Abstract
APOBEC1 (A1) is a cytidine deaminase involved in the regulation of lipids in the small intestine. Herpes simplex virus 1 (HSV-1) is a ubiquitous pathogen that is capable of infecting neurons in the brain, causing encephalitis. Here, we show that A1 is induced during encephalitis in neurons of rats infected with HSV-1. In cells stably expressing A1, HSV-1 infection resulted in significantly reduced virus replication compared to that in control cells. Infectivity could be restored to levels comparable to those observed for control cells if A1 expression was silenced by specific A1 short hairpin RNAs (shRNA). Moreover, cytidine deaminase activity appeared to be essential for this inhibition and led to an impaired accumulation of viral mRNA transcripts and DNA copy numbers. The sequencing of viral gene UL54 DNA, extracted from infected A1-expressing cells, revealed G-to-A and C-to-T transitions, indicating that A1 associates with HSV-1 DNA. Taken together, our results demonstrate a model in which A1 induction during encephalitis in neurons may aid in thwarting HSV-1 infection.
INTRODUCTION
The human apolipoprotein B-editing catalytic polypeptide (ABOBEC) family is a group of zinc-dependent DNA and RNA cytidine deaminases and consists of AID, APOBEC1 (A1), APOBEC2 (A2), seven APOBEC3s (APOBEC3A [A3A] to A3H), and APOBEC4 (A4). A1, the first APOBEC to be discovered, is known to introduce a premature stop codon into host apolipoprotein B mRNA in the gastrointestinal tract, an event critical for lipid metabolism (17, 40, 61). The editing by A1 is highly precise and specifically converts C to U at position 6666 of the apolipoprotein B mRNA substrate (46). Along with APOBEC1 complementation factor (ACF), these two proteins constitute the minimal required components necessary for the editing of apolipoprotein B mRNA in vitro (37).
Cytidine deaminases first came into the limelight as antiviral factors after A3G was identified as a cellular restriction factor capable of inhibiting HIV-1 dissemination in the absence of HIV-1 virus infectivity factor (Vif) (56). This molecule was later shown to inhibit retrovirus infection by inducing a massive hypermutation of the murine leukemia virus (MLV) genome (23). Further detailed studies revealed that APOBEC molecules are packaged into HIV-1 virions in virus producer cells via a specific interaction with Gag and viral RNA and then exert their deaminase activity in subsequent target cells on a single-stranded DNA (ssDNA) intermediate synthesized by reverse transcriptase (3, 28, 55). Editing can lead to nonsynonymous mutations, such as premature stop codons, in critical proteins (e.g., reverse transcriptase) necessary for virus replication and infectivity, severely impairing the next round of infection (54, 64). Extensive studies to assess the antiviral nature of these APOBEC enzymes have been performed across a broad range of retroviruses and hepatitis B virus (HBV) (7, 35, 36, 42, 43, 50, 56, 58).
Herpes simplex virus (HSV) is an enveloped, double-stranded DNA (dsDNA) virus and a member of the genus Alphaherpesviridae. One in every 250,000 to 500,000 individuals infected with HSV-1 experiences a devastating disease known as HSV encephalitis (HSE), characterized by acute inflammation and/or hemorrhaging in the central nervous system (CNS) (62). HSV-1 is the predominant causative agent of sporadic encephalitis in western countries, and it is estimated to be responsible for over 90% of HSE cases (62).
In this study, we report a novel finding that A1 expression is induced during HSV-1 infection in neurons of infant rat brains, more than 30% of which were able to recover from HSE. Moreover, an investigation into the potential antiviral role of A1 in vitro revealed that A1 inhibited virus replication directly by targeting mainly viral DNA in a deaminase-dependent manner, resulting in a stalling of virus replication. To the best of our knowledge, this is the first report to indicate the potential antiviral function of A1 against herpesviruses in the context of HSE.
MATERIALS AND METHODS
Cells, plasmids, viruses, and infection.
Vero cells and rabbit skin cells (RSCs) were maintained in Dulbecco's modified Eagle medium (DMEM) and supplemented with fetal calf serum (FCS) and antibiotics as previously described (4). Rat A1 cDNA was obtained by reverse transcription (RT)-PCR of mRNA derived from HSV-1-infected rat brain tissue and inserted into pcDNA3.1/Zeo(+) (Invitrogen). An A1 mutant, A1E63A, was generated by site-directed mutagenesis using a QuikChange II site-directed mutagenesis kit (Stratagene). Hemagglutinin (HA)-tagged empty vector, wild-type A1 (A1WT) or A1E63A mutant DNAs were transfected into RSCs and then cultured for 10 days in the presence of zeocin. Colonies were then screened protein expression, and expression was confirmed by Western blotting with an anti-HA monoclonal antibody (MAb). Green fluorescent protein (GFP)-expressing replication-competent HSV-1 strain YK333 (60) was used as previously described (4). HSV-1 YK333 was created by the intergenic insertion of a GFP-expressing cassette between UL3 and UL4 of the virus genome. Green fluorescence can be detected after HSV infects host cells and starts viral gene expression (60); however, since this event occurs before viral DNA replication, the GFP signal detected in these cells is not indicative of HSV-1 progeny production. The titers were determined by determining the standard 50% tissue culture infective dose (TCID50) or PFU on Vero cells. For the inhibition of HSV-1, we added 25 μg/ml of acyclovir (Sigma-Aldrich, St. Louis, MO) into the culture medium. UV-inactivated HSV-1 was prepared by exposing the virus to a total of 3 J/cm2 using a CL-1000 UV cross-linker for 10 min.
Generation of shRNA and shRNA-expressing cells.
Short hairpin RNA (shRNA) plasmids were generated by the cloning of A1-specific or luciferase control open reading frame (ORF)-targeted sense and antisense sequences into a pBAsi-hU6 puromycin vector (TaKaRa), via BamHI and XbaI restriction sites. The targeted A1 and luciferase sequences are as follows: A1/736 (5′-GATCCCCGACGCTCCGTTACCCGGTTACGTGTGCTGTCCGTAACCAGGTAATGGAGCATCTTTTTGGAAAT-3′), A1/1119 (5′-GATCCCCGTTCTTCAAGGCTGCCGTTACGTGTGCTGTCCGTAATGGCAGCTTTGAAGAGCTTTTTGGAAAT-3′), and Luciferase (5′-GATCCCCGTGCGTTGTTGGTGTTAATACGTGTGCTGTCCGTATTGGCACCAGCAGCGCACTTTTTGGAAAT-3′). A1-expressing RSCs were transfected with the shRNA plasmids by using Lipofectamine 2000, selected with puromycin for individual clones, and finally analyzed by Western blotting for the suppression of A1.
Animal models and tissue collection.
Wistar Hannover GALAS (Global Alliance for Laboratory Animal Standardization) rats (Clea Japan, Inc.) were anesthetized and then intracranially inoculated with 1.0 × 106 TCID50 HSV-1 (YK333) or phosphate-buffered saline (PBS) as a negative control. For most studies, 14-day-old rats were used unless indicated otherwise. Rats exhibiting a range of symptoms were classified into four groups, as described in Table 1. For histological examinations, tissue samples were fixed by immersion in 4% paraformaldehyde (PFA) and processed as described previously (4). All animal experiments were carried out according to the guidelines for animal experimentation at Kyoto University.
Table 1.
Classification of rats exhibiting various degrees of HSE
| Classificationa | No. of rats (%) | No. of rats showing neurological symptom ofb: |
Pathological change score |
GFP-positive region scoref | HSV-1-positive region scoref | ||||
|---|---|---|---|---|---|---|---|---|---|
| Monoplegia | Quadriplegia | Seizure | Tissue damagec | CD3-positive cellsd | CD68-positive cellse | ||||
| Severe | 17 (27) | 1 | 15 | 11 | + | ++ | ++ | ++ | ++ |
| Mild | 14 (22.2) | 7 | 7 | 4 | ++ | +++ | +++ | +++ | +++ |
| Survived | 24 (38.1) | 22 | 0 | 0 | − | + | + | + | + |
| Healthy | 8 (12.7) | 0 | 0 | 0 | − | − | − | − | − |
| Total | 63 | ||||||||
HSV-1-infected rats were classified as follows: severe, which died within 5 days after showing symptoms of severe encephalitis; mild, which died between 5 and 7 days after showing symptoms of mild encephalitis; survived, which recovered after transient encephalitis; and healthy, which did not display HSE symptoms.
The number of rats exhibiting neurological symptoms such as monoplegia, quadriplegia, and seizures.
The percentage of damaged areas including cell loss, degeneration of cells, and/or hemorrhaging in the coronal section is presented as follows: −, <5%; +, 5 to 10%; ++, >15%.
The number of CD3-positive cells in one infected area of the coronal section was quantified and is presented as follows: −, not detected; +, <5 cells; ++, 5 to 20 cells; +++, >20 cells.
The number of CD68-positive cells in one infected area of the coronal section was quantified and is presented as follows: −, not detected; +, <5 cells; ++, 5 to 50 cells; +++, >50 cells.
The percentage of GFP- or HSV-1-positive areas in the coronal section is presented as follows: −, not detected; +, <5%; ++, 5 to 20%; +++, >20%.
Transcriptome analysis.
RNA was prepared from rat brains by using an RNeasy extraction kit (Qiagen), and transcriptome analysis was performed by using a GeneChip Rat Genome 230 2.0 array (Affymetrix) according to the manufacturer's instructions. Microarray data were analyzed by using Gene Spring software (Agilent).
Real-time PCR.
Nucleic acids were extracted as previously described (4). For the analyses of HSV-1 mRNA products, 1 μg of RNA was reverse transcribed by using a QuantiTect reverse transcription kit (Qiagen) according to the manufacturer's instructions. The cDNA (50 ng) was used as a template to amplify the UL54 (immediate-early [IE]), UL30 (early [E]), and UL27 (late [L]) genes by real-time PCR using Power SYBR green PCR master mix (Applied Biosystems). The threshold was set to a value of 1.00, and the number of cycles required to reach the threshold cycle (CT) was determined and then normalized to the CT of the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The HSV-1 DNA copy number was measured as previously described (4). Thermocycler conditions for the real-time PCR were as follows: 95°C for 10 min and 95°C for 15s plus 60°C for 1 min for 40 cycles. The primers used for real-time PCR are as follows: A1 Forward (5′-ACCACGCAGATCCTCGAAAT-3′), A1 Reverse (5′-TCTTGCTCCGTCATGATCTGG-3′), HSV-1 UL54 Forward (5′-CCGCGACGACCTGGAATCGG-3′), HSV-1 UL54 Reverse (5′-GGCGAGCGGCGTCGAGTATC-3′), HSV-1 UL30 Forward (5′-AGAGGGACATCCAGGACTTTGT-3′), HSV-1 UL30 Reverse (5′-CAGGCGCTTGTTGGTGTAC-3′), HSV-1 UL27 Forward (5′-TCGCCTTTCGCTACGTCAT-3′), HSV-1 UL27 Reverse (5′-GGTTCTTGAGCTCCTTGGTGG-3′), GAPDH Forward (5′-ACTAAAGGGCATCCTGGGCTA-3′), and GAPDH Reverse (5′-TGGAAGAATGGGAGTTGCTGT-3′).
Immunohistochemistry and antibodies.
Brain sections and cultured cells were treated as described previously (4, 31). The following primary and secondary antibodies were used: goat anti-APOBEC1 polyclonal antibody (Pab) (Santa Cruz Biotechnology), rabbit anti-HSV-1 Pab (DakoCytomation), mouse anti-MAP2 MAb (Upstate), anti-HA MAb (Roche), Alexa Fluor 594-conjugated anti-goat IgG (Invitrogen), and Cy5-conjugated anti-mouse IgG or Alexa Fluor 647-conjugated streptavidin (Invitrogen). Nuclei were stained by using Hoechst 33342 dye (Invitrogen). Each sample was examined under a confocal laser microscope (TCS SP2 AOBS; Leica Microsystems), using 405-, 543-, and 633-nm excitations with 10×, 20×, and 40× objectives.
Sequencing analysis of UL54.
Nested PCR of UL54 was carried out with Pfu Ultra II Fusion DNA polymerase (Stratagene). In the first round of PCR, 100 ng of template cDNA or DNA, derived from either HSV-1-infected A1-expressing or control cells, was used with the appropriate primers in a total volume of 25 μl under the following thermocycler conditions: 1 min at 98°C; 20 s at 98°C, 20 s at 57°C, and 30 s at 72°C for 35 cycles; and 3 min at 72°C. For the second round of PCR, 2.5 μl of the first PCR mixture was used as a template in a total volume of 25 μl under the following thermocycler conditions: 1 min at 93.4°C; 20 s at 93.4°C, 20 s at 57°C, and 30 s at 72°C for 35 cycles; and 3 min at 72°C. The UL54 amplicon was cloned into a pUC19 vector, and the plasmid DNA was amplified. A BigDye Terminator cycle sequencing kit (v3.1) was used next, and samples were read on an ABI Prism 3130 sequencer. Sequences were analyzed by using Sequencher 4.9 software. The primers used for the nested PCR are as follows: First PCR UL54 Forward (5′-AGCTTTGGCCGCAGCGCACA-3′), First PCR UL54 Reverse (5′-GAGTTGCAATAAAAATATTTGCCGTGCAC-3′), Second PCR UL54 Forward (5′-GGTCTAGAAGCTTTGGCCGCAGCGCACA-5′), and Second PCR UL54 Reverse (5′-ATCAAGCTTCCTCGCGCCTTCAGGTAGCA-3′).
Statistical analysis.
Statistical significance was determined by using the Student t test and χ2 analysis. A P value of less than 0.05 was considered statistically significant.
RESULTS
Rat model for survival of encephalitis with intracranial HSV-1 inoculation.
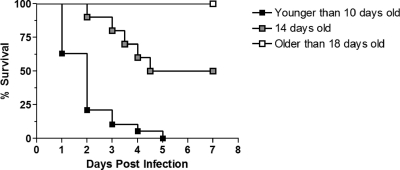
In the past, rats have been reported to be useful models for examining HSE induced by both HSV-1 and HSV-2, and, depending on the age of the animal, there appears to be an age-dependent resistance to infection for survival (5, 15). In line with those reports, our preliminary studies revealed that infant GALAS rats before the age of 14 days suffered from 100% mortality when infected by an intracranial injection of GFP-expressing HSV-1 (YK333) (Fig. 1). On the other hand, rats 18 days of age or older displayed 100% survival after HSV-1 inoculation. Interestingly, the infection of 14-day-old rats resulted in half of the animals surviving (Fig. 1). These results indicated that possible intrinsic factors expressed in rats from this age may help to protect them against HSV.
Fig. 1.
Age-dependent death induced by the intracranial inoculation of 1.0 × 106 TCID50 HSV-1 YK333 in Wistar GALAS rats. The inoculation of rats younger than 10 days old resulted in 100% mortality (groups composed of 5-day-old rats [n = 7], 7-day-old rats [n = 7], and 10-day-old rats [n = 5]). The inoculation of 14-day-old rats resulted in a survival rate of 50% (n = 40). Rats older than 18 days of age showed a survival rate of 100% (groups composed of 18-day-old rats [n = 7], 21-day-old rats [n = 7], and 28-day-old rats [n = 7]).
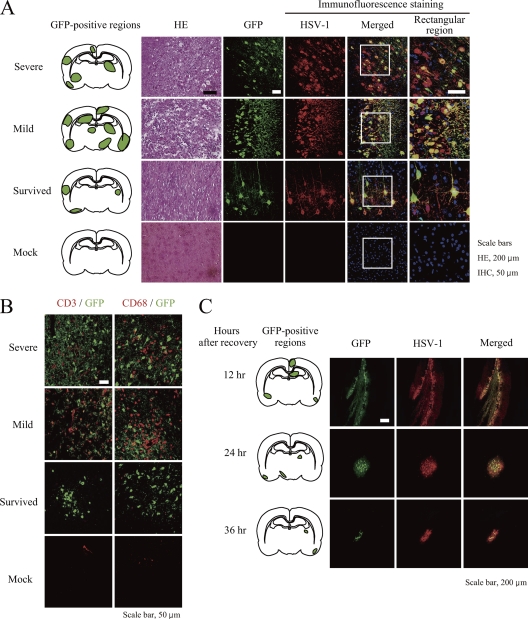
In order to the elucidate factors contributing to the survival of the HSV-1-infected 14-day-old rats, we focused on the observed phenotype for this age group. We proceeded to inoculate 63 animals with HSV-1 for a more in-depth assessment of HSE. We found that 17 HSV-1-injected rats (27%) (indicated as severe in Table 1 and Fig. 2A) died after showing severe signs of encephalitis, such as weight loss and quadriplegia, within 5 days after inoculation. Fourteen injected rats (22%) (indicated as mild in Table 1 and Fig. 2A) died after showing a milder degree of weight loss and quadriplegia at between 5 and 7 days. Interestingly, 24 injected rats (38.1%) (indicated as survived in Table 1 and Fig. 2A) recovered from HSE after showing temporary paralysis; HSV-1 infection in these mice was confirmed by GFP-positive (GFP+) and HSV-1-positive (HSV-1+) regions. The final group of 8 injected rats did not exhibit symptoms of encephalitis and did not possess GFP+ and HSV-1+ regions (indicated as healthy in Table 1). The range of symptoms displayed by the rats indicated that some were able to cope with HSV-1 infection better than others, which we speculated to be due to the containment of virus infection.
Fig. 2.
HSE model induced by GFP-HSV-1 inoculation of infant rats. Fourteen-day-old infant rats were intracranially injected with GFP-expressing HSV-1 (YK333), and brain tissues were collected from rats exhibiting severe or mild encephalitis as described in Table 1 or from surviving rats 36 h after the disappearance of paralysis. PBS (mock)-inoculated rats were used as negative controls, and brain tissues were collected at the end of the experiment. (A) GFP+ (green) regions in the coronal plane, with hematoxylin and eosin (HE)-stained, GFP+ (green), and HSV antigen-positive (red) brain cells in the tissue section. Nuclei were also stained with Hoechst 33342 dye (blue) and are shown in the merged image. A zoom-in view of the rectangular region in the merged image is shown to the right (immunofluorescence staining). (B) GFP+ (green), CD3+ (red), or CD68+ (red) cells in the tissue section. (C) GFP+ regions in the coronal plane (right) and GFP+ and HSV antigen-positive cells in tissue sections (left) from surviving rats at the indicated hours after the disappearance of paralysis. Representative results are shown. Scale bars are 200 μm for hematoxylin-and-eosin-stained sections and 50 μm for the other sections.
In the brain tissues of rats with severe and mild symptoms, taken at the time of death from HSE, we found vast hemorrhagic and necrotic damage (Fig. 2A). Furthermore, disseminated GFP+ regions, which were confirmed to be HSV-1+ cells by immunostaining with an anti-HSV antibody, indicated massive spreading and a multifocally distributed infection in the brain, typical of encephalitis in human infants (Table 1 and Fig. 2A) (62). Severe and mild symptoms in rats were also associated with extensive leukocyte infiltration by CD3+ T cells and CD68+ macrophages (Fig. 2B, red); however, brain samples from recovered rats taken 36 h after the disappearance of paralysis showed less infiltration in the parenchyma and fewer GFP+ cells (Table 1 and Fig. 2B, third row). Meanwhile, we could not detect pathological changes or GFP+ or HSV+ cells in the brains of mock-infected rats (Fig. 2A, bottom), confirming that the observed results were indeed dependent on HSV-1 infection. These data also indicated that limited but clear HSV-1 infection occurred in the brain of the rats that survived.
To investigate whether our initial observation of reduced HSV-1 infection in recovered rats was due to the control of HSV-1 dissemination over the course of infection or due simply to an inadequate initial infection, brain samples of rats that survived were taken 12, 24, and 36 h after the disappearance of paralysis, and the extent of GFP+ regions was determined. The results showed that the sizes of the HSV-1-infected regions diminished over a span of 36 h, suggesting that the virus was being contained and cleared (Fig. 2C). These data also suggest the existence of an anti-HSV factor(s) induced upon HSV-1 infection, which appears to be expressed in CNS parenchymal cells, including neurons or glia, since the number of invading lymphocytes and macrophages did not seem to correlate with the clearance of the virus.
Apolipoprotein B-editing catalytic subunit 1 is induced during HSV-1 infection in brain tissue.
In order to identify potential candidate molecules which are induced in the brain upon HSV-1 infection, we isolated RNA from GFP+ and neighboring regions of brains from two surviving rats at 4 and 6 days postinfection (corresponding to 12 and 36 h after the disappearance of paralysis, respectively) and performed microarray analysis of mRNA from both survived and mock-infected rat brains. When we compared the levels of mRNA from these rats, 47 genes showed augmented expression levels (see Table S1 in the supplemental material). Among them, the levels of A1 mRNA were clearly increased by 4.59- and 41.6-fold at 4 and 6 days postinfection, respectively.
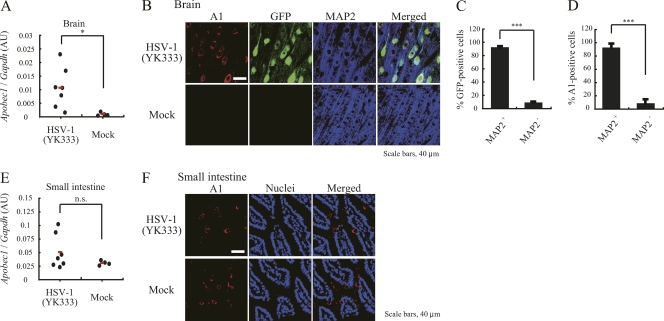
Since A1 and related A3 family proteins are known to be strong inhibitors of HIV-1 infection, we examined the possibility of an association between A1 and HSV-1 infection. In rat brain tissue, we were able to detect significantly elevated levels of A1 cDNA exclusively in HSV-1-infected rats by real-time PCR (Fig. 3A). As expected, we confirmed high levels of A1 cDNA expression in the small intestine of mock- and HSV-1-infected rats, where A1 is constitutively expressed (Fig. 3E). Immunofluorescence staining of brain tissue with an A1-specific antibody demonstrated A1-positive (A1+) staining only after HSV-1 infection but not after mock infection (Fig. 3B), corroborating the PCR findings.
Fig. 3.
Expression of A1 in HSV-1-infected rat brains. (A and E) Levels of A1 cDNA expression, quantified by real-time PCR, in the brain (A) and small intestine (E) of HSV-1-inoculated rats, which consisted of 4 rats with mild symptoms and 2 recovered rats, and of 4 PBS-inoculated rats. AU, arbitrary units. (B and F) A1+ (red), GFP+ (green), and MAP2+ (blue) neuronal cells of an HSV-1- and mock-infected rat brain (B, top) and A1+ cells in the small intestine (F) of HSV-1- and mock-infected rats (bottom). Representative results are shown. Scale bars are 200 μm. (C and D) Quantification data presented as a percentage of MAP2 antigen-positive cells in GFP+ (C) or A1+ (D) cells. *, P < 0.05 compared to mock-infected cells; ***, P < 0.005 compared to MAP2 antigen-negative cells.
To further investigate the cell type-specific expression of A1 after HSV-1 infection, we performed coimmunostaining with an anti-MAP2 (neuron-specific) antibody and an anti-A1 antibody. A1+ staining was observed predominantly in MAP2+ cells that were HSV-1 infected but not in MAP2-negative (MAP2−) cells, indicating that A1 expression was induced in neurons within the HSV-1-infected brain (Fig. 3B to D). On the other hand, the expression levels of A1 in the small intestine were high regardless of HSV-1 infection (Fig. 3E and F). These data suggested that A1 may serve as an intrinsic neuronal tissue factor for the control of HSV-1 infection in the CNS.
A1 inhibits HSV-1 infection in a deaminase-dependent and -independent manner.
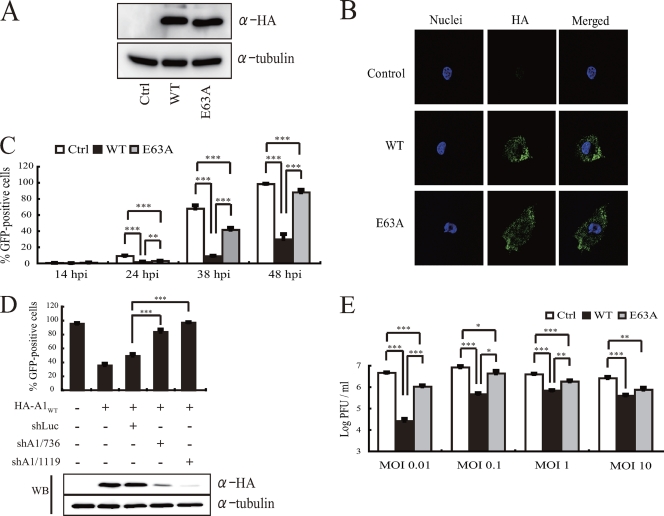
To examine the potential anti-HSV-1 activity of A1, we generated RSCs stably expressing N-terminally HA-tagged wild-type rat A1 (A1WT). A1 is a zinc-dependent deaminase with 3 residues (His61, Cys93, and Cys96) needed for coordinating zinc binding and a glutamate residue at position 63 in the catalytic domain essential for cytidine deaminase activity (39). Hence, we also generated RSCs stably expressing an N-terminally HA-tagged A1 deaminase-deficient mutant (A1E63A). Control A1 (A1Ctrl)-expressing RSCs were generated by using an empty HA vector. Western blotting using an anti-HA antibody confirmed that the expression levels of A1WT and A1E63A were equivalent in these cell lines (Fig. 4A). Immunofluorescence staining of A1WT and A1E63A a showed similar localization, which was comparable to that in A1+ neuronal cells of HSV-1-infected brain tissue (Fig. 3B and 4B, respectively).
Fig. 4.
A1 inhibits HSV-1 infection. (A) Western blotting of cells stably transfected with HA-tagged wild-type, E63A mutant, and control A1 proteins were probed with an anti-HA antibody for A1 expression or an antitubulin antibody as an internal standard. (B) Immunofluorescence staining of HA-tagged A1WT-, A1E63A-, or A1Ctrl-expressing RSCs with an anti-HA antibody (green). The nucleus was stained with Hoechst 33342 dye (blue). (C) Longitudinal analysis of GFP-expressing HSV-1 infection in cells stably transfected with wild-type A1, E63A mutant, or control A1. **, P < 0.05; ***, P < 0.005. (D) Percentage of GFP-positive cells in HA-tagged A1WT-expressing RSCs stably expressing A1- or luciferase-targeted shRNAs 48 h after YK333 infection. ***, P < 0.005 compared to cells treated with luciferase-targeted shRNA (shLuc). A Western blot was probed with an anti-HA antibody for the expression of A1 and antitubulin as an internal standard. (E) Level of PFU in the supernatant of A1Ctrl-, A1WT-, and A1E63A-expressing RSCs 48 h after infection by YK333 at various MOIs. *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
We next proceeded to assess the potential antiviral activity of A1 against HSV-1 by infecting A1WT- or A1E63A-expressing RSCs with GFP-expressing HSV-1 at a multiplicity of infection (MOI) of 0.01. As shown in Fig. 4C, the level of GFP expression was significantly attenuated in A1WT-expressing RSCs compared to RSCs expressing A1E63A and A1Ctrl at 48 h postinfection (hpi). Interestingly, A1E63A-expressing cells also had a modest inhibitory effect on HSV-1, although this activity did not appear to be as potent as that in wild-type-expressing RSCs, suggesting a somewhat deaminase-independent inhibition mechanism (Fig. 4C). When two different shRNAs were used to specifically target A1WT, the level of infectivity of HSV-1 was restored and comparable to that observed with A1Ctrl (Fig. 4D). Overall, the low and high levels of A1 protein expression resulting from specific shRNAs (A1/736 and A1/1119) against A1 and control luciferase, respectively, correlated inversely with HSV-1 infectivity (Fig. 4D) and suggested that A1 expression is essential for the inhibition of HSV-1 infection.
In order to analyze the level of attenuation of HSV-1 replication, we looked at the virus titers of supernatants taken from A1Ctrl-, A1WT-, and AE63A-expressing cells infected at higher MOIs. The virus titer of supernatants from HSV-1-infected A1WT-expressing cells was significantly attenuated compared to that of supernatants from empty-vector-expressing cells or cells stably expressing A1E63A (Fig. 4E). Similarly to what was observed for the inhibition of HSV-1 infection and GFP expression described above, A1E63A also inhibited HSV-1 titers in a statistically significant fashion (Fig. 4E). However, cytidine deaminase activity was crucial for potent inhibition to take place. Taken together, HSV-1 replication was being significantly inhibited by A1 in a deaminase-dependent and -independent manner.
A1 inhibits HSV-1 gene transcription and DNA replication.
HSV-1 gene expression is highly ordered and occurs in a cascade-dependent manner, which is divided into three stages, with the transcription products subdivided into 3 broad groups termed α, β, and γ, also known as the immediate-early (IE), early (E), and late (L) genes, respectively (27). The α genes, produced soon after infection, promote β gene transcription and increase viral protein synthesis. Thereafter, β genes are transcribed and enhance DNA replication, which, in turn, signals γ gene transcription for the production of viral proteins necessary for virion assembly (12).
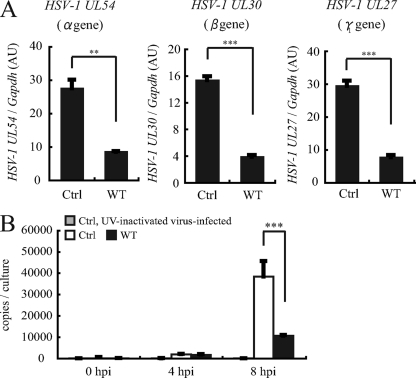
To better understand the mechanism by which A1 inhibits HSV-1 replication, we first examined the expression levels of representative genes from the α, β, and γ groups. UL54, an α gene that plays a role in the shutoff of host protein synthesis and also enhances viral gene expression; UL30, a β gene and the DNA polymerase responsible for DNA replication; and UL27, a γ gene and envelope glycoprotein, were analyzed by real-time PCR, and their expression levels were normalized to GAPDH levels. As can be seen from the expression level of HSV-1 cDNA extracted from cells stably expressing A1Ctrl and A1WT at 8 hpi at an MOI of 0.01, the levels of all three genes analyzed were significantly lower in A1WT-expressing cells than in A1Ctrl-expressing cells (Fig. 5A).
Fig. 5.
Inhibition of HSV-1 mRNA and DNA. (A) HSV-1 UL54, UL30, and UL27 gene expression levels were measured by real-time PCR and normalized to GAPDH levels. Nucleic acids were extracted from HSV-1-infected wild-type A1-expressing and control RSCs at 8 hpi. (B) DNA copy number was measured by real-time PCR in A1-expressing and control RSCs 4 and 8 hpi. As a control, UV-inactivated virus was also used to infect control RSCs. **, P < 0.05; ***, P < 0.005.
Given that α gene synthesis precedes DNA replication, we expected that the observed low level of expression of the immediate-early genes would affect HSV-1 DNA replication and, thus, the viral DNA copy number. Indeed, at 8 hpi, the HSV-1 DNA copy number was nearly 4 times lower in the A1WT-expressing cells than in the A1Ctrl-expressing cells (Fig. 5B). As an additional control, the infection of control RSCs with a UV-inactivated virus did not result in viral DNA replication (Fig. 5B). Altogether, our results indicated that A1 affected viral gene expression and also DNA replication.
A1 induces mutations in HSV-1 DNA.
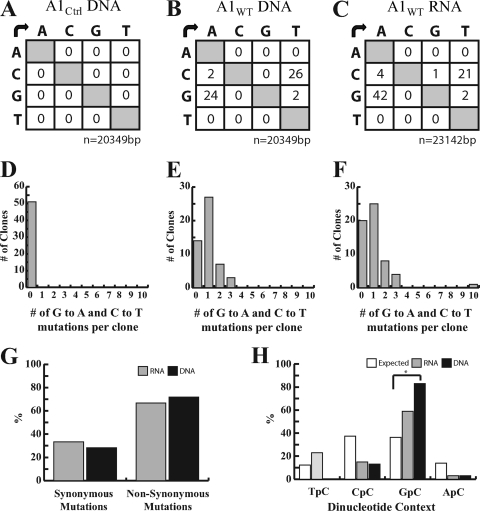
A1 homologues are putative inducers of mutations in retrovirus DNA and also in DNA of A1-expressing bacteria (8, 24, 29, 44, 49). Thus, to investigate the mechanism of the A1-dependent inhibition of HSV-1 gene expression, we looked at the potential editing of HSV-1 viral DNA. The UL54 DNA extracted from HSV-1-infected A1WT-expressing RSCs showed evidence of both G→A and C→T transitions, indicating that plus and minus strands were being edited (Fig. 6B). The majority of the 51 clones analyzed from A1WT-expressing RSCs contained 1 mutation within the sequenced region; however, a few clones harbored 2 or 3 mutations (Fig. 6E). On the other hand, A1Ctrl-derived clones showed no signs of mutations (Fig. 6A and D), suggesting that the observed nucleotide changes were induced by A1.
Fig. 6.
Sequencing analysis of UL54 DNA and cDNA from A1WT- and A1Ctrl-expressing HSV-1-infected RSCs. A 399-bp region of UL54 DNA or cDNA, extracted at 8 hpi, was amplified by nested PCR and cloned for sequencing. (A to C) Mutation matrices for DNA extracted from A1Ctrl-expressing RSCs (51 clones) (A), DNA extracted from A1WT-expressing RSCs (51 clones) (B), and cDNA extracted from A1WT-expressing RSCs (58 clones). (D to F) Numbers of clones harboring G-to-A and C-to-T transitions were assessed and are represented for DNA extracted from A1Ctrl-expressing RSCs (D), DNA extracted from A1WT-expressing RSCs (E), and cDNA extracted from A1WT-expressing RSCs (F). (G) Mutations from UL54 cDNA and DNA were narrowed down to 39 and 30 commonly mutated positions, respectively. The percentages of synonymous or nonsynonymous mutations are depicted in the bar graph. (H) Rat A1 had a tendency toward a dinucleotide context for 5′GpC in viral UL54 DNA and cDNA and was statistically significant for the DNA. A χ2 analysis showed that compared to expected values, 5′GpC was significantly favored on UL54 DNA, as indicated by the asterisk (P < 0.001).
Given that rat A1 is also known to edit retroviral RNA, we looked at cDNA extracted from A1WT-expressing RSCs at 8 hpi at an MOI of 0.01 for evidence of additional deaminase activities (8). Similar to the DNA sequencing results, G→A and C→T transitions in viral cDNA were observed, with the majority of clones containing 1 mutation, while clones with 2 or 3 mutations were also observed (Fig. 6C and F). Interestingly, 1 clone contained 10 G→A mutations, suggesting that a second round of editing may be occurring on mRNA transcribed from the mutated DNA (Fig. 6F), yet the main editing appeared to be occurring on the viral DNA, which was mirrored by the mutations observed for viral cDNA (Fig. 6B and C). Looking at the types of mutations arising from 39 commonly mutated positions in UL54 cDNA and 30 commonly mutated positions in UL54 DNA, approximately 70% of the mutations were found to lead to nonsynonymous substitutions in the analyzed gene (Fig. 6G).
APOBECs such as A3G are known to have an editing preference for 5′CpC and 5′TpC sequences, and we attempted to assess the dinucleotide preference of rat A1 (59). As can be seen in Fig. 6H, we found the preferred context to be 5′GpC on both viral DNA and RNA (Fig. 6H), although previous studies indicated a tendency of rodent A1 to prefer 5′TpC (11, 29, 49). Altogether, these results indicate that rat A1 acts directly on DNA for editing.
A1 inhibits HSV-1 transcription independently of viral mRNA editing.
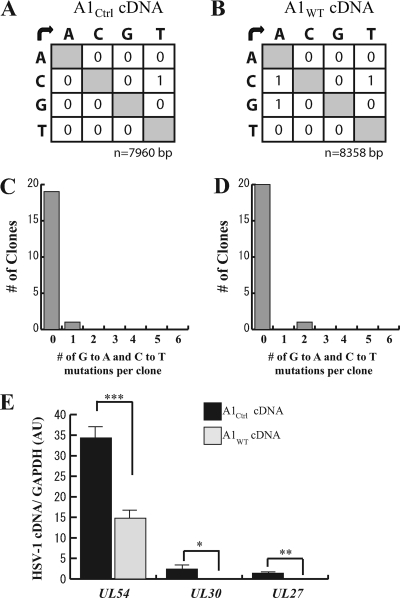
To further elucidate the mechanism by which A1 inhibits HSV-1 infection, we focused on the initial transcription of viral genes in the presence of the nucleoside analogue inhibitor acyclovir, which effectively inhibits nascent DNA synthesis. If A1 acts mainly on ssDNA substrates for mutagenesis, then we expected to see a reduced deamination of HSV-1 cDNA during transcription and few clones harboring signature A1-mediated mutations. At an MOI of 1, the sequencing of UL54 mRNA extracted at 2 hpi revealed a low frequency of C→U and G→A transitions in samples taken from A1WT-expressing cells similar to that of nucleic acids obtained from A1Ctrl-expressing cells, further indicating that A1 is acting on DNA and not mRNA (Fig. 7A and B). In addition, only 1 clone containing these mutations was observed for each A1Ctrl- and A1WT-expressing RSC (Fig. 7C and D). However, when we looked at the levels of viral gene transcripts at this time point, α, β, and γ gene expression levels were still significantly lower in A1WT- than in A1Ctrl-expressing RSCs (Fig. 7E). Altogether, these data suggest that although A1 did not induce mutations in viral RNA, its association with ssDNA may serve to block and inhibit transcription.
Fig. 7.
A1 inhibition of HSV-1 RNA transcripts is independent of RNA mutagenesis. A1WT- and A1Ctrl-expressing RSCs were infected with YK333 at an MOI of 1 for 2 h in the presence of 25 μg/ml of acyclovir. (A and B) Mutation matrices for UL54 cDNA extracted from A1Ctrl-expressing RSCs (A) and A1WT-expressing RSCs (B). (C and D) Numbers of clones harboring G-to-A and C-to-T transitions from cDNA extracted from A1Ctrl-expressing RSCs (20 colonies) (C) and cDNA extracted from A1WT-expressing RSCs (21 colonies) were assessed and are represented in the bar graphs. (E) Levels of HSV-1 mRNA for UL54, UL30, and UL27 in A1Ctrl- and A1WT-expressing RSCs were measured by real-time PCR and normalized to GAPDH levels. *, P < 0.05; **, P < 0.0005; ***, P < 0.00005.
DISCUSSION
From our findings, we propose a novel role for A1 as an inhibitor of HSV-1 infection in the context of HSE in rats. Although the induction mechanism remains to be elucidated in future studies, we posit that after HSV-1 infection, the expression of A1 in neuronal cells inhibits the early stages of the virus life cycle. The molecule may edit viral DNA in the nucleus and/or serve as a physical blockade by binding to DNA during transcription, resulting in reduced expression and dysfunction of the edited viral genes. The stalling of the HSV-1 gene cascade may prevent the virus from ramping up to full-scale production and allow for a controlled immune response.
We found that both viral DNA and RNA harbored G→A transitions; however, DNA was the main target of A1 during HSV-1 infection (Fig. 6B and C). Furthermore, when DNA synthesis was blocked, cDNA isolated from virus-infected cells displayed few mutations, further reinforcing the assumption that A1 is acting on a DNA substrate (Fig. 7A and B). HSV-1 DNA is known to replicate within the nucleus, and while A1 has been reported to shuttle between the cytosol and the nucleus in the presence of APOBEC1 complementation factor (ACF) for a nuclear distribution, if ACF is mutated or absent, then A1 will have a primarily cytoplasmic localization (9, 25). Here, we show a similar distribution of A1 in the cytosol in vivo in HSV-1-infected neurons and in vitro in A1-expressing RSCs (Fig. 3B and 4B). However, because we observed mutations in HSV-1 DNA in vitro (Fig. 6B), this suggests that a small pool of A1 also exists in the nucleus and is involved in the direct interaction with and mutation of genomic viral DNA.
Although we obtained evidence of cytidine deaminase-dependent mutations in HSV-1 nucleic acids, it is possible that our observed results could be an underestimate of the actual degree of mutagenesis that is taking place inside A1-expressing cells. HSV encodes a uracil DNA glycosylase, which is able to remove uracil bases from DNA, allowing the subsequent repair of the damaged DNA (32, 38, 63). This activity may be masking the true extent of deamination taking place by A1.
It is worth noting that the editing of HSV-1 DNA alone may not completely account for the inhibition of HSV-1. We found 33% of mutated positions in the sequenced region of UL54 DNA and 28% of UL54 RNA to be synonymous mutations, meaning that their function would remain intact (Fig. 6G). Furthermore, mutations alone do not account entirely for the observed inhibition of α gene transcription (Fig. 5A), since the transcription of these genes is highly dependent on the incoming tegument protein VP16, which acts as a transactivator (1, 2). Therefore, it is possible that the editing of nucleic acids by A1 does not play the major antiviral role against HSV-1. Instead, an alternative explanation for the mechanism may be that A1 is inhibiting gene transcription through an undefined mechanism similar to the A3G deaminase-independent inhibition of HIV-1 reverse transcription products, perhaps through a physical block of transcription or DNA replication (6, 18, 19, 41, 57). Supporting this assumption are data from our deaminase-deficient mutant AE63A, which showed a partial inhibition of HSV-1 infection although not to the same extent as that of the A1WT protein (Fig. 4C and E). Moreover, diminished HSV-1 transcript levels were observed for A1WT-expressing RSCs when DNA synthesis was inhibited, even in the absence of RNA cytidine deamination (Fig. 7E). However, because of the presence of repair mechanisms encoded by HSV-1, such as the uracil DNA glycosylase described above, we were not able to rule out the possibility that viral ssDNA is mutated during transcription.
If mutated viral transcripts are synthesized and make their way to the cytoplasm for translation, we suspect that the functionality of the synthesized viral protein may be affected. As mentioned above, the gene expression of HSV-1 is tightly regulated, and defects in certain genes can have detrimental effects on the virus life cycle. For instance, studies of temperature-sensitive HSV-1 mutants revealed that defects in the IE genes ICP4 and UL54 render the virus replication deficient (16, 47, 51). Furthermore, IE genes have pleiotropic functions as transcriptional activators of viral genes and suppressors of host protein synthesis. In the case of UL54, other functions include the inhibition of host mRNA splicing, an increase in viral mRNA levels, and the export of viral mRNA from the nucleus (10, 21, 22, 30, 52, 53). Given that HSV-1 proteins have essential regulatory roles, it is conceivable that disruptions of these genes can cause a stall in the virus replication cycle. Our results also support this possibility, as evidenced by the reduced HSV-1 mRNA and DNA levels, GFP expression, and virus titers in infected A1-expressing RSCs (Fig. 4C and E and 5A and B, respectively). This effect was dependent on A1 expression, as the silencing of A1 by specific shRNAs restored HSV-1 infection (Fig. 4D).
Our in vitro results raise the issue of whether the expression of A1 induced in rat brains during HSE aids in the inhibition of virus infection or not. Rat A1 is ubiquitously expressed in virtually all tissues, such as the spleen and liver, and while A1 functions within an editing complex in the small intestine for the deamination of apolipoprotein B mRNA, it is also expressed in tissues where apolipoprotein B mRNA is not present, calling into question whether there is an additional role for this enzyme (26). Indications that A1 also has antiviral activity on top of its already known physiological function have been shown by mouse models of infection with either MLV or HBV. Splenocytes taken from MLV-infected mice or hepatocytes taken from HBV-infected transgenic mice both displayed signs of an A1-specific editing of viral genomes, indicating a direct role in virus inhibition (45, 49). From our in vivo findings, recovered rats and rats with mild symptoms showed an increase in A1 mRNA expression levels, which was not observed for mock-infected rats (Fig. 3A and B). Moreover, rat brain tissue infected by HSV-1 revealed that the induction of A1 occurred predominantly in HSV-1-infected neuronal cells, which suggests a possible antiviral function for this molecule in the brain.
The consequence of the increased cytidine deamination of the HSV-1 genome for virus infectivity has been exemplified by a previous report showing that HSV-1 mutants containing a defective uracil DNA glycosylase were 100,000 times less neuroinvasive in mice intracranially inoculated with HSV-1 than their wild-type counterparts (48). On the other hand, uracil DNA glycosylase-deficient HSV-1 did not have issues replicating in vitro, suggesting that this enzyme is necessary for virus infectivity in rodent neurons because HSV-1 is more prone to cytidine deamination in these cells (48). It is tempting to speculate that A1 in rats may also be playing a role in mutating the HSV-1 DNA in vivo to inhibit HSV-1 infection in the brain in our model.
A1 may also help to reduce HSE in vivo by slowing down virus dissemination long enough to allow an appropriate immune response to be mounted, thus eliminating the virus without causing excessive damage from infiltrating macrophages during encephalitis or lytic damage resulting from massive viral infection. In fact, it was speculated previously that an overzealous immune response may be causing more harm than good in the CNS (13). For instance, the depletion of macrophages during HSV-1 infection in mice was demonstrated previously to lead to a higher survival rate (34). On the other hand, uncontrolled virus dissemination can also lead to a fatal outcome in mice missing the type I interferon receptor (14, 33). Needless to say, a balance must be struck between the host immune response and the extent of virus infection. It is plausible that A1 helps to achieve both of these feats by inhibiting virus infection, resulting in a toned-down immune response, as was shown in vitro (Fig. 4C and E). Here, we observed diminished leukocyte infiltration in surviving rats compared to that in rats with mild and severe symptoms, which died within 5 to 7 days after infection in our in vivo model (Fig. 2A). Moreover, the sizes of the HSV-1-infected areas diminished over time in surviving rats, suggesting the containment of virus dissemination (Fig. 2C).
Given that rat A1 has the potential to inhibit HSV-1 replication, it would be intriguing to investigate whether human A1 plays a role during encephalitis in humans. However, we believe that this is unlikely, since the expression profile of human A1 is quite distinct from that of rodent A1. Rat A1 has at least three clusters of different transcriptional promoter sites within the gene giving rise to its diverse tissue expression, whereas human A1 appears to be confined to the small intestine, with no evidence of its expression in other tissues (20, 26). Nonetheless, it will be interesting to explore whether human A1 is expressed in neurons during HSV-1 infection.
In conclusion, we identified rat A1 as a novel anti-HSV-1 molecule induced during virus infection in rat brains. The mechanism by which A1 inhibits virus replication appears to be through the interference of the HSV-1 gene cascade in a predominantly deaminase-dependent manner involving the editing of viral DNA. In order to assess the extent of the A1 inhibition of HSV-1 infection during encephalitis, future studies using knockout animals may provide insights into the contribution of A1 to the control of HSE.
Supplementary Material
ACKNOWLEDGMENTS
We thank Atsushi Koito, Youichi Suzuki, and Kei Sato for their generous support of our study and Bernard Roizman for providing us with RSCs.
This work was supported in part by a grant-in-aid for scientific research (S and B) and a grant-in-aid for young scientists (B) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan and a Research on HIV/AIDS grant (200932025A to Y.K.) from the Ministry of Health, Labor, and Welfare of Japan; P.G. was supported by the foreign student program of the MEXT.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 20 July 2010.
REFERENCES
- 1. Ace C. I., Dalrymple M. A., Ramsay F. H., Preston V. G., Preston C. M. 1988. Mutational analysis of the herpes simplex virus type 1 trans-inducing factor Vmw65. J. Gen. Virol. 69(Pt. 10):2595–2605 [DOI] [PubMed] [Google Scholar]
- 2. Ace C. I., McKee T. A., Ryan J. M., Cameron J. M., Preston C. M. 1989. Construction and characterization of a herpes simplex virus type 1 mutant unable to transinduce immediate-early gene expression. J. Virol. 63:2260–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alce T. M., Popik W. 2004. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J. Biol. Chem. 279:34083–34086 [DOI] [PubMed] [Google Scholar]
- 4. Ando Y., Kitayama H., Kawaguchi Y., Koyanagi Y. 2008. Primary target cells of herpes simplex virus type 1 in the hippocampus. Microbes Infect. 10:1514–1523 [DOI] [PubMed] [Google Scholar]
- 5. Bergstrom T., Svennerholm B., Conradi N., Horal P., Vahlne A. 1991. Discrimination of herpes simplex virus types 1 and 2 cerebral infections in a rat model. Acta Neuropathol. 82:395–401 [DOI] [PubMed] [Google Scholar]
- 6. Bishop K. N., Holmes R. K., Malim M. H. 2006. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 80:8450–8458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bishop K. N., et al. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14:1392–1396 [DOI] [PubMed] [Google Scholar]
- 8. Bishop K. N., Holmes R. K., Sheehy A. M., Malim M. H. 2004. APOBEC-mediated editing of viral RNA. Science 305:645. [DOI] [PubMed] [Google Scholar]
- 9. Blanc V., Henderson J. O., Kennedy S., Davidson N. O. 2001. Mutagenesis of apobec-1 complementation factor reveals distinct domains that modulate RNA binding, protein-protein interaction with apobec-1, and complementation of C to U RNA-editing activity. J. Biol. Chem. 276:46386–46393 [DOI] [PubMed] [Google Scholar]
- 10. Chen I. H., Sciabica K. S., Sandri-Goldin R. M. 2002. ICP27 interacts with the RNA export factor Aly/REF to direct herpes simplex virus type 1 intronless mRNAs to the TAP export pathway. J. Virol. 76:12877–12889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen Z., et al. 2010. Hypermutation induced by APOBEC-1 overexpression can be eliminated. RNA 16:1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Conley A. J., Knipe D. M., Jones P. C., Roizman B. 1981. Molecular genetics of herpes simplex virus. VII. Characterization of a temperature-sensitive mutant produced by in vitro mutagenesis and defective in DNA synthesis and accumulation of gamma polypeptides. J. Virol. 37:191–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Conrady C. D., Drevets D. A., Carr D. J. 2010. Herpes simplex type I (HSV-1) infection of the nervous system: is an immune response a good thing? J. Neuroimmunol. 220:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Conrady C. D., Thapa M., Wuest T., Carr D. J. 2009. Loss of mandibular lymph node integrity is associated with an increase in sensitivity to HSV-1 infection in CD118-deficient mice. J. Immunol. 182:3678–3687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Crawford J. P., Percy D. H., Hatch L. A. 1979. Experimental encephalitis in the newborn rat due to herpes simplex virus type 2. Exp. Mol. Pathol. 31:44–55 [DOI] [PubMed] [Google Scholar]
- 16. DeLuca N. A., Courtney M. A., Schaffer P. A. 1984. Temperature-sensitive mutants in herpes simplex virus type 1 ICP4 permissive for early gene expression. J. Virol. 52:767–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Driscoll D. M., Zhang Q. 1994. Expression and characterization of p27, the catalytic subunit of the apolipoprotein B mRNA editing enzyme. J. Biol. Chem. 269:19843–19847 [PubMed] [Google Scholar]
- 18. Gaddis N. C., Chertova E., Sheehy A. M., Henderson L. E., Malim M. H. 2003. Comprehensive investigation of the molecular defect in vif-deficient human immunodeficiency virus type 1 virions. J. Virol. 77:5810–5820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guo F., Cen S., Niu M., Saadatmand J., Kleiman L. 2006. Inhibition of formula-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J. Virol. 80:11710–11722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hadjiagapiou C., Giannoni F., Funahashi T., Skarosi S. F., Davidson N. O. 1994. Molecular cloning of a human small intestinal apolipoprotein B mRNA editing protein. Nucleic Acids Res. 22:1874–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hardwicke M. A., Sandri-Goldin R. M. 1994. The herpes simplex virus regulatory protein ICP27 contributes to the decrease in cellular mRNA levels during infection. J. Virol. 68:4797–4810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hardy W. R., Sandri-Goldin R. M. 1994. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J. Virol. 68:7790–7799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harris R. S., et al. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803–809 [DOI] [PubMed] [Google Scholar]
- 24. Harris R. S., Petersen-Mahrt S. K., Neuberger M. S. 2002. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol. Cell 10:1247–1253 [DOI] [PubMed] [Google Scholar]
- 25. Henderson J. O., Blanc V., Davidson N. O. 2001. Isolation, characterization and developmental regulation of the human apobec-1 complementation factor (ACF) gene. Biochim. Biophys. Acta 1522:22–30 [DOI] [PubMed] [Google Scholar]
- 26. Hirano K., Min J., Funahashi T., Davidson N. O. 1997. Cloning and characterization of the rat apobec-1 gene: a comparative analysis of gene structure and promoter usage in rat and mouse. J. Lipid Res. 38:1103–1119 [PubMed] [Google Scholar]
- 27. Honess R. W., Roizman B. 1974. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J. Virol. 14:8–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huthoff H., Malim M. H. 2007. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and virion encapsidation. J. Virol. 81:3807–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ikeda T., et al. 2008. The antiretroviral potency of APOBEC1 deaminase from small animal species. Nucleic Acids Res. 36:6859–6871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jean S., LeVan K. M., Song B., Levine M., Knipe D. M. 2001. Herpes simplex virus 1 ICP27 is required for transcription of two viral late (gamma 2) genes in infected cells. Virology 283:273–284 [DOI] [PubMed] [Google Scholar]
- 31. Kitayama H., Miura Y., Ando Y., Koyanagi Y. 2008. Human immunodeficiency virus type-1 vulnerates nascent neuronal cells. Microbiol. Immunol. 52:78–88 [DOI] [PubMed] [Google Scholar]
- 32. Lindahl T. 1979. DNA glycosylases, endonucleases for apurinic/apyrimidinic sites, and base excision-repair. Prog. Nucleic Acid Res. Mol. Biol. 22:135–192 [DOI] [PubMed] [Google Scholar]
- 33. Luker G. D., Prior J. L., Song J., Pica C. M., Leib D. A. 2003. Bioluminescence imaging reveals systemic dissemination of herpes simplex virus type 1 in the absence of interferon receptors. J. Virol. 77:11082–11093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lundberg P., et al. 2008. The immune response to herpes simplex virus type 1 infection in susceptible mice is a major cause of central nervous system pathology resulting in fatal encephalitis. J. Virol. 82:7078–7088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mahieux R., et al. 2005. Extensive editing of a small fraction of human T-cell leukemia virus type 1 genomes by four APOBEC3 cytidine deaminases. J. Gen. Virol. 86:2489–2494 [DOI] [PubMed] [Google Scholar]
- 36. Mangeat B., et al. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99–103 [DOI] [PubMed] [Google Scholar]
- 37. Mehta A., Driscoll D. M. 1998. A sequence-specific RNA-binding protein complements apobec-1 to edit apolipoprotein B mRNA. Mol. Cell. Biol. 18:4426–4432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mullaney J., Moss H. W., McGeoch D. J. 1989. Gene UL2 of herpes simplex virus type 1 encodes a uracil-DNA glycosylase. J. Gen. Virol. 70(Pt. 2):449–454 [DOI] [PubMed] [Google Scholar]
- 39. Navaratnam N., et al. 1995. Evolutionary origins of apoB mRNA editing: catalysis by a cytidine deaminase that has acquired a novel RNA-binding motif at its active site. Cell 81:187–195 [DOI] [PubMed] [Google Scholar]
- 40. Navaratnam N., et al. 1993. The p27 catalytic subunit of the apolipoprotein B mRNA editing enzyme is a cytidine deaminase. J. Biol. Chem. 268:20709–20712 [PubMed] [Google Scholar]
- 41. Newman E. N., et al. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15:166–170 [DOI] [PubMed] [Google Scholar]
- 42. Noguchi C., et al. 2005. G to A hypermutation of hepatitis B virus. Hepatology 41:626–633 [DOI] [PubMed] [Google Scholar]
- 43. Okeoma C. M., Lovsin N., Peterlin B. M., Ross S. R. 2007. APOBEC3 inhibits mouse mammary tumour virus replication in vivo. Nature 445:927–930 [DOI] [PubMed] [Google Scholar]
- 44. Petersen-Mahrt S. K., Neuberger M. S. 2003. In vitro deamination of cytosine to uracil in single-stranded DNA by apolipoprotein B editing complex catalytic subunit 1 (APOBEC1). J. Biol. Chem. 278:19583–19586 [DOI] [PubMed] [Google Scholar]
- 45. Petit V., et al. 2009. Murine APOBEC1 is a powerful mutator of retroviral and cellular RNA in vitro and in vivo. J. Mol. Biol. 385:65–78 [DOI] [PubMed] [Google Scholar]
- 46. Powell L. M., et al. 1987. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell 50:831–840 [DOI] [PubMed] [Google Scholar]
- 47. Preston C. M. 1979. Control of herpes simplex virus type 1 mRNA synthesis in cells infected with wild-type virus or the temperature-sensitive mutant tsK. J. Virol. 29:275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pyles R. B., Thompson R. L. 1994. Evidence that the herpes simplex virus type 1 uracil DNA glycosylase is required for efficient viral replication and latency in the murine nervous system. J. Virol. 68:4963–4972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Renard M., Henry M., Guetard D., Vartanian J. P., Wain-Hobson S. 2010. APOBEC1 and APOBEC3 cytidine deaminases as restriction factors for hepadnaviral genomes in non-humans in vivo. J. Mol. Biol. 400:323–334 [DOI] [PubMed] [Google Scholar]
- 50. Rosler C., et al. 2005. APOBEC-mediated interference with hepadnavirus production. Hepatology 42:301–309 [DOI] [PubMed] [Google Scholar]
- 51. Sacks W. R., Greene C. C., Aschman D. P., Schaffer P. A. 1985. Herpes simplex virus type 1 ICP27 is an essential regulatory protein. J. Virol. 55:796–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sandri-Goldin R. M. 1998. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 12:868–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sandri-Goldin R. M. 1994. Properties of an HSV-1 regulatory protein that appears to impair host cell splicing. Infect. Agents Dis. 3:59–67 [PubMed] [Google Scholar]
- 54. Sato K., et al. 2010. Remarkable lethal G-to-A mutations in vif-proficient HIV-1 provirus by individual APOBEC3 proteins in humanized mice. J. Virol. 84:9546–9556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schafer A., Bogerd H. P., Cullen B. R. 2004. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology 328:163–168 [DOI] [PubMed] [Google Scholar]
- 56. Sheehy A. M., Gaddis N. C., Choi J. D., Malim M. H. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646–650 [DOI] [PubMed] [Google Scholar]
- 57. Shindo K., et al. 2003. The enzymatic activity of CEM15/Apobec-3G is essential for the regulation of the infectivity of HIV-1 virion but not a sole determinant of its antiviral activity. J. Biol. Chem. 278:44412–44416 [DOI] [PubMed] [Google Scholar]
- 58. Suspene R., et al. 2005. Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc. Natl. Acad. Sci. U. S. A. 102:8321–8326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Suspene R., et al. 2004. APOBEC3G is a single-stranded DNA cytidine deaminase and functions independently of HIV reverse transcriptase. Nucleic Acids Res. 32:2421–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tanaka M., Kodaira H., Nishiyama Y., Sata T., Kawaguchi Y. 2004. Construction of recombinant herpes simplex virus type I expressing green fluorescent protein without loss of any viral genes. Microbes Infect. 6:485–493 [DOI] [PubMed] [Google Scholar]
- 61. Teng B., Burant C. F., Davidson N. O. 1993. Molecular cloning of an apolipoprotein B mRNA editing protein. Science 260:1816–1819 [DOI] [PubMed] [Google Scholar]
- 62. Whitley R. J. 2006. Herpes simplex encephalitis: adolescents and adults. Antiviral Res. 71:141–148 [DOI] [PubMed] [Google Scholar]
- 63. Worrad D. M., Caradonna S. 1988. Identification of the coding sequence for herpes simplex virus uracil-DNA glycosylase. J. Virol. 62:4774–4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yu Q., et al. 2004. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 11:435–442 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.