Abstract
Geminiviruses are small DNA viruses that replicate in nuclei of infected plant cells by using plant DNA polymerases. These viruses encode a protein designated AL1, Rep, or AC1 that is essential for viral replication. AL1 is an oligomeric protein that binds to double-stranded DNA, catalyzes the cleavage and ligation of single-stranded DNA, and induces the accumulation of host replication machinery. It also interacts with several host proteins, including the cell cycle regulator retinoblastoma-related protein (RBR), the DNA replication protein PCNA (proliferating cellular nuclear antigen), and the sumoylation enzyme that conjugates SUMO to target proteins (SUMO-conjugating enzyme [SCE1]). The SCE1-binding motif was mapped by deletion to a region encompassing AL1 amino acids 85 to 114. Alanine mutagenesis of lysine residues in the binding region either reduced or eliminated the interaction with SCE1, but no defects were observed for other AL1 functions, such as oligomerization, DNA binding, DNA cleavage, and interaction with AL3 or RBR. The lysine mutations reduced or abolished virus infectivity in plants and viral DNA accumulation in transient-replication assays, suggesting that the AL1-SCE1 interaction is required for viral DNA replication. Ectopic AL1 expression did not result in broad changes in the sumoylation pattern of plant cells, but specific changes were detected, indicating that AL1 modifies the sumoylation state of selected host proteins. These results established the importance of AL1-SCE1 interactions during geminivirus infection of plants and suggested that AL1 alters the sumoylation of selected host factors to create an environment suitable for viral infection.
INTRODUCTION
Geminiviruses constitute a large family of plant viruses with circular, single-stranded DNA (ssDNA) genomes packaged within geminate particles (77, 82). They infect a broad range of plants and cause devastating crop diseases (57, 63). The family Geminiviridae is classified into four genera, Begomovirus, Curtovirus, Topocuvirus, and Mastrevirus, based on their genome organizations, host ranges, and insect vectors (25, 26). The largest genus corresponds to the begomoviruses, which can have bipartite genomes (A and B components), like Tomato golden mosaic virus (TGMV), or monopartite genomes, like Tomato yellow leaf curl Sardinia virus (TYLCSV).
Geminiviruses replicate through double-stranded DNA (dsDNA) intermediates (34). Begomoviruses encode two proteins involved in viral replication. AL1 (also called AC1, C1, and Rep) is essential for replication (23), while AL3 (also called AC3, C3, and REn) enhances viral DNA accumulation (86). AL1 is a multifunctional protein that mediates the virus-specific recognition of its cognate origin (28), is required for the initiation and termination of viral DNA synthesis (28, 49, 70), and acts as a DNA helicase (18, 19). A variety of protein interactions have been demonstrated for TGMV AL1 and other geminivirus replication proteins, including the formation of homomultimers (72) and interactions with AL3/REn (83, 84) and coat protein (CP) (56).
Geminiviruses do not encode their own DNA polymerases and rely on host nuclear DNA replication machinery, like many mammalian DNA tumor viruses. However, they are able to replicate in nuclei of mature plant cells that are inactive for DNA replication. Accumulating evidence strongly supports the notion that geminivirus proteins have a significant impact on a variety of host pathways (reviewed in references 31 and 35), including differentiation, cell cycle control, DNA replication, plasmodesma function, and RNA silencing. The AL1 protein binds to several host factors (1, 4, 5, 13–15, 46) and reprograms mature plant cells to create a permissive environment for viral replication. These factors include interactions with essential components of the DNA replisome, like proliferating cell nuclear antigen (PCNA) (62) and the host retinoblastoma-related protein (RBR), which regulates cell division and differentiation in plants (22).
Posttranslational modifications of proteins play critical roles in many cellular processes because they cause rapid changes in the function of preexisting proteins, multiprotein complexes, and subcellular structures. Their versatility in regulating protein function and cell behavior makes them particularly attractive targets for viruses. Sumoylation is a posttranslational process that involves the covalent attachment of a 10-kDa ubiquitin-like polypeptide (Ubl) called SUMO (also known as sentrin, Smt3 UPL, and PIC1) to a target protein (reviewed in references 2, 29, 36, and 87). Posttranslational modification by SUMO employs ATP-dependent reaction cascades that are mechanistically similar to ubiquitination, involving sequential activation and conjugation. SUMO activation is driven by an E1 enzyme (SUMO-activating enzyme SAE1/SAE2 heterodimer). SUMO conjugation is mediated by a single E2 enzyme (SUMO-conjugating enzyme [SCE1], also known as Ubc9) that is essential for cell viability and sumoylation in yeast, animals, and plants (37, 81, 85). The final transfer of SUMO from SCE1 to the substrate can occur directly or can be enhanced by SUMO ligases, such as the PIAS family proteins. Lysine-conjugated SUMO can be specifically cleaved by SUMO proteases (SENPs), making this a dynamic process (reviewed in references 10 and 65).
The addition of SUMO occurs exclusively at lysine residues, most commonly in the acceptor motif ΨKx(E/D), where Ψ is a large hydrophobic amino acid, K is the target lysine, x is any amino acid, and E/D is glutamic or aspartic acid. Two different extensions of the simple SUMO acceptor consensus site were identified recently. The phosphorylation-dependent sumoylation motif (PDSM) consists of a conventional sumoylation motif followed by a phosphorylated Ser and a Pro residue (ΨKxexxpSP) (39). The second acceptor consensus includes the negatively charged amino acid-dependent sumoylation motif (NDSM) (90).
Sumoylation is associated with diverse outcomes, ranging from changes in localization to altered activity and, in some cases, the stabilization of the modified protein. All of these effects might be the result of changes in the molecular interactions of sumoylated proteins (29). Sumoylation can mask a binding site in its target, inhibiting interactions with other proteins; increase the number of binding sites on its target, facilitating the binding of other molecules, such as proteins or DNA; or produce a conformational change that modulates activity.
The core components for sumoylation have been identified in Arabidopsis thaliana (48, 69, 81). The Arabidopsis genome encodes eight full-length SUMO genes (AtSUMO genes), a single gene for a SUMO-conjugating enzyme homolog of SCE1/Ubc9 (AtSCE1a), and a large number of SUMO proteases (20). Only three SUMO E3 ligases (SIZ1, HPY2, and MMS21) have been identified in Arabidopsis (41, 42, 61). SUMO influences a variety of plant responses to the environment (24, 60). It is involved in tolerance to cold, heat, drought, and salt stress (16, 21, 48, 59, 61, 92); modulates abscisic acid and cytokinin responses (41, 54); and has an important role in phosphate homeostasis (61). The loss of the SUMO, E1, or E2 enzyme leads to embryonic lethality (81), indicating that sumoylation is essential for normal plant development. Sumoylation controls the time of flower initiation (43, 67) and meristem and root development via cell cycle regulation (41, 42). Recent studies have identified SUMO target host proteins involved in DNA-related or RNA-dependent processes, such as the regulation of chromatin structure, splicing, and translation (11, 24, 58).
Several observations, including the pathogen manipulation of SUMO conjugation (40, 45, 76), the modification of SUMO levels altering pathogen infection in plants (15, 32), and sumoylation influencing innate immunity (52), indicate that SUMO also plays an important role in the plant defense response. In animal systems, an increasing number of proteins from both RNA and DNA viruses have been shown to modify the sumoylation status of host proteins either by preventing de novo sumoylation or by enhancing desumoylation. Many of these viral proteins are also targets of sumoylation (9, 78). In sharp contrast, only the interaction between AL1 and the sumoylation machinery has been described for plants (15). Here we map the SCE1 interaction motif in AL1 and present evidence that the interaction is required for viral infection and replication. We show that the AL1-SCE1 interaction does not alter the general sumoylation pattern in plant cells but may specifically influence the SUMO conjugation of selected host proteins.
MATERIALS AND METHODS
General methods.
Manipulations of Escherichia coli and Saccharomyces cerevisiae strains and nucleic acids were performed according to standard methods (79). Plant DNA gel blots were performed as described previously (15). E. coli strain DH5α was used for subcloning. All PCR-amplified fragments cloned in this work were fully sequenced. Agrobacterium tumefaciens strain LBA 4404 was used for the agroinfiltration assays.
Plasmids and cloning.
Cloning details are provided in the supplemental material. Table S1 in the supplemental material summarizes the engineering of the plasmids used in this work. Table S2 in the supplemental material contains all the oligonucleotides used in this study.
Yeast two-hybrid GAL4 system.
Yeast two-hybrid assays were performed as described previously (15). Yeast strain PJ696, which contains the three reporter genes lacZ, HIS3, and ADE2, was used in the two-hybrid screens (27). Yeast cells were cotransformed with bait and prey plasmids as described previously by Gietz (30). The transformation mixture was plated onto the yeast selective dropout (SD) selection medium lacking Trp and Leu (SD/−Trp−Leu) (19a). Transformants were recovered during a period of 3 to 5 days and checked for growth on three selection media: SD/−Trp−Leu−Ade, SD/−Trp−Leu−His, or SD/−Trp Leu−His−Ade. Quantitative β-galactosidase assays were performed in liquid cultures as described previously (14). Immunoblotting of the AL1 proteins expressed in yeasts was monitored with the ECL enhanced chemiluminescence detection system (Amersham Pharmacia Biotech AB). The primary antibody was a polyclonal anti-TGMV AL1 antibody (33). All constructs were tested in at least three independent experiments, with each experiment including four independent transformants per construct.
Infection and replication assays.
Nicotiana benthamiana plants were infected by bombardment or agroinoculation. For bombardment, the wild type or the mutant replicon (10 μg) for TGMV DNA-A was precipitated onto 1-mm gold microprojectiles in the presence or absence of the corresponding wild-type DNA-B replicon. The wild-type TGMV A and B plasmids were pMON1564 (modified as described in the supplemental material) and pTG1.4B (28), respectively. Agroinoculation was performed by using wild-type or mutant constructs for TGMV A and pGB2 and a TGMV B dimer cloned into pGA482 (7). Total DNA was extracted from young leaf tissue from individual plants. The DNA (2.5 μg/lane) was digested with XhoI, resolved on 1% agarose gels, transferred onto a nylon membrane, and hybridized with an α-32P-radiolabeled probe specific for DNA-A isolated as an EcoRI fragment (2,588 bp) from pMON1565 (70). The AL1 coding region recovered from total DNA extracted from infected plants was amplified by using primers AL1For and AL1Rev and sequenced.
Transient-replication assays were performed as described previously (28). Total DNA was extracted 72 h after transfection, digested with DpnI/XhoI, and examined for double- and single-stranded viral DNA accumulation by agarose gel blot analysis using α-32P-radiolabeled virus-specific probes against TGMV A. Each replication assay was performed in at least three independent experiments.
Bimolecular fluorescence complementation (BiFC) assays.
The reconstitution of yellow fluorescent protein (YFP) fluorescence was determined by the transient coexpression of selected protein pairs. N. benthamiana leaves were infiltrated with A. tumefaciens LBA4404 cells carrying the corresponding binary plasmids according to a previously described protocol (64). The reconstitution of YFP fluorescence was analyzed as described previously (3). Confocal laser scanning microscopy was performed as described previously (91), using a TCS SP5 2 (Leica) microscope. Fluorescence was quantified from four independent experiments by using ImageJ software (1997 to 2009; W. S. Rasband, U.S. National Institutes of Health, Bethesda, MD [http://rsb.info.nih.gov/ij/]).
Recombinant protein purification and AL1 DNA-binding and DNA cleavage assays.
The glutathione S-transferase (GST) and GST-AL1 (wild-type and mutant versions) expression cassettes from pNSB547 and pNSB314 were integrated into the parent bacmid vector bMON14272 in E. coli (55). Recombinant bacmid DNA was purified from E. coli and transfected into Sf9 cells (Bac-to-Bac baculovirus expression system; Invitrogen). The baculovirus-mediated expression of GST and the GST-AL1 protein was confirmed by immunoblot analysis with anti-GST (Sigma-Aldrich) and anti-AL1 (33) polyclonal antibodies. High-titer lysates were prepared and used for subsequent infections and protein production as described previously (70).
DNA electrophoretic mobility shift assays were performed as described previously (73), using the wild-type and mutant versions of the GST-AL1 protein produced in Sf9 cells. Wild-type and mutant versions of the His10-tagged AL11-180 protein were expressed in E. coli, purified by Ni affinity chromatography, and used for DNA cleavage assays as described previously by Nash et al. (68).
RESULTS
Mapping of the SCE1-interacting domain of AL1.
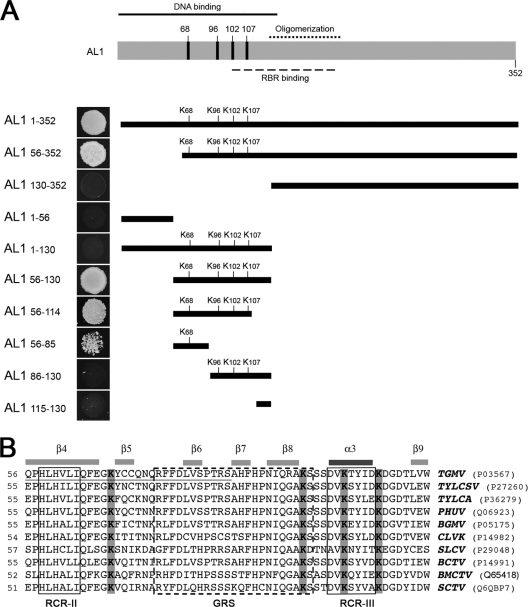
Previous work from our laboratory showed that TGMV AL1 interacts with Nicotiana benthamiana SCE1 (NbSCE1) through its N-terminal half (15). To map this interaction further, we examined the abilities of a series of truncated AL1 proteins to bind to SCE1. The TGMV AL1 truncations were expressed as fusions with the GAL4 DNA-binding domain (DBD), and the interaction with an activation domain (AD)-NbSCE1 fusion was analyzed by using yeast two-hybrid growth assays (Fig. 1A). When the first 130 residues of AL1 (AL1130-352) were removed, the interaction with NbSCE1 was abolished. However, when residues 1 to 56 (AL156-352) were deleted, no reduction in yeast growth was detected. Furthermore, AL1 fragments comprising amino acids 56 to 130 (AL156-130) or amino acids 56 to 114 (AL156-114) were able to induce yeast growth with an efficiency similar to that of full-length AL1. To further characterize the SCE1-binding motif, we analyzed the interaction of AL1 fragments comprising residues 56 to 85 (AL156-85), residues 86 to 130 (AL186-130), and residues 115 to 130 (AL1115-130). An interaction was detected only with AL156-85, although its strength was significantly reduced relative to that of the full-length protein. The interaction differences between the various baits were not due to variations in expression or stability, because immunoblot analysis using an anti-GAL4 DBD antibody showed that all of the truncated AL1-DBD fusions accumulated to similar levels in yeast (data not shown).
Fig. 1.
(A) Interaction between TGMV AL1 deletions and NbSCE1. Diagram of AL1 showing the position of the DNA-binding (71), oligomerization (72), and RBR-binding (46) domains. Boxes below the diagram indicate the sizes of truncated AL1 proteins designated by their N- and C-terminal amino acids. Lysine residues located in the SCE1-binding domain of AL1 are marked. Shown are data for the growth of yeast cells cotransformed with NbSCE1 and one of the AL1 fusion proteins (partial or complete AL1 clones) in media lacking histidine and containing 2 mM 3-amino-1,2,4-triazole (3-AT). Yeast growth images are on the left of each bar, representing the different AL1 protein deletions. (B) Comparison of the amino acid sequences of AL156-114 fragments from TGMV with the equivalent regions of AL1/Rep homologs from other geminiviruses (TYLCA, Tomato yellow leaf curl Australia virus; PHYVV, Pepper huasteco yellow vein virus; BGMV, Bean golden mosaic virus; CLVK, Cassava latent virus Kenya; SLCV, Squash leaf curl virus; BCTV, Beet curly top virus; BMCTV, Beet mild curly top virus; STCV, Spinach curly top virus). GenBank accession numbers corresponding to the Rep/AL1 protein sequences are indicated in parentheses. Lysine residues are highlight in gray. Predicted beta-sheets and alpha-helix structures are indicated. GRS (dotted box), RCR-II, and RCR-III (box) domains are also marked.
Together, these results showed that the SCE1-binding domain in AL1 is located between amino acids 56 and 114. Amino acids 56 to 85 are likely to form the core SCE1-binding domain, while sequences located between residues 86 and 114 may stabilize or enhance the interaction. We constructed a three-dimensional (3D) model for the amino-terminal region (amino acids 7 to 122) of AL1, using the ProMod method from Swiss-Model and homology with TYLCSV AL1/Rep (79.3% identity within this region) (12). As shown in Fig. 1B, the core SCE1-interacting region of AL1 (residues 56 to 85) corresponds to a conserved structural motif with three beta-sheets (β4, β5, and β6), while residues 86 to 114 correspond to three beta-sheets and one alpha-helix (β7, β8, α3, and β9). The complete sequence contains two of the three conserved motifs (RCR-II and RCR-III) characteristic of enzymes mediating rolling-circle replication (RCR) and the intervening geminivirus Rep sequence (GRS) element found in all geminivirus AL1/Rep proteins (47, 68).
Previous results located the SCE1-binding domain of AL1 between amino acids 130 and 180, as a truncated AL1 protein corresponding to residues 1 to 180 supported yeast growth, while the fragment encompassing amino acids 1 to 130 (AL11-130) did not (15) (Fig. 1). Since both truncated proteins accumulate at similar levels, the lack of an interaction between SCE1 and AL11-130 must be due to alterations in protein structure, possibly because of incorrect protein folding in yeast.
Mutation of lysine residues of the AL1-SCE1-interacting domain alter binding to SCE1.
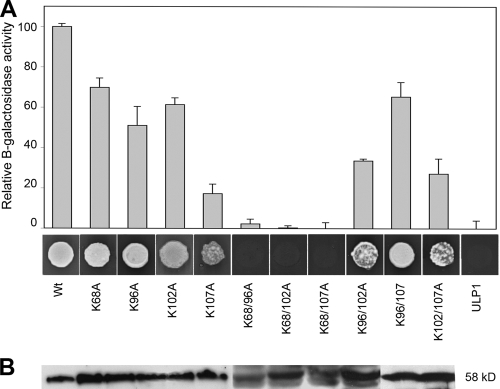
Previous results suggested that sumoylation substrates interact with SCE1 primarily through the sumoylation motif Y-K-xD/E (8, 53), while other studies with mammalian SCE1/UBC9-binding proteins indicated that SCE1 binds preferentially to hydrophobic regions containing LK and/or KL dipeptides (75). TGMV AL1 contains three putative sumoylation sites (positions 23 to 28, 141 to 147, and 297 to 301) identified by prediction programs (SUMOsp 2.0 [http://sumosp.biocuckoo.org/] and SUMOplot [Abgent]). It also has two conserved LK pairs (positions 270 to 271 and 327 to 328). However, all of these motifs are located outside the AL1-SCE1-binding region identified in Fig. 1A. Specific lysine residues also play crucial roles in most SCE1 interactions. The AL1-SCE1-binding domain (residues 56 to 114) contains four lysine residues (K68, K98, K102, and K107) that are conserved in most AL1/Rep homologs from begomoviruses, curtoviruses, and topocuviruses (Fig. 1B). Although previous results showed that K107 is required for DNA binding and cleavage (46, 71), no functions have been assigned to K68, K96, or K102. We generated alanine substitution mutations at the four lysine residues and determined the abilities of the corresponding AL1 mutant proteins to interact with SCE1. We produced single-site mutants (K68A, K98A, K102A, and K107A) and all combinations of double-site mutants (K68/96A, K68/102A, K68/107A, K96/102A, K96/107, and K102/107A) in TGMV AL1. The mutant AL1 open reading frames (ORFs) fused to the GAL4 DBD were expressed in yeast and analyzed for binding to AD-SCE1 in the two-hybrid system (Fig. 2A).
Fig. 2.
(A) Single and double TGMV AL1 mutant interactions with NbSCE1. Interactions were assayed by measuring the β-galactosidase activity in total protein extracts and are expressed as a percentage of the interaction between the wild-type (Wt) AL1 protein and NbSCE1. The negative control corresponds to yeast coexpressing S. cerevisiae ULP1 (ubiquitin-like protein) and NbSCE1. The bars correspond to data from an average of three independent experiments, each assayed for four independent transformants. The error bars correspond to two standard errors. (B) Immunoblot analysis of protein extracts from yeast cells coexpressing NbSCE1 and AL1 using an anti-AL1 polyclonal antibody. The molecular mass for the observed band is 58 kDa.
No obvious differences in growth were detected in yeast cells cotransformed with any of the single mutants. However, the strength of the interaction with SCE1 relative to that of wild-type AL1 was reduced for all the mutants, ranging from an 82% reduction for the K107A mutant to 32% for the K68A mutant, suggesting that the lysine residues are required for full binding activity. Double mutants containing the K68A substitution and one of the other lysine residues (K68A/K96A, K68A/K102A, or K68A/K107A) lost the ability to bind SCE1 almost completely. A minor, but noticeable, reduction was also detected for the K96A/K102A double mutant. However, the binding activities of double mutants containing any combination of K107A (K96A/K107A and K102A/K107A) were reduced significantly compared to those of the corresponding single mutants. All mutant proteins accumulated to levels comparable to those of a DBD-wild-type AL1 fusion (Fig. 2B), showing that the variations in the interaction efficiency were not due to differences in expression levels. Together, these results suggested that K68 plays a major role in SCE1 binding and that K96, K102, and K107 have redundant roles in the interaction.
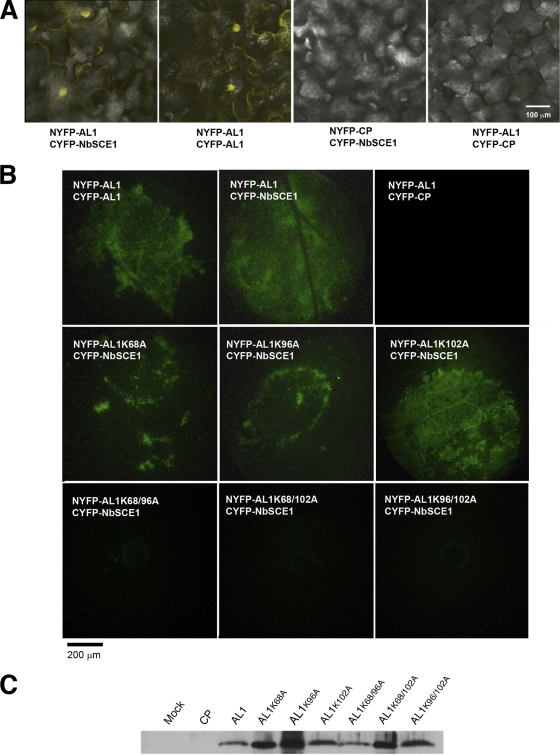
AL1-SCE1 interactions were confirmed in planta by using bimolecular fluorescence complementation (BiFC) with a split yellow fluorescent protein (YFP) reporter (44). To validate this experimental approach for the analysis of AL1 interactions, we first confirmed the ability of the BiFC assay to detect AL1 oligomerization in vivo. When N-terminal fusions of wild-type AL1 to both the N terminus of YFP (NYFP) and the C terminus of YFP (CYFP) were transiently coexpressed in N. benthamiana, YFP fluorescence was observed mainly in nuclei of the infiltrated leaf, which is indicative of AL1-AL1 interactions (Fig. 3A). We also observed YFP fluorescence mostly in nuclei of cells coinfiltrated with constructs corresponding to NYFP-AL1 and CYFP-NbSCE1 (Fig. 3A). In contrast, the expression of NYFP-AL1 or CYFP-NbSCE1 alone (data not shown) or the coexpression of NYFP-AL1 or CYFP-NbSCE1 with the coat protein of Prunus necrotic ringspot virus (PNRV) fused to NYFP or CYFP, respectively, did not generate fluorescence (Fig. 3A). Taken together, these results demonstrated that AL1 and SCE1 can associate in plant cells.
Fig. 3.
BiFC analyses showing in vivo oligomerization of AL1 and interaction with NbSCE1. N. benthamiana leaves were coagroinfiltrated with constructs fused to the C terminus (CYFP) or N terminus (NYFP) of YFP. Reconstituted YFP fluorescence was monitored 3 days after infiltration with a confocal microscope (A) or an epifluorescence binocular microscope with a Leica 10446364 filter for YFP emission (B). (A) Interaction of AL1 (wild type, AL1), NbSCE1, and CP from PNRV. (B) BiFC of AL1 (wild type and mutants) with NbSCE1. (C) Immunoblot analysis of protein extracts from leaves coagroinfiltrated with CYFP-NbSCE1 and wild-type or mutant AL1 or PNRV CP. Mock samples correspond to extracts from leaves agroinfiltrated with constructs expressing CYPF or NYFP. Fusion AL1 proteins were detected by using a polyclonal antibody against AL1. The molecular mass for the observed band is 58 kDa.
Since the K107A mutation affects other AL1 functions in addition to SCE1 binding (46, 71), we did not analyze this mutant further. To evaluate the role played by the K68, K96, and K102 residues in the AL1-SCE1 interaction in planta, we performed BiFC assays by coexpressing CYFP-NbSCE1 and NYFP fused to single and double mutant AL1 proteins. In these assays, the AL1 K102A mutant showed no significant difference in fluorescence compared to that of wild-type AL1. However, the substitution of either K68A or K96A reduced fluorescence in the infiltrated tissue 74% and 58%, respectively, relative to that of wild-type AL1. Furthermore, when a second AL1 mutation was introduced (K68A/K96A, K68A/K102A, or K96A/K102A), no fluorescence was detected (Fig. 3B). These results indicated that the K68 and K96 residues may be involved in SCE1 binding in planta, as observed for the two-hybrid assays. However, the K102A mutation alone did not alter the strength of the interaction. Immunoblot analysis using an anti-AL1 polyclonal antibody to probe total protein extracts from infiltrated leaves indicated that the mutant AL1 proteins accumulated to similar or higher levels than those of the wild-type protein, indicating that variations in interactions were not due to poor expression (Fig. 3C).
The K68A mutation reduces viral DNA accumulation and symptom severity in plants.
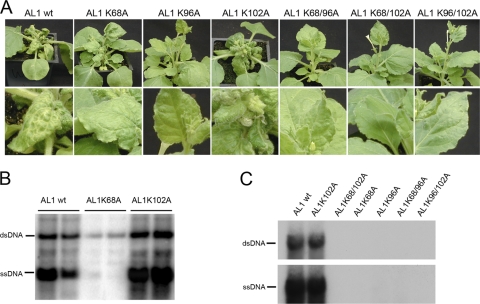
To determine whether the K68A, K96A, or K102A mutation affects viral infection or symptom development, we generated viral replicons containing these AL1 mutations and used them in plant infection and transient-replication assays. Plant infections were carried out on N. benthamiana plants by the cobombardment of TGMV B and either wild-type or mutant TGMV A replicons, with the latter carrying single or double AL1 point mutations. Plants inoculated with wild-type virus developed symptoms by 6 to 7 days postinoculation, exhibiting leaf curling, general chlorosis, and stunted growth (Fig. 4A). The K102A mutant virus caused symptoms that were indistinguishable from those caused by the wild-type virus, indicating that this mutation does not visibly alter the infection process. In contrast, plants inoculated with the K68A mutant virus developed only mild stunting and leaf curling and never displayed chlorosis. The K96A single mutant and all the double mutants (K68A/K96A, K68A/K102A, and K96A/K102A) did not produce detectable symptoms at 6 weeks postinoculation (Fig. 4A).
Fig. 4.
(A) Symptoms of N. benthamiana cobombarded with TGMV B and wild-type (wt) or mutant AL1 TGMV A replicons. Leaf details for each plant are shown in the bottom panels. (B) Total DNA (2.5 mg/lane) was isolated from systemically infected leaves from two plants (wild type and the K68A and K102A mutants) at 19 days postinfection and analyzed on DNA gel blots. TGMV DNA was detected by using a radiolabeled probe specific for the A component. The two intense bands for each line represent double- and single-stranded forms of TGMV A. (C) Tobacco protoplasts were transfected with TGMV A and B replicons with either wild-type or mutant AL1 ORFs. Total DNA was isolated from cells at 72 h posttransfection and analyzed on DNA gel blots by using a radiolabeled probe specific for the A component.
We examined TGMV DNA levels in N. benthamiana plants inoculated with either wild-type or mutant viral constructs. Samples from infected leaves at 14 days postinoculation were analyzed by the hybridization of a TGMV A-specific probe to leaf tissue prints (see Fig. S4 in the supplemental material). Viral DNA accumulation correlated with the intensity of the symptoms. All plants infected with wild-type and K102A mutant viruses showed equivalent amounts of viral DNA accumulation. A reduction in viral DNA levels was observed for plants infected with the K68A mutant, suggesting that lower levels of viral DNA are associated with mild symptoms. No viral DNA was detected in any of the plants infected with the K96A mutant or any of the three double mutants. Viral ssDNA and dsDNA accumulation patterns were examined on DNA gel blots of total DNA from systemically infected leaves hybridized with a TGMV A-specific probe. Plants infected with the K102A mutant contained essentially wild-type levels of viral ssDNA and dsDNA (Fig. 4B). In contrast, levels of both DNA forms were reduced in K68A mutant-inoculated plants relative to wild-type-inoculated plants. The decrease was more apparent for ssDNA than for dsDNA.
To confirm that the viral DNA in plants infected with the K68A or K102A mutant did not result from the replication of revertants, we extracted DNA from symptomatic young leaves collected at 19 days postinfection from plants inoculated with the corresponding mutant viruses (three plants per infection). A 471-bp fragment encoding AL1 amino acids 42 to 199 was amplified with primers AL1For and AL1Rev and fully sequenced. All analyzed fragments contained the mutations, confirming that the alanine replacement of K68 and K102 is stable in infected plants.
To determine whether the reduced viral DNA levels in plants infected with the mutants was due to a defect in replication and/or dissemination through the plant, we monitored viral DNA accumulation in Nicotiana tabacum BY-2 protoplasts transfected with the TGMV A replicons. In these assays, only the K102A mutant supported viral replication (Fig. 4C). No nascent viral DNA corresponding to the K96A mutant and the double mutants was detected in the replication assays, indicating that their inability to infect plants is due to a replication defect. The replication of the K68 mutant was also not observed in transient assays, even though it causes attenuated symptoms and accumulates to low levels in plants. This discrepancy most likely reflects the ability of the K68 mutant to move into adjacent cells and replicate in infected plants but not in protoplasts, thereby facilitating the detection of very low levels of viral replication in planta.
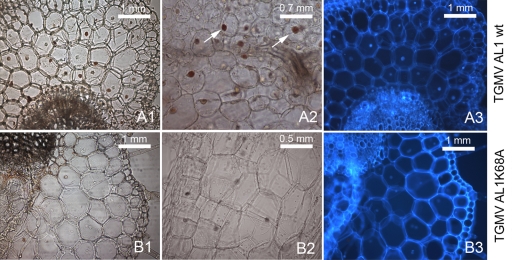
Tissue sections from infected N. benthamiana plants showed that even though the intensity of the signal per cell was lower for the K68A AL1 mutant than for the wild-type virus, there was no difference in the numbers of infected nuclei (Fig. 5B1 and B2), indicating that variations in viral DNA accumulation are consistent with a decrease in viral production rather than a reduction in the number of infected cells in the plant (Fig. 5).
Fig. 5.
Immunolocalization of TGMV in infected N. benthamiana plants. Panels A1, A2, and A3 correspond to wild-type TGMV A- and B-inoculated plants, while panels B1, B2, and B3 correspond to plants infected with TGMV B and the TGMV A K68A mutant. Panels A3 and B3 are the same sections as those in panels A1 and B1, respectively, but with 4′,6-diamidino-2-phenylindole (DAPI)-stained nuclei. Sectioning was performed 19 days after plant infection, and a polyclonal AL1 primary antibody was used. Arrows indicate examples of infected nuclei.
Mutations in K68 and K102 do not alter other AL1 functions.
To rule out a potential role of the mutated lysine residues in other protein interactions, we first asked if the AL1 mutants can form oligomers in two-hybrid assays. Oligomerization was assayed by the cotransformation of yeast cells with constructs expressing the mutant version of the protein fused to the Gal4 DBD and AD. No significant differences were observed for any of the mutant fusions and the wild-type AL1 fusions in reporter gene assays (see Fig. S1A in the supplemental material).
We then examined the abilities of the AL1 mutants to interact with AL3 and RBR because their binding regions (amino acids 101 to 180) overlap with the SCE1-binding region in AL1 (1, 46, 83). We coexpressed TGMV AL1 and AL3 as GAL4 DBD and AD fusions, respectively. The K68A and K102A single mutants and the K68A/K102A double mutant showed AL3-binding activities similar to those of wild-type AL1 (see Fig. S1C in the supplemental material). In contrast, the AL1 K96A mutant displayed reduced AL3-binding activity. Although TGMV AL1 interacts with RBR proteins from both maize and Arabidopsis in the yeast two-hybrid assays, binding to maize RBR is stronger and easier to detect (1, 46). Thus, we assayed AL1 binding to a fusion to the GAL4 DBD of a truncated version (Zm214C) of maize RBR1, comprising the pocket-C-terminal region of the protein (amino acids 214 to 866) (1, 46). Neither the single nor the double mutations altered AL1-RBR binding significantly (Fig. S1B).
AL1 also specifically binds to DNA direct repeats in the 5′-intergenic region of the TGMV genome (28) through a domain located in the first 130 amino acids of the protein (73) (Fig. 1A). To determine whether the lysine substitutions that affect the AL1-SCE1 interaction also alter the AL1 DNA-binding activity, we used GST fusions of wild-type and mutated AL1. GST-AL1 fusion proteins were expressed in insect cells and partially purified by binding to glutathione resin. The DNA-binding activities of the fusion proteins were tested in electrophoretic mobility shift assays by titration with a radiolabeled dsDNA fragment (comprising TGMV A nucleotides 62 to 92) that includes the AL1-binding site. Shifted complexes were observed with wild-type AL1 and all the mutant proteins. No significant differences were detected in either the intensity or the number of the retarded complexes between wild-type AL1 and the mutant versions (see Fig. S2 in the supplemental material).
It was shown recently that an AL1 mutant encompassing the K96A mutation is not competent for DNA cleavage (68), which could account for the failure of this mutant to infect plants and support viral replication. In contrast, recombinant His10-tagged proteins corresponding to wild-type AL11-180 and the K68A or K68A/K102A mutant displayed similar cleavage activities in assays using a fluorescently labeled ssDNA containing the origin cleavage site (see Fig. S3 in the supplemental material). Based on these results and those described above, we concluded that the K68A and K68A/K102A mutations do not significantly alter activities known to be mediated by regions that overlap or contain the SCE1-binding region in AL1.
The AL1-SCE1 interaction does not depend on K68 or K102 sumoylation.
It was shown previously that SCE1-binding motifs often include lysine residues that are sumoylated (80). To determine if the role of K68 and K102 in the AL1-SCE1 interaction depends on their sumoylation, we generated single and double mutants of K68 and K102 by replacing the lysine residue with arginine, which maintains the positive charge but eliminates any chance of conjugation to SUMO or ubiquitin. Yeast two-hybrid assays showed that single K68R and double K68/102R mutants were altered in NbSCE1 binding (see Fig. S5 in the supplemental material), indicating that the AL1-SCE1 interaction does not depend on the sumoylation of K68 or K102.
Recombinant viruses, including single K68R or K102R or double K68/102R mutations, produced symptoms in N. benthamiana plants that were indistinguishable from those caused by the wild-type virus (see Fig. S5A in the supplemental material). Furthermore, all plants infected with wild-type and mutant viruses showed equivalent amounts of viral DNA accumulation (Fig. S5B). The stability of the K68R and K102R mutations during infection was confirmed as described above for the corresponding alanine mutations. Consistent with this result, all attempts using in vitro and in vivo assays failed to detect AL1 sumoylation (data not shown).
AL1 modulates sumoylation of host targets.
There is evidence that animal viruses can alter the sumoylation status of host proteins (9). To gain insight into whether AL1-SCE1 interactions affect host cell sumoylation, we expressed AL1 in plants and analyzed changes in the overall host sumoylation pattern. Leaves from young N. benthamiana plants were infiltrated with A. tumefaciens cultures containing a binary plasmid that expresses TGMV AL1 from a 35S Cauliflower mosaic virus (CaMV) promoter (pBINX′-AL1). Total protein extracts from agroinfiltrated leaves were analyzed by immunoblotting using an anti-SUMO1 antibody (Fig. 6A). The numbers and the intensities of cross-reacting bands in AL1-expressing and control leaf samples were similar, indicating that AL1 does not alter the general sumoylation pattern of plant proteins. However, the intensities of two bands migrating at 50 and 82 kDa were enhanced in the AL1-expressing samples. Although the size of the 50-kDa band matches that expected for AL1-SUMO (51 kDa), it does not correspond to the viral protein, because immunoblots of the same extracts using an anti-AL1 polyclonal antibody did not cross-react with the 50-kDa band (Fig. 6B). It is unlikely that the sumoylation of AL1 would preclude its detection when a polyclonal antibody is used. Taken together, these results indicated that AL1 expression does not have a global effect on the host sumoylation system. Nevertheless, the two changes observed for AL1-expressing N. benthamiana leaves suggested that the viral protein might modulate the sumoylation state of the specific host target(s). However, since AL1 is sufficient to induce host transcription (35), we cannot rule out that the changes correspond to sumoylated proteins that are strongly induced in response to AL1 and are not detected in the control plants due to low expression levels.
Fig. 6.
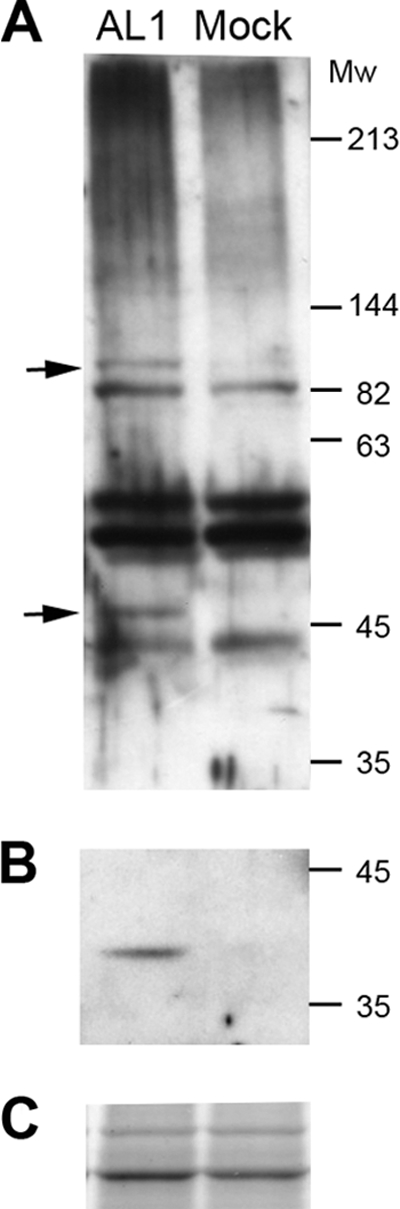
Accumulation of SUMO protein conjugates in N. benthamiana leaves ectopically expressing AL1. Protein extracts from agroinfiltrated N. benthamiana leaves expressing AL1 were analyzed by protein gel blots using an anti-NbSUMO1 polyclonal antibody (N. benthamiana) (A) or an anti-AL1 polyclonal antibody (B). Each lane contained 35 mg (N. benthamiana) of total protein extract. Similar results were obtained in three independent experiments. Arrows indicate bands that accumulated differentially when AL1 was expressed. (C) Loading controls. Mw, molecular weight (in thousands).
DISCUSSION
Given the small size of geminivirus DNA genomes, their replication and movement are dependent largely on host factors. Small DNA viruses typically encode a few multifunctional proteins that mediate replication, regulate the expression of their own genes, and manipulate the cell machinery to create an appropriate cell environment for viral reproduction. Protein interactions between viral and host proteins play key roles in all these processes. Geminivirus AL1/Rep is a multifunctional protein that interacts with many cellular factors, including SCE1, the conjugating enzyme (E2) of the sumoylation system (15). In the last few years, this posttranslational modification system has emerged as a central theme in the regulation of protein function. Consequently, it is no surprise that viral proteins were among the first described to exploit the host SUMO system. To better understand the biological role of the interaction between AL1/Rep and SCE1, we mapped the SCE1-binding domain of AL1, constructed viral mutants affected in the interaction, and analyzed the effect of these mutations on viral infection and replication.
Two-hybrid assays of truncated AL1 proteins located the SCE1-binding site in the N terminus of the viral protein to a region that spans residues 56 to 114. The lower level of SCE1-binding activity of AL156-85 than that of AL156-114 and the inability of AL186-130 to bind to SCE1 suggest that the region consists of a core region encompassing residues 56 to 85 that is essential for the SCE1-AL1-binding interaction and a supporting region including residues 86 to 114 that enhances or stabilizes the interaction. This model is also supported by the observation that the combination of an AL1 K68A mutation with a K96A, K102A, or K107A mutation abolishes SCE1 binding in yeast. The SCE1-interacting region is located within the DNA-binding and cleavage/ligation domains of AL1 (71, 73) and overlaps with the AL3- and RBR-binding regions (46, 83) but is outside the oligomerization core. In a three-dimensional model of AL1 based on the previously reported structure of the homolog TYLCSV Rep (12), the lysines analyzed in this work are located on the surface of the protein (Fig. 7), where they are accessible for interactions with other proteins.
Fig. 7.
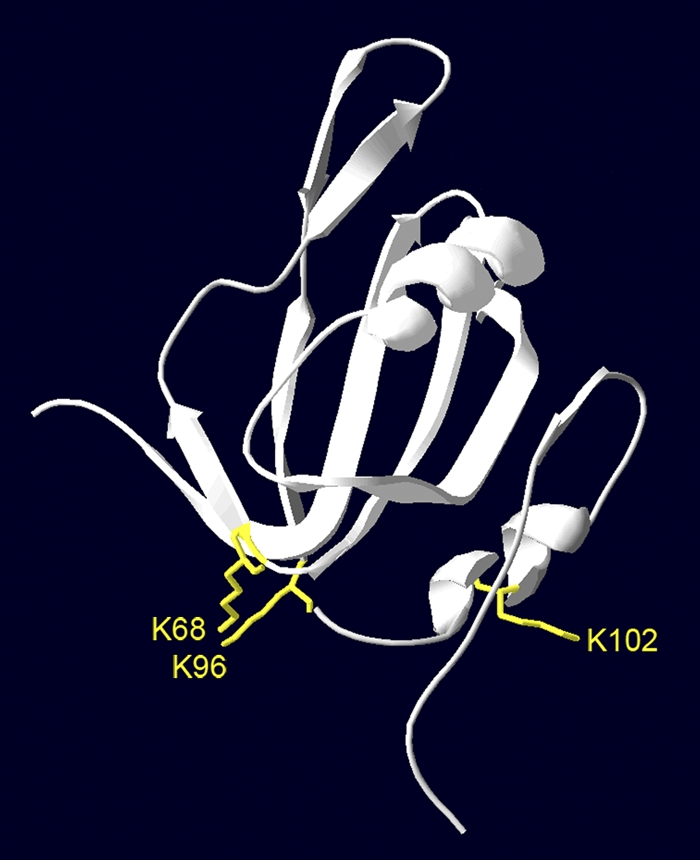
Three-dimensional model of the AL1 N terminus (residues 4 to 122). Lysine residues analyzed in this work are highlighted in yellow. The model is based on the previously reported structure of the homolog TYLCSV AL1/Rep (12).
BiFC results established key roles for K68 and K96 in the AL1-SCE1 interaction in planta, since a clear reduction in such interactions was detected with both the K68A and K96A mutant versions of AL1. BiFC assays also indicated that K102 is not essential for the interaction in the context of the plant but plays a supporting role by enhancing the effect of the K68A mutation, since there were no significant differences between wild-type AL1 and the K102 single mutant, while leaves agroinfiltrated with the K68A/K102A and K96A/K102A double mutants did not display any fluorescent signal. Although the interaction was reduced to a similar extent in both double mutants, suggesting an equivalent contribution to the AL1-SCE1 interaction for both lysine residues (K68 and K96A), the quantitative yeast two-hybrid analysis indicated that these residues do not play an equal role in SCE1 binding. The difference in the reduction of the interaction shown by the K96A/K102A mutant in yeast and in planta could also be due to differences in the cell type. The position predicted by the AL1 three-dimensional model for the three lysine residues supports the results. Lysines 68 and 96 are located close together and point in the same direction, while K102 is positioned on opposite sides of the structure (Fig. 7). SCE1-AL1 binding is likely to be mediated by the protein region where K68 and K96 are located, while additional interactions that enhance binding may involve residues on the opposite side of the protein. The three lysines involved in SCE1 binding are highly conserved among all begomoviruses and curtoviruses. K68 and K96 are also conserved in mastreviruses, while K102 is either absent or has been replaced by an arginine.
Infection analyses and viral replication assays of tobacco protoplasts with the lysine mutants suggested a direct link between the AL1-SCE1 interaction and efficient viral replication. The mutants with a diminished capacity to interact with SCE1 (the K68A and K96A single mutants and all the double mutants) were affected in their ability to infect the plant, while the K102A single mutant was indistinguishable from the wild-type virus. Likewise, the K68A and K96A single mutants and all the double mutants displayed no detectable replication in protoplast assays, while the K102A mutant replicated to wild-type levels. However, while the K96A mutation and all double mutations seemed to disable viral replication completely, the severe restriction of K68A replication was not complete, because this mutant was still able to infect the plants.
We showed that single or double mutants of lysines 68, 96, and 102 are active for DNA binding, oligomerization, and interactions with RBR, and it is therefore unlikely that the mutations alter the general structure of the AL1 protein. However, although the K68A and K96A single mutants exhibited a similar reduction in SCE1 interactions, they displayed clear differences in DNA replication and infectivity, with the latter displaying an almost complete failure to replicate. The radical replication defect of the K96A mutant may reflect the involvement of this residue in additional AL1 activities other than SCE1 binding, such as AL3 binding (see Fig. S1C in the supplemental material) or DNA cleavage (68). In contrast, the K68A mutant seemed to be affected exclusively by the SCE1-AL1 interaction. Although the K102A mutation does not affect viral replication or SCE binding, the additive effect on both activities observed for the K68A/K102A double mutant further suggests a functional correlation between SCE1 binding and replication. This notion is also supported by the results obtained with the arginine substitutions for K68 and K102, which do not impair AL1-SCE1 binding or affect the ability of the mutant viruses to infect and replicate in plants.
Interactions with the sumoylation system have been described for several proteins encoded by mammalian DNA viruses that replicate in the nucleus. Among these DNA viruses, all viral proteins known to interact with the sumoylation system are immediate-early or early proteins, like AL1/Rep (reviewed in references 9, 17, 88, and 89). The biological effects of these interactions are in most cases the sumoylation of the viral protein and/or the interference with the host sumoylation program. Although TGMV AL1 has three putative sumoylation sites, we have not detected a sumoylation of the viral protein using many different experimental approaches. This negative result does not exclude the possibility that AL1 is a target for SUMO modification, especially if only a small fraction of the protein is modified.
In addition to taking advantage of their host modification system to sumoylate their own proteins, viruses can alter the sumoylation of host proteins either by preventing or inducing de novo sumoylation or by enhancing desumoylation, as long as the outcome is a more favorable environment for viral propagation. For example, the adenovirus Gam1 and papillomavirus E6 proteins cause widespread changes in the sumoylation level of host proteins by inhibiting E1/E2 sumoylation enzymes (17, 38, 74). In other cases, the effect on host sumoylation is not general but restricted to specific host factors. This specific effect can result in a reduction of sumoylation, as exemplified by herpes simplex virus (HSV) ICPO and human cytomegalovirus (HCMV) IE1, which reduce the sumoylation of the promyelocytic leukemia protein (PML) (6, 51), and the papillomavirus E7 and E1A proteins, which abrogate the SUMO modification of the pRB tumor suppressor (50). Conversely, viral proteins can increase the sumoylation of specific proteins, as illustrated by the adenovirus E1B-55K stimulation of p53 sumoylation (66). The overexpression of AL1 in plant leaves does not produce an overall alteration of host protein sumoylation, as would be expected if the interaction with AL1 modified SCE1 activity. Thus, it seems more likely that by interacting with SCE1, AL1 modulates the sumoylation level of a subset of host targets, creating a suitable environment for viral replication. Since this multifunctional viral protein is able to interact with host proteins that are sumoylated (for example, PCNA and RBR), we speculate that AL1/Rep stimulates or impairs SUMO attachment to one or more of its host protein partners. The determination of the number and identity of these plant targets will require a broad proteomic analysis, with prime candidates being host proteins that are sumoylated and also interact with AL1/Rep.
From the results generated in the last 10 years, it is clear that SUMO is important for animal viruses. The results presented in this paper demonstrate that this is also true for plant DNA viruses. Although many putative effects have been attributed to sumoylation, the precise biological role that this process plays in terms of viral fitness remains to be determined. Future experiments that identify host proteins whose sumoylation is modulated by AL1/Rep will clarify the role of the AL1-SCE1 interaction during infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank Vicente Pallas for providing the bimolecular fluorescent complementation vectors. We thank M. A. Botella, C. Gutiérrez, R. Flores, and J. Jiménez for helpful suggestions and discussions.
This research was supported by grants from the Spanish Ministerio de Ciencia y Tecnología (AGL2007-66062-C02-02/AGR) and the National Science Foundation (MCB-0110536) to L.H.-B. M.A.S.-D. was awarded a predoctoral fellowship from the Junta de Andalucia and an EMBO short-term fellowship (ASTF no. 240-05).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 20 July 2011.
REFERENCES
- 1. Ach R. A., et al. 1997. RRB1 and RRB2 encode maize retinoblastoma-related proteins that interact with a plant D-type cyclin and geminivirus replication protein. Mol. Cell. Biol. 17:5077–5086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anckar J., Sistonen L. 2007. SUMO: getting it on. Biochem. Soc. Trans. 35:1409–1413 [DOI] [PubMed] [Google Scholar]
- 3. Aparicio F., Sanchez-Navarro J. A., Pallas V. 2006. In vitro and in vivo mapping of the Prunus necrotic ringspot virus coat protein C-terminal dimerization domain by bimolecular fluorescence complementation. J. Gen. Virol. 87:1745–1750 [DOI] [PubMed] [Google Scholar]
- 4. Arguello-Astorga G., et al. 2004. A novel motif in geminivirus replication proteins interacts with the plant retinoblastoma-related protein. J. Virol. 78:4817–4826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bagewadi B., Chen S., Lal S. K., Choudhury N. R., Mukherjee S. K. 2004. PCNA interacts with Indian mung bean yellow mosaic virus Rep and downregulates Rep activity. J. Virol. 78:11890–11903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bailey D., O'Hare P. 2002. Herpes simplex virus 1 ICP0 co-localizes with a SUMO-specific protease. J. Gen. Virol. 83:2951–2964 [DOI] [PubMed] [Google Scholar]
- 7. Bejarano E. R., Lichtnstein C. P. 1994. Expression of TGMV antisense RNA in transgenic tobacco inhibits replication of BCTV but not ACMV geminiviruses. Plant Mol. Biol. 24:241–248 [DOI] [PubMed] [Google Scholar]
- 8. Bernier-Villamor V., Sampson D. A., Matunis M. J., Lima C. D. 2002. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108:345–356 [DOI] [PubMed] [Google Scholar]
- 9. Boggio R., Chiocca S. 2006. Viruses and sumoylation: recent highlights. Curr. Opin. Microbiol. 9:430–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bossis G., Melchior F. 2006. SUMO: regulating the regulator. Cell Div. 1:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Budhiraja R., et al. 2009. Substrates related to chromatin and to RNA-dependent processes are modified by Arabidopsis SUMO isoforms that differ in a conserved residue with influence on desumoylation. Plant Physiol. 149:1529–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Campos-Olivas R., Louis J. M., Clerot D., Gronenborn B., Gronenborn A. M. 2002. The structure of a replication initiator unites diverse aspects of nucleic acid metabolism. Proc. Natl. Acad. Sci. U. S. A. 99:10310–10315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Castillo A., et al. 2007. Identification of plants genes involved in TYLCV replication, p. 207–222 In Czosnek H. (ed.), Tomato yellow leaf curl virus diseases. Springer, Dordrecht, Netherlands [Google Scholar]
- 14. Castillo A. G., Collinet D., Deret S., Kashoggi A., Bejarano E. R. 2003. Dual interaction of plant PCNA with geminivirus replication accessory protein (Ren) and viral replication protein (Rep). Virology 312:381–394 [DOI] [PubMed] [Google Scholar]
- 15. Castillo A. G., Kong L. J., Hanley-Bowdoin L., Bejarano E. R. 2004. Interaction between a geminivirus replication protein and the plant sumoylation system. J. Virol. 78:2758–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Catala R., et al. 2007. The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell 19:2952–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chiocca S. 2007. Viral control of the SUMO pathway: Gam1, a model system. Biochem. Soc. Trans. 35:1419–1421 [DOI] [PubMed] [Google Scholar]
- 18. Choudhury N. R., et al. 2006. The oligomeric Rep protein of Mungbean yellow mosaic India virus (MYMIV) is a likely replicative helicase. Nucleic Acids Res. 34:6362–6377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Clerot D., Bernardi F. 2006. DNA helicase activity is associated with the replication initiator protein Rep of tomato yellow leaf curl geminivirus. J. Virol. 80:11322–11330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a. Clontech Laboratories, Inc. July 2009. Yeast protocols handbook, PT3024-1 (PR973283) Clontech Laboratories, Inc., Mountain View, CA [Google Scholar]
- 20. Colby T., Matthai A., Boeckelmann A., Stuible H. P. 2006. SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol. 142:318–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Conti L., et al. 2008. Small ubiquitin-like modifier proteases overly tolerant to SALT1 and -2 regulate salt stress responses in Arabidopsis. Plant Cell 20:2894–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Desvoyes B., Ramirez-Parra E., Xie Q., Chua N. H., Gutierrez C. 2006. Cell type-specific role of the retinoblastoma/E2F pathway during Arabidopsis leaf development. Plant Physiol. 140:67–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Elmer J. S., et al. 1988. Genetic analysis of the tomato golden mosaic virus. II. The product of the AL1 coding sequence is required for replication. Nucleic Acids Res. 16:7043–7060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Elrouby N., Coupland G. 2010. Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc. Natl. Acad. Sci. U. S. A. 107:17415–17420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fauquet C. M., et al. 2003. Revision of taxonomic criteria for species demarcation in the family Geminiviridae, and an updated list of begomovirus species. Arch. Virol. 148:405–421 [DOI] [PubMed] [Google Scholar]
- 26. Fauquet C. M., et al. 2008. Geminivirus strain demarcation and nomenclature. Arch. Virol. 153:783–821 [DOI] [PubMed] [Google Scholar]
- 27. Fields S., Song O. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245–246 [DOI] [PubMed] [Google Scholar]
- 28. Fontes E. P., Eagle P. A., Sipe P. S., Luckow V. A., Hanley-Bowdoin L. 1994. Interaction between a geminivirus replication protein and origin DNA is essential for viral replication. J. Biol. Chem. 269:8459–8465 [PubMed] [Google Scholar]
- 29. Geiss-Friedlander R., Melchior F. 2007. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8:947–956 [DOI] [PubMed] [Google Scholar]
- 30. Gietz R. D. 2006. Yeast two-hybrid system screening. Methods Mol. Biol. 313:345–371 [DOI] [PubMed] [Google Scholar]
- 31. Gutierrez C., et al. 2004. Geminivirus DNA replication and cell cycle interactions. Vet. Microbiol. 98:111–119 [DOI] [PubMed] [Google Scholar]
- 32. Hanania U., Furman-Matarasso N., Ron M., Avni A. 1999. Isolation of a novel SUMO protein from tomato that suppresses EIX-induced cell death. Plant J. 19:533–541 [DOI] [PubMed] [Google Scholar]
- 33. Hanley-Bowdoin L., Elmer J. S., Rogers S. G. 1990. Expression of functional replication protein from tomato golden mosaic virus in transgenic tobacco plants. Proc. Natl. Acad. Sci. U. S. A. 87:1446–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hanley-Bowdoin L., Settlage S. B., Orozco B. M., Nagar S., Robertson D. 2000. Geminiviruses: models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Biochem. Mol. Biol. 35:105–140 [PubMed] [Google Scholar]
- 35. Hanley-Bowdoin L., Settlage S. B., Robertson D. 2004. Reprogramming plant gene expression: a prerequisite to geminivirus DNA replication. Mol. Plant Pathol. 5:149–156 [DOI] [PubMed] [Google Scholar]
- 36. Hay R. T. 2005. SUMO: a history of modification. Mol. Cell 18:1–12 [DOI] [PubMed] [Google Scholar]
- 37. Hayashi N., Shirakura H., Uehara T., Nomura Y. 2006. Relationship between SUMO-1 modification of caspase-7 and its nuclear localization in human neuronal cells. Neurosci. Lett. 397:5–9 [DOI] [PubMed] [Google Scholar]
- 38. Heaton P. R., Deyrieux A. F., Bian X. L., Wilson V. G. 2011. HPV E6 proteins target Ubc9, the SUMO conjugating enzyme. Virus Res. 158:199–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hietakangas V., et al. 2006. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Natl. Acad. Sci. U. S. A. 103:45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hotson A., Chosed R., Shu H., Orth K., Mudgett M. B. 2003. Xanthomonas type III effector XopD targets SUMO-conjugated proteins in planta. Mol. Microbiol. 50:377–389 [DOI] [PubMed] [Google Scholar]
- 41. Huang L., et al. 2009. The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root. Plant J. 60:666–678 [DOI] [PubMed] [Google Scholar]
- 42. Ishida T., et al. 2009. SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell 21:2284–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jin J. B., et al. 2008. The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J. 53:530–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kerppola T. K. 2008. Bimolecular fluorescence complementation: visualization of molecular interactions in living cells. Methods Cell Biol. 85:431–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim J. G., et al. 2008. XopD SUMO protease affects host transcription, promotes pathogen growth, and delays symptom development in Xanthomonas-infected tomato leaves. Plant Cell 20:1915–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kong L. J., et al. 2000. A geminivirus replication protein interacts with the retinoblastoma protein through a novel domain to determine symptoms and tissue specificity of infection in plants. EMBO J. 19:3485–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Koonin E. V., Ilyina T. V. 1993. Computer-assisted dissection of rolling circle DNA replication. Biosystems 30:241–268 [DOI] [PubMed] [Google Scholar]
- 48. Kurepa J., et al. 2003. The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. J. Biol. Chem. 278:6862–6872 [DOI] [PubMed] [Google Scholar]
- 49. Laufs J., et al. 1995. Geminivirus replication: genetic and biochemical characterization of Rep protein function, a review. Biochimie 77:765–773 [DOI] [PubMed] [Google Scholar]
- 50. Ledl A., Schmidt D., Muller S. 2005. Viral oncoproteins E1A and E7 and cellular LxCxE proteins repress SUMO modification of the retinoblastoma tumor suppressor. Oncogene 24:3810–3818 [DOI] [PubMed] [Google Scholar]
- 51. Lee H. R., et al. 2004. Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J. Virol. 78:6527–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lee J., Miura K., Bressan R. A., Hasegawa P. M., Yun D. J. 2007. Regulation of plant innate immunity by SUMO E3 ligase. Plant Signal. Behav. 2:253–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lin D., et al. 2002. Identification of a substrate recognition site on Ubc9. J. Biol. Chem. 277:21740–21748 [DOI] [PubMed] [Google Scholar]
- 54. Lois L. M., Lima C. D., Chua N. H. 2003. Small ubiquitin-like modifier modulates abscisic acid signaling in Arabidopsis. Plant Cell 15:1347–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Luckow V. A., Lee S. C., Barry G. F., Olins P. O. 1993. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J. Virol. 67:4566–4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Malik P. S., Kumar V., Bagewadi B., Mukherjee S. K. 2005. Interaction between coat protein and replication initiation protein of Mung bean yellow mosaic India virus might lead to control of viral DNA replication. Virology 337:273–283 [DOI] [PubMed] [Google Scholar]
- 57. Mansoor S., Briddon R. W., Zafar Y., Stanley J. 2003. Geminivirus disease complexes: an emerging threat. Trends Plant Sci. 8:128–134 [DOI] [PubMed] [Google Scholar]
- 58. Miller M. J., Barrett-Wilt G. A., Hua Z., Vierstra R. D. 2010. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 107:16512–16517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Miura K., et al. 2007. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19:1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Miura K., Lee J., Miura T., Hasegawa P. M. 2010. SIZ1 controls cell growth and plant development in Arabidopsis through salicylic acid. Plant Cell Physiol. 51:103–113 [DOI] [PubMed] [Google Scholar]
- 61. Miura K., et al. 2005. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc. Natl. Acad. Sci. U. S. A. 102:7760–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Moldovan G. L., Pfander B., Jentsch S. 2007. PCNA, the maestro of the replication fork. Cell 129:665–679 [DOI] [PubMed] [Google Scholar]
- 63. Morales F. J., Anderson P. K. 2001. The emergence and dissemination of whitefly-transmitted geminiviruses in Latin America. Arch. Virol. 146:415–441 [DOI] [PubMed] [Google Scholar]
- 64. Morilla G., Castillo A. G., Preiss W., Jeske H., Bejarano E. R. 2006. A versatile transreplication-based system to identify cellular proteins involved in geminivirus replication. J. Virol. 80:3624–3633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mukhopadhyay D., Dasso M. 2007. Modification in reverse: the SUMO proteases. Trends Biochem. Sci. 32:286–295 [DOI] [PubMed] [Google Scholar]
- 66. Muller S., Dobner T. 2008. The adenovirus E1B-55K oncoprotein induces SUMO modification of p53. Cell Cycle 7:754–758 [DOI] [PubMed] [Google Scholar]
- 67. Murtas G., et al. 2003. A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of small ubiquitin-related modifier conjugates. Plant Cell 15:2308–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nash T. E., et al. 2011. Functional analysis of a novel motif conserved across geminivirus Rep proteins. J. Virol. 85:1182–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Novatchkova M., Budhiraja R., Coupland G., Eisenhaber F., Bachmair A. 2004. SUMO conjugation in plants. Planta 220:1–8 [DOI] [PubMed] [Google Scholar]
- 70. Orozco B. M., Hanley-Bowdoin L. 1996. A DNA structure is required for geminivirus replication origin function. J. Virol. 70:148–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Orozco B. M., Hanley-Bowdoin L. 1998. Conserved sequence and structural motifs contribute to the DNA binding and cleavage activities of a geminivirus replication protein. J. Biol. Chem. 273:24448–24456 [DOI] [PubMed] [Google Scholar]
- 72. Orozco B. M., Kong L. J., Batts L. A., Elledge S., Hanley-Bowdoin L. 2000. The multifunctional character of a geminivirus replication protein is reflected by its complex oligomerization properties. J. Biol. Chem. 275:6114–6122 [DOI] [PubMed] [Google Scholar]
- 73. Orozco B. M., Miller A. B., Settlage S. B., Hanley-Bowdoin L. 1997. Functional domains of a geminivirus replication protein. J. Biol. Chem. 272:9840–9846 [DOI] [PubMed] [Google Scholar]
- 74. Pozzebon M. 2009. Inhibition of the SUMO pathway by Gam1. Methods Mol. Biol. 497:285–301 [DOI] [PubMed] [Google Scholar]
- 75. Rangasamy D., Wilson V. G. 2000. Bovine papillomavirus E1 protein is sumoylated by the host cell Ubc9 protein. J. Biol. Chem. 275:30487–30495 [DOI] [PubMed] [Google Scholar]
- 76. Roden J., Eardley L., Hotson A., Cao Y., Mudgett M. B. 2004. Characterization of the Xanthomonas AvrXv4 effector, a SUMO protease translocated into plant cells. Mol. Plant Microbe Interact. 17:633–643 [DOI] [PubMed] [Google Scholar]
- 77. Rojas M. R., Hagen C., Lucas W. J., Gilbertson R. L. 2005. Exploiting chinks in the plant's armor: evolution and emergence of geminiviruses. Annu. Rev. Phytopathol. 43:361–394 [DOI] [PubMed] [Google Scholar]
- 78. Rosas-Acosta G., Russell W. K., Deyrieux A., Russell D. H., Wilson V. G. 2005. A universal strategy for proteomic studies of SUMO and other ubiquitin-like modifiers. Mol. Cell. Proteomics 4:56–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 80. Sampson D. A., Wang M., Matunis M. J. 2001. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 276:21664–21669 [DOI] [PubMed] [Google Scholar]
- 81. Saracco S. A., Miller M. J., Kurepa J., Vierstra R. D. 2007. Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol. 145:119–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Seal S. E., Jeger M. J., Van den Bosch F. 2006. Begomovirus evolution and disease management. Adv. Virus Res. 67:297–316 [DOI] [PubMed] [Google Scholar]
- 83. Settlage S. B., Miller A. B., Gruissem W., Hanley-Bowdoin L. 2001. Dual interaction of a geminivirus replication accessory factor with a viral replication protein and a plant cell cycle regulator. Virology 279:570–576 [DOI] [PubMed] [Google Scholar]
- 84. Settlage S. B., Miller A. B., Hanley-Bowdoin L. 1996. Interactions between geminivirus replication proteins. J. Virol. 70:6790–6795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Seufert W., Futcher B., Jentsch S. 1995. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature 373:78–81 [DOI] [PubMed] [Google Scholar]
- 86. Sunter G., Hartitz M. D., Hormuzdi S. G., Brough C. L., Bisaro D. M. 1990. Genetic analysis of tomato golden mosaic virus: ORF AL2 is required for coat protein accumulation while ORF AL3 is necessary for efficient DNA replication. Virology 179:69–77 [DOI] [PubMed] [Google Scholar]
- 87. Ulrich H. D. 2009. The SUMO system: an overview. Methods Mol. Biol. 497:3–16 [DOI] [PubMed] [Google Scholar]
- 88. Wilson V. G., Rangasamy D. 2001. Viral interaction with the host cell sumoylation system. Virus Res. 81:17–27 [DOI] [PubMed] [Google Scholar]
- 89. Wilson V. G., Rosas-Acosta G. 2005. Wrestling with SUMO in a new arena. Sci. STKE 2005:pe32. [DOI] [PubMed] [Google Scholar]
- 90. Yang S. H., Galanis A., Witty J., Sharrocks A. D. 2006. An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J. 25:5083–5093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Yang X., et al. 2007. Functional modulation of the geminivirus AL2 transcription factor and silencing suppressor by self-interaction. J. Virol. 81:11972–11981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yoo C. Y., et al. 2006. SIZ1 small ubiquitin-like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid. Plant Physiol. 142:1548–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.