Abstract
That pigs may play a pivotal role in the emergence of pandemic influenza was indicated by the recent H1N1/2009 human pandemic, likely caused by a reassortant between viruses of the American triple-reassortant (TR) and Eurasian avian-like (EA) swine influenza lineages. As China has the largest human and pig populations in the world and is the only place where both TR and EA viruses have been reported to cocirculate, it is potentially the source of the H1N1/2009 pandemic virus. To examine this, the genome sequences of 405 swine influenza viruses from China were analyzed. Thirty-six TR and EA reassortant viruses were identified before and after the occurrence of the pandemic. Several of these TR-EA reassortant viruses had genotypes with most segments having the same lineage origin as the segments of the H1N1/2009 pandemic virus. However, these viruses were generated from independent reassortment events throughout our survey period and were not associated with the current pandemic. One TR-EA reassortant, which is least similar to the pandemic virus, has persisted since 2007, while all the other variants appear to be transient. Despite frequent reassortment events between TR and EA lineage viruses in China, evidence for the genesis of the 2009 pandemic virus in pigs in this region is still absent.
INTRODUCTION
Human pandemic influenza strains of the last century appear to have been generated by reassortment between previously circulating human and avian viruses (9, 12). Domestic pigs are often considered intermediate hosts that have facilitated these reassortment events, given that most pandemic or genetically related viruses have become established in swine (12). For example, classical H1N1 swine influenza (CS) and H3N2 human origin viruses have been prevalent in the swine population since soon after their emergence (11).
The only successful introduction of a wholly avian H1N1 influenza virus into mammals occurred in pigs, establishing the H1N1 “Eurasian avian-like” (EA) swine influenza lineage, which prevails across Europe and Asia (2). Triple-reassortant (TR) H3N2 and H1N2 swine viruses, which combine avian origin PB2 and PA genes with CS and human origin genes, provide further evidence that pigs act as “mixing vessels” to facilitate reassortment events (26). Indeed, pigs may be the only host in which both human and avian influenza viruses can persist (17). Thus, in pigs, avian viruses or their genes can become mammalian adapted, and reassortment of viruses of different origins will be facilitated.
The recent human H1N1/2009 pandemic (pdm/09) virus, which emerged in Mexico and the United States in April 2009 and subsequently spread throughout the world, was caused by a reassortant virus that had its NA and M segments derived from the EA lineage while its remaining segments were derived from the TR lineage (6, 19). This is the first clear evidence that reassortment involving only swine viruses could facilitate the emergence of a human pandemic virus and that avian-origin genes, after becoming “mammalian adapted” in pigs, could pass on to humans and be transmitted readily from human to human. In North America, H3N2 and H1N2 TR viruses have been maintained in pigs for the last decade. However, EA viruses are prevalent in Eurasia and have never been reported from North America.
Southern China is the only region where both TR and EA viruses have been reported to cocirculate (19, 25), creating an apparently ideal opportunity for reassortment events like that which led to the emergence of the pdm/09 virus. Consequently, the precursor of the pdm/09 virus could have been generated in southern China (19) and spread unnoticed to North America before the outbreak in humans. Molecular-clock dating has suggested there is a 9- to 17-year window of uncertainty in the time of origin of the virus, and no samples of precursor pdm/09-related viruses are available from this time (19). However, the lack of systematic influenza surveillance in pigs in many regions creates additional uncertainty as to the origin of the pdm/09 virus.
Surveillance of swine influenza will help in understanding the genesis of the H1N1/2009 pandemic. Recently, hundreds of swine influenza viruses isolated in Hong Kong were sequenced and analyzed. The findings revealed extensive reassortment among CS, TR, and EA viruses (19, 21, 22). Some H1N1 TR-EA (Sw/HK/72/2007-like) reassortants have persisted since 2007, establishing a genetic lineage in the pig population of this region (19, 21, 22). However, the key events in the reassortment between the TR and EA virus lineages that could have led to the genesis of the pdm/09 virus have not yet been defined.
In this study, we infer the evolutionary history of swine influenza viruses sampled in Hong Kong from 1998 to 2010 and in Guangdong from 2009 to 2010, together with other complete influenza A genome sequences taken from GenBank. All TR-EA reassortant viruses generated in China were identified, and their evolutionary pathways and reassortment patterns were analyzed in detail. These TR-EA reassortant viruses, which include two groups of highly similar viruses, were generated from independent reassortment events and had different evolutionary pathways. Except for the Sw/HK/72/07 sublineage, none of the viruses could be isolated from more than one sampling occasion, and they are likely transient. Although TR-EA reassortment events occurred in the swine population in China before and after the pandemic, direct precursors of the pdm/09 virus were not recognized.
MATERIALS AND METHODS
Virus isolation and sequencing.
Swine influenza surveillance has been conducted since May 1998 in Hong Kong and since December 2009 in Guangdong. Tracheal or nasal swabs were collected fortnightly and weekly from slaughtered swine. Swab materials were inoculated into 9- to 10-day-old embryonated chicken eggs and Madin-Darby canine kidney (MDCK) cells. Virus isolates were identified and subtyped as previously described (13). The genomes of four isolates from Guangdong and one from Hong Kong were sequenced and verified following previously described procedures (14, 19, 21).
Sequence preparation and alignment.
The nucleotide sequences of all influenza A viruses with complete genomes were downloaded from GenBank (1) in November 2010. They were aligned, together with the sequences of the influenza virus strains isolated in this study and from earlier surveys (22, 27) using MUSCLE v3.5 (5). Sequences with >10 ambiguous nucleotides were excluded from the alignment, and columns were removed from the alignment if gaps occurred in >90% of the sequences. Manual editing and refinement of alignments was done in MEGA4 (20). Sequences of selected avian, human, and equine influenza viruses were combined with the complete genome data of all swine influenza viruses (see Table S1 in the supplemental material), resulting in data sets of eight gene segments that comprise a total of 20,168 sequences.
Phylogenetic analyses.
Maximum-likelihood (ML) phylogenies were inferred for each of the eight genome segments, using a heuristic tree search algorithm implemented in PhyML v2.4.5 (8). The GTR + I + Γ4 (general time-reversible model with invariant sites and 4 Γ distributed heterogeneous substitution rates) nucleotide substitution model was used in all cases. One thousand pseudoreplicates were generated for bootstrapping analyses using the same ML method in PhyML. The robustness of the ML topology was further evaluated with the topologies sampled by the Bayesian Monte Carlo Markov chain (BMCMC) method as implemented in MrBayes v3.2 (16) using a chain length of 5 × 106 steps, with sampling every 1,000 steps.
Genotype classification.
Individual host-specific lineages were identified from the phylogenies. Establishment of human- and swine-specific lineages in the phylogenies of different genome segments indicate their introduction from either the avian reservoir or established mammalian virus populations and has been reported previously (2, 11, 24, 26). Sublineages were defined as monophyletic lineages with >70% bootstrap support and >0.9 posterior probability. A small number of sublineages in some gene phylogenies (especially those of shorter genes) have lower support. However, they remain the best groupings identified from the ML estimates, as well as the bootstrap and Bayesian analyses.
Molecular-clock dating of reassortment events.
The time and order of reassortment events giving rise to a particular genotype were inferred from the time of divergence (tDIV) of the reassorted gene segments within their windows of uncertainty (10). tDIV was estimated using a uncorrelated log-normal relaxed molecular clock (3) in a BMCMC joint estimation framework as implemented in BEAST v1.6.1 (4). A Bayesian skyline demographic model and the GTR + I + Γ4 nucleotide substitution model with codon partitioning (18) were used. A chain length of 108 steps, sampled every 10,000 steps, was used. The convergence of the tDIV estimates was assessed using Tracer v1.4.1 (15).
Nucleotide sequence accession numbers.
The nucleotide sequences obtained in the present study are available from GenBank under accession numbers JN375282 to JN375321.
RESULTS
Virus data set.
All complete influenza A genome sequences from our surveillance of pigs in Hong Kong (1998 onward) and in southern China (2009 onward) (19, 21, 22, 27, and this work), were combined with all complete influenza A genome sequences from GenBank and subjected to phylogenetic analysis.
Genotype identification.
Swine influenza genotypes were characterized by the lineage of origin of each of their gene segments based on phylogenetic analyses (Fig. 1 and 2; see Fig. S1 to S3 in the supplemental material) (see Materials and Methods). A total of 36 viruses with genotypes comprised of TR and EA lineage segments were isolated in China (Table 1). They formed eight distinct genotype patterns, five of which had a majority of TR origin segments. Only two of these genotypes were represented by more than one virus. Four closely related viruses (Sw/GD/2693/2010-like; isolated on one sampling occasion) and two other distinct viruses (HK78/03 and HK915/04) had seven TR origin segments and an EA origin M segment. Twenty-four viruses similar to Sw/HK/72/2007 with seven EA origin segments and a TR origin NS segment were isolated on nine occasions. Four of the genotype patterns (nine viruses) had at least five internal gene segments that were from the same lineage as those of the pdm/09 virus. To trace the phylogenetic relationships of the gene segments in these TR-EA reassortants, the sublineage structure of swine influenza viruses from southern China was analyzed.
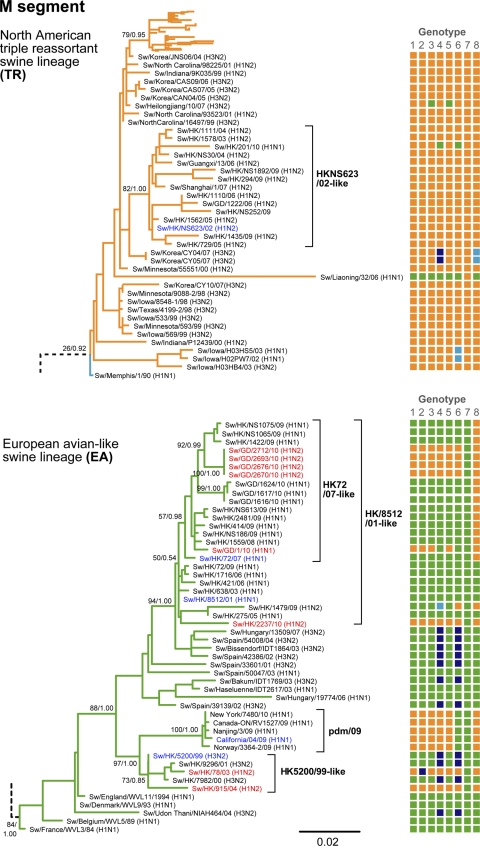
Fig. 1.
TR and EA phylogenies of the M gene of the influenza A virus. Shown is the ML phylogeny of the M gene, with numbers showing bootstrap support (left) and posterior probability (right) for selected nodes. The genotypes of these swine influenza viruses are shown on the right as an array of eight blocks representing each gene segment (numbers 1 to 8 indicate PB2, PB1, PA, HA, NP, NA, M, and NS, respectively), and the color of a block indicates the lineage in which that segment is located in the gene phylogeny (orange, TR; green, EA; blue, CS; purple, HuH3N2). The names of the TR-EA reassortant viruses are highlighted in red, and those of representative viruses for the sublineages are in blue. Only reference viruses relevant to the TR-EA reassortant viruses are included. The phylogenies of the complete data sets are shown in Fig. S1 to S3 in the supplemental material.
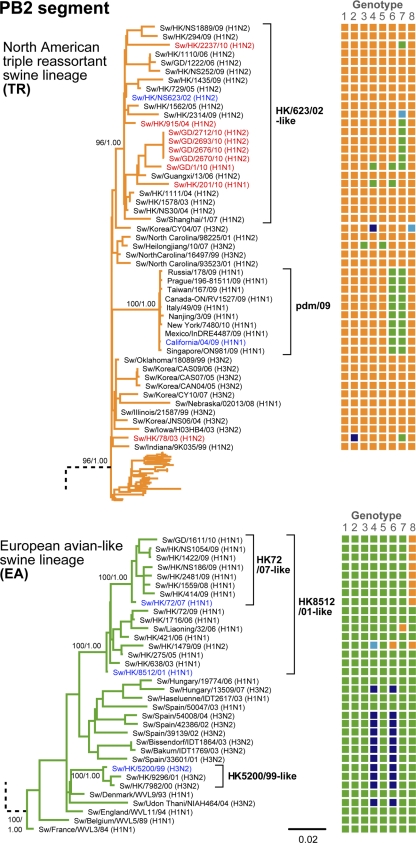
Fig. 2.
TR and EA phylogenies of the PB2 gene of the influenza A virus. See the legend to Fig. 1 for details. The names of the TR-EA reassortant viruses are highlighted in red, and those of representative viruses for the sublineages are in blue.
Table 1.
Genotypes of the TR-EA reassortant swine influenza viruses
| Strain name | Lineage assigned to gene segment |
Source | |||||||
|---|---|---|---|---|---|---|---|---|---|
| PB2 | PB1 | PA | HA | NP | NA | M | NS | ||
| CA/04/09 (H1N1) pdm/09 | TR | TR | TR | TR | TR | EA | EA | TR | 6 |
| Sw/HK/78/03 (H1N2) | TR | HU3a | TR | TR | TR | TR | EA | TR | 19 |
| Sw/HK/915/04 (H1N2) | TR | TR | TR | TR | TR | TR | EA | TR | 19 |
| Sw/HK/2237/10 (H1N2) | This study | ||||||||
| Sw/GD/2670/10 (H1N2) | This study | ||||||||
| Sw/GD/2676/10 (H1N2) | This study | ||||||||
| Sw/GD/2693/10 (H1N2) | This study | ||||||||
| Sw/GD/2712/10 (H1N2) | This study | ||||||||
| Sw/GD/1/10 (H1N1) | TR | TR | TR | EA | TR | EA | EA | TR | 23 |
| Sw/HK/201/10 (H1N1) | TR | TR | TR | EA | TR | pdm | TR | TR | 21 |
| Sw/Heilongjiang/10/07 (H3N2) | TR | TR | EA | TR | EA | TR | TR | TR | GenBank |
| Sw/HK/1479/09 (H1N2) | EA | EA | EA | CS | EA | TR | EA | TR | 21 |
| Sw/Liaoning/32/06 (H1N1) | EA | EA | EA | EA | EA | EA | TR | EA | GenBank |
| Sw/HK/72/07-likeb (H1N1) | EA | EA | EA | EA | EA | EA | EA | TR | 22, 27 |
HU3, H3N2 human lineage.
Sw/HK/72/07-like represents 24 related isolates.
Phylogenies of different swine influenza virus lineages from southern China.
Major groupings of North American and Eurasian avian, human, and CS influenza viruses were identified as basal lineages throughout the phylogenetic trees, with generally strong bootstrap support (Fig. 1 and 2; see Fig. S1 to S3 in the supplemental material). Distinct groups of the more recently emerged swine influenza lineages, EA and TR, could also be distinguished, as previously observed (2, 11, 24, 26). All major swine influenza lineages, CS, EA, and TR, were cocirculating in the pigs in southern China during our survey period.
Three major sublineages of Chinese swine viruses (HK103/93-like, HK86/79-like, and HK25/77-like) were identified in the CS lineage (see Fig. S1 to S3 in the supplemental material). It is possible there were multiple introductions of CS viruses into China, because these sublineages are intercalated with North American CS isolates in polytomic nodes in some gene phylogenies (see Fig. S1 to S3 in the supplemental material). Diversity in the HK103/93-like sublineage might have been caused by local evolution since the initial introduction of the parent virus around 1993 (22).
In the EA lineage, two monophyletic groups of swine isolates from Hong Kong and China represent regional viral circulation from 1999 to 2002 (HK5200/99-like; first isolate, Sw/HK/5200/99) and 2001 to 2010 (HK8512/01-like; first isolate, Sw/HK/8512/01) (Fig. 1 and 2; see Fig. S1 to S3 in the supplemental material). These sublineages indicate two separate introductions from Europe, with the second subsequently replacing the first. The HK5200/99-like sublineage was derived from an H3N2 reassortant variant (surface genes from human H3N2 virus and internal genes from an EA H1N1 virus) (Fig. 1). The HK8512/01-like sublineage evolved from a purely EA virus. Through further reassortment with TR viruses, a subgroup of viruses with TR NS segments emerged in 2007 (the HK72/07-like subgroup; first isolate, Sw/HK/72/07). The HK72/07-like subgroup became the first established group of TR-EA reassortant viruses detected in the pigs in southern China.
All TR viruses isolated in Hong Kong and Guangdong form a single monophyletic sublineage (HK623/02-like; first isolate, Sw/HK/NS623/02), except for one isolate that was a TR-EA reassortant, Sw/HK/78/03.
Evolutionary pathways of TR-EA reassortant viruses.
Detailed evolutionary pathways of the nine TR-EA reassortants (six distinct virus groups of four genotypes) that had at least five internal genes of similar lineage origin to the pdm/09 virus are shown in Fig. 3. Two of these viruses (HK78/03 and HK915/04) were isolated in Hong Kong before the 2009 pandemic outbreak (19). The M segments of both viruses were from the HK5200/99-like sublineage, while the PB1 segment of HK78/03 was derived from a contemporary human H3N2 influenza virus (closest to A/New York/336/99). All remaining segments were of TR origin, but those of HK915/04 belonged to the HK623/02-like sublineage while those of HK78/03 were more similar to TR viruses from North America and diverged close to the most recent common ancestor of all TR viruses (see Fig. S1 to S3 in the supplemental material).
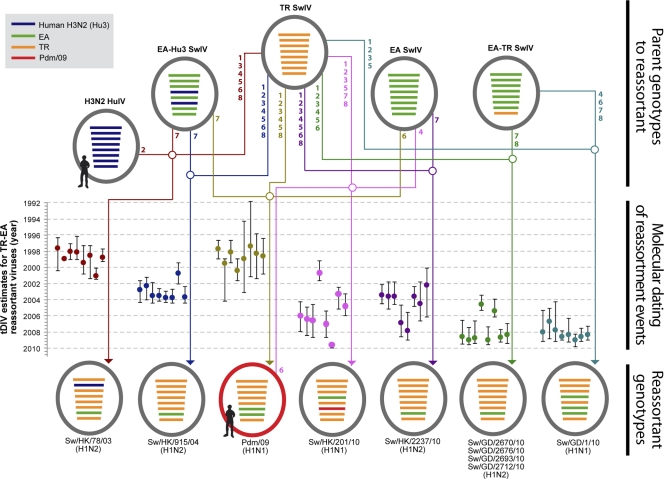
Fig. 3.
Genotypes and reassortment of TR-EA swine viruses. Each horizontal bar represents a gene segment (from top to bottom, PB2, PB1, PA, HA, NP, NA, M, and NS), and its color indicates the lineage from which it originated (see the legend). The arrows indicate the recombination sources (arrow tails) and the resulting reassortant (arrowheads). The numbers at the arrow tails indicate the segments that were contributed to the reassortant virus. Only genotypes relevant to the TR-EA reassortant viruses are shown. HuIV, AvIV, and SwIV are human, avian, and swine influenza viruses, respectively. Divergence time estimates (tDIV) are shown for each segment (PB2 to NS, left to right) of each reassortant virus. The sublineages of the parent viruses are H3N2 HuIV (closest to NY/336/99), EA-Hu3 (HK5200/99-like sublineage), TR SwIV (HK623/02-like sublineage and North American isolates), EA SwIV (HK8512/01-like sublineage and European isolates), and EA-TR SwIV (HK72/07-like subgroup).
The remaining seven viruses were isolated after the 2009 pandemic outbreak. Four of these viruses are very closely related to each other (GD2693/10-like). The other three viruses—GD1/10, HK2237/10, and HK201/10—were isolated separately. All TR origin segments of these viruses originate from the HK623/02-like sublineage, with the NS segment of the GD1/10 and GD2693/10-like viruses coming from the HK72/07-like subgroup of this sublineage. The EA origin segments of these viruses were mostly from the HK72/07-like subgroup of the HK8512/01-like sublineage. The M segment of HK2237/10 was from the larger HK8512/01-like sublineage, as was the hemagglutinin (HA) segment of HK201/10. The NA segment of HK201/10 was from the pdm/09 lineage. None of these virus groups clustered together in the phylogenies of all eight segments, indicating they were generated by individual reassortment events.
All remaining TR-EA reassortant viruses identified here (HLJ10/07, HK1479/09, LN32/06, and the HK72/07-like viruses) have fewer internal gene segments of common lineage origin with the pdm/09 virus. Apart from HLJ10/07, which has only PA and NP segments of EA origin, the majority of the segments of these viruses are of EA origin. All of the EA origin segments in these viruses are from the HK8512/01-like sublineage. LN32/06 has only the M segment of TR origin, and HK72/07-like viruses have only the NS segment of TR origin. Their remaining gene segments are from the EA lineage. HK1479/09 is a reassortant with the HA segment from the CS lineage and the NA and NS segments from the TR lineage. All other segments are from the EA lineage. The TR origin segments of HLJ10/07 and LN32/06 were derived from North American lineages, but those of HK1479/09 were from the HK623/02-like sublineage.
Time of emergence of TR-EA reassortant viruses.
The tDIV from the parent viruses was estimated for each of the segments of the pdm/09 virus and the nine TR-EA reassortants with at least five internal genes of similar lineage origin (Fig. 3). HK78/03 and the pdm/09 viruses appear to have been generated at approximately the same time, i.e., 1997 to 2001, whereas HK915/04, HK2237/10, GD2693/10-like, GD1/10, and HK201/10 formed subsequently. In HK201/10, the tDIV estimate of its pdm/09 origin NA segment was more recent than those of its TR and EA origin segments, suggesting a TR-EA reassortant precursor of HK201/10 was generated prior to the introduction of the pdm/09 gene segment. The TR origin genes of the pdm/09 and HK78/03 viruses are located outside the HK623/02-like TR sublineage, which was established after their estimated divergence times (1997 to 2001). In the other reassortant viruses, the TR origin segments are from the HK623/02-like sublineage.
Both early TR-EA reassortant viruses, HK78/03 and HK915/04, were estimated to have acquired their EA origin M segments from the HK5200/99-like viruses around 2000 to 2001 (Fig. 1 and 3), when the HK5200/99-like sublineage predominated in southern China. EA origin segments of the other viruses were acquired from the HK8512/01-like and HK72/07-like viruses, which have essentially replaced the HK5200/99-like sublineage.
DISCUSSION
Previous phylogenetic analyses revealed that the 2009 pandemic H1N1 (pdm/09) virus was a reassortant with two or three parents. It is generally accepted that all these parents belong to either the TR H1N2 swine influenza virus lineage originally present in pigs in North America or the EA H1N1 swine virus lineage circulating in pigs in European countries (6, 7, 19). However, cocirculation of both parent virus lineages in pigs, which could facilitate the reassortment events leading to the genesis and emergence of this TR-EA pandemic virus, is only known, from accessible epidemiological data, to have occurred in China since 2001.
In this study, all TR-EA reassortant viruses isolated in pigs in this region during the last decade were characterized, and their genotypes and evolutionary patterns were identified. TR-EA reassortants have been repeatedly isolated in China since 2003 and were also found on recent sampling occasions after the occurrence of the 2009 influenza pandemic (Table 1). However, these TR-EA reassortant viruses arose from independent reassortment events. The reassortants detected before the pandemic outbreak, HK915/04 and HK78/03, have not been isolated again in southern China, suggesting they were transient infections. Their initial transmissions might have been confined to a small swine population unable to sustain sufficient reinfections for effective adaptation to the host to occur.
Viruses with genotypes identical to that of the pdm/09 virus, in terms of the lineage origin of the segments, were not found in this region. Of the eight TR-EA reassortant genotypes detected in pigs in China, none had a TR origin H1 HA gene combined with an EA origin N1 NA gene (Table 1). Except for the pdm/09 virus itself, the TR-like H1 may not be naturally compatible with an EA-like N1. Incomplete compatibility of the surface genes might also be the reason why the reassortants identified here, apart from HK72/07-like viruses, could not become established in pigs. The TR-like NS gene was found in seven of the eight reassortant genotypes identified here. In reassortment with an EA virus, this segment contributed to the only persistent TR-EA genotype observed in pigs (HK72/07-like viruses) (Table 1).
Repeated emergence of TR-EA reassortant viruses over the last decade in China appears consistent with the proposal that the pdm/09 virus might have arisen in southern China and spread unnoticed to Mexico and the United States. However, the failure of any of these reassortants, except the mainly EA origin HK72/07-like viruses, to persist in pigs suggests that they may lack the characteristics of a pandemic virus or that the combinations of segments are insufficiently compatible to produce a stable virus.
All major swine influenza virus lineages were cocirculating in pigs in southern China, and reassortment between different virus lineages is not a rare event. However, our analyses revealed that none of the gene segments of pdm/09 virus, except the M segment, fall into the sublineages of the EA and TR viruses established in southern China. The pdm/09 M segment is either within or a neighbor of the HK5200/99 sublineage, which also contributed the M segment to the early TR-EA reassortants (HK78/03 and HK915/04). Most of the pdm/09 gene segments diverge from the clusters of North American TR and European EA isolates. Therefore, the probability of the pdm/09 virus having arisen in China is very low. As our knowledge of how to identify the pandemic potential of an influenza virus is incomplete, it is still unknown why the TR-EA reassortants identified here, despite having genotype origins related to the pdm/09 virus, failed to cause a pandemic outbreak.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the National Institutes of Health (National Institute of Allergy and Infectious Diseases contract HSN266200700005C), the Li Ka Shing Foundation, and the Area of Excellence Scheme of the UGC of the Hong Kong SAR (grant AoE/M-12/06) for financial support.
We thank the staff from the International Institute of Infection and Immunity (Shantou, China) and the State Key Laboratory of Emerging Infectious Diseases (Shenzhen and Hong Kong SAR, China) for technical assistance. We acknowledge the support of HPCPower projects for bioinformatics and computational services from the Computer Centre of The University of Hong Kong. We also thank W. K. Kwan and Frankie Cheung for technical assistance.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 27 July 2011.
REFERENCES
- 1. Bao Y., et al. 2008. The influenza virus resource at the National Center for Biotechnology Information. J. Virol. 82:596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brown I. H. 2000. The epidemiology and evolution of influenza viruses in pigs. Vet. Microbiol. 74:29–46 [DOI] [PubMed] [Google Scholar]
- 3. Drummond A. J., Ho S. Y., Phillips M. J., Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4:e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Drummond A. J., Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Edgar R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Garten R. J., et al. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gibbs A. J., Armstrong J. S., Downie J. C. 2009. From where did the 2009 ‘swine-origin’ influenza A virus (H1N1) emerge? Virol. J. 6:207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guindon S., Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 9. Kawaoka Y., Krauss S., Webster R. G. 1989. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J. Virol. 63:4603–4608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lam T. Y., et al. 2008. Evolutionary analyses of European H1N2 swine influenza A virus by placing timestamps on the multiple reassortment events. Virus Res. 131:271–278 [DOI] [PubMed] [Google Scholar]
- 11. Olsen C. W. 2002. The emergence of novel swine influenza viruses in North America. Virus Res. 85:199–210 [DOI] [PubMed] [Google Scholar]
- 12. Parrish C. R., Kawaoka Y. 2005. The origins of new pandemic viruses: the acquisition of new host ranges by canine parvovirus and influenza A viruses. Annu. Rev. Microbiol. 59:553–586 [DOI] [PubMed] [Google Scholar]
- 13. Peiris J. S., et al. 2001. Cocirculation of avian H9N2 and contemporary “human” H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? J. Virol. 75:9679–9686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Poon L. L., et al. 2010. Rapid detection of reassortment of pandemic H1N1/2009 influenza virus. Clin. Chem. 56:1340–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rambaut A., Drummond A. J. 2009, posting date Tracer v1.5. http://tree.bio.ed.ac.uk/software/tracer
- 16. Ronquist F., Huelsenbeck J. P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574 [DOI] [PubMed] [Google Scholar]
- 17. Scholtissek C., Hinshaw V. S., Olsen C. W. 1998. Influenza in pigs and their role as the intermediate host, p. 137–145In Nicholson K. G., Webster R. G., Hay A. J.(ed.), Textbook of influenza. Blackwell Science, Oxford, United Kingdom [Google Scholar]
- 18. Shapiro B., Rambaut A., Drummond A. J. 2006. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol. Biol. Evol. 23:7–9 [DOI] [PubMed] [Google Scholar]
- 19. Smith G. J., et al. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459:1122–1125 [DOI] [PubMed] [Google Scholar]
- 20. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 21. Vijaykrishna D., et al. 2010. Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science 328:1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vijaykrishna D., et al. 2011. Long-term evolution and transmission dynamics of swine influenza A viruses. Nature 473:519–522 [DOI] [PubMed] [Google Scholar]
- 23. Xu M., et al. 2011. Isolation and genetic analysis of a novel triple-reassortant H1N1 influenza virus from a pig in China. Vet. Microbiol. 147:403–409 [DOI] [PubMed] [Google Scholar]
- 24. Yu H., et al. 2008. Genetic evolution of swine influenza A (H3N2) viruses in China from 1970 to 2006. J. Clin. Microbiol. 46:1067–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yu H., et al. 2009. Isolation and genetic characterization of avian-like H1N1 and novel ressortant H1N2 influenza viruses from pigs in China. Biochem. Biophys. Res. Commun. 386:278–283 [DOI] [PubMed] [Google Scholar]
- 26. Zhou N. N., et al. 1999. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J. Virol. 73:8851–8856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhu H., et al. Novel reassortment of Eurasian avian-like and pandemic/2009 influenza viruses in swine: infectious potential for humans. J. Virol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.