Abstract
The double-stranded DNA genome of vaccinia virus (VACV), the prototype poxvirus, contains approximately 200 open reading frames (ORFs) that are transcribed at early, intermediate, and late stages of infection. Previous high-throughput deep RNA sequencing allowed us to map 118 VACV early genes that are expressed before viral DNA replication and 93 postreplicative genes. However, the intermediate- and late-stage postreplicative genes could not be differentiated. Here, we synchronized infections with a reversible inhibitor of DNA replication and used a VACV mutant that conditionally transcribes late genes to sequence the two classes of mRNAs. In addition, each postreplicative ORF was individually expressed under conditions that distinguished intermediate and late classes. We identified 38 VACV genes that belong to the late class and 53 that belong to the intermediate class, with some of the latter continuing to be expressed late. These data allowed us to prepare a genome-wide early, intermediate, and late transcription map. Inspection of sequences upstream of these ORFs revealed distinctive characteristics of intermediate and late promoters and suggested that some promoters have intermediate and late elements. The intermediate genes encoded many DNA binding/packaging and core-associated proteins in addition to late transcription factors; the late genes encoded many morphogenesis and mature virion membrane proteins, including those involved in entry, in addition to early transcription factors. The top-ranked antigens for CD4+ T cells and B cells were mainly intermediate rather than late gene products. The differentiation of intermediate and late genes may enhance understanding of poxvirus replication and lead to improvements in expression vectors and recombinant vaccines.
INTRODUCTION
Poxviruses are of general interest because of their wide distribution in nature, responsibility for causing human and animal diseases, and use as expression vectors for vaccines and investigational purposes (18). Poxviruses are unusual in that they replicate and transcribe their large double-stranded DNA genomes in the cytoplasm rather than the nucleus of infected cells. Vaccinia virus (VACV), the prototype member of the poxvirus family, has a DNA genome of approximately 200 kbp that encodes more than 200 proteins. The open reading frames (ORFs) are closely spaced and temporally regulated. Early-, intermediate-, and late-class mRNAs can be detected in 20, 100, and 140 min, respectively, after synchronous infection of HeLa cells with VACV (3). The enzymes necessary for transcription of early genes are packaged in the infectious particle and expressed immediately after entry of the core into the cytoplasm, whereas de novo RNA, protein, and DNA synthesis is required for transcription of intermediate and late genes. The virus-encoded multisubunit RNA polymerase, in conjunction with class-specific transcription factors, transcribes the genes of each class. The intermediate transcription factor genes have early promoters, late transcription factor genes have intermediate promoters, and early transcription factor genes have late promoters, providing a cascade mechanism of regulation (4, 9, 10, 15, 22).
It has been difficult to distinguish intermediate and late genes because expression of both classes is dependent on viral DNA replication. One approach was to construct a recombinant VACV in which the late transcription factor G8 was conditionally regulated (26). SDS-polyacrylamide gel electrophoresis resolved polypeptides that had been labeled with [35S]methionine in cells infected in the presence or absence of inducer. Based on the relative intensities of the bands under the two conditions, at least 13 intermediate proteins and an equal number of late ones were resolved. Since this method can detect only the most abundant proteins, the potential number of intermediate proteins was considered to be much greater (26). Thus far, however, only a few genes have been characterized as intermediate (15, 17, 22, 23, 26), and similarities between known intermediate and late promoters make it difficult to distinguish them based on sequence alone (4, 10, 17).
Recent high-throughput deep RNA sequencing (RNA-seq) studies led to the identification of 118 early ORFs that are expressed prior to viral DNA replication and 93 that are expressed after DNA replication (25). However, due to the small difference in temporal expression levels, the close spacing of ORFs, and the extensive read-through of transcripts, neither RNA-seq nor tiling methodologies were able to distinguish intermediate and late mRNAs (1, 25). For the present study, we sequenced RNAs from synchronously infected cells in which the late transcription factor G8 was conditionally expressed in order to discriminate intermediate and late transcripts. To further identify intermediate and late genes, we cloned and expressed each postreplicative ORF separately. Together, these analyses revealed 38 late genes and 53 intermediate genes, some of which continue to be expressed late. This information was used to construct the first genome-wide early, intermediate, and late transcription map; characterize intermediate and late promoter sequences; and group early, intermediate, and late genes according to functions.
MATERIALS AND METHODS
Viruses and cells.
The Western Reserve (WR) strain of VACV (American Type Culture Collection VR-1354) and recombinant forms were used. The recombinant virus vRO-G8R with isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible late transcriptional factor G8R was described previously (24, 26). HeLa S3 cells were maintained in minimum essential medium with spinner modification (Quality Biological, Gaithersburg, MD) supplemented with 5% equine serum (HyClone, Logan, UT). Infection of HeLa S3 cells was as described previously (24). BS-C-1 cells were cultured in minimum essential medium with Earle's balanced salts (Quality Biological) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and 100 μg/ml penicillin-streptomycin. All cells were cultured at 37°C in a 5% CO2 atmosphere. Preparation, purification, and titration of VACV were described previously (11, 24). Where indicated in the text and figures, 100 μg/ml cycloheximide (CHX) or 40 μg/ml cytosine arabinoside (AraC) was added to the medium.
Plasmids.
VACV postreplicative genes with a C-terminal or N-terminal Flag tag or three copies of the Flag tag (3×Flag), including 80 to 100 bp upstream of the start codon, were PCR amplified from VACV WR DNA using primers containing promoter- and tag-encoding sequences and cloned into Zero Blunt TOPO PCR cloning vector (Invitrogen, Carlsbad, CA). The enhanced green fluorescent protein (EGFP) ORF with adjacent VACV promoter sequences was amplified by PCR and cloned into Zero Blunt TOPO PCR vector.
mRNA isolation, RNA-seq, and computational analyses.
Polyadenylated RNA isolation, purification, and cDNA sequencing were carried out as described previously (25) with modifications. Briefly, polyadenylated RNA was isolated and purified using a Dynabeads mRNA kit (Invitrogen). Strand-specific cDNA libraries were prepared using a Whole Transcriptome Analysis Kit (Ambion, Austin, TX) and a SOLiD Transcriptome Multiplexing Kit (Applied Biosystems, Foster City, CA) for multiplexed sequencing with an Applied Biosystems SOLiD 3+ system. An Applied Biosystems Whole Transcriptome Analysis Pipeline was used to process the reads against the VACV genome (NCBI accession number NC_006998) with four mismatches allowed. The transcriptome maps were visualized with Mochiview (14).
Transfection and Western blotting.
BS-C-1 cells were transfected with plasmids and Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The cells were lysed at 16 to 18 h after transfection. For Western blotting, cell lysates were resolved by SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes using an iBlot apparatus (Invitrogen). The membranes were blocked with 5% nonfat milk in Tris-buffered saline with 0.05% Tween 20 for 1 h, incubated with primary antibody for 1 to 2 h at room temperature or overnight at 4°C, washed with Tris-buffered saline with 0.05% Tween 20, incubated with horseradish peroxidase-conjugated secondary antibody or IRDye 800-conjugated secondary antibody for 1 h at room temperature, washed with Tris-buffered saline with 0.05% Tween 20, and developed using chemiluminescent substrate (Pierce, Rockford, IL) or a Li-Cor Odyssey infrared imager (Li-Cor Biosciences, Lincoln, NE). Anti-Flag M2 antibody was purchased from Agilent Technologies (Santa Clara, CA). Anti-EGFP antibody was purchased from Roche (Basel, Switzerland). Anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was purchased from Covance (Princeton, NJ).
Nucleotide sequence accession number.
The cDNA sequences were deposited under accession number SRA037996 in the Sequence Read Archive of the National Library of Medicine.
RESULTS
Deep sequencing of viral mRNAs.
Our strategy to distinguish intermediate and late mRNAs employed the recombinant VACV vRO-G8R, in which expression of the late transcription factor G8 is regulated by the Escherichia coli lac operator and repressor (26). Previous studies had shown that in the absence of the inducer IPTG, expression of G8 was reduced 99% compared to wild-type virus, early and intermediate gene expression was unaffected, and synthesis of late mRNA and protein was reduced about 90%. Since late expression is not completely inhibited under these conditions, we added a synchronization protocol (Fig. 1) in which early mRNA synthesis was allowed to occur for 4 h in the presence of hydroxyurea (HU), a reversible ribonucleotide reductase inhibitor that blocks DNA synthesis. Under these conditions, RNA polymerase, intermediate transcription factors, and DNA replication proteins accumulate in cells infected with vRO-G8R in the presence or absence of IPTG (+IPTG or −IPTG, respectively). At the end of 4 h, HU was removed, and the protein synthesis inhibitor CHX was added to the cells without IPTG. The purpose of CHX was to further ensure that late transcription factors were not synthesized and to enhance the stringency of the repression. After removal of HU, it was anticipated that DNA replication would occur rapidly, followed by successive synthesis of intermediate and late mRNAs in cells infected in the presence of IPTG. In the absence of IPTG and presence of CHX, however, the transcription program would be arrested after synthesis of intermediate mRNAs.
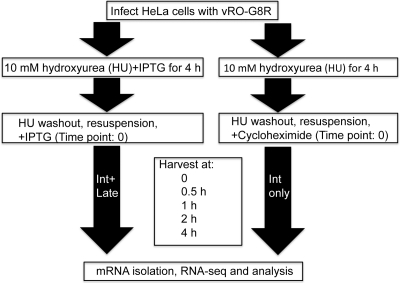
Fig. 1.
Flowchart for intermediate (Int) and late mRNA analyses. HeLa S3 cells infected with vRO-G8R at a multiplicity of infection of 20 PFU per cell were incubated for 4 h in the presence of 10 mM HU to inhibit DNA replication with or without 5 mM IPTG. Synchronization of the infection was achieved by washing the cells to remove HU and resuspending them in the presence of CHX to prevent late gene expression or in the presence of IPTG to allow late gene expression. The cells were harvested at 0, 0.5, 1, 2, and 4 h for RNA isolation and sequencing.
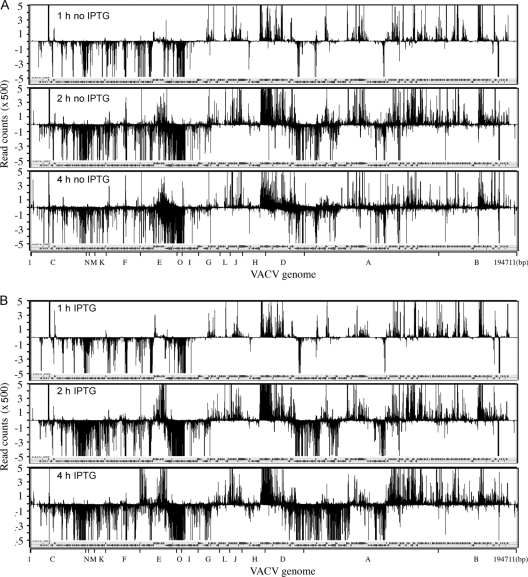
Cells infected in the presence or absence of IPTG were harvested at 0, 0.5, 1, 2, and 4 h after HU removal according to the protocol shown in Fig. 1. Polyadenylated RNAs were purified, cDNAs were synthesized, and deep sequencing was performed. Because the sequencing method involves PCR amplification of short cDNAs that are then aligned on the genome, the reads even within ORFs are not uniform and hence give a fragmented appearance. The 50-nucleotide (nt) sequence reads that mapped to the VACV genome increased over time and ranged from 1.2 to 7.2 million per sample. The sequence reads, as counts per nucleotide, were superimposed over the annotated VACV genome. At 0, 0.5, and 1 h, the transcription patterns under both +IPTG and −IPTG conditions were similar (Fig. 2; see also Fig. S1 in the supplemental material) and corresponded to the early mRNA pattern previously described (25). The up and down lines represent RNAs that were synthesized rightward from the top strand and leftward from the bottom strand, respectively. At 2 h, additional peaks were evident for both the +IPTG and −IPTG samples, presumably representing intermediate mRNAs made under both conditions (Fig. 2). The patterns of the 2- and 4-h samples without IPTG were similar, whereas the pattern continued to evolve in the 4-h +IPTG sample as late mRNAs were made (Fig. 2). The transcripts tended to occur in clusters on the bottom and top strands corresponding to the direction of the ORFs.
Fig. 2.
Temporal expression of VACV mRNAs following removal of HU. HeLa cells were infected with vRO-G8R in the absence or presence of IPTG as outlined in Fig. 1. After removal of HU, polyadenylated RNA was isolated at 0, 0.5, 1, 2, and 4 h, and cDNAs were made and sequenced. Read counts per nucleotide were plotted along the complete VACV ORF map. Counts above and below the line represent RNAs transcribed from the top DNA strand in the rightward direction and from the bottom strand in the leftward direction, respectively. Only the 1-, 2-, and 4-h data are shown here; the 0- and 0.5-h data are shown in Fig. S1 in the supplemental material. ORFs are indicated by arrows; the letters at the bottom represent HindIII fragments. (A) Cells without IPTG and with CHX. (B) Cells with IPTG and without CHX.
To quantify the differences between the samples with and without IPTG, we first excluded the reads that mapped to known early ORFs (25) and normalized the number of reads that mapped to each postreplicative ORF by the total number of reads from that sample. The +IPTG/−IPTG ratios of the read numbers from cells infected in the presence and absence of IPTG were plotted (Fig. 3). From 0 to 2 h, the +IPTG/−IPTG ratios for many ORFs were close to 1, consistent with similar patterns of expression of intermediate mRNAs in the presence and absence of IPTG. The +IPTG/−IPTG ratios increased between 2 and 4 h, as indicated by the decrease in the correlation coefficient (R) and P value, with high ratios representing ORFs with late transcripts, low ratios likely representing ORFs with downregulated intermediate transcripts, and ORFs with ratios near 1 consistent with maintenance of some intermediate transcripts.
Fig. 3.
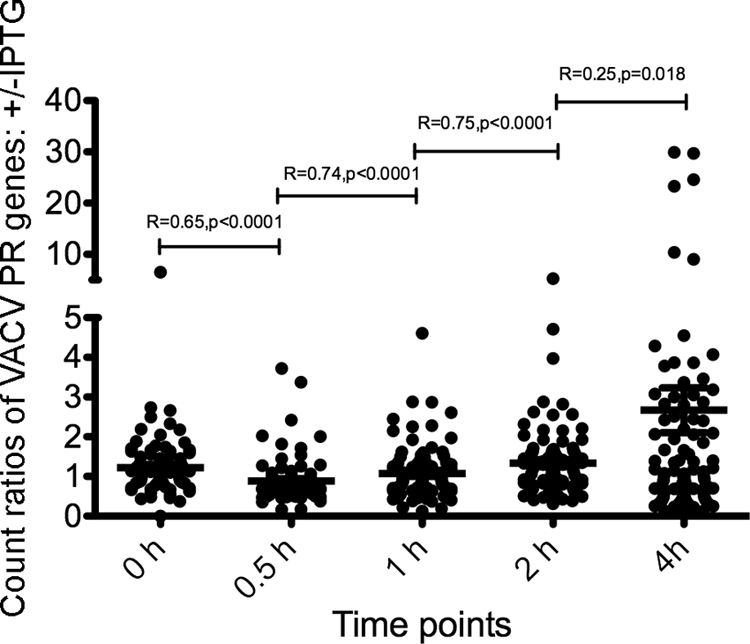
Ratios (+IPTG/−IPTG) of read counts mapping to VACV postreplicative ORFs. For each time point, the reads mapping to early mRNAs were removed, and the reads mapping to postreplicative ORFs were normalized by the total read counts. The ratios (+IPTG/−IPTG) of read counts were plotted. The horizontal lines are the mean ratios with the standard error of the mean at each time. R is the correlation coefficient between two neighboring sets of ratios; a smaller R signifies a change in ratios and correlates with the presence of late mRNAs in the sample with IPTG at 4 h. The P values are the chance that random ratios would result in a correlation coefficient as far from zero (or further) as observed if there really is no correlation.
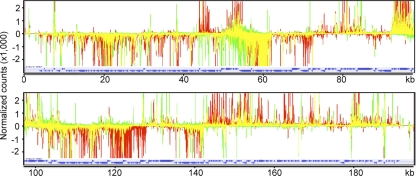
To better visualize differences, the 4-h transcription maps for the +IPTG and −IPTG samples were superimposed (Fig. 4). Peaks unique to the +IPTG infection were considered to be late mRNAs. One clear example is F17R, a well-characterized gene with a strong late promoter that is well separated from other genes transcribed from the same DNA strand, thus providing an opportunity to look at the emergence of a late mRNA. The F17R mRNA, which starts at approximately 44 kbp on the upper strand and apparently extends antisense to ORFs on the lower strand, was seen only in the +IPTG sample. The reduction in intermediate mRNAs following late mRNA synthesis (3) could account for the predominance of some RNAs in the −IPTG sample compared to the +IPTG sample. Intermediate and late RNAs were transcribed from both DNA strands but occurred in clusters on the same strand. Overall, the results suggested similar numbers of intermediate and late mRNAs, as predicted from studies of the defect in protein synthesis resulting from repression of a late transcription factor (24). However, due to the close spacing of ORFs, the extensive read-through of transcripts, and the possibility that many ORFs have hybrid promoters with early, intermediate, or late elements, it was difficult to classify individual ORFs with high confidence from the RNA-seq data alone.
Fig. 4.
Superimposed VACV genome-wide transcriptome maps at 4 h after HU removal. The data obtained at 4 h after HU removal, as described in the legend of Fig. 2, were plotted as the number of reads per nucleotide and expanded. The color code is as follows: red, reads with IPTG; green, reads without IPTG; yellow, superimposed reads.
Identification of intermediate and late genes by expression and Western blotting.
Having determined that there are a large number of intermediate genes, we employed an independent strategy to identify them based on the following insights. Although RNA polymerase and intermediate transcription factors are synthesized in cells infected in the presence of a DNA replication inhibitor, the absence of template prevents intermediate gene expression (15). However, the templates for individual genes can be introduced into infected cells by transfection. Transfected genes with intermediate promoters are transcribed in the presence of a DNA replication inhibitor, whereas plasmids with late promoters and early promoters are not (4, 22). This procedure was previously used in individual cases to confirm intermediate expression or analyze promoter mutations but had not been applied systematically. The general scheme is shown in Fig. 5. We cloned each putative intermediate and late ORF, with a C-terminal Flag epitope tag, together with 80 to 100 bp of upstream DNA. The 93 plasmids constructed were individually transfected into cells that had been infected 1 h earlier with VACV in the absence or presence of AraC, an inhibitor of DNA replication. Subsequently, the cells were lysed and analyzed by Western blotting using an antibody to the epitope tag. A nonspecific band that reacted with the anti-Flag antibody even in untransfected cells was used as a loading control (data not shown). We detected proteins derived from 80 of the 93 ORFs under at least one of the conditions. To increase the sensitivity of Western blotting, C-terminal 3×Flag-tagged constructs of the remaining 13 ORFs were made allowing detection of 11 more proteins. The products of two pseudogene ORFs, VACWR003 and VACWR145, were barely detectable even with the 3×Flag tag and were considered not expressed.
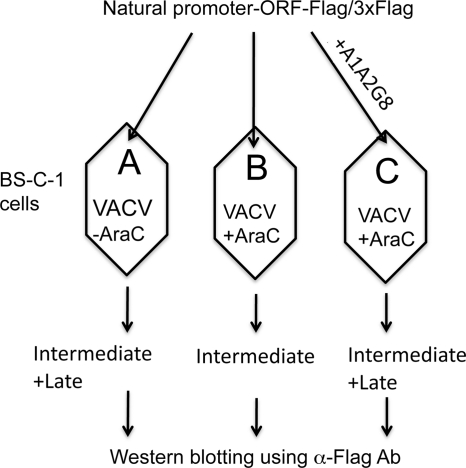
Fig. 5.
Scheme for distinguishing intermediate and late genes by transfection. BS-C-1 cells were infected with VACV in the absence (A) or presence (B and C) of AraC and transfected with a single plasmid containing a Flag-tagged ORF under its natural intermediate or late promoter (A and B) or cotransfected with the latter plasmid and plasmid A1A2G8 (C) containing the three late transcription factors under their natural intermediate promoters. The lysates were analyzed by Western blotting with antibody to the Flag epitope (α-Flag Ab). In the absence of AraC, both intermediate and late genes can be expressed; in the presence of AraC only intermediate genes can be expressed since late transcription factors are not made. However, late gene expression in the presence of AraC can be rescued by cotransfection of a plasmid expressing the three late transcription factors. In a related assay (not shown here) the VACV ORF was replaced by EGFP under the control of the putative intermediate or late promoter.
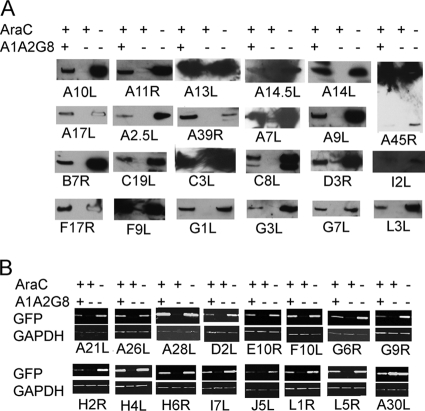
Based on protein expression patterns (Fig. 6), the genes were divided into three categories: category I, 39 ORFs with no or trace expression in the presence of AraC and robust expression in the absence of AraC; category IIA, 31 ORFs with robust expression in the presence or absence of AraC; category IIB, 21 ORFs with robust expression in the presence of AraC and with much less expression in the absence of AraC. The simple interpretation is that category I comprises late genes and that categories IIA and IIB comprise intermediate genes.
Fig. 6.
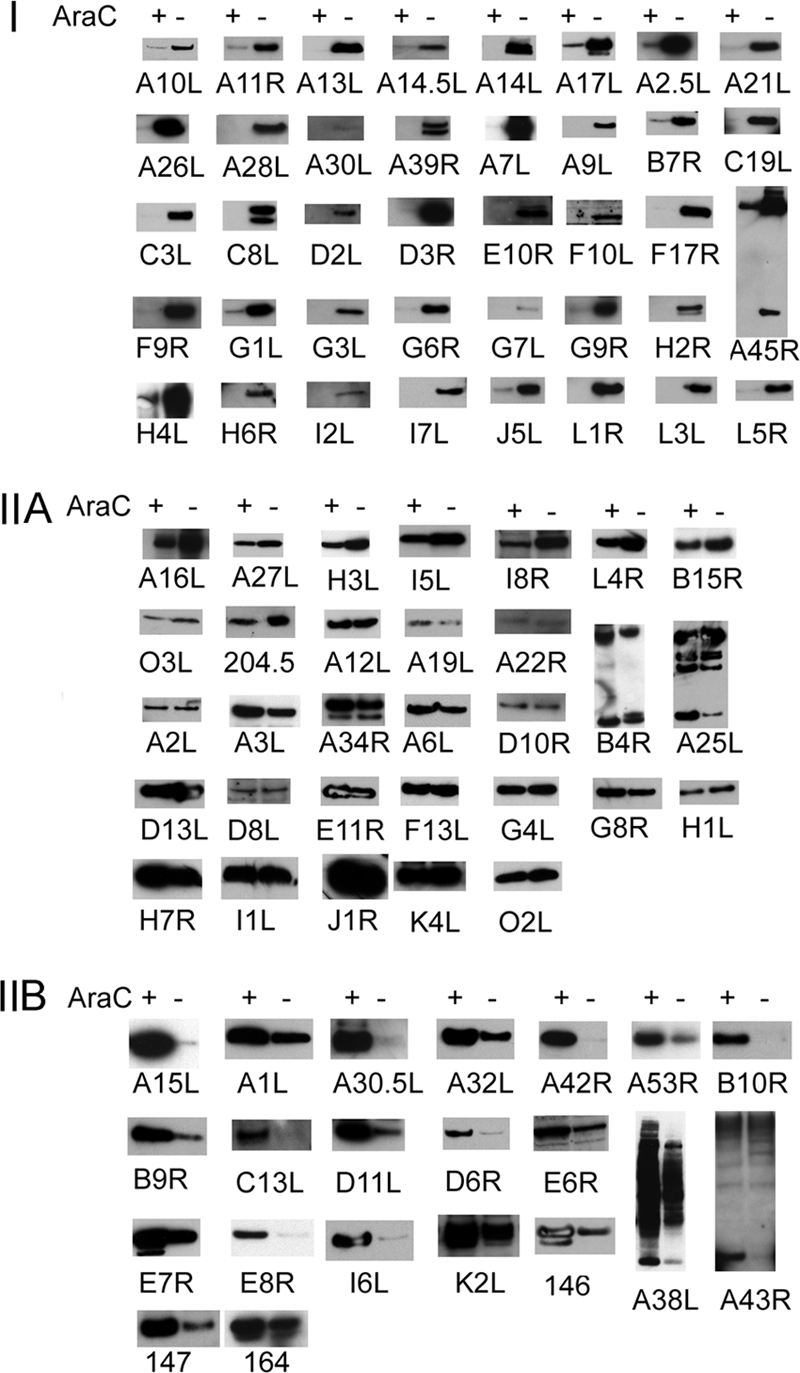
Expression of individual genes in the presence and absence of AraC. Employing the scheme shown in Fig. 5, each Flag- or 3×Flag-tagged VACV postreplicative gene in a plasmid was transfected into VACV-infected BS-C-1 cells in the presence (+) or absence (−) of AraC. Western blots were probed with anti-Flag M2 antibody. In most cases, a single band of the expected size was discerned, and only a small part of the Western blot is shown. An expanded segment of the Western blot is shown in cases where more than one band is present due to protein processing, glycosylation, or SDS-resistant aggregates. Where applicable, the common (Copenhagen) HindIII fragment letter/number was used as the ORF name; otherwise the WR number was used. The protein expression patterns are as follows: I, robust expression only in the absence of AraC; IIA, robust expression in the absence and presence of AraC; IIB, higher expression in the presence of AraC than in the absence.
Before concluding that all category I proteins belonged to the late class, we had to prove that their failure to be expressed in the presence of AraC was due to the absence of the late transcription factors A1, A2, and G8. An alternative hypothesis was that some category I proteins were really intermediate but unstable in the absence of an unexpressed partner protein. To test these possibilities, we infected cells in the presence and absence of AraC and cotransfected each putative late ORF plasmid with the plasmid A1A2G8 encoding the three late transcription factors under their natural intermediate promoters, as indicated in Fig. 5C. Protein expression was fully or partially rescued by cotransfection of the A1A2G8 plasmid in the presence of AraC for more than half of the ORFs, proving that they were late genes (Fig. 7 A).
Fig. 7.
Rescue of late gene expression by cotransfection of late transcription factors. As described in the scheme of Fig. 5, late gene expression was rescued by expressing late transcription factors in the presence of AraC. (A) BS-C-1 cells were infected with VACV in the presence (+) or absence (−) of AraC and in each case transfected with a plasmid containing a Flag- or 3×Flag-tagged VACV postreplicative gene with its natural promoter. The cells were cotransfected with A1A2G8 (+) or the vector plasmid (−). The cells were lysed, and Western blots were probed with anti-Flag M2 antibody. (B) The protocol was similar to that of panel A except that the plasmid encoded EGFP under the VACV promoter instead of the Flag-tagged ORF. The Western blot was probed with anti-EGFP antibody and with anti-GAPDH antibody as a loading control. In both panels, the common (Copenhagen) HindIII fragment letter/number was used as the ORF name.
The ORFs that did not respond to A1A2G8 rescue were A21L, A26L, A28L, A30L, D2L, E10R, F10L, G9R, H2R, H4L, H6R, I7L, J5L, L1R, and L5R. According to the alternative hypothesis, the proteins expressed from these ORFs might be unstable in the absence of a partner protein. In addition, we considered yet another possibility that failure of rescue by the A1A2G8 plasmid was because the 15 ORFs belonged to a novel expression class with different or additional transcription factor requirements. To address the possibilities of specific protein instability and additional transcription factor requirements, we constructed 15 new plasmids that contained the promoter regions attached to the EGFP coding sequence. Cells were infected in the presence and absence of AraC and cotransfected with the EGFP plasmids and the A1A2G8 plasmid. In each case (except for A30L) expression of EGFP was fully or partially rescued by the late transcription factors, indicating that they are true late promoters (Fig. 7B). In the case of the A30L promoter, EGFP was expressed robustly with or without cotransfection of the A1A2G8 plasmid (Fig. 7B), indicating that A30L contains an intermediate promoter. Therefore, it seems likely that the polypeptides encoded by the 15 ORFs listed above were unstable in the presence of AraC due to the absence of other VACV proteins. Supporting this idea, the proteins encoded by the A21L, A28L, G9R, H2R, J5L, L1R, L5R, and J5L ORFs interact and are associated with the entry-fusion complex (5, 20), and the proteins encoded by A30L, D2L, and F10L interact and are part of the seven-protein complex required for interaction of viral membranes and viroplasm (21).
The genes in categories IIA and IIB must be intermediate because of their expression in the presence of AraC. However, we were curious regarding the differences in their expression levels in the absence of AraC. The lower expression of category IIB ORFs in the absence of AraC would be consistent with previous findings that intermediate mRNAs decline sharply after late genes are expressed during a normal infection (3). An alternative possibility, that the epitope tags were lost by processing when other VACV postreplicative genes including proteinases were expressed, was investigated by placing the epitope tag at the N terminus instead of the C terminus of several ORFs (A15L, A30.5L, B10R, B9R, and C13L) in category IIB. However, the proteins with N-terminal tags were also greatly decreased in the absence of AraC (not shown), suggesting either downregulation of expression or extensive proteolysis. Expression of the ORFs in category IIA in the absence of AraC could be due to their resistance to repression by late proteins or because they have late as well as intermediate promoter elements, as discussed below.
DISCUSSION
VACV genome-wide transcription map.
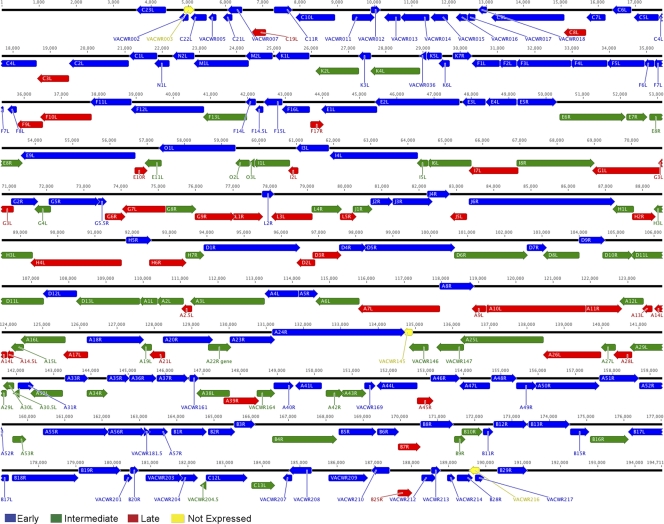
In Table S1 in the supplemental material, we listed 53 ORFs classified as intermediate and 38 classified as late. Taking the above data together with the 118 ORFs previously classified as early (25), we constructed a genome-wide transcription map (Fig. 8). The clustering of ORFs transcribed in the same direction and the preponderance of early genes near the two ends of the genome were previously noted (25). The intermediate and late ORFs were dispersed mostly in the central part of the genome with nominal stage-specific clustering. Adjacent RNAs with opposite polarities were uncommon, and only one example of convergent intermediate transcripts and no example of divergent intermediate transcripts was found. There have been studies of oppositely oriented late promoters and late promoters oppositely oriented to intermediate promoters (16). Because of the experimental methods used, the assigned promoter class is the one with earliest expression. Thus, a gene that is expressed by VACV before DNA replication was classified as early regardless of whether it might also be transcribed by intermediate or late transcription complexes. As an example, previous studies had indicated that I3L has both early and intermediate promoters (22); however, this is not shown in Table S1 or in the transcription map (Fig. 8). Similarly, non-early genes that were expressed in the absence of late transcription factors were classified as intermediate even though they may also have late promoter elements. To remedy this, we plan to systematically investigate all candidate dual promoter genes and refine the map.
Fig. 8.
VACV transcriptome map. VACV WR ORFs are shown as colored arrows indicating the direction of transcription. When applicable, the common (Copenhagen) HindIII fragment letter/number name was used to identify ORFs; otherwise the VACV WR name was provided. The numbers from 1 to 194711 indicate the nucleotide positions on the VACV genome. Note that each ORF has been assigned the stage at which its earliest expression can be detected and that the presence of additional promoter elements that potentially contribute to later stages of gene expression are not indicated.
Promoters of VACV intermediate and late genes.
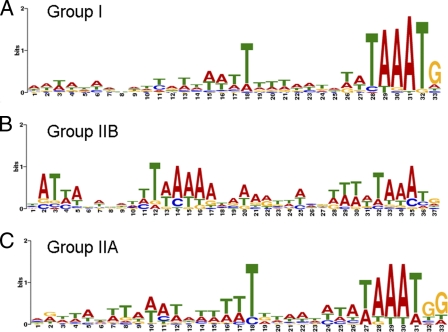
Having classified the postreplicative genes into intermediate and late categories, we examined stage-specific promoter elements. The possibility of mixed promoter elements led us to divide the genes into three groups. The first group contained the 38 genes, excluding A30L, in protein expression category I (Fig. 6). Genes in this group (except for A30L) were predicted to contain only late promoter elements since there was little or no expression in the absence of DNA replication. The second group contains 21 intermediate genes in category IIB (Fig. 6) that have higher protein levels in the absence of DNA replication, implying that they contain only intermediate promoter elements. The third group contains 31 intermediate genes from category IIA (Fig. 6) (plus A30L) with relatively high protein levels in the presence and absence of DNA replication, raising the possibility that many of them have mixed intermediate and late promoter elements or resist repression. In each group, the sequences between 50 nt upstream and 4 nt downstream of start codons were analyzed with the motif discovery program MEME (2). The TAAAT motif usually followed by a G was a signature of late promoters (Fig. 9 A). In addition, late promoters had a conserved T at the 10th nucleotide upstream of TAAAT with several less-conserved T residues surrounding it (Fig. 9A). The intermediate-only promoters invariably contained TAAA upstream of the ORF, but only a subset had TAAAT, and even fewer had TAAATG (Fig. 9B). In addition, the intermediate-only promoters lacked the T at the 10th nucleotide upstream of TAAAT but contained a higher AT-rich sequence 15 to 19 nucleotides upstream, followed by a predominant T (Fig. 9B). Many of the third-category promoters appeared to have mixed intermediate and late elements: the T following the TAAA motif was more frequent than in intermediate-only promoters as was the T residue 9 or 10 nucleotides upstream of TAAAT (Fig. 9C). In addition, A residues were more frequent further upstream than in late-only promoters. The promoter of the previously described intermediate gene I1L (17), which was assigned to category IIA containing potential intermediate-late promoters, contains the two signatures of late genes, i.e., TAAATG and T at the 9th or 10th nucleotide upstream of TAAAT in addition to upstream AT residues present in intermediate genes.
Fig. 9.
Motif logos of VACV promoters. Motifs were generated by the program MEME using sequences spanning 50 nt upstream and 4 nt downstream of the start codons. The search parameters were one motif per sequence and a motif length of 33 to 38 nt. Motifs represent late genes (A), intermediate genes (B), and intermediate genes with late expression (C), as shown in Fig. 6.
The present results generally confirmed and extended previous analyses of intermediate and late promoters based on smaller sample sets and mutagenesis (4, 10, 12, 13, 17). More precise mapping of the transcriptional start sites and additional mutagenesis studies are needed to further explore the differences between intermediate and late pro- moters.
Functions of intermediate and late genes.
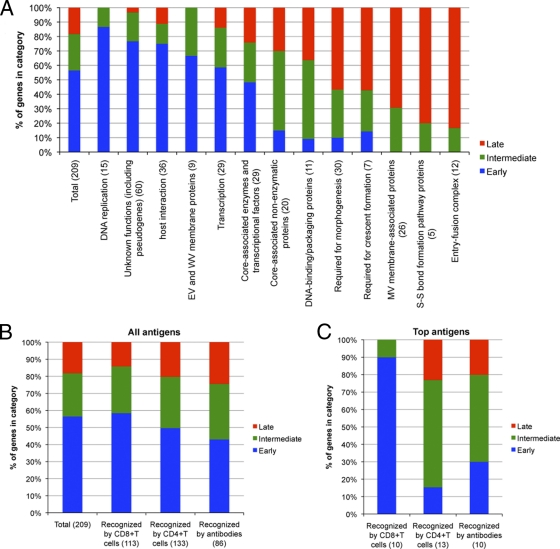
VACV genes were grouped into 13 partially overlapping categories in order to compare functional and temporal expression relationships. The categories with predominant early genes are DNA replication, overlapping transcription and core-associated enzymes, host interactions, and extracellular virion membrane proteins (Fig. 10 A). The partially overlapping categories of transcription and core-associated enzymes also contain intermediate and late genes (Fig. 10A). The two groups with the largest percentage of intermediate genes are DNA binding/packaging and core-associated nonenzymatic proteins (Fig. 10A). Groups with a preponderance of late genes encode redox disulfide bond enzymes, overlapping morphogenesis and crescent formation proteins, and mature virion membrane proteins and components of the entry-fusion complex. It appears that the intermediate proteins are more involved with interacting with the newly synthesized viral genome, and the late proteins are involved with the formation of the primary viral membrane. One theme is that proteins that need to interact are frequently synthesized at the same time. For example, most of the entry-fusion proteins have intramolecular disulfide bonds that are formed by redox proteins, and both the entry and redox proteins belong to the late class. On the other hand, the finding of proteins synthesized at different times, such as those for formation of the extracellular viral membrane and the mature virion membrane, supports independent pathways of membrane morphogenesis. It is possible that synthesis of some proteins at the wrong time would have a deleterious effect. This was suggested by previous experiments with the intermediate A32 DNA packaging protein though overexpression as well as late synthesis could have contributed to decreased virus replication (7). We have planned studies to more critically examine the effects of altering the timing of gene expression.
Fig. 10.
Gene functions and antigenicity of different expression classes. (A) The individual genes in each functional category are listed in Table S1 in the supplemental material. The numbers of genes in each category are indicated. (B) CD8+, CD4+, and B cell antigens were obtained from a recent review (19) and were grouped by expression class according to Table S1. The numbers following each category indicate the number of genes in that category. (C) Highly recognized CD8+ T cell (G5.5R, G5R, E2L, A19L, D12L, F12L, C7L, A47L, A55R, and A48R), CD4+ T cell (D13L, H3L, L4R, A2.5L, B2R, A9L, A26L, D8L, F13L, H7R, I1L, J6R, and O3L), and B cell (A10L, H3L, B5R, A33R, A27L, A56R, A25L, D13L D8L, and A13L) antigens, from the same review as in panel B, were grouped by expression class.
Antigenic properties of intermediate and late gene products.
A recent review summarized the VACV antigens recognized by CD8+ T cells, CD4+ T cells, and B cells (19). The distribution of these antigenic proteins in different expression classes is shown in Fig. 10B, and the top-ranked proteins are shown in Fig. 10C. The genes for top-ranked CD8+ T cell antigens are predominantly in the early class, as noted previously (6, 8, 19). The genes for top-ranked CD4+ T cell and B cell antigens are mostly synthesized after DNA replication but with a higher proportion in the intermediate than the late class (Fig. 10C).
Additional thoughts.
Like many other aspects of poxvirus biology, the transcription system is highly complex, and its conservation reflects development early in the evolution of the family. Intermediate and late promoters have some similar features, suggesting that they diverged from a common sequence. The differentiation of intermediate and late genes should allow us to investigate the functional consequences of swapping promoters and even melding the intermediate and late transcription systems by replacing the promoters of the late transcription factors. The latter mutants might recapitulate a putative early stage of poxvirus evolution.
Supplementary Material
ACKNOWLEDGMENTS
We thank P. S. Satheshkumar for providing the A1A2G8 plasmid and helpful suggestions, Catherine Cotter for cell culture, and other laboratory members for additional assistance.
The study was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 27 July 2011.
REFERENCES
- 1. Assarsson E., et al. 2008. Kinetic analysis of a complete poxvirus transcriptome reveals an immediate-early class of genes. Proc. Natl. Acad. Sci. U. S. A. 105:2140–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bailey T. L., Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28–36 [PubMed] [Google Scholar]
- 3. Baldick C. J., Jr., Moss B. 1993. Characterization and temporal regulation of mRNAs encoded by vaccinia virus intermediate stage genes. J. Virol. 67:3515–3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baldick C. J., Keck J. G., Moss B. 1992. Mutational analysis of the core, spacer and initiator regions of vaccinia virus intermediate class promoters. J. Virol. 66:4710–4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bisht H., Weisberg A. S., Moss B. 2008. Vaccinia virus L1 protein is required for cell entry and membrane fusion. J. Virol. 82:8687–8694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bronte V., et al. 1997. Antigen expression by dendritic cells correlates with the therapeutic effectiveness of a model recombinant poxvirus tumor vaccine. Proc. Natl. Acad. Sci. U. S. A. 94:3183–3188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cassetti M. C., Merchlinsky M., Wolffe E. J., Weisberg A. S., Moss B. 1998. DNA packaging mutant: repression of the vaccinia virus A32 gene results in noninfectious, DNA-deficient, spherical, enveloped particles. J. Virol. 72:5769–5780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Coupar B. E. H., Andre M. E., Both G. W., Boyle D. B. 1986. Temporal regulation of influenza hemagglutinin expression in vaccinia virus recombinants and effects on the immune response. Eur. J. Immunol. 16:1479–1487 [DOI] [PubMed] [Google Scholar]
- 9. Davison A. J., Moss B. 1989. The structure of vaccinia virus early promoters. J. Mol. Biol. 210:749–769 [DOI] [PubMed] [Google Scholar]
- 10. Davison A. J., Moss B. 1989. The structure of vaccinia virus late promoters. J. Mol. Biol. 210:771–784 [DOI] [PubMed] [Google Scholar]
- 11. Earl P. L., Cooper N., Wyatt L. S., Moss B., Carroll M. W. 2001. Preparation of cell cultures and vaccinia virus stocks. Curr. Protoc. Mol. Biol. Chapter 16, unit 16.6. doi: 10.1002/0471142727.mb1616s43 [DOI] [PubMed] [Google Scholar]
- 12. Hänggi M., Bannwarth W., Stunnenberg H. G. 1986. Conserved TAAAT motif in vaccinia virus late promoters: overlapping TATA box and site of transcription initiation. EMBO J. 5:1071–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hirschmann P., Vos J. C., Stunnenberg H. G. 1990. Mutational analysis of a vaccinia virus intermediate promoter in vivo and in vitro. J. Virol. 64:6063–6069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Homann O. R., Johnson A. D. 2010. MochiView: versatile software for genome browsing and DNA motif analysis. BMC Biol. 8:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Keck J. G., Baldick C. J., Moss B. 1990. Role of DNA replication in vaccinia virus gene expression: a naked template is required for transcription of three late transactivator genes. Cell 61:801–809 [DOI] [PubMed] [Google Scholar]
- 16. Knutson B. A., Drennan M., Liu X., Broyles S. S. 2009. Bidirectional transcriptional promoters in the vaccinia virus genome. Virology 385:198–203 [DOI] [PubMed] [Google Scholar]
- 17. Knutson B. A., Liu X., Oh J., Broyles S. S. 2006. Vaccinia virus intermediate and late promoter elements are targeted by the TATA-binding protein. J. Virol. 80:6784–6793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moss B. 2007. Poxviridae: the viruses and their replication, p. 2905–2946 In Knipe D. M., et al., 5th ed vol. 2 Lippincott Williams and Wilkins, Philadelphia, PA [Google Scholar]
- 19. Moutaftsi M., et al. 2010. Uncovering the interplay between CD8, CD4 and antibody responses to complex pathogens. Future Microbiol. 5:221–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Senkevich T. G., Ojeda S., Townsley A., Nelson G. E., Moss B. 2005. Poxvirus multiprotein entry-fusion complex. Proc. Natl. Acad. Sci. U. S. A. 102:18572–18577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Szajner P., Jaffe H., Weisberg A. S., Moss B. 2004. A complex of seven vaccinia virus proteins conserved in all chordopoxviruses is required for the association of membranes and viroplasm to form immature virions. Virology 330:447–459 [DOI] [PubMed] [Google Scholar]
- 22. Vos J. C., Stunnenberg H. G. 1988. Derepression of a novel class of vaccinia virus genes upon DNA replication. EMBO J. 7:3487–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xiang Y., et al. 1998. The vaccinia virus A18R DNA helicase is a postreplicative negative transcription elongation factor. J. Virol. 72:7012–7023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang Z., Bruno D. P., Martens C. A., Porcella S. F., Moss B. 2011. Genome-wide analysis of the 5′ and 3′ ends of vaccinia virus early mRNAs delineates regulatory sequences of annotated and anomalous transcripts. J. Virol. 85:5897–5909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang Z., Bruno D. P., Martens C. A., Porcella S. F., Moss B. 2010. Simultaneous high-resolution analysis of vaccinia virus and host cell transcriptomes by deep RNA sequencing. Proc. Natl. Acad. Sci. U. S. A. 107:11513–11518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang Y. F., Keck J. G., Moss B. 1992. Transcription of viral late genes is dependent on expression of the viral intermediate gene G8R in cells infected with an inducible conditional-lethal mutant vaccinia virus. J. Virol. 66:6470–6479 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.