of plasmids pNL4-3, pNL4-3 Udel, pNL4-3 Urd, and pSHIVDH12-CL7 has been previously described (1, 26, 43, 46). pSHIVAD8-MV was constructed in two steps: (i) insertion of the 8,556-bp KasI to HindIII fragment (containing viral gag through env gene sequences) from a molecular clone of unintegrated DNA obtained from rhesus peripheral blood mononuclear cell (PBMC)-infected SHIVAD8PBMC (38) into the genetic background of pSHIVAD8 (38) to generate SHIVAD8-KH17 and (ii) insertion of the 3,029-bp EcoRI to HindIII fragment (containing vpr through env gene sequences) obtained by reverse transcriptase (RT) PCR cloning of the RNA present in the week 42 plasma of macaque CK15, previously inoculated with SHIVAD8#2 (38), into similarly digested SHIVAD8-KH17. The vpu-defective SHIVDH12-CL7/Udel and SHIVAD8-MV/Udel mutants were constructed by first subcloning the respective EcoRI to KpnI fragments and then deleting the 5′ 163 bp of the vpu gene (up to the 5′ terminus of the env gene) by PCR-based mutagenesis. SHIVDH12-CL7/Urd was constructed by randomizing the TM domain of the DH12-CL7 Vpu using oligonucleotide-based mutagenesis. The resulting sequence is shown in Fig. 1B. The SHIVDH12-CL7/Urd TM domain sequence was subsequently transferred into SHIVAD8-MV, resulting in SHIVAD8-MV/Urd (Fig. 1B). A nef-deficient variant, SHIVDH12-CL7/Nef−, was constructed using PCR-based site-directed mutagenesis by mutating the Nef initiation codon (ATG) to TAG, followed by a second in-frame stop codon (TGA) at Nef position 3. A Vpu/Nef double mutant, SHIVDH12-CL7/Udel/Nef−, was created by transferring the mutated nef gene sequence into the backbone of SHIVDH12-CL7/Udel. pNL4-3/TM-CL7 carries the Vpu TM domain of SHIVDH12-CL7 and was constructed using standard PCR techniques. All constructs were verified by sequence analysis.
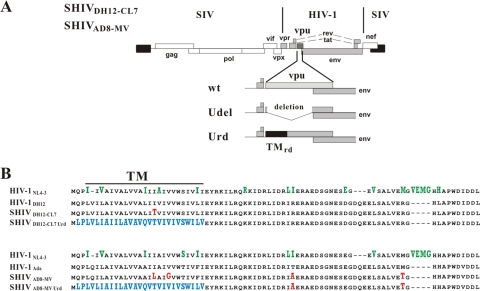
Fig. 1.
Constructs used in the current study. (A) Schematic representation of the SHIV constructs. SHIVDH12-CL7 and SHIVAD8-MV are chimeric viruses expressing HIV-1 Vpr, Tat, Rev, Vpu, and Env products (shaded areas) in a SIVmac239 genetic background. The region encompassing the vpu gene is expanded, and vpu mutants are shown schematically. TMrd, randomized TM domain. (B) Amino acid alignment of Vpu variants employed in this study. Amino acid differences between parental HIV-1 Vpu proteins and those present in SHIV isolates are highlighted in red. The randomized TM sequences in the Urd mutants are shown in blue. Amino acid differences between HIV-1NL4-3 Vpu and HIV-1DH12 or HIV-1Ada Vpu are shown in green.
Cell culture.
HEK293T (293T) is a human kidney cell line lacking expression of endogenous BST-2. 293T cells were cultured in Dulbecco's modified Eagle's minimal essential medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum. Rhesus monkey PBMCs were prepared and cultured as described previously (22).
Transfection, infection, and reverse transcriptase assays.
Virus stocks were prepared by transfecting 293T cells with each molecular clone using Lipofectamine 2000 (Invitrogen, Carlsbad, CA); culture supernatants were collected 48 h later and stored at −80°C until use. Virion-associated RT activity was measured as described previously (60). Concanavalin A (ConA)-stimulated rhesus PBMCs (5 × 106) were infected with the indicated viruses (normalized by particle-associated RT activity) by spinoculation (39) for 1 h and maintained for 12 days. Tissue culture medium was replaced daily; these supernatant samples were monitored for RT activity.
Virus release assay.
Cells were transfected as described in the text with constant amounts of proviral vectors and increasing amounts of BST-2. Twenty-four hours later, cells were washed with phosphate-buffered saline (PBS), scraped, and resuspended in 3 ml labeling medium lacking methionine (Millipore Corp., Billerica, MA). Cells were then incubated for 10 min at 37°C to deplete the endogenous methionine pool. Cells were then suspended in 400 μl of labeling medium together with 150 μCi of Express35S35S protein labeling mix (Perkin Elmer, Shelton, CT). Cells were labeled for 90 min at 37°C. Cells and virus-containing supernatants were then separated by centrifugation and processed separately for immunoprecipitation, as follows. To identify HIV-1-specific proteins, cells were lysed with 150 μl of Triton lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.5% Triton X-100) and incubated on ice for 5 min. To identify SHIV-specific proteins, cells were lysed in NP-40-deoxycholate (DOC) lysis buffer (20 mM Tris, pH 7.5, 2 mM EDTA, 20 mM NaCl, 1% Igepal detergent, 0.5% sodium deoxycholate) and incubated on ice for 5 min. After lysis, the cells were pelleted at 13,000 × g for 2 min to remove insoluble material. The supernatants were used for immunoprecipitation. Virus-containing supernatants were treated with 150 μl of Triton lysis buffer (HIV-1) and NP-40-DOC lysis buffer (SHIV) to disrupt viral membranes. Cell and virus lysates were adjusted to a 1.1-ml total volume with PBS containing bovine serum albumin (BSA; final concentration of BSA, 0.1%) and incubated on a rotating wheel for 1 h at 4°C with protein A-Sepharose coupled with an HIV-positive patient serum sample or plasma from SHIV-infected macaques. Beads were washed twice with wash buffer (50 mM Tris, pH 7.4, 300 mM NaCl, 0.1% Triton X-100). Bound proteins were eluted by heating in sample buffer for 10 min at 95°C, separated by SDS-PAGE, and visualized by fluorography. Virus release was quantified by PhosphoImage analysis using a Fujifilm FLA7000 system.
Animals.
Rhesus macaques were maintained in accordance with the guidelines of the Committee on the Care and Use of Laboratory Animals (8) and were housed in a biosafety level 2 facility; biosafety level 3 practices were followed. Phlebotomies, intravenous virus inoculations, and euthanasias were performed as described previously (12, 19).
Plasma viral RNA quantitation.
Viral RNA levels in plasma were determined by real-time RT-PCR (Prism 7700 sequence detection system; Applied Biosystems, Foster City, CA) as described previously (12).
Flow cytometric analysis.
To detect BST-2 expression on the surface of rhesus macaque T cells, CD4+ T cells were isolated from rhesus PBMCs and stimulated with concanavalin A (25 μg/ml) and interleukin-2 (20 units/ml) as described previously (22, 25). BST-2 expression was analyzed 72 h later using a cross-reactive rabbit polyclonal antibody against human BST-2 (34). Cells were washed twice in ice-cold PBS containing 1% BSA. Cells were blocked for 10 min with mouse IgG (1.25 μg/ml; Millipore Corp., Billerica, MA). Cells were incubated with primary antibody (anti-BST-2; 1:100) for 30 min at 4°C, washed twice with ice-cold PBS containing 1% BSA, followed by the addition of allophycocyanin (APC)-conjugated anti-rabbit IgG secondary antibody (Jackson ImmunoResearch Laboratories Inc., West Grove, PA) in PBS containing 1% BSA. Incubation was for 30 min at 4°C in the dark. Cells were then washed twice with ice-cold PBS containing 1% BSA and fixed with 1% paraformaldehyde in PBS. For in vivo lymphocyte immunophenotyping, EDTA-treated blood samples were stained for flow cytometric analysis as described previously (37) using combinations of the following fluorochrome-conjugated monoclonal antibodies (obtained from BD Biosciences Pharmingen, San Diego, CA): CD3 (phycoerythrin [PE]), CD4 (PE, peridinin chlorophyll protein [PerCP]-Cy5.5]), CD8 (APC), CD20 (fluorescein isothiocyanate [FITC]), CD28 (FITC), and CD95 (APC). Finally, cells were analyzed on a FACSCalibur flow cytometer (BD Biosciences). Data analysis was performed using CellQuest Pro (BD Biosciences) and Flow Jo (Tree Star Inc., Ashland, OR) software.
RESULTS
Vpu from HIV-1 DH12 can antagonize rhesus BST-2.
SHIVs have been used as challenge viruses in vaccine experiments to assess the potency of candidate immunogens designed to elicit antiviral neutralizing antibodies because they carry the HIV-1 env gene. They are usually constructed by inserting a gene segment, which may encode a portion of Vpr and all of the Tat, Rev, Vpu, and Env proteins, from a particular HIV-1 isolate into the genetic background of SIVmac239 (20, 43). In this study, we have used X4-tropic SHIVDH12 and R5-tropic SHIVAD8, which are phenotypically distinct. The intravenous inoculation of rhesus macaques with large doses (>2,500 50% tissue culture infective doses [TCID50s]) of the X4-tropic molecular clone SHIVDH12-CL7 (43), derived from SHIVDH12 (19, 20, 52), results in high sustained levels of plasma viremia; the rapid, unrelenting, and systemic depletion of naïve and memory CD4+ T cells; and death from immunodeficiency in 3 to 6 months. In contrast, a new pathogenic R5-tropic SHIV clone (SHIVAD8-MV), derived from the recently described SHIVAD8 (38), maintains variable plasma viral loads (102 to 105 RNA copies/ml), exclusively targets memory CD4+ T lymphocytes, and causes clinical symptoms of AIDS at 1 to 2 years postinfection (p.i.).
To ascertain whether the SHIV vpu gene might contribute to its replication phenotype in macaque cells, wild-type (wt) X4- and R5-utilizing SHIVs and their Vpu mutant derivatives were used to infect rhesus monkey PBMCs. The X4-tropic molecular clone SHIVDH12-CL7 (43), derived from SHIVDH12 (20, 52), and a new pathogenic R5-tropic SHIV clone (SHIVAD8-MV), derived from the recently described SHIVAD8 (38), were used in these studies. Two types of Vpu mutants were constructed for each SHIV molecular clone (Fig. 1A). The Vpu Udel mutants retain the Vpu initiation codon but carry an out-of-frame deletion eliminating vpu gene sequences upstream of the env gene. These mutants were modeled on a natural deletion present in the vpu gene of the HIV-1NY5 isolate (4, 26). The Urd mutants express a Vpu protein carrying a randomized Vpu TM domain. These mutants were constructed to specifically inactivate Vpu-mediated particle-releasing activity but not the CD4 downregulation function (46). As shown in Fig. 1B, the amino acid sequences of the parental HIV-1DH12 and HIV-1Ada Vpu TM regions differ from one another as well as from the sequence of the HIV-1NL4-3 Vpu prototype. With respect to the latter comparison, the HIV-1DH12 Vpu differs from the HIV-1NL4-3 Vpu at 20 amino acid (aa) positions, including 6 located in the TM region. HIV-1AD8 Vpu differs from HIV-1NL4-3 at 17 aa residues, 6 of which map to the TM domain. In addition, 1 and 2 amino acid changes in the Vpu TM region, associated with the animal passaging used to generate pathogenic SHIVs, were present in the SHIVDH12-CL7 and SHIVAD8-MV Vpu proteins, respectively.
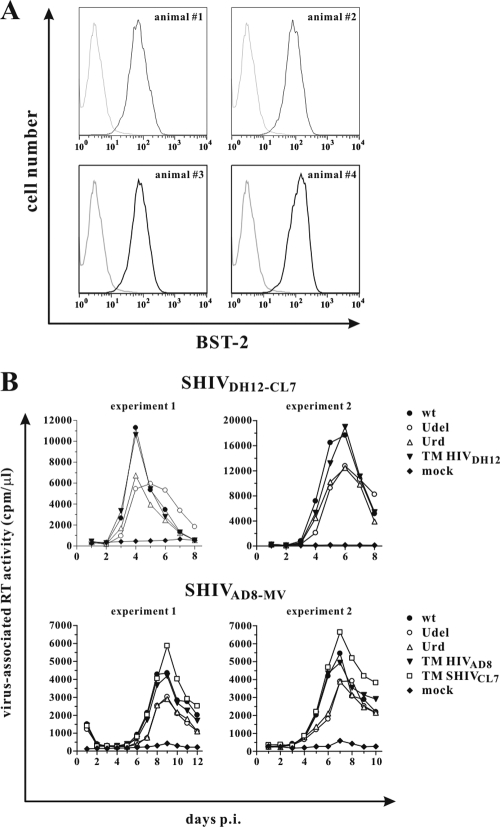
Prior to infecting macaque PBMCs, we assessed the BST-2 expression by fluorescence-activated cells sorter (FACS) analysis and found that CD4+ T lymphocytes from four rhesus monkey donors expressed cell surface BST-2 (Fig. 2A). To determine whether the expression of Vpu might modulate virus production during spreading infections in macaque T cells, rhesus PBMCs were infected with wt SHIVDH12-CL7 or Vpu mutant derivatives. The results of two of three independent experiments are shown in Fig. 2B (top panels). The results indicated that both of the Vpu mutant viruses (Udel, Urd) released less progeny virions into the supernatant medium than the wt SHIV, suggesting that the Vpu protein was functioning in monkey cells. Of note, the difference in peak RT activities of wt and Vpu-defective viruses was relatively small and in the range of 2-fold. These results are consistent with the reported small 2- to 3-fold effects of Vpu on HIV-1 release during virus spread in human PBMCs and likely reflect an inherently inefficient inhibitory effect of BST-2 on virus release in these cells (46, 48). A similar result was obtained with wt and Vpu mutants of the R5-tropic SHIVAD8-MV, although the differences between the amounts of wt and Vpu-minus viruses in the medium were not as great (Fig. 2, bottom panels). To ascertain whether the in vivo passaging, which attended the generation of the pathogenic SHIVs and had resulted in minor alterations in their Vpu TM domains (shown by red letters in Fig. 1B), was responsible for the Vpu-mediated enhanced replication in rhesus PBMCs, SHIV derivatives containing the parental HIV-1 Vpu TM regions were constructed (TM HIVDH12 and TM HIVAD8). Interestingly, these SHIV derivatives released similar amounts of progeny virions as the passaged wt SHIV, indicating that the vpu gene in the original HIV-1 isolates was also active in rhesus cells. Because each SHIV isolate encoded a distinct Vpu TM domain and the SHIVDH12-CL7 Vpu appeared to have higher activity in macaque PBMCs, a SHIVAD8-MV derivative bearing a chimeric vpu gene consisting of the SHIVDH12-CL7 Vpu TM region and the SHIVAD8-MV Vpu cytoplasmic domain (TM SHIVCL7) was constructed. The Vpu TM domain of SHIVDH12-CL7 differs from that of AD8-MV in six positions (A16L, L18T, A19V, G21V, T24S, F27L; Fig. 1B). As shown in Fig. 2B, the SHIV isolate carrying the chimeric TM SHIVCL7 vpu gene released higher levels of virus than wt SHIVAD8-MV. Thus, the differences between SHIVDH12-CL7 and SHIVAD8-MV with respect to their relative resistance to rhesus BST-2 could be due to differences in the TM domains of their Vpu proteins. Taken together, these results indicate that two SHIVs carrying vpu genes from primary HIV-1 isolates release larger amounts of progeny virions during spreading infections in rhesus PBMCs than their Vpu-minus derivatives. This augmented phenotype appears to map to the Vpu TM domain.
Fig. 2.
Vpu-deficient SHIVs produce less cell-free virus in rhesus PBMCs than wt SHIVs. (A) CD4+ T cells were isolated from PBMCs of four rhesus macaques and processed for FACS analysis as described in Materials and Methods. Black lines, BST-2 staining; gray lines, isotype controls. (B) ConA-activated rhesus PBMCs were infected with equal reverse transcriptase units of SHIVDH12-CL7 variants (top) or SHIVAD8-MV variants (bottom). All virus stocks were produced in transfected 293T cells. Virus replication was monitored by measuring the virus-associated RT activity in the culture supernatants over time.
SIV Nef and DH12 CL7 Vpu contribute equally to functional inhibition of rhesus BST2.
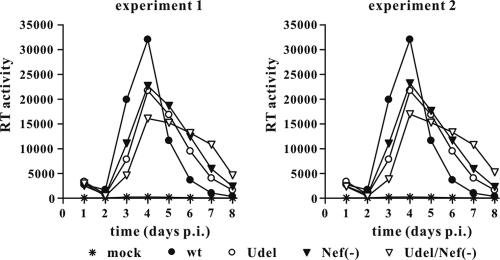
Several recent studies have reported that the Nef proteins of some SIV strains, including SIVmac239, have Vpu-like activity because they can antagonize monkey BST-2 and facilitate virus release from BST-2-positive monkey cells (24, 45, 63). Our SHIV constructs are based on SIVmac239 and are expected to express a functional Nef protein. To assess the relative contribution of SIVmac239 Nef and HIV-1 Vpu to virus replication in rhesus PBMCs, we constructed two SHIV variants carrying a defective nef gene in the context of either SHIVDH12-CL7 or SHIVDH12-CL7/Udel, as described in Materials and Methods. Virus stocks of wt SHIVDH12-CL7 and the Vpu-defective variant (Udel), as well as a Nef-deficient (Nef−) variant and the Vpu/Nef double mutant (Udel/Nef−), were produced in 293T cells and virus replication in rhesus PBMCs was monitored for 8 days (Fig. 3). Two independent infections by Nef− and Udel/Nef− viruses are shown (experiment 1 and experiment 2). Interestingly, inactivation of HIV-1 Vpu or SIVmac239 Nef had very similar effects on virus replication and resulted in reduced peak RT values in both experiments. Of note, inactivation of both Vpu and Nef led to a further reduction in peak RT values, indicating that the antagonistic effects of Vpu and Nef on rhesus BST-2 are additive. The knockout of Nef did not affect viral replication kinetics, and the Nef/Vpu double mutant revealed only a slight delay in the replication profiles. This suggests that SIV Nef, like its HIV-1 Vpu counterpart, inhibits an activity of rhesus BST-2 that impairs the release of cell-free virions but does not inhibit cell-to-cell spread of the virus. We conclude that in the context of SHIVDH12-CL7, Vpu and Nef contribute equally and in an additive manner to the release of particles from rhesus PBMCs.
Fig. 3.
SIV Nef and HIV-1 Vpu contribute equally to the antagonism of rhesus BST-2. ConA-activated rhesus PBMCs were infected with equal reverse transcriptase units of SHIVDH12-CL7 wt or variants defective in Vpu (Udel) or Nef [Nef(−)] or lacking both Vpu and Nef [Udel/Nef(−)]. All virus stocks were produced in transfected 293T cells. Virus replication was monitored by measuring the virus-associated RT activity in the culture supernatants over time.
SHIV Vpu proteins can counteract human and rhesus BST-2.
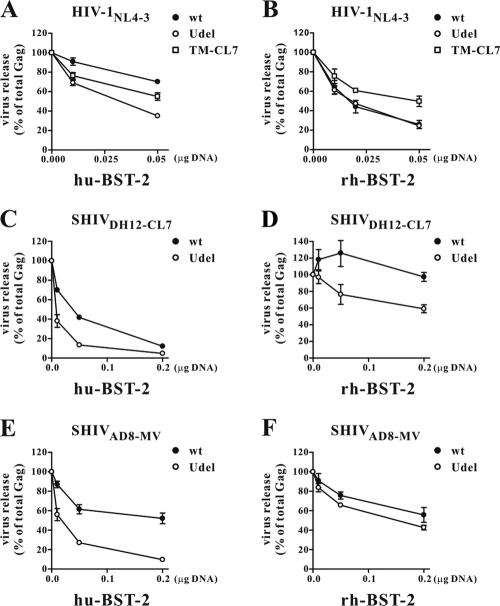
To examine whether Vpu-mediated SHIV production in macaque cells was associated with antagonism of rhesus BST-2, the BST-2-null human cell line 293T was cotransfected with HIV-1 or SHIV carrying wt, mutant, or chimeric vpu genes plus increasing amounts of plasmids expressing human or rhesus BST-2. Cells were labeled for 90 min with [35S]methionine at 24 h posttransfection, and cell lysates or cell-free virus-containing supernatants were immunoprecipitated, using an HIV-1-positive patient serum sample or plasma from SHIV-infected macaques. Immunoprecipitated viral proteins were separated by SDS-PAGE and quantitated by PhosphoImage analysis, and virus release was calculated as the percentage of Gag protein in the supernatant medium compared to the total amount of intra- plus extracellular Gag protein.
Not unexpectedly, wt HIV-1NL4-3 Vpu was able to counteract human BST-2 but not rhesus BST-2 (Fig. 4A and B), as previously reported by several laboratories (10, 14, 15, 24, 32, 41, 45, 58, 61, 62). Interestingly, and consistent with the results of the PBMC infectivity assays, SHIVDH12-CL7, bearing the wt vpu gene, released considerably more progeny virions than the Vpu mutant SHIV in the presence of large amounts of rhesus BST-2 (Fig. 4D). Since the Vpu TM domain confers virus-releasing activity, we wondered whether HIV-1NL4-3, carrying a chimeric Vpu containing the TM region from SHIVDH12-CL7, would better antagonize rhesus BST-2. As shown in Fig. 4B, HIVNL4-3(TM-CL7) did, in fact, generate augmented levels of supernatant particles compared to wt HIV-1NL4-3 in the presence of rhesus BST-2. The capacity of SHIVAD8-MV Vpu to counteract rhesus BST-2 was also evaluated. Although the wt vpu gene in SHIVAD8-MV consistently exhibited more activity against rhesus BST-2 than the mutant vpu gene, it was clearly not as potent as the SHIVDH12-CL7 Vpu in counteracting rhesus BST-2 (compare Fig. 4D and F). This difference was also previously observed in rhesus PBMC infectivity assays. The higher basal particle-releasing activities observed with both SHIV Vpu Udel mutants in the presence of rhesus BST-2 are worth noting. This could reflect an SIV Nef (present in both SHIV plasmids)-mediated anti-rhesus BST-2 effect in this assay system.
Fig. 4.
SHIV Vpu proteins can counteract human (hu) and rhesus (rh) BST-2. (A and B) Analyses of HIV-1NL4-3 derivatives against human BST-2 (A) and rhesus BST-2 (B); (C and D) analyses of SHIVDH12 CL7 derivatives against human BST-2 (C) and rhesus BST-2 (D); (E and F) analyses of SHIVAD8-MV derivatives against human BST-2 (E) and rhesus BST-2 (F). Cells were metabolically labeled for 90 min with [35S]methionine, and cell lysates and cell-free supernatants were subjected to immunoprecipitation by an HIV-positive patient serum sample or serum from an SHIV-infected monkey. Immunoprecipitates were subjected to SDS-PAGE, and virus release was quantified by PhosphoImage analysis using a Fujifilm FLA7000 system. Virus release was calculated independently for each sample by determining the percentage of cell-free CA protein relative to the total amount of intra- and extracellular Gag protein. Virus release in the absence of BST-2 (0 μg) was defined as 100% for each sample.
Vpu-deficient SHIVs are less pathogenic in rhesus macaques than their wt counterparts.
The attenuated replicative phenotype observed with SHIVDH12-CL7 and SHIVAD8-MV mutants in rhesus PBMCs raised the possibility that these HIV-1 vpu genes might enhance infectivity and disease progression in macaques inoculated with these SHIVs. It should be noted that pathogenic X4-tropic SHIV infections of rhesus monkeys are typically dose dependent: (i) with large inoculum sizes (e.g., 1,000 to 5,000 TCID50s), plasma viral loads are sustained (105 to 107 RNA copies/ml), CD4+ T cell numbers rapidly and irreversibly fall to low levels, and animals succumb to immunodeficiency at 12 to 20 weeks p.i.; and (ii) following exposure to small inoculum sizes, peak levels of plasma viremia are durably controlled, CD4+ T cell counts return to or remain at preinfection levels, and infected animals rarely develop symptomatic disease (21, 43). Normally progressing R5-tropic SHIVAD8-infected macaques usually generate high peak virus loads, lower sustained levels of plasma viremia, and a gradual loss of CD4+ T lymphocytes and disease development (38).
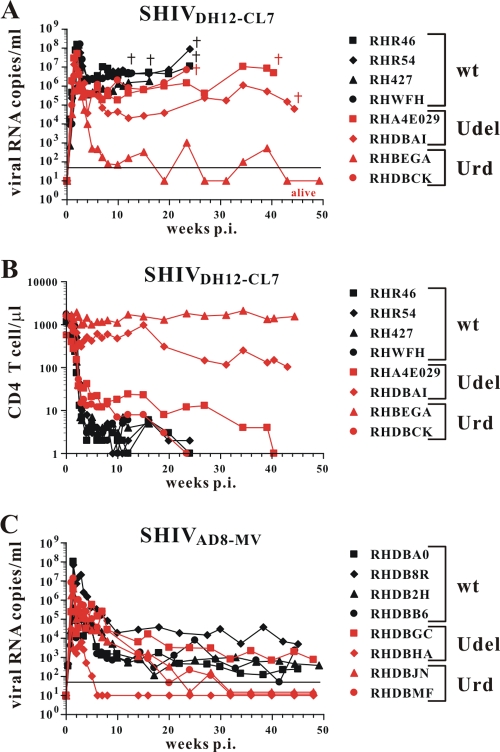
As shown in Fig. 5A and B, four historic wt SHIVDH12-CL7-infected control animals (30) inoculated intravenously with 5,000 TCID50s of virus experienced the expected high levels of plasma viremia and rapid depletion of their CD4+ T cells. Four other macaques were newly inoculated with 10,000 TCID50s of Vpu-defective SHIVDH12-CL7 (Udel and Urd, shown in red in Fig. 5A and B). One of these monkeys (RHDBCK) exhibited a clinical course indistinguishable from that of the four animals inoculated with the wt SHIVDH12-CL7 (shown in black in Fig. 5A and B). Two of the three remaining macaques (RHA4E029 and RHDBAI) experienced somewhat lower levels of set-point viremia, more gradual losses of CD4+ T lymphocytes, and delayed death from AIDS at weeks 40 and 44 p.i., respectively. The third monkey (RHBEGA), also infected with Vpu-deficient X4 SHIVDH12-CL7, is asymptomatic at week 74 p.i. with undetectable plasma viral RNA loads and little change in the number of CD4+ T cells measured prior to inoculation. Of the four macaques inoculated with the Vpu-defective R5-tropic SHIVAD8-MV, three currently have undetectable levels of plasma viremia and one (RHDBGC) has viral RNA loads similar to those of animals infected with wt virus (Fig. 5C). CD4+ T cell numbers in macaque RHDBGC are lower than preinoculation values (data not shown). Taken together, these results indicate that X4- and R5-tropic SHIVs carrying mutant vpu genes exhibit an attenuated replication phenotype in vivo compared to wt viruses.
Fig. 5.
Vpu-deficient SHIVs are less pathogenic in rhesus macaques than their wt counterparts. A group of four rhesus macaques each was infected with the X4-tropic wt or Udel SHIVDH12-CL7 (A and B) or the R5-tropic SHIVAD8-MV (C). The levels of plasma viremia were monitored over a period of 45 weeks (A and C), and absolute numbers of peripheral CD4+ T cells in animals infected by the X4-tropic viruses are shown (B). Euthanized monkeys are indicated (†).
DISCUSSION
Like other lentiviral accessory proteins, HIV-1 Vpu is multifunctional and has evolved to regulate several independent activities important for HIV replication and disease induction. Initially, Vpu was shown to enhance virion release from infected cells, a property that likely facilitates the dissemination of virus systemically (54, 55). It was subsequently noted that Vpu was also able to induce the degradation of the receptor for HIV-1 (CD4), a property that may augment the production of infectious virus particles by preventing the formation of intracellular, trafficking-incompetent, CD4-Env complexes (59). Finally, Vpu was found to induce host cell apoptosis (2). The role of Vpu-mediated host cell apoptosis for optimal HIV-1 replication is not immediately obvious, and host cell apoptosis may represent an unintended side effect of Vpu-induced CD4 degradation since both mechanisms involve binding to and sequestration of β-TrCP (6, 31). While there is still an ongoing debate about which of the reported Vpu functions is most relevant to virus replication and disease induction in vivo, the recent identification of BST-2 as a Vpu substrate has refocused attention on the regulation of virion release.
The precise mechanism responsible for the Vpu antagonism of BST-2, thereby enhancing virus release, is still unresolved. However, there is increasing evidence indicating that Vpu functions intracellularly to prevent the surface transport of BST-2 (3, 10, 11, 17, 33, 53). It is interesting to note that Vpu is not ubiquitously expressed by all primate lentiviruses but is unique to HIV-1 and some SIVs (SIVcpz, SIVgor, and SIVgsn). HIV-2 and most other SIVs do not encode a Vpu protein to counter BST-2. Instead, these viruses express Vpu-like activities in their env and nef genes, respectively (5, 7, 16, 17, 24, 45, 47, 49, 63). This suggests that regulation of virus release is a conserved property of lentiviruses and as such must be critical for virus replication. This is supported by the observation that compensatory changes in the cytoplasmic tail of gp41 confer resistance to BST-2 in a pathogenic SIV strain from which nef is deleted (49). Up to now it had been generally presumed that HIV-1 Vpu could antagonize only human and great ape BST-2 variants, a premise that was primarily based on studies conducted with the Vpu encoded by the prototypic HIV-1NL4-3 isolate (14, 24, 32, 45, 58, 61, 62). In this regard, HIV-1NL4-3 was originally constructed by ligating the 5′ half (long terminal repeat [LTR]-gag-pol-vif-vpr sequences) of the HIV-1NY5 isolate to the 3′ half (vpr-tat-rev-vpu-env-nef-LTR sequences) of HIV-1LAV at the single EcoRI site located in the vpr gene (1). Thus, the source of HIV-1 NL4-3 Vpu is HIV-1LAV. Interestingly, Ruiz et al. recently reported that SHIVKU-2MC4, encoding the vpu gene from HIV-1HXB2, also a derivative of HIV-1LAV, failed to antagonize pig-tailed macaque BST-2 in virion release assays or TZM-bl infectivity assays (42). This result is consistent with our failure to show any inhibition of rhesus macaque BST-2 using the Vpu from HIV-1NL4-3 (derived from the HIV-1LAV Vpu) (Fig. 3B). Taken together, these results reiterate the failure of the HIV-1NL4-3 Vpu to antagonize monkey BST-2, whether it is evaluated as a component of HIV-1 or SHIV.
It is remarkable that Vpu encoded by SHIVDH12-CL7 enhanced virus release, despite the presence of a functional nef gene. Indeed, the results shown in Fig. 3 indicate that neither SIVmac239 Nef nor DH12 Vpu expressed in the context of SHIVDH12-CL7 is capable of fully antagonizing rhesus BST-2 in primary rhesus PBMCs. Instead, the two proteins appear to function additively to provide maximal virus release. It should be pointed out that the potent antagonism of the SHIVDH12-CL7 Vpu against both simian and human BST-2 may reflect the way in which the parental HIV-1DH12 was identified and selected for use in animal studies. In an attempt to generate an HIV-1 chimpanzee disease model in 1995, 23 primary isolates from AIDS patients were screened for their capacity to rapidly spread and release high levels of progeny virions from chimpanzee PBMCs (51). HIV-1DH12 was unique among the three chimpanzee-tropic isolates obtained because of its ability to infect cultured PBMCs from 25 of 25 chimpanzee donors tested. Although HIV-1DH12 failed to maintain detectable levels of plasma viremia or induce disease in inoculated chimpanzees, it was the source of the HIV-1 vpu gene sequences present in SHIVDH12 (52) and its molecularly cloned and pathogenic derivative SHIVDH12-CL7 (43). The relationship between replication capacity in chimpanzee cells and inhibition of macaque BST-2 is not presently understood.
There have been very few studies evaluating Vpu function during nonhuman primate lentivirus infections in vivo. The earliest, using first-generation nonpathogenic SHIVs carrying wt Vpu (from HIV-1BH10) or mutant Vpu (from SHIVHXBc2) to inoculate cynomolgus monkeys, reported somewhat lower levels of circulating virus in animals infected with Vpu-defective SHIVs (29). Nearly a decade later, Hout et al. observed profoundly attenuated replication for a SHIV Vpu mutant (with a scrambled TM region) compared to the wt SHIVKU-1 (derived from SHIVHXBc2) both in vitro (human C8166 cells) and in vivo (pig-tailed macaques) (18). In monkeys inoculated with the Vpu-defective SHIV, peak levels of plasma viremia at week 2 p.i. were reduced 50-fold and viral set points were 2 to 3 log units lower at week 24 p.i. than those measured for wt SHIVKU-1-infected macaques. In a recent follow-up study, Ruiz et al. reported that the wt Vpu present in SHIVKU-2MC4 failed to counteract pig-tailed macaque BST-2 (42). In addition, deletion of Vpu in SHIVKU-2MC4 did not affect virus release due to the presence of SIV Nef, which antagonizes monkey BST-2 (24, 45, 61, 63). On the basis of the profound replication defect previously observed with the Vpu-minus SHIVKU-2MC4 (18), Ruiz et al. proposed that the Vpu TM domain may possess functional activities unrelated to BST-2 antagonism, which facilitates virus replication in vivo (42). Because full-length sequencing of the wt and Vpu mutant SHIVDH12-CL7 and SHIVAD8-MV genomes confirmed that each virus was isogenic except for the TM regions of Vpu, we believe that the attenuated phenotype observed in 6 of 8 animals inoculated with Vpu TM mutants in the present study, despite the presence of an intact SIV nef gene, is physiologically relevant. Given that replication and disease induction by primate lentiviruses are multigenic and Nef antagonism of simian BST-2 continues in the absence of a functional Vpu in the inoculated monkeys, the subtle defects in maintaining plasma viremia, depleting CD4+ T cells, and disease induction observed could represent the loss of auxiliary particle release activity mediated by Vpu or recently described immunomodulatory functions attributed to Vpu, such as downregulation of Cd1d or NK T- and B-cell antigen (35, 50). While a contribution of Vpu-induced degradation of rhesus CD4 cannot formally be ruled out, the fact that our experiments involving Urd—a Vpu mutant capable of degrading human CD4—yielded results very similar to those observed with a variant lacking Vpu entirely (i.e., Udel) argues against a significant contribution of Vpu-induced CD4 degradation to the observed phenotypic differences.
ACKNOWLEDGMENTS
We very much appreciate Reza Sadjadpour's help with constructing Nef-deficient variants of our molecular SHIV clones. We thank Yoshiaki Nishimura for critical comments on the manuscript. We further thank Ranjini Iyengar for determining viral RNA levels and Alicia Buckler-White and Ronald Plishka for conducting nucleotide sequence analyses. We appreciate the contributions of Boris Skopits and Rahel Petros for diligently assisting in the care and maintenance of our animals.
Pooled HIV-1-positive patient serum was obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: HIV-IG was from NABI and NHLBI (catalog no. 3957).
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Published ahead of print on 20 July 2011.
REFERENCES
- 1. Adachi A., et al. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Akari H., Bour S., Kao S., Adachi A., Strebel K. 2001. The human immunodeficiency virus type 1 accessory protein Vpu induces apoptosis by suppressing the nuclear factor kappaB-dependent expression of antiapoptotic factors. J. Exp. Med. 194:1299–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Andrew A. J., Miyagi E., Strebel K. 2011. Differential effects of human immunodeficiency virus type 1 Vpu on the stability of BST-2/tetherin. J. Virol. 85:2611–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benn S., et al. 1985. Genomic heterogeneity of AIDS retroviral isolates from North America and Zaire. Science 230:949–951 [DOI] [PubMed] [Google Scholar]
- 5. Bour S., Akari H., Miyagi E., Strebel K. 2003. Naturally occurring amino acid substitutions in the HIV-2 ROD envelope glycoprotein regulate its ability to augment viral particle release. Virology 309:85–98 [DOI] [PubMed] [Google Scholar]
- 6. Bour S., Perrin C., Akari H., Strebel K. 2001. The human immunodeficiency virus type 1 Vpu protein inhibits NF-kappa B activation by interfering with beta TrCP-mediated degradation of Ikappa B. J. Biol. Chem. 276:15920–15928 [DOI] [PubMed] [Google Scholar]
- 7. Bour S., Strebel K. 1996. The human immunodeficiency virus (HIV) type 2 envelope protein is a functional complement to HIV type 1 Vpu that enhances particle release of heterologous retroviruses. J. Virol. 70:8285–8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Committee on the Care and Use of Laboratory and Animals 1985. Guide for the care and use of laboratory animals. U.S. Department of Health and Human Services publication no. NIH 85-23 National Institutes of Health, Bethesda, MD [Google Scholar]
- 9. Douglas J. L., et al. 2009. Vpu directs the degradation of the human immunodeficiency virus restriction factor BST-2/tetherin via a {beta}TrCP-dependent mechanism. J. Virol. 83:7931–7947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dube M., et al. 2010. Antagonism of tetherin restriction of HIV-1 release by Vpu involves binding and sequestration of the restriction factor in a perinuclear compartment. PLoS Pathog. 6:e1000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dube M., et al. 2009. Suppression of tetherin-restricting activity upon human immunodeficiency virus type 1 particle release correlates with localization of Vpu in the trans-Golgi network. J. Virol. 83:4574–4590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Endo Y., et al. 2000. Short- and long-term clinical outcomes in rhesus monkeys inoculated with a highly pathogenic chimeric simian/human immunodeficiency virus. J. Virol. 74:6935–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gendelman H. E., et al. 1988. Efficient isolation and propagation of human immunodeficiency virus on recombinant colony-stimulating factor 1-treated monocytes. J. Exp. Med. 167:1428–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goffinet C., et al. 2009. HIV-1 antagonism of CD317 is species specific and involves Vpu-mediated proteasomal degradation of the restriction factor. Cell Host Microbe 5:285–297 [DOI] [PubMed] [Google Scholar]
- 15. Gupta R. K., et al. 2009. Mutation of a single residue renders human tetherin resistant to HIV-1 Vpu-mediated depletion. PLoS Pathog. 5:e1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gupta R. K., et al. 2009. Simian immunodeficiency virus envelope glycoprotein counteracts tetherin/BST-2/CD317 by intracellular sequestration. Proc. Natl. Acad. Sci. U. S. A. 106:20889–20894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hauser H., et al. 2010. HIV-1 Vpu and HIV-2 Env counteract BST-2/tetherin by sequestration in a perinuclear compartment. Retrovirology 7:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hout D. R., et al. 2005. Scrambling of the amino acids within the transmembrane domain of Vpu results in a simian-human immunodeficiency virus (SHIV(TM)) that is less pathogenic for pig-tailed macaques. Virology 339:56–69 [DOI] [PubMed] [Google Scholar]
- 19. Igarashi T., et al. 2002. Rapid and irreversible CD4+ T-cell depletion induced by the highly pathogenic simian/human immunodeficiency virus SHIV(DH12R) is systemic and synchronous. J. Virol. 76:379–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Igarashi T., et al. 1999. Emergence of a highly pathogenic simian/human immunodeficiency virus in a rhesus macaque treated with anti-CD8 mAb during a primary infection with a nonpathogenic virus. Proc. Natl. Acad. Sci. U. S. A. 96:14049–14054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Igarashi T., et al. 2003. Early control of highly pathogenic simian immunodeficiency virus/human immunodeficiency virus chimeric virus infections in rhesus monkeys usually results in long-lasting asymptomatic clinical outcomes. J. Virol. 77:10829–10840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Imamichi H., et al. 2002. Amino acid deletions are introduced into the V2 region of gp120 during independent pathogenic simian immunodeficiency virus/HIV chimeric virus (SHIV) infections of rhesus monkeys generating variants that are macrophage tropic. Proc. Natl. Acad. Sci. U. S. A. 99:13813–13818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Iwabu Y., et al. 2009. HIV-1 accessory protein Vpu internalizes cell-surface BST-2/tetherin through transmembrane interactions leading to lysosomes. J. Biol. Chem. 284:35060–35072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jia B., et al. 2009. Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLoS Pathog. 5:e1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kamada K., et al. 2006. Generation of HIV-1 derivatives that productively infect macaque monkey lymphoid cells. Proc. Natl. Acad. Sci. U. S. A. 103:16959–16964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Klimkait T., Strebel K., Hoggan M. D., Martin M. A., Orenstein J. M. 1990. The human immunodeficiency virus type 1-specific protein Vpu is required for efficient virus maturation and release. J. Virol. 64:621–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kobayashi T., et al. 2011. Identification of amino acids in the human tetherin transmembrane domain responsible for HIV-1 Vpu interaction and susceptibility. J. Virol. 85:932–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Le Tortorec A., Neil S. J. 2009. Antagonism to and intracellular sequestration of human tetherin by the human immunodeficiency virus type 2 envelope glycoprotein. J. Virol. 83:11966–11978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li J. T., et al. 1995. Persistent infection of macaques with simian-human immunodeficiency viruses. J. Virol. 69:7061–7067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mao H., et al. 2005. CD8+ and CD20+ lymphocytes cooperate to control acute simian immunodeficiency virus/human immunodeficiency virus chimeric virus infections in rhesus monkeys: modulation by major histocompatibility complex genotype. J. Virol. 79:14887–14898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Margottin F., et al. 1998. A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol. Cell 1:565–574 [DOI] [PubMed] [Google Scholar]
- 32. McNatt M. W., et al. 2009. Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLoS Pathog. 5:e1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mitchell R. S., et al. 2009. Vpu antagonizes BST-2-mediated restriction of HIV-1 release via beta-TrCP and endo-lysosomal trafficking. PLoS Pathog. 5:e1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miyagi E., Andrew A. J., Kao S., Strebel K. 2009. Vpu enhances HIV-1 virus release in the absence of Bst-2 cell surface down-modulation and intracellular depletion. Proc. Natl. Acad. Sci. U. S. A. 106:2868–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moll M., Andersson S. K., Smed-Sorensen A., Sandberg J. K. 2010. Inhibition of lipid antigen presentation in dendritic cells by HIV-1 Vpu interference with CD1d recycling from endosomal compartments. Blood 116:1876–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Neil S. J., Zang T., Bieniasz P. D. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430 [DOI] [PubMed] [Google Scholar]
- 37. Nishimura Y., et al. 2004. Highly pathogenic SHIVs and SIVs target different CD4+ T cell subsets in rhesus monkeys, explaining their divergent clinical courses. Proc. Natl. Acad. Sci. U. S. A. 101:12324–12329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nishimura Y., et al. 2010. Generation of the pathogenic R5-tropic simian/human immunodeficiency virus SHIVAD8 by serial passaging in rhesus macaques. J. Virol. 84:4769–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. O'Doherty U., Swiggard W. J., Malim M. H. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74:10074–10080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Perez-Caballero D., et al. 2009. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell 139:499–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rong L., et al. 2009. The transmembrane domain of BST-2 determines its sensitivity to down-modulation by HIV-1 Vpu. J. Virol. 83:7536–7546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ruiz A., et al. 2010. BST-2 mediated restriction of simian-human immunodeficiency virus. Virology 406:312–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sadjadpour R., et al. 2004. Induction of disease by a molecularly cloned highly pathogenic simian immunodeficiency virus/human immunodeficiency virus chimera is multigenic. J. Virol. 78:5513–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sakai H., Tokunaga K., Kawamura M., Adachi A. 1995. Function of human immunodeficiency virus type 1 Vpu protein in various cell types. J. Gen. Virol. 76Pt 11:2717–2722 [DOI] [PubMed] [Google Scholar]
- 45. Sauter D., et al. 2009. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6:409–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schubert U., et al. 1996. The two biological activities of human immunodeficiency virus type 1 Vpu protein involve two separable structural domains. J. Virol. 70:809–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schubert U., Bour S., Willey R. L., Strebel K. 1999. Regulation of virus release by the macrophage-tropic human immunodeficiency virus type 1 AD8 isolate is redundant and can be controlled by either Vpu or Env. J. Virol. 73:887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schubert U., Clouse K. A., Strebel K. 1995. Augmentation of virus secretion by the human immunodeficiency virus type 1 Vpu protein is cell type independent and occurs in cultured human primary macrophages and lymphocytes. J. Virol. 69:7699–7711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Serra-Moreno R., Jia B., Breed M., Alvarez X., Evans D. T. 2011. Compensatory changes in the cytoplasmic tail of gp41 confer resistance to tetherin/BST-2 in a pathogenic nef-deleted SIV. Cell Host Microbe 9:46–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shah A. H., et al. 2010. Degranulation of natural killer cells following interaction with HIV-1-infected cells is hindered by downmodulation of NTB-A by Vpu. Cell Host Microbe 8:397–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shibata R., et al. 1995. Isolation and characterization of a syncytium-inducing macrophage/T-cell line-tropic human immunodeficiency virus type 1 isolate that readily infects chimpanzee cells in vitro and in vivo. J. Virol. 69:4453–4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shibata R., et al. 1997. Infection and pathogenicity of chimeric simian-human immunodeficiency viruses in macaques: determinants of high virus loads and CD4 cell killing. J. Infect. Dis. 176:362–373 [DOI] [PubMed] [Google Scholar]
- 53. Skasko M., et al. 2011. BST-2 is rapidly down-regulated from the cell surface by the HIV-1 protein Vpu: evidence for a post-ER mechanism of Vpu-action. Virology 411:65–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Strebel K., Klimkait T., Martin M. A. 1988. A novel gene of HIV-1, vpu, and its 16-kilodalton product. Science 241:1221–1223 [DOI] [PubMed] [Google Scholar]
- 55. Terwilliger E. F., Cohen E. A., Lu Y. C., Sodroski J. G., Haseltine W. A. 1989. Functional role of human immunodeficiency virus type 1 vpu. Proc. Natl. Acad. Sci. U. S. A. 86:5163–5167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Theodore T. S., et al. 1996. Construction and characterization of a stable full-length macrophage-tropic HIV type 1 molecular clone that directs the production of high titers of progeny virions. AIDS Res. Hum. Retroviruses 12:191–194 [DOI] [PubMed] [Google Scholar]
- 57. Van Damme N., et al. 2008. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3:245–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Van Damme N., Guatelli J. 2008. HIV-1 Vpu inhibits accumulation of the envelope glycoprotein within clathrin-coated, Gag-containing endosomes. Cell. Microbiol. 10:1040–1057 [DOI] [PubMed] [Google Scholar]
- 59. Willey R. L., Maldarelli F., Martin M. A., Strebel K. 1992. Human immunodeficiency virus type 1 Vpu protein regulates the formation of intracellular gp160-CD4 complexes. J. Virol. 66:226–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Willey R. L., et al. 1988. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J. Virol. 62:139–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yang S. J., et al. 2010. Anti-tetherin activities in Vpu-expressing primate lentiviruses. Retrovirology 7:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yoshida T., Kao S., Strebel K. 2011. Identification of residues in the BST-2 TM domain important for antagonism by HIV-1 Vpu using a gain-of-function approach. Front. Microbiol. 2:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang F., et al. 2009. Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe 6:54–67 [DOI] [PMC free article] [PubMed] [Google Scholar]