Abstract
The recombination rate in Newcastle disease virus (NDV) was as high as 10% in RDP analysis with full-length NDV genome sequences available in GenBank. We found that two NDV strains, China/Guangxi09/2003 and NDV/03/018, previously reported as recombinants, failed to show any evidence of recombination upon complete genome resequencing. Furthermore, we were able to reproduce artificial recombination by amplification of the M gene in a mixed sample of strains LaSota and ZJ1. It appears that the recombination of NDV is not as common as has been reported. NDV sequences in GenBank should be analyzed with caution during bioinformatic analyses for natural recombination events.
TEXT
Newcastle disease (ND) is one of the most devastating diseases in poultry. The causative agent, Newcastle disease virus (NDV), is a member of the Avulavirus genus in the Paramyxoviridae family (2, 9, 11, 24). The NDV genome consists of approximately 15-kb-long nonsegmented single-stranded negative-sense RNA that codes for six proteins, including nucleoprotein (NP), phosphoprotein (P), matrix (M) protein, fusion (F) protein, hemagglutinin neuraminidase (HN), and polymerase protein (L) (2, 6, 28). Although NDV has only one serotype, substantial antigenic and genetic diversity have been previously recognized (2, 19). According to earlier reports, at least 10 genotypes (genotypes I to X) have been described for NDVs (4, 29).
The main dynamics of evolution in nonsegmented RNA viruses either are due to the inherent error rate of the RNA-dependent RNA polymerase or occur as a result of recombination (30). While polymerase error is believed to be the main driving force for NDV evolution (30), it has been established that recombination in nonsegmented negative-sense RNA viruses, including NDV, is rare (1, 7). Indeed, during 2005 to 2009, we characterized more than 100 NDV strains in our laboratory (20, 43), but no recombinant strains were detected on the basis of the analysis of F and HN gene sequences. However, in recent years, more and more recombination events have been reported for NDVs, with the recombination occurring throughout the whole genome (7, 8, 15, 30, 36, 47, 44, 46). Han et al. and Zhang et al. even reported recombinants of NDVs that involved multiple genotypes (15, 46).
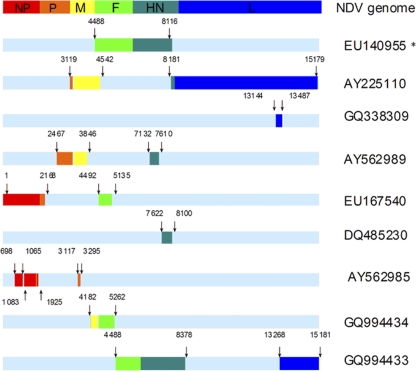
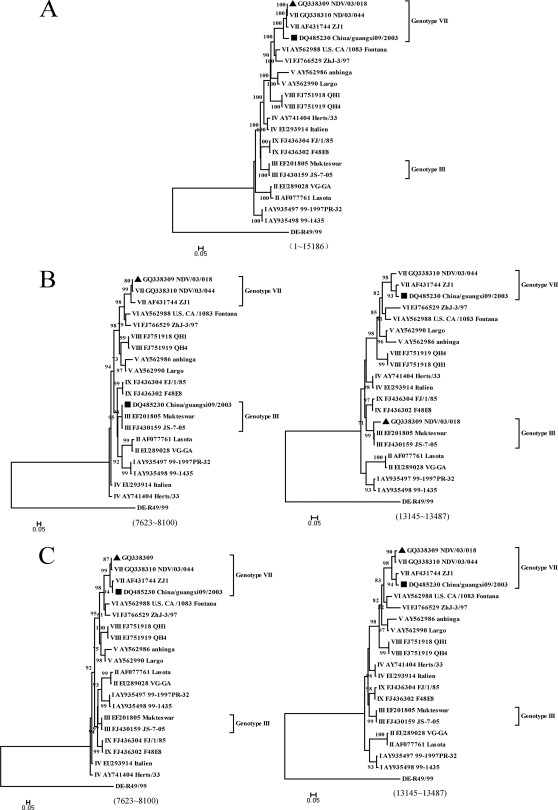
This controversy has prompted us to investigate whether the recombination events in NDVs are as common as has been reported. Eighty complete genomic sequences of NDV retrieved from GenBank (Table 1 ) were edited using BioEdit version 7.0.0 and aligned using ClustalX version 1.83 software (41). To detect recombination events over the whole genome of NDVs, we used different statistical methods included in the RDP3.42 software package (17): RDP (21), Geneconv (33), Bootscan (22), Maxchi (40), Chimaera (35), SiScan (12) and 3Seq (5). As different algorithms might not be completely consistent with each other, any breakpoint supported by five or more methods with P values ≤ 10−5 was set as a positive recombination signature. KBNP, a chimeric vaccine strain (GenBank accession number EU140955), was used as a control to evaluate the prediction capability of the program. Our analysis indicated a recombination rate as high as 10%. There are at least 8 recombinants in the 80 NDVs, involving 15 recombination events (Fig. 1). These events were detected throughout the whole genome but more frequently in M, F, and HN genes. Moreover, 5 of the 8 recombinants were involved in multirecombination events. Interestingly, these recombinants were mainly isolated in China and the recombination events were concentrated in viruses of genotypes I, II, III (three genotypes of viruses widely used as vaccines), and VII (a predominant genotype currently circulating worldwide). Among these recombinants, virus strain China/Guangxi09/2003 (DQ485230) was a putative daughter virus of genotype VII virus FWM (GU564399) and genotype III virus Mukteswar (EF201805), as detected by RDP (P = 2.528 × 10−70), Geneconv (1.478 × 10−68), BootScan (1.730 × 10−70), MaxChi (2.539 × 10−13), Chimaera (1.628 × 10−13), and SiScan (2.267 × 10−17). Another virus, NDV/03/018 (GQ338309), appeared to have arisen from a recombination event between genotype VII virus NDV/03/044 (GQ338310) and genotype III virus JS-7-05 (FJ430159), as identified accordingly to analysis by RDP (P = 1.862 × 10−35), Geneconv (3.859 × 10−21), Bootscan (7.608 × 10−35), MaxChi (1.044 × 10−08), Chimaera (3.401 × 10−07), and SiScan (3.344 × 10−08). The maximum-likelihood (ML) trees were constructed using PhyML and the GTR+I+G model and selected by jModelTest (13, 14, 34), and the approximate-likelihood-ratio test values were examined for branch support (3, 14). The tree based on the full-length genomes indicated that these two potential recombinants were affiliated with genotype VII (Fig. 2 A), while the putative recombination regions clustered into genotype III (Fig. 2B).
Table 1.
Sequences of NDVs used in this study
| Accession no. | Strain | Country | Genotype | Referencea |
|---|---|---|---|---|
| AY562991 | Ulster-67 | Ireland | I | |
| AY935489 | 01-1108 | AUS | I | 18a |
| AY935490 | 02-1334 | AUS | I | 18a |
| AY935491 | 98-1154 | AUS | I | 18a |
| AY935492 | 98-1249 | AUS | I | 18a |
| AY935493 | 98-1252 | AUS | I | 18a |
| AY935494 | 99-0655 | AUS | I | 18a |
| AY935496 | 99-0868lo | AUS | I | 18a |
| AY935497 | 99-1997PR-32 | AUS | I | 18a |
| AY935498 | 99-1435 | AUS | I | 18a |
| AY935500 | I-2 progenitor | AUS | I | 18a |
| DQ097394 | PHY-LMV42 | Germany | I | 8a |
| GQ918280 | BHG | Sweden | I | 30b |
| HM063422 | D3 | China | I | |
| HM063424 | R8 | China | I | |
| HM125898 | WDK/JX/7793/2004 | China | I | |
| AF077761 | La Sota | II | 9 | |
| AF309418 | B1 | USA | II | |
| AF375823 | B1 isolate Takaaki | USA | II | 30c |
| AY225110 | HB94 isolate V4 | China | II | |
| DQ060053 | A11-ND026 | China | II | |
| EU140955 | KBNP | II | 7b | |
| EU289028 | VG-GA | USA | II | 33b |
| EU546165 | JL-1 | China | II | |
| FJ386392 | NDV01 | China | II | |
| FJ386393 | NDV02 | China | II | |
| FJ386394 | NDV03 | China | II | |
| FJ386395 | NDV04 | China | II | |
| FJ386396 | NDV05 | China | II | |
| FJ939313 | NDV-Chicken | Egypt | II | 30a |
| GQ994433 | XD/Shandong/08 | China | II | 46 |
| GU978777 | APMV-1-U.S.-GB | USA | II | 33a |
| Y18898 | Clone30 | II | 37b | |
| EF201805 | Mukteswar | China | III | |
| FJ430159 | JS-7-05 | China | III | 37a |
| FJ430160 | JS-9-05 | China | III | 37a |
| AY741404 | Herts/33 | Holland | IV | 9a |
| EU293914 | Italien | China | IV | 42a |
| AY562986 | Anhinga | USA | V | |
| AY562987 | U.S.(CA)/211472/02 | USA | V | |
| AY562990 | Largo | USA | V | |
| AJ880277 | Pigeon paramyxvirus-1 | Hunga | VI | 41a |
| AY562988 | U.S.(CA)/1083 | USA | VI | |
| AY562989 | Italy/2736/00 | Italy | VI | |
| FJ410145 | PPMV-1/New York/84 | USA | VI | |
| FJ410147 | PPMV-1/Maryland/84 | USA | VI | |
| FJ766526 | JS/07/22 | China | VI | |
| FJ766527 | JS/07/16 | China | VI | |
| FJ766528 | NDV05-029 | China | VI | |
| FJ766529 | ZhJ-3/97 | China | VI | |
| FJ766530 | JS-07-04 | China | VI | |
| FJ766531 | JS-07-03 | China | VI | |
| GQ338311 | ND/05/028 | China | VI | |
| GQ429293 | Italy/2736/00 | Italy | VI | 10 |
| HM063425 | P4 | China | VI | |
| HM063423 | W4 | China | VI | |
| AF431744 | ZJ1 | China | VII | 17a |
| AF473851 | SF02 | China | VII | 48 |
| AY562985 | Indonesia/14698/90 | Indonesia | VII | |
| DQ485229 | China/Guangxi7/2002 | China | VII | |
| DQ485230 | China/Guangxi09/2003 | China | VII | |
| DQ485231 | China/Guangxi11/2003 | China | VII | |
| DQ486859 | GM strain | China | VII | |
| DQ659677 | NA-1 | China | VII | |
| DQ839397 | KBNP/Korea | Korea | VII | 7a |
| EU167540 | SRZ03 | China | VII | 36 |
| FJ872531 | China(Fujian)/FP1/02 | China | VII | |
| GQ338309 | NDV/03/018 | China | VII | |
| GQ338310 | NDV/03/044 | China | VII | |
| GQ849007 | JSD0812 | China | VII | |
| GU143550 | Go/CH/HLJ/LL01/08 | China | VII | |
| GU564399 | FMW | China | VII | |
| GQ994434 | QG/Hebei/07 | China | VII | 46 |
| FJ751918 | QH1 | China | VII | |
| FJ751919 | QH4 | China | VII | |
| FJ436302 | F48E8 | China | IX | 37 |
| FJ436303 | ZJ/1/86 | China | IX | 37 |
| FJ436304 | FJ/1/85 | China | IX | 37 |
| FJ436305 | JS/1/97 | China | IX | 37 |
| FJ436306 | JS/1/02 | China | IX | 37 |
A reference number is given if available.
Fig. 1.
Recombination events detected simultaneously by five or more methods using the RDP3.42 program with P values ≤ 10−5. The nucleotide positions of breakpoints in the whole genome are shown above or below the bars. Red, orange, yellow, green, cyan, and blue bars indicate NP, P, M, F, HN, and L genes, respectively. The asterisk indicates KBNP (EU140955), a chimerical vaccine strain that has the genotype II Lasota vaccine strain backbone and the F and HN genes from a genotype VII virus, which was used as a control to evaluate the prediction capability of the program.
Fig. 2.
Maximum-likelihood trees based on whole-genome and putative regions of NDVs. The scale bars represent the numbers of substitutions per site, and the numbers at each node represent values from approximate-likelihood-ratio tests for branch (Shimodaira-Hasegawa-like) support. Support values under 70 were removed. Putative recombinants are marked by either a black square for China/Guangxi09/2003 or a black triangle for NDV/03/018. (A) Phylogenetic trees of NDV strains based on complete genomic sequences showed that the two putative recombinants are of genotype VII. (B) Phylogenetic trees based on the putative recombination regions showed that they belong to genotype III. (C) Phylogenetic trees based on the revised sequences showed that the putative recombinant regions are affiliated with genotype VII, indicating that the two old versions of sequences in GenBank represent artificial recombinants.
Using strains China/Guangxi09/2003 and NDV/03/018 as examples, we attempted to validate whether the unexpected high recombination rate was due to inaccuracy of some of the NDV sequences deposited in GenBank. The two viruses were resequenced and analyzed in this study. Viruses were subjected to plaque purification using primary chicken embryo fibroblasts (16). Viral RNAs were extracted from infective allantoic fluid by the use of TRIzol reagent (Invitrogen, Carlsbad, CA). The putative recombination regions of both viruses were amplified using specific primers (available upon request). PCR products were sequenced using an ABI Prism BigDye Terminator version 3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA). ML trees based on the putative recombination regions of our newly obtained sequences were constructed. Their positions in the new trees were consistent with those in the trees constructed from full-length sequences (Fig. 2A and C). However, the putative recombinants detected with the old versions of sequences in GenBank (DQ485230 and GQ338310) were not observed, indicating that these recombinants are artificial. As both viruses were detected as recombinants of genotype VII and genotype III viruses, we then tried to identify the presence of genotype III viruses in original samples. Using genotype III-specific primers (available upon request), we were able to amplify genotype III sequences from both samples. These genotype III sequences are identical to the putative recombination regions in the old versions of sequences in GenBank. These data demonstrated that the original China/Guangxi09/2003 and NDV/03/018 isolates were mixtures of genotype VII and genotype III viruses and that the genomic sequences deposited in GenBank previously might be a reflection of artificial recombination. It is very likely that these two recombinants were generated through RNA template switching between genotype VII and III components.
To provide evidence for a potential artificial recombination resulting in a mosaic sequence, we performed reverse transcription-PCR (RT-PCR) amplification of the M gene from a mixed sample of LaSota (allantoic fluid; HA titer, 512) and ZJ1 (allantoic fluid; HA titer, 128) viruses, which represent genotypes II and VII, respectively. The volume ratio of the former to the latter was 1,000:1. Parental LaSota and ZJ1 viruses were used as controls. A pair of primers, 5′-AGGGCAGAGCCAARACARTAC-3′ and 5′-CGCRGTTTGRCTCCAGAGTAT-3′, were used for the amplification. The amplification was performed in a 25-μl total reaction volume with 1.5 U Taq DNA polymerase (Fermentas, CA). PCR products were cloned into pGEM-T vector (Promega), and multiple clones were sequenced. Clones carrying either genotype II or VII sequences were identified. Notably, the two genotype sequences could also be identified in the same clone, e.g., the clone designated L1000Z1M-4 (Fig. 3), which was identified as a recombinant by all of the seven statistical methods with P values < 10−5 (data not shown). These results suggest that artificial recombination events can be easily induced with samples containing mixed virus genotypes or strains, probably through polymerase template switching during the PCR procedure.
Fig. 3.
Artificial recombinant fragments were amplified from a mixed-NDV sample. LaSotaM and ZJ1M represent M gene sequences of Lasota and ZJ1. LaSota sequences and ZJ1 sequences are indicated by green and red bars, respectively. LaSota and ZJ1 genome sequences were identified in one clone (L1000Z1M-4). The fragment between the green and red arrows denotes a potential template switching position.
Our study clearly demonstrated that some NDV sequences in GenBank are inaccurate and may affect the bioinformatic analysis for NDV evolution. Consistent with these findings, we noticed that another NDV strain, Italy/2736/00, possesses two accession numbers (AY562989 and GQ429293) in GenBank. The virus was characterized as a recombinant based on the originally uploaded sequence (10). Later in 2009, the author submitted a new version of the sequence (GQ429293) and indicated the absence of recombination in the virus and that the earlier sequence represented an artificial recombination. In 2003, F and HN gene sequences of JS2/98/Go were deposited in GenBank with assigned accession numbers AF456439 and AF456430, respectively. The phylogenetic analysis of these sequences indicated that this virus was a recombinant of F and HN genes from genotypes VI and VII. However, subsequent resequencing confirmed this virus to be an entirely genotype VII strain (data not shown). We believe that the actual recombination rate in NDVs is lower than that deduced by using NDV sequences from GenBank.
Currently, lentogenic NDV strains Hitchner B1, Australia V4, and LaSota and mesogenic strain Mukteswar are used as live vaccines worldwide (19, 25, 26, 42), especially in Asia. However, infections caused by virulent NDV strains frequently occur in poultry despite vaccination, providing a good opportunity for the coexistence of virulent and vaccine NDV strains. Due to the propensity of Taq DNA polymerase to slip during the elongation step in PCR (18, 23, 27, 31, 32, 38, 39, 45), molecular work using RNA templates extracted from a field virus sample (which may contain nucleic acids from various NDV strains) is likely to produce artificial recombination fragments by template switching during the process of PCR. On the other hand, the cross-contamination of PCR products, which was pointed out earlier as a frequent occurrence in NDV research laboratories (1), might also cause artificial recombination. We suggest careful verification of NDV sequences while performing bioinformatic analysis for evaluation of virus evolution. In addition, field samples should be subjected to plaque purification before sequencing to ensure the accuracy of NDV sequence data deposited in GenBank.
Nucleotide sequence accession numbers.
We have recently updated sequences of China/Guangxi09/2003 and NDV/03/018 in GenBank under accession numbers JF343539 and JF343538, respectively.
Acknowledgments
We thank Xie Zhixun of Guangxi Veterinary Research Institute for offering NDV strain China/Guangxi09/2003. We are grateful to Subbiah Elankumaran from Virginia Polytechnic Institute and State University for his help in editing the manuscript.
This work was supported by the National Natural Science Foundation of China (grant 30630048) and the Earmarked Fund for Modern Agro-industry Technology Research System in China (nycytx-41-G07).
Footnotes
Published ahead of print on 20 July 2011.
REFERENCES
- 1. Afonso C. L. 2008. Not so fast on recombination analysis of Newcastle disease virus. J. Virol. 82:9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alexander D. J., Senne D. A. 2008. Newcastle disease virus, other avian paramyxovirus, and pneumovirus infections, p. 75–115 In Saif Y. M. (ed.), Diseases of poultry, 12th ed. Blackwell Publishing, Oxford, United Kingdom [Google Scholar]
- 3. Anisimova M., Gascuel O. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55:539–552 [DOI] [PubMed] [Google Scholar]
- 4. Ballagi-Pordány A., Wehmann E., Herczeg J., Belak S., Lomniczi B. 1996. Identification and grouping of Newcastle disease virus strains by restriction site analysis of a region from the F gene. Arch. Virol. 141:243–261 [DOI] [PubMed] [Google Scholar]
- 5. Boni M. F., Posada D., Feldman M. W. 2007. An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics 176:1035–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chambers P., Millar N. S., Bingham R. W., Emmerson P. T. 1986. Molecular cloning of cDNA to Newcastle disease virus, and nucleotide sequence analysis of the junction between the genes encoding the haemagglutinin-neuraminidase and the large protein. J. Gen. Virol. 67(Pt. 3):475–486 [DOI] [PubMed] [Google Scholar]
- 7. Chare E. R., Gould E. A., Holmes E. C. 2003. Phylogenetic analysis reveals a low rate of homologous recombination in negative-sense RNA viruses. J. Gen. Virol. 84:2691–2703 [DOI] [PubMed] [Google Scholar]
- 7a. Cho S. H., Kim S. J., Kwon H. J. 2007. Genomic sequence of an antigenic variant Newcastle disease virus isolated in Korea. Virus Genes. 35:293–302 [DOI] [PubMed] [Google Scholar]
- 7b. Cho S. H., et al. 2008. Characterization of a recombinant Newcastle disease virus vaccine strain. Clin. Vaccine Immunol. 15:1572–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chong Y. L., Padhi A., Hudson P. J., Poss M. 2010. The effect of vaccination on the evolution and population dynamics of avian paramyxovirus-1. PLoS Pathog. 6:e1000872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a. Czeglédi A., et al. 2006. Third genome size category of avian paramyxovirus serotype 1 (Newcastle disease virus) and evolutionary implications. Virus Res. 120:36–48 [DOI] [PubMed] [Google Scholar]
- 9. de Leeuw O., Peeters B. 1999. Complete nucleotide sequence of Newcastle disease virus: evidence for the existence of a new genus within the subfamily Paramyxovirinae. J. Gen. Virol. 80(Pt. 1):131–136 [DOI] [PubMed] [Google Scholar]
- 9a. de Leeuw O. S., koch G., Hartog L., Ravenshorst N., Peeters B. P. 2005. Virulence of Newcastle disease virus is determined by the cleavage site of the fusion protein and by both the stem region and globular head of the haemagglutinin-neuraminidase protein. J. Gen. Virol. 86:1759–1769 [DOI] [PubMed] [Google Scholar]
- 10. Dortmans J. C., Koch G., Rottier P. J., Peeters B. P. 2009. Virulence of pigeon paramyxovirus type 1 does not always correlate with the cleavability of its fusion protein. J. Gen. Virol. 90:2746–2750 [DOI] [PubMed] [Google Scholar]
- 11. Fauquet C. M., Fargette D. 2005. International committee on taxonomy of viruses and the 3,142 unassigned species. Virol. J. 2:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gibbs M. J., Armstrong J. S., Gibbs A. J. 2000. Sister-scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 16:573–582 [DOI] [PubMed] [Google Scholar]
- 13. Guindon S., et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321 [DOI] [PubMed] [Google Scholar]
- 14. Guindon S., Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 15. Han G. Z., He C. Q., Ding N. Z., Ma L. Y. 2008. Identification of a natural multi-recombinant of Newcastle disease virus. Virology 371:54–60 [DOI] [PubMed] [Google Scholar]
- 16. Harper D. R. 1989. A novel plaque assay system for paramyxoviruses. J. Virol. Methods 25:347–350 [DOI] [PubMed] [Google Scholar]
- 17. Heath L., van der Walt E., Varsani A., Martin D. P. 2006. Recombination patterns in aphthoviruses mirror those found in other picornaviruses. J. Virol. 80:11827–11832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a. Huang Y., Wan H. Q., Liu H. Q., Wu Y. T., Liu X. F. 2004. Genomic sequence of an isolate of Newcastle disease virus isolated from an outbreak in geese: a novel six nucleotide insertion in the non-coding region of the nucleoprotein gene. Arch. Virol. 149:1445–1457 [DOI] [PubMed] [Google Scholar]
- 18. Jansen R., Ledley F. D. 1990. Disruption of phase during PCR amplification and cloning of heterozygous target sequences. Nucleic Acids Res. 18:5153–5156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a. Kattenbelt J. A., Stevens M. P., Gould A. R. 2006. Sequence variation in the Newcastle disease virus genome. Virus Res. 116:168–184 [DOI] [PubMed] [Google Scholar]
- 19. Kim L. M., et al. 2007. Phylogenetic diversity among low-virulence Newcastle disease viruses from waterfowl and shorebirds and comparison of genotype distributions to those of poultry-origin isolates. J. Virol. 81:12641–12653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu X., et al. 2009. Surveillance for avirulent Newcastle disease viruses in domestic ducks (Anas platyrhynchos and Cairina moschata) at live bird markets in Eastern China and characterization of the viruses isolated. Avian Pathol. 38:377–391 [DOI] [PubMed] [Google Scholar]
- 21. Martin D., Rybicki E. 2000. RDP: detection of recombination amongst aligned sequences. Bioinformatics 16:562–563 [DOI] [PubMed] [Google Scholar]
- 22. Martin D. P., Williamson C., Posada D. 2005. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21:260–262 [DOI] [PubMed] [Google Scholar]
- 23. Marton A., Delbecchi L., Bourgaux P. 1991. DNA nicking favors PCR recombination. Nucleic Acids Res. 19:2423–2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mayo M. A. 2002. Virus taxonomy—Houston 2002. Arch. Virol. 147:1071–1076 [DOI] [PubMed] [Google Scholar]
- 25. McFerran J. B., Dane D. S., Briggs E. M., Connor T., Nelson R. 1968. Further investigations on enterovirus-neutralising substances in human and animal sera. J. Pathol. Bacteriol. 95:93–99 [DOI] [PubMed] [Google Scholar]
- 26. McFerran J. B., Nelson R. 1971. Some properties of an avirulent Newcastle disease virus. Arch. Gesamte Virusforsch. 34:64–74(In German.) [DOI] [PubMed] [Google Scholar]
- 27. Meyerhans A., Vartanian J. P., Wain-Hobson S. 1990. DNA recombination during PCR. Nucleic Acids Res. 18:1687–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Millar N. S., Chambers P., Emmerson P. T. 1988. Nucleotide sequence of the fusion and haemagglutinin-neuraminidase glycoprotein genes of Newcastle disease virus, strain Ulster: molecular basis for variations in pathogenicity between strains. J. Gen. Virol. 69(Pt. 3):613–620 [DOI] [PubMed] [Google Scholar]
- 29. Miller P. J., Decanini E. L., Afonso C. L. 2010. Newcastle disease: evolution of genotypes and the related diagnostic challenges. Infect. Genet. Evol. 10:26–35 [DOI] [PubMed] [Google Scholar]
- 30. Miller P. J., Kim L. M., Ip H. S., Afonso C. L. 2009. Evolutionary dynamics of Newcastle disease virus. Virology 391:64–72 [DOI] [PubMed] [Google Scholar]
- 30a. Mohamed M. H., et al. 2009. Complete genome sequence of a virulent Newcastle disease virus isolated from an outbreak in chickens in Egypt. Virus Genes 39:234–237 [DOI] [PubMed] [Google Scholar]
- 30b. Munir M., et al. 2010. Complete genome analysis of an avian paramyxovirus type 1 strain isolated in 1994 from an asymptomatic black-headed gull (Larus ridibundus) in southern Sweden. Avian Dis. 54:923–930 [DOI] [PubMed] [Google Scholar]
- 30c. Nakaya T., et al. 2001. Recombinant Newcastle disease virus as a vaccine vector. J. Virol. 75:11868–11873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Odelberg S. J., Weiss R. B., Hata A., White R. 1995. Template-switching during DNA synthesis by Thermus aquaticus DNA polymerase I. Nucleic Acids Res. 23:2049–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pääbo S., Irwin D. M., Wilson A. C. 1990. DNA damage promotes jumping between templates during enzymatic amplification. J. Biol. Chem. 265:4718–4721 [PubMed] [Google Scholar]
- 33. Padidam M., Sawyer S., Fauquet C. M. 1999. Possible emergence of new geminiviruses by frequent recombination. Virology 265:218–225 [DOI] [PubMed] [Google Scholar]
- 33a. Paldurai A., Kumar S., Nayak B., Samal S. K. 2010. Complete genome sequence of highly virulent neurotropic Newcastle disease virus strain Texas GB. Virus Genes 41:67–72 [DOI] [PubMed] [Google Scholar]
- 33b. Perozo F., Villegas P., Dolz R., Afonso C. L., Purvis L. B. 2008. The VG/GA strain of Newcastle disease virus: mucosal immunity, protection against lethal challenge and molecular analysis. Avian Pathol. 37:237–245 [DOI] [PubMed] [Google Scholar]
- 34. Posada D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25:1253–1256 [DOI] [PubMed] [Google Scholar]
- 35. Posada D., Crandall K. A. 2001. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc. Natl. Acad. Sci. U. S. A. 98:13757–13762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Qin Z., et al. 2008. F gene recombination between genotype II and VII Newcastle disease virus. Virus Res. 131:299–303 [DOI] [PubMed] [Google Scholar]
- 37. Qiu X., et al. 2011. Entire genome sequence analysis of genotype IX Newcastle disease viruses reveals their early-genotype phylogenetic position and recent-genotype genome size. Virol. J. 8:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a. Qiu X., et al. 2009. Full-length genome analysis of two genotype III velogenic Newcastle disease virus strains reveals their close relationship with vaccine Mukteswar. Wei Sheng Wu Xue Bao. 49:302–308(In Chinese.) [PubMed] [Google Scholar]
- 37b. Römer-Oberdörfer A., Mundt E., Mebatsion T., Buchholz U. J., Mettenleiter T. C. 1999. Generation of recombinant lentogenic Newcastle disease virus from cDNA. J. Gen. Virol. 80:2987–2995 [DOI] [PubMed] [Google Scholar]
- 38. Saiki R. K., et al. 1988. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239:487–491 [DOI] [PubMed] [Google Scholar]
- 39. Shafikhani S. 2002. Factors affecting PCR-mediated recombination. Environ. Microbiol. 4:482–486 [DOI] [PubMed] [Google Scholar]
- 40. Smith J. M. 1992. Analyzing the mosaic structure of genes. J. Mol. Evol. 34:126–129 [DOI] [PubMed] [Google Scholar]
- 41. Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41a. Ujvári D. 2006. Complete nucleotide sequence of IT-227/82, an avian paramyxovirus type-1 strain of pigeons (Columba livia). Virus Genes 32:49–57 [DOI] [PubMed] [Google Scholar]
- 42. Waterson A. P., Pennington T. H., Allan W. H. 1967. Virulence in Newcastle disease virus. A preliminary study. Br. Med. Bull. 23:138–143 [DOI] [PubMed] [Google Scholar]
- 42a. Wei D., Yang B., Li Y. L., Xue C. F., Chen Z. N., Bian H. 2008. Characterization of the genome sequence of an oncolytic Newcastle disease virus strain Italien. Virus Res. 135:312–319 [DOI] [PubMed] [Google Scholar]
- 43. Wu S., et al. 9 November 2010, posting date. Genetic diversity of Newcastle disease viruses isolated from domestic poultry species in Eastern China during 2005-2008. Arch. Virol. [Epub ahead of print.] doi:10.1007/s00705-010-0851-5 [DOI] [PubMed] [Google Scholar]
- 44. Yin Y., et al. 2011. Molecular characterization of Newcastle disease viruses in Ostriches (Struthio camelus L.): further evidences of recombination within avian paramyxovirus type 1. Vet. Microbiol. 149:324–329 [DOI] [PubMed] [Google Scholar]
- 45. Zaphiropoulos P. G. 1998. Non-homologous recombination mediated by Thermus aquaticus DNA polymerase I. Evidence supporting a copy choice mechanism. Nucleic Acids Res. 26:2843–2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang R., Wang X., Su J., Zhao J., Zhang G. 2010. Isolation and analysis of two naturally occurring multi-recombination Newcastle disease viruses in China. Virus Res. 151:45–53 [DOI] [PubMed] [Google Scholar]
- 47. Zhang R., et al. 2010. Phylogenetic characterization of Newcastle disease virus isolated in the mainland of China during 2001-2009. Vet. Microbiol. 141:246–257 [DOI] [PubMed] [Google Scholar]
- 48. Zou J., Shan S., Yao N., Gong Z. 2005. Complete genome sequence and biological characterizations of a novel goose paramyxovirus-SF02 isolated in China. Virus Genes. 30:13–21 [DOI] [PubMed] [Google Scholar]