Abstract
Since the demonstration that almost 80% of human immunodeficiency virus type 1 (HIV-1) infections result from the transmission of a single variant from the donor, biological features similar to those of HIV mucosal transmission have been reported for macaques inoculated with simian immunodeficiency virus (SIV). Here we describe the early diversification events and the impact of challenge doses on viral kinetics and on the number of variants transmitted in macaques infected with the chimeric simian/human immunodeficiency virus SHIVsf162p4. We show that there is a correlation between the dose administered and the number of variants transmitted and that certain inoculum variants are preferentially transmitted. This could provide insight into the viral determinants of transmission and could aid in vaccine development. Challenge through the mucosal route with high doses results in the transmission of multiple variants in all the animals. Such an unrealistic scenario could underestimate potential intervention measures. We thus propose the use of molecular evolution analysis to aid in the determination of challenge doses that better mimic the transmission dynamics seen in natural HIV-1 infection.
INTRODUCTION
A vaccine for the prevention of human immunodeficiency virus type 1 (HIV-1) infection is a global health priority. The most recent phase III clinical trial, RV144, reported a 31% rate of protection from infection after immunization with a combination of a recombinant canarypox virus vector (vCP1521) encoding HIV-1 Env, Gag, protease, and envelope proteins from HIV-1 subtypes B and E (43). Efforts to further develop improved clinical vaccine candidates based on combinations with HIV-1 Env antigens are in progress. Macaque species, including rhesus macaques (Macaca mulatta), cynomolgus macaques (Macaca fascicularis), and pig-tailed macaques (Macaca nemestrina), are frequently used to evaluate the efficacy of HIV-1 vaccine candidates. Simian immunodeficiency virus (SIV) derived from sooty mangabeys (SIVsm) causes an AIDS-like disease in rhesus macaques, with a pattern of disease progression that closely resembles the development of AIDS in humans. In this setting, surrogate SIV antigens are used primarily to evaluate T-cell-based HIV-1 vaccine candidates. However, the envelope structure of SIV differs significantly from that of HIV-1, and neutralizing monoclonal antibodies (Nabs) specific for HIV-1 do not neutralize SIVmac. Chimeric SIVmac viruses expressing the Env of HIV-1 have been demonstrated to be infectious for macaques and have been instrumental in demonstrating the in vivo efficacy of anti-HIV-1 Env Nabs in passive immunization studies (3, 9, 17, 30, 34, 49). Furthermore, the SHIV macaque model has been utilized with at least 3 different species of macaques and has proven invaluable in evaluating the efficacy of HIV-1 Env-based vaccine candidates in various active HIV-1 vaccine protocols in vivo. However, following the failure of the Merck STEP trial (55), in which the protective effects of an adenovirus type 5-based vector expressing HIV proteins for nonhuman primates were not reproduced in humans, the use of these models as gatekeepers for the advancement of vaccine candidates to human clinical trials has been controversial. Although the results generated from nonhuman primate challenges are informative, it has been suggested that they should not be considered the benchmark for the advancement of vaccine candidates into clinical trials until better immune correlates of protection in humans can be identified (48). These concerns indicate that reevaluation and refinement of the available nonhuman primate models are of critical importance.
Traditionally, nonhuman primates have been challenged with a single high dose (between 1,000 and 5,000 50% tissue culture infective doses [TCID50]) of the challenge virus either intravenously or mucosally. The use of high doses ensures that all naïve control animals become infected after a single exposure. However, in light of the finding that HIV transmission is a low-probability event that increases with multiple exposures (5, 11, 14, 19), repeated-low-dose exposure models in macaques have been introduced in an attempt to better mimic natural HIV transmission (2, 5, 8, 11, 18, 19, 32, 33, 35, 36, 42, 51). Here, animals are challenged multiple times with virus doses ranging from 10 to 100 TCID50. Although repeated low-dose challenges use inoculum doses that better reflect the viral concentrations found in semen (37) and lead to a more realistic dynamic of exposure, a larger number of animals is required per group to compensate for the fact that some animals are naturally resistant to low-dose exposures (24); longer follow-up times are necessary; and an “immunization” effect may occur.
Current data suggest that approximately 80% of HIV-1 clade A, B, C, and D infections result from the transmission of a single viral variant from the donor (22). Detailed analysis of low-dose SIV administered to macaques intrarectally and vaginally has revealed biological features similar to those of HIV mucosal transmission (23, 28, 50). In general, one or a few viral variants were transmitted after challenge, indicating the presence of an extreme genetic bottleneck upon transmission and reflecting similarities with HIV mucosal transmission (22). Furthermore. the rate of transmission was dose related, supporting the observation that the rate of HIV transmission is associated with the donor's virus titer (40). More recently, it was reported that high-dose SIV challenges result in a shorter eclipse phase (the time elapsed between infection and the detection of the virus in blood), a higher number of transmitted variants, and greater innate immune activation in macaques exposed to repeated low doses of SIV (28). These findings with SIV suggest that the inoculum dose has a strong impact on the number of variants transmitted as well as on early diversification events, underscoring the importance of defining a challenge dose that more closely reflects natural HIV infection. The use of high-dose challenge settings in early preclinical studies may have underestimated the potential efficacy of certain vaccine candidates, given the unnaturally high variety of genetically diverse variants that were transmitted, a scenario that conflicts with natural HIV transmission.
Whereas the early diversification events and the impact of challenge doses on viral kinetics and on the number of variants transmitted have been investigated in macaques infected with SIV (23, 28, 50), no information is available for macaques infected with SHIV. Here we describe for the first time the early diversification events occurring upon mucosal transmission of the chimeric R5-tropic virus SHIVsf162p4. We also describe how the inoculum dose affects the number of variants transmitted. Our analysis suggests that the use of molecular evolution tools can aid in the determination of challenge doses that better mirror natural HIV transmission.
MATERIALS AND METHODS
Study design and animals.
Peripheral blood mononuclear cells (PBMC) and plasma specimens were collected from Indian rhesus macaques (Macaca mulatta) that had received a single intrarectal inoculation of 1 ml of a 1:2, 1:10, 1:20, or 1:40 dilution of the cell-free challenge stock SHIVsf162p4 (4, 16), corresponding to 1,800, 360, 180, and 90 TCID50. All study animals were purpose bred in captivity, were mature, weighed more than 4 kg, and were housed at the Biomedical Primate Research Center, Rijswijk, The Netherlands, according to international guidelines for nonhuman primate care and use (9a, 9b). During the experiment, the animals were housed separately in cages equipped to give them the ability to express their physiological and behavioral needs. All animals were healthy, had no previous immunosuppressive treatment, were negative for all known simian retroviruses, and had no antigen cross-reactivity with SIV or simian T-cell leukemia virus (STLV).
Viral nucleic acid determination.
Plasma virus load was determined by quantitative competitive reverse transcription-PCR as described previously (53).
DNA extraction.
DNA was extracted from PBMC using the QIAamp DNA Mini kit and QIAamp Blood Mini kit (Qiagen) according to the manufacturer's recommendations.
Viral RNA extraction and cDNA synthesis.
A 200-μl volume of plasma was used to extract viral RNA by using the QIAamp viral RNA kit (Qiagen) according to the manufacturer's instructions. Columns were eluted with 65 μl of a 1:5 dilution of AVE buffer (Qiagen)–water plus carrier RNA. RNA either was used immediately for cDNA synthesis or was aliquoted and stored at −80°C. Viral RNA was reverse transcribed using Superscript III (Invitrogen) according to the manufacturer's instructions. First, 30 μl of RNA was mixed with 0.5 mM each deoxynucleoside triphosphate (dNTP) and 0.25 μM reverse primer SHIVR2 (5′-GCCTCACTGATACCCCTACC-3′) to a final volume of 39 μl, and the mixture was incubated at 65°C for 5 min. Then 5× buffer, dithiothreitol (DTT), RnaseOUT, and Superscript III enzyme were added to final concentrations of 1×, 5 mM, 2 μg/μl, and 10 U/μl, respectively. The reaction mixture was then incubated at 50°C for 1 h, followed by an extra hour at 55°C. cDNA was treated with RNase H and either was used immediately for PCR or was aliquoted and stored at −80°C.
Single-genome amplification (SGA).
In order to amplify env from a single DNA template, DNA was serially diluted to find the appropriate dilution that gave <30% positive reactions in 94 reactions. According to a Poisson distribution, this appropriate DNA dilution results in amplicons for each positive reaction that are derived from a single DNA molecule more than 80% of the time (45).
First-round PCR was carried out under the following conditions: 1× buffer, 2 mM magnesium sulfate (MgSO4), 0.2 mM dNTPs, 0.025 U/pl High-Fidelity Platinum Taq DNA polymerase (Invitrogen), 0.2 μM forward (BFow-out; 5′-GCAATAGTTGTGTGGTCCATAGTAATCATAG-3′) and reverse (SHIVR2; 5′-GCCTCACTGATACCCCTACC-3′) primers, and nuclease-free water up to 20 μl. One microliter of the appropriate cDNA dilution was added as the template. The thermocycler conditions were as follows: 1 cycle of 94°C for 10 min; 35 cycles of a denaturing step at 94°C for 15 s, an annealing step at 43°C for 30 s, and an extension step at 68°C for 4 min; and a final extension at 68°C for 20 min.
Second-round PCRs were performed under the same conditions using the following primers: BFW162-AC (5′-AATAGACCGGTTAATCGATAGAATAACAG-3′) and SHIVp4RW (5′-TCCTCTAGACCCTGATTGTATTTCTGTCC-3′). For second-round PCRs, 1 μl of the first-round PCR product was used as a template. Thermocycler conditions were identical to the first-round conditions, but with an initial cycle of 94°C for 2 min and 45 cycles.
For each 96-well plate, two negative controls were included to detect contamination.
To screen for positive PCRs, PCR products were run on precast 1% agarose gels (E-Gel 96 1% agarose gels; Invitrogen). To avoid cross-contamination, the preparation of the PCR master mix and the addition of DNA were carried out in separate rooms. Only dedicated equipment was used, and the whole PCR procedure was performed using a unidirectional flow.
Evolutionary analysis.
Individual env sequence reads were assembled using the Lasergene SeqMan (DNAStar) package. Individual chromatograms were visually inspected for the presence of multiple peaks at single base positions, which would indicate amplification from multiple templates or a Taq polymerase error, which occurred in the early rounds of amplification. Sequences with multiple or ambiguous peaks were excluded from analysis. The remaining sequences were aligned manually using Se-Al, version 2.0a11 Carbon (http://tree.bio.ed.ac.uk/software/seal/).
All sequences were tested for hypermutation by APOBEC3G/F with Hypermut, version 2.0 (www.hiv.lanl.gov).
Maximum-likelihood (ML) trees were estimated using PAUP, version 4.0 beta, under the best-fit substitution model calculated by Modeltest, version 3.7 (39), using the Akaike information criterion (AIC). Sequences with deletions and insertions were excluded.
To assess env diversity, the observed mean pairwise distances (p-distances) were calculated using MEGA, version 4 (52), using pairwise deletions and uniform rates among sites. Hypermutated sequences and sequences with insertions and deletions were excluded from diversity calculations.
Mean numbers of nonsynonymous (dN) and synonymous (dS) substitutions per site (dN/dS ratios) were estimated using the single-likelihood ancestor counting (SLAC) algorithm available in the Datamonkey Web interface of the HyPhy software package (38). Alignments stripped of sequences with insertions, deletions, and hypermutations were used for this analysis.
Recombination was screened by Genetic Algorithms for Recombination Detection (GARD) (25), available in the Datamonkey Web interface of the HyPhy software package, using the REV nucleotide substitution bias model and beta-gamma site-to-site variation (4 rate classes).
To address the number of potential N-linked glycosylation sites, nucleotide sequences were translated into amino acid sequences using Se-Al, version 2.0a11 Carbon, and were then screened by N-GlycoSite (www.hiv.lanl.gov) (58).
All statistical calculations were carried out using R, version 2.10.1 (41).
Poisson-Fitter was used to test if the Hamming distances followed a Poisson distribution and a star-like phylogeny (12). Alignments were stripped of hypermutated sequences for this analysis.
Enumeration of transmissions.
The transmitted variants were identified and enumerated using SeqTrack (20). To test for the nonuniform probability of transmission of particular variants, we fitted a mixture of two binomial distributions to the number of times (out of 7) a variant was transmitted, and we compared the goodness of fit using Akaike's information criterion (where lower scores indicate a better fit). This was fitted using the gamlss.mx function in R.
Neutralization assays.
Neutralizing titers of sera were assessed using single-round competent viruses expressing either an envelope gene of the SHIVsf162p4 inoculum or a composite of envelope genes present in each animal at week 2 postchallenge and TZM-bl cells as a target as described previously (27). Briefly, the virus was incubated with serially diluted antisera for 1 h at 37°C before being placed in wells of 96-well microplates seeded with TZM-bl cells. After 48 h, cells were lysed, and the luciferase signal in the lysate was developed with the britelite plus substrate (Perkin-Elmer) and was read on a luminometer.
Nucleotide sequence accession numbers.
All the sequences determined in this study have been deposited in the EMBL data bank under accession numbers JN205462 to JN205754.
RESULTS
Study design and infection kinetics.
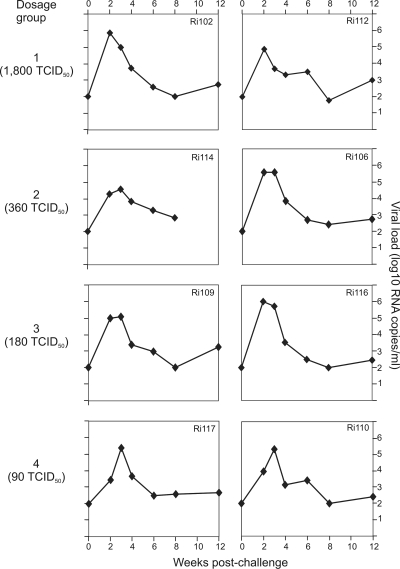
Four groups, each consisting of two rhesus monkeys (Macaca mulatta), were inoculated with descending infectious doses of the R5-tropic SHIVsf162p4 challenge stock (see Table 1). The infection kinetics of the challenged animals is illustrated in Fig. 1. There were no differences in the overall infection rate across the dosage groups; all animals became infected after a single atraumatic rectal mucosal inoculation. Viral loads were measured at 2, 3, 4, 6, 8, and 12 weeks postchallenge and ranged from 2.6 × 103 to 1 × 106 viral copies (median, 1 × 105 viral copies), peaking at 2 or 3 weeks postchallenge. By use of one-way analysis of variance (ANOVA), there were no statistically significant differences between the groups at any of the time points (P = 0.35).
Table 1.
Summary of sequences analyzed
| Animal IDa and time postchallenge | Dosage group | Dose (TCID50) | No. of sequences: |
|||||
|---|---|---|---|---|---|---|---|---|
| From proviral DNA | From viral RNA | With indels | With stop codons | Hypermutated | ||||
| Inoculum | 22 | 0 | 1 | 0 | ||||
| Ri112 | 1 | 1,800 | ||||||
| Wk 2 | 22 | 12 | 3 | 1 | 2 | |||
| Wk 12 | 29 | 0 | 0 | 0 | 2 | |||
| Ri102 | 1 | 1,800 | ||||||
| Wk 2 | 12 | 15 | 1 | 1 | 0 | |||
| Wk 12 | 17 | 0 | 0 | 0 | 3 | |||
| Ri114 | 2 | 360 | ||||||
| Wk 2 | 23 | 0 | 1 | 0 | 1 | |||
| Wk 12 | 0 | |||||||
| Ri106 | 2 | 360 | ||||||
| Wk 2 | 20 | 0 | 0 | 1 | 1 | |||
| Wk 12 | 17 | 0 | 1 | 0 | 1 | |||
| Ri116 | 3 | 180 | ||||||
| Wk 2 | 22 | 0 | 0 | 1 | 1 | |||
| Wk 12 | 27 | 0 | 1 | 0 | 0 | |||
| Ri109 | 3 | 180 | ||||||
| Wk 2 | 17 | 0 | 2 | 0 | 1 | |||
| Wk 12 | 17 | 0 | 0 | 0 | 0 | |||
| Ri117 | 4 | 90 | ||||||
| Wk 2 | 17 | 0 | 0 | 1 | 0 | |||
| Wk 12 | 15 | 0 | 1 | 0 | 1 | |||
| Ri110 | 4 | 90 | N/Ab | N/A | N/A | N/A | N/A | |
| Total | 255 | 49 | 10 | 6 | 13 | |||
ID, identification code.
N/A, not available.
Fig. 1.
Infection kinetics. Four groups of two animals each were challenged intrarectally with different doses of a SHIVsf162p4 virus stock. Blood samples were collected every 2 weeks for a total period of 12 weeks. The group numbers and 50% tissue culture infectious doses are given on the left. The detection level of the assay was 100 viral copies/ml of plasma.
Viral diversity of the SHIVsf162p4 inoculum.
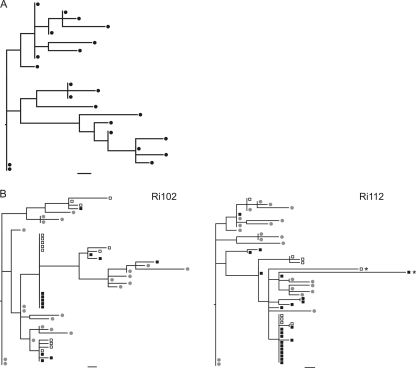
A total of 22 full-length proviral env sequences were derived from the challenge virus stock by single-genome amplification (SGA) followed by direct sequencing. This sample size allowed the detection of variants present in 12.8% or more of the within-host viral population with 95% confidence. The phylogenetic relationships among sequences were estimated using maximum likelihood (Fig. 2 A). The heterogeneity of the inoculum was illustrated by the lack of a majority variant. Indeed, only three variants exhibited a frequency higher than 1, and all the other variants were present at a frequency of 1 out of 22. Randomly distributed unique mutations as well as shared polymorphisms were detected. The overall genetic distance of the stock was 0.293%, which is consistent with the env diversity estimated for another SHIVsf162 stock (54).
Fig. 2.
(A) ML phylogenetic tree of the envelope gene of the virus inoculum. (B) ML trees of envelope sequences derived from proviral DNA, plasma viral RNA, and the inoculum. Envelope genes were amplified using SGA and were sequenced directly from animals Ri112 and Ri102 at 2 weeks postchallenge. Filled squares, sequences derived from proviral DNA; open squares, viral RNA sequences derived from plasma; circles, sequences derived from the challenge stock. Bars, 1 nucleotide substitution. Asterisks indicate hypermutated sequences.
Comparison of sequences derived from proviral DNA and viral RNA.
To compare the viral population circulating in plasma with that integrated into the host DNA, we examined paired PBMC-derived DNA and RNA samples collected from animals Ri102 and Ri112 at 2 weeks postchallenge. The reconstructed phylogenies showed that most lineages were represented by sequences deriving from both viral RNA and proviral DNA (Fig. 2B). It is noteworthy that identical sequences were found for all predominant env variants in both RNA and DNA populations. Only a few minor variants were traced in one of the two sources (data not shown). To formally test for the presence of compartmentalization between circulating and integrated virus, we performed an analysis of molecular variance (AMOVA). No statistically significant differences were found between compartments when a Monte Carlo test was performed with 1,000 replicates (P > 0.05), in agreement with previous observations for HIV-1-infected patients (15, 47). Given that no major differences between sequences derived from plasma viral RNA and proviral DNA were found, sequences derived from cell-associated DNA were used for subsequent analysis or both viral RNA and proviral DNA when available.
Viral diversity in SHIV-infected macaques.
Full-length env genes were amplified by SGA and were sequenced directly from proviral DNA extracted from PBMC isolated at week 2 and week 12 postchallenge. A total of 255 full-length proviral env sequences derived from PBMC were analyzed (23 to 51 sequences per animal; median, 34). In addition, for animals Ri102 and Ri112, 27 full-length env sequences were obtained from viral RNA extracted from plasma at week 2 postchallenge. It was not possible to amplify full-length env using SGA or to perform bulk amplification for animal Ri110. A total of 13 hypermutated sequences were found in all seven animals (Table 1). For the overall diversity calculations, hypermutated sequences and variants with insertions and deletions were excluded.
The observed mean env diversity ranged from 0.03 to 0.31% for sequences obtained 2 weeks postchallenge and from 0.05 to 0.21% for week 12 postchallenge (Table 2). According to the env sequence diversity, animals could be divided into two distinct groups: a high-diversity group and a low-diversity group. Animals Ri102, Ri112, and Ri114 were classified as high diversity and had a mean env sequence diversity of 0.22%, whereas animals Ri106, Ri109, Ri116, and Ri117 were classified as low diversity and had a mean env diversity of 0.04%. Further analysis indicated that animals belonging to the high-diversity group were infected by multiple variants, while the low-diversity animals were infected by a few variants (see below) (Fig. 3).
Table 2.
Analysis of within-host variation
| Animal IDa and time postchallenge | Dosage group | No. of mutations |
Global dN/dS ratio | env diversity (%) | Diversity group | |
|---|---|---|---|---|---|---|
| Synonymous | Nonsynonymous | |||||
| Inoculum | 0.29 | N/Ab | ||||
| Ri112 | 1 | High | ||||
| Wk 2 | 44 | 25 | 0.40 | 0.15 | ||
| Wk 12 | 45 | 30 | 0.47 | 0.18 | ||
| Ri102 | 1 | High | ||||
| Wk 2 | 41 | 55 | 0.48 | 0.24 | ||
| Wk 12 | 19 | 25 | 0.38 | 0.21 | ||
| Ri114 (wk 2) | 2 | 64 | 65 | 0.51 | 0.31 | High |
| Ri106 | 2 | Low | ||||
| Wk 2 | 3 | 9 | 0.87 | 0.049 | ||
| Wk 12 | 6 | 12 | 0.58 | 0.09 | ||
| Ri116 | 3 | Low | ||||
| Wk 2 | 3 | 5 | 0.82 | 0.03 | ||
| Wk 12 | 6 | 8 | 0.59 | 0.05 | ||
| Ri109 | 3 | Low | ||||
| Wk 2 | 3 | 3 | 0.41 | 0.034 | ||
| Wk 12 | 4 | 8 | 0.63 | 0.05 | ||
| Ri117 | 4 | Low | ||||
| Wk 2 | 3 | 8 | 1.39 | 0.05 | ||
| Wk 12 | 2 | 7 | 1.49 | 0.1 | ||
| Ri110 | 4 | N/A | N/A | N/A | N/A | N/A |
ID, identification code.
N/A, not available.
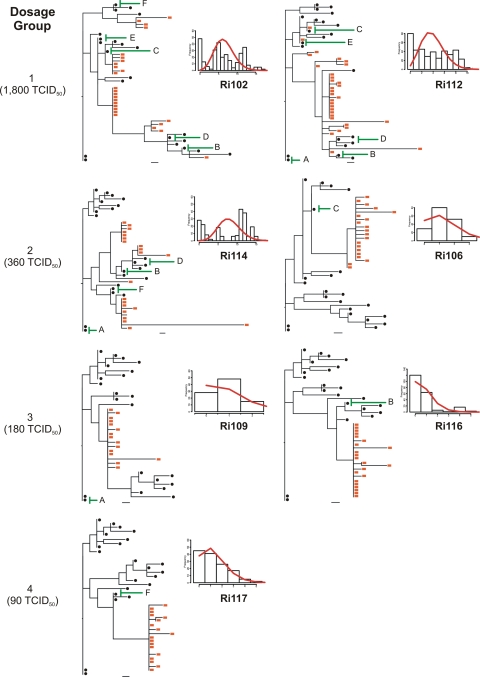
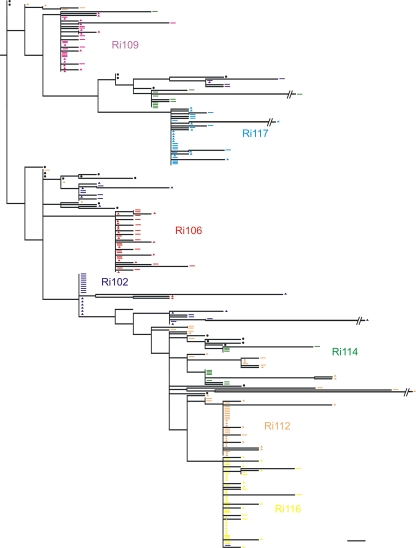
Fig. 3.
Relationship between the virus dose administered and the number of variants transmitted. ML phylogenetic trees were reconstructed with envelope sequences amplified from samples obtained at week 2 postchallenge, together with sequences from the inoculum. Red squares, animal-derived sequences; black circles, inoculum-derived sequences. Bars, 1 nucleotide substitution. Green lines indicate the transmitted inoculum variants identified by SeqTrack. Plots show the Hamming distance distributions (calculated after the removal of hypermutated sequences). The red curves on the plots represent the model predictions of the Hamming distance distribution.
Characterization of early diversification events in SHIV-infected macaques.
Sequence analysis of several HIV-1-infected cohorts, including those infected by subtypes A, B, C, and D, has shown that the majority of mucosal infections result from the transmission of single variants (60 to 90%) (1, 15, 21, 22, 45, 46). The remaining infections result from the transmission of multiple variants and are associated with high-risk behaviors (22). These results indicate that there is a severe genetic bottleneck upon transmission. Under the assumptions of no selection pressure and a lack of recombination, it has been shown that each HIV-1 founder variant diversifies following a Poisson distribution into a lineage composed of identical and/or near-identical sequences. In this model, monophyletic low-diversity lineages are expected to show a star-like phylogeny, since the founder genotypes accumulate mutations at random that are unlikely to be shared. The consensus sequence of each lineage is that of the most recent common ancestor (MRCA) and thus constitutes the genotype of the founder variant (22). Moreover, experimental vaginal and rectal transmission of SIV in rhesus macaques follows the same early diversification pattern (23, 50). We thus asked if intrarectal SHIV infection also recapitulates early HIV-1 diversification events. To this end, we reconstructed the phylogenies for all animals by using envelope gene sequences from week 2 postchallenge, and we used Poisson-Fitter to determine whether the distribution of mutations followed a Poisson distribution and a star-like phylogeny (12). Poisson-Fitter computes the best Poisson distribution using maximum likelihood on Hamming distance frequencies (the number of base positions at which two genomes differ). It also determines whether a given distribution deviates from a Poisson distribution using a χ2 goodness-of-fit test and tests for star-like phylogeny.
The phylogenies of all individual animals containing sequences from week 2 postchallenge, along with the sequences from the inoculum, were inferred (Fig. 3). Sequences derived from animals belonging to the low-diversity group showed a single env lineage with a star-like phylogeny (animals Ri106, Ri109, Ri116, and Ri117). Moreover, for animals Ri106, Ri109, and Ri117, the calculated Hamming distances did not differ significantly from the Poisson distribution, since the goodness-of-fit P values were high (Fig. 3 and Table 3). These results suggested that the infections in animals Ri117, Ri109, and Ri106 likely resulted from the transmission of a single or a few viral variants. The phylogeny of animal Ri117, which received the lowest dose and included 17 sequences derived from week 2 postchallenge along with 22 inoculum-derived sequences, displayed a star-like pattern, with all the mutations being unique. A total of 10 sequences were identical; 4 sequences showed single mutations; and 3 sequences differed by 2 to 3 nucleotides from the founder variant. All sequences coalesced with the same founder genotype, which differed by 5 nucleotides from the closest inoculum sequence. Both animals of dosage group 3, as well as animal Ri106, of dosage group 2, displayed similar early diversification patterns except that three sequences differed by 4 to 12 nucleotides from the corresponding founder sequence. Indeed, all of these sequences were presumably enriched with APOBEC-related G-to-A hypermutations. As stated above, the phylogeny of animal Ri116 displayed a single env lineage with star-like phylogeny. However, the calculated frequencies of the Hamming distances differed significantly from a Poisson distribution. When the number of transmissions was estimated for this animal using SeqTrack (see the next section), a single viral variant was identified. These data strongly suggest that infection of this animal resulted from the transmission of a single variant and that the lack of fit to a Poisson distribution was due to the presence of high diversity within the lineage.
Table 3.
Analysis of early diversification events in SHIV-infected macaques using Poisson-Fitter
| Animal IDa | Lambda (SD) | Days (CI)b | χ2 | df | Goodness-of-fit P value | Poisson distribution | Star-like phylogeny |
|---|---|---|---|---|---|---|---|
| Stock | 7.455 (0.4919) | 123 (107, 139) | 14.8432452409528 | 9 | 0.0953 | No | No |
| Ri112 | 3.892 (0.5363) | 64 (47, 82) | 539.667893696136 | 9 | 0 | No | No |
| Ri102 | 6.52 (0.7562) | 108 (83, 133) | 102362.007295707 | 12 | 0 | No | No |
| Ri114 | 7.971 (0.4948) | 132 (115, 148) | N/Ac | 11 | 0 | No | No |
| Ri106 | 1.251 (0.2002) | 21 (14, 27) | 1.26632991655766 | 2 | 0.531 | Yes | Yes |
| Ri116 | 0.7524 (0.2845) | 13 (3, 22) | 53.5951151999382 | 4 | 6.4e−11 | No | Yes |
| Ri109 | 0.8571 (0.1924) | 14 (8, 21) | 1.91557932984410 | 1 | 0.166 | Yes | Yes |
| Ri117 | 1.279 (0.3262) | 21 (11, 32) | 0.392539076564358 | 3 | 0.942 | Yes | Yes |
| Ri110 | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
ID, identification code.
CI, confidence interval.
N/A, not available.
In contrast to animal Ri106, the second animal of dosage group 2 (Ri114) showed a heterogeneous infection. In this animal, three discrete low-diversity env lineages with similar representation and two separate unique sequences were observed (Fig. 3). The facts that the phylogeny inferred from the sequences obtained from Ri114 did not display a star-like pattern and that the Hamming distances did not follow a Poisson distribution (Table 3) suggest that this animal was infected by multiple variants.
Animals Ri102 and Ri112, who received the highest challenge dose, displayed distinct phylogenies that contrasted with those observed in animals from the low env diversity group (Fig. 3). For both animals, a predominant env lineage consisting of 11 and 18 sequences, respectively, was detected along with several other, less prevalent lineages, again indicating the transmission of multiple variants. Moreover, the distribution of Hamming distances in these two animals did not follow a Poisson distribution. Indeed, the fit to the model was so poor that the retrieved P value for the goodness of fit was extremely low and was reported as zero (Table 3). However, the distribution of Hamming distances did follow a Poisson distribution when each lineage was analyzed separately. It is noteworthy that two sequences from animal Ri112 were identical to two inoculum sequences.
Lineages with minor representation were found in all three animals belonging to the high env diversity group (Ri114, Ri102, and Ri112). This could be explained by a lower replicative fitness of these variants. However, it should be noted that in multivariant infections, the likelihood of identifying at least one sequence of each founder lineage becomes dependent on the number of sequences analyzed.
Enumeration of transmitted variants.
To formally quantify and identify the transmissions that occurred from the inoculum to individual animals upon challenge, we applied the recently developed algorithm SeqTrack (20). SeqTrack infers ancestry relationships among sample sequences. Since it considers that ancestors and descendants are sampled together, it is able to infer direct and indirect ancestries between the isolates sampled. The total numbers of transmissions and the inoculum-transmitted variants are given in Table 4. One variant was transmitted to animals Ri109, Ri116, and Ri117 (inoculum variants A, B, and F, respectively); two variants were transmitted to animal Ri106 (inoculum variants C and E); and multiple variants were transmitted to animals Ri102, Ri112, and Ri114. Only 6 of 22 isolates from the stock were identified by SeqTrack as transmitted variants, and each was transmitted to three or four macaques. We fitted a mixture of two binomial distributions to the number of times (out of 7) a variant was transmitted. There was a significant improvement in the fit of this model (AIC, 46.8) over that of the null model that each variant in the stock had an equal probability of being transmitted (AIC, 74.4), indicating that some inoculum variants were preferentially transmitted.
Table 4.
Viral variants transmitted from the inoculum to individual animals identified by SeqTrack
| Transmitted inoculum variant | No. of transmissions in: |
|||||||
|---|---|---|---|---|---|---|---|---|
| Group 1 (1,800 TCID50) |
Group 2 (360 TCID50) |
Group 3 (180 TCID50) |
Group 4 (90 TCID50) |
|||||
| Ri102 | Ri112 | Ri106 | Ri114 | Ri109 | Ri116 | Ri117 | Ri110 | |
| A | 0 | 1 | 0 | 1 | 1 | 0 | 0 | N/A |
| B | 1 | 1 | 0 | 1 | 0 | 1 | 0 | N/A |
| C | 1 | 1 | 1 | 0 | 0 | 0 | 0 | N/A |
| D | 1 | 1 | 0 | 1 | 0 | 0 | 0 | N/A |
| E | 1 | 1 | 1 | 0 | 0 | 0 | 0 | N/A |
| F | 1 | 0 | 0 | 1 | 0 | 0 | 1 | N/A |
| Total | 5 | 5 | 2 | 4 | 1 | 1 | 1 | N/A |
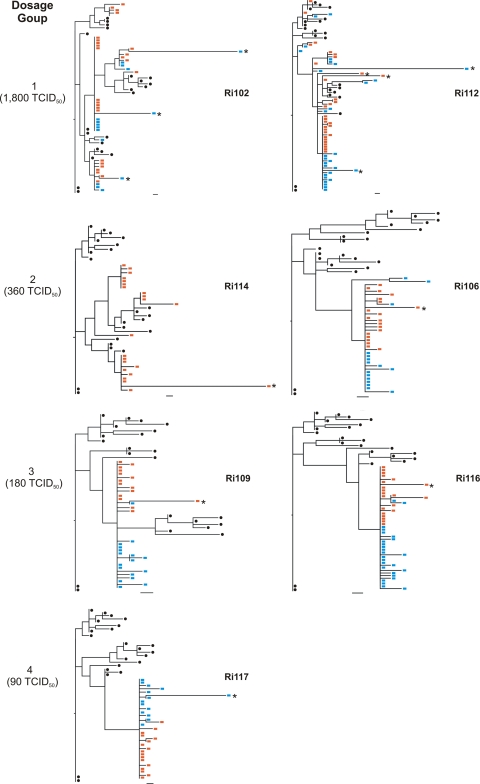
As expected, the ancestors of all the sequences from week 2 were traced back to the virus inoculum sample (not shown). The ancestors of the week 12 sequences were traced back to a week 2 sequence or, for three particular animals (Ri106, Ri102, and Ri112), to a sequence in the inoculum that was already detected as a transmitted variant at week 2 (not shown). However, one week 12 variant of animal Ri112 had an ancestor in the virus inoculum that was not detected as a transmitted variant at week 2. This result was also supported by the phylogenies reconstructed with combined week 2 and week 12 sequences (Fig. 4), where it is evident that a further transmission had occurred in this animal but was revealed only when week 12 sequences were analyzed. This is in agreement with the findings of Felber et al. (10) showing that transmissions occurring in the acute phase of infection, but not detected early on, emerge in the chronic phase of infection in SIV-infected macaques.
Fig. 4.
ML trees of envelope sequences amplified from samples obtained at week 2 and week 12 postchallenge along with sequences from the inoculum. Red squares, week 2 animal-derived sequences; blue squares, week 12 animal-derived sequences; black circles, inoculum-derived sequences. Bars, 1 nucleotide substitution. Asterisks indicate hypermutated sequences.
Importantly, we found a correlation between the exposure dose and the number of transmitted variants (Spearman correlation coefficient, 0.96; P = 0.00054). This indicates that the challenge dose can be used to determine an infectious-dynamics scenario that better suits the preclinical intervention strategy under study.
Selection analysis and single-site mutations.
For the majority of samples. the number of nonsynonymous mutations was higher than the number of synonymous mutations (Table 2). To estimate the selection pressures acting on the HIV-1 envelope in the SHIV/macaque model, the global ratio of nonsynonymous to synonymous mutations (dN/dS) was calculated. The estimated global dN/dS ratio for the complete data set was 0.61, with two codons under positive selection (codons 164 and 824). Interestingly, codon 164 lies within the V2 loop, which is known to harbor neutralizing antibody epitopes, and thus, mutations in V2 are likely to confer escape from the host's antibody responses (44, 57). Six other sites were found to be negatively selected (codons 46, 250, 252, 435, 584, and 692), indicative of positions that are constrained to undergo change, since they might be functionally or structurally important for replicative fitness. For example, codons 250 and 252 are located within the CD4 binding site, and thus, mutations at these positions are most likely deleterious. The global dN/dS ratio was also estimated for each individual animal and time point (Table 2); in general, these were similar to the global dN/dS ratio. The estimated dN/dS ratio for animal Ri117 was remarkably higher, above 1. However, no positively selected sites were detected for this animal.
We applied a recently developed Bayesian technique in an attempt to identify single nucleotide sites that showed evidence of a change in the distribution of bases (relative to the inoculum), at either week 2 or week 12, that was unlikely to have arisen purely due to random mutation events (29). Using this approach, we identified 23 nucleotide sites of interest (Table 5). Interestingly, the two positively selected codons identified by selection analysis were also identified by this method, since they showed changes in the distribution of bases in at least one of the samples (at week 2 or week 12) compared to the inoculum. Along the same lines, by use of this model, the third base in four of the six negatively selected codons showed strong evidence of change in comparison with the inoculum, resulting in synonymous changes at weeks 2 and 12. Moreover, eight nucleotide sites were identified as exhibiting changes in more than one animal, which could be indicative of positions that, upon transmission, have been favored to increase in frequency, possibly because they confer increased fitness (see below).
Table 5.
Nucleotide sites of interest resulting from the implementation of a Bayesian model
| Nucleotide site | Animal | Selection analysis | Location |
|---|---|---|---|
| 360 | Ri106 | C1 | |
| 393 | Ri112 | V1 | |
| 406 | Ri102 | V1 | |
| 436 | Ri117 | V1 | |
| 417 | Ri114 | V1 | |
| 491 | Ri106 | Positively selected site | V2 |
| Ri117 | |||
| 672 | Ri117 | C2 | |
| 750 | Ri102 | Negatively selected site | CD4 binding site |
| Ri106 | |||
| 756 | Ri109 | Negatively selected site | CD4 binding site |
| Ri116 | |||
| 771 | Ri106 | C2 | |
| 836 | Ri117 | C2 | |
| 945 | Ri116 | V3 | |
| 994 | Ri112 | C3 | |
| Ri116 | |||
| Ri117 | |||
| 1006 | Ri112 | C3 | |
| Ri116 | |||
| Ri117 | |||
| 1285 | Ri106 | C4 | |
| Ri117 | |||
| 1305 | Ri116 | Negatively selected site | C4 |
| 1353 | Ri114 | V5 | |
| 1479 | Ri102 | C5 | |
| 1518 | Ri114 | N terminus of gp41 | |
| 1525 | Ri116 | N terminus of gp41 | |
| 1752 | Ri109 | Negatively selected site | gp41 upstream of MPERa |
| Ri112 | |||
| Ri116 | |||
| Ri117 | |||
| 2188 | Ri106 | Cytoplasmic tail/C terminus of gp41 | |
| 2470 | Ri109 | Positively selected site | C terminus of gp41 |
| Ri116 | |||
| Ri117 |
MPER, membrane-proximal external region.
Genetic variation within the host during the course of infection.
To understand the patterns of virus evolution within the first 12 weeks of infection, we reconstructed the phylogenies of all animals for which we had both week 2 and week 12 postchallenge sequences (Fig. 4). For all the animals infected by a single variant, most of the sequences obtained at week 12 postchallenge were identical to the predominant variant present at week 2 or harbored one or two nucleotide changes. For animals Ri109, Ri106, and Ri116, a few nucleotide changes that were first detected at week 2 postchallenge were also detected at week 12, although they were a minority of the week 12 sequences. In particular, a nucleotide change at position 1525 first appeared at week 2 postchallenge in two sequences and was again detected at week 12 in only one sequence from animal Ri106. The same nucleotide change was first detected at week 12 postchallenge in three sequences from animal Ri109. This nonsynonymous mutation changes a methionine to a valine immediately downstream of the gp120–gp41 cleavage site without producing a change in the glycosylation pattern.
For animals Ri102 and Ri112, which were infected with multiple variants, the predominant lineage that was detected at week 2 postchallenge remained predominant at week 12. Indeed, most of the clusters that were first detected at week 2 persisted until week 12. No sequences were amplified from animal Ri114 at week 12, since this animal was found dead in his cage at week 9 postchallenge.
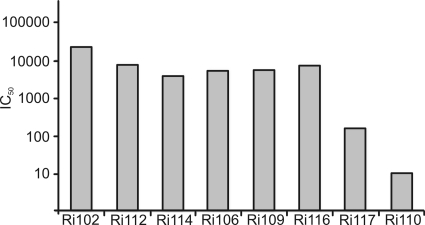
In general, the genetic composition of the viral variants detected at week 12 displayed greater complexity, manifested as an increase in diversity and increases in the numbers of synonymous and nonsynonymous mutations, than the viral composition at week 2. Overall, both at week 2 and at week 12 postchallenge, mutations were uniformly distributed along the entire envelope gene. This increase in the complexity of the genetic composition of the viral variants at week 12 is expected and could be attributed in part to a response of the virus to the development of adaptive immune responses. To address this point, we assayed for the development of neutralizing antibodies (Nabs). Sera collected from each animal at week 8 postchallenge were tested against single-round competent virus expressing an envelope gene derived from the SHIVsf162p4 challenge inoculum (Fig. 5). All the pseudoviruses were tested against monoclonal antibody b12 as an internal control (not shown). By week 8 postchallenge, all animals had developed Nabs. Interestingly, most of the animals developed Nabs as early as week 4 postchallenge against autologous envelope genes present at week 2 postchallenge (not shown), in agreement with the findings of Kraft et al. (26).
Fig. 5.
Neutralizing antibody responses. Sera collected from each animal at week 8 postchallenge were tested against single-round competent virus expressing an envelope gene derived from the SHIVsf162p4 challenge inoculum. The reciprocals of serum dilutions at which 50% inhibition of viral infection occurred (IC50) are reported.
Phylogenetic distribution of the founder genotypes within the inoculum.
We compiled the sequences obtained from all the animals included in this study together with the sequences generated from the inoculum and inferred a phylogenetic tree (Fig. 6). Animals infected with multiple variants could be easily identified, since their sequences were spread widely throughout the tree, again indicating multiple transmission events. Most of the sequences from these three animals (Ri112, Ri102, and Ri114) differed by two or more nucleotides from a sequence in the inoculum, with the exception of animal Ri112, from which two sequences were identical to two other sequences in the inoculum. Sequences derived from animals infected with single variants were characterized by the presence of single low-diversity lineages that differed in all cases by three or more nucleotides from a given variant present in the inoculum.
Fig. 6.
Composite phylogenetic tree of all envelope sequences together with inoculum sequences. Rectangles and triangles indicate animal-derived sequences from weeks 2 and 12, respectively. Animal-derived sequences are color coded. Black circles, inoculum-derived sequences. Bar, 1 nucleotide substitution.
At first glance, this composite phylogeny seemed to show that no distinct genotype of the inoculum was preferentially transmitted, since the sequences of all animals were equally dispersed among the inoculum sequences. However, analysis of the transmitted variants using SeqTrack indicated that some variants were transmitted to multiple animals. Since these variants were present at a frequency of 1 within the viral population of the inoculum, we can exclude the possibility that they were consistently transmitted, since they were overrepresented in the challenge inoculum, as suggested by the study of Stone et al. (50). Therefore, we provide here, for the first time, formal evidence for the preferential transmission of variants within the inoculum. The Bayesian technique described above aided in the identification of nucleotide positions that could be important for transmission. As mentioned above, 8 of the 23 sites of interest found by this method were identified in more than one animal (Table 4). All the transmitted variants possessed a mutation at least in one of those sites. Those mutations were transmitted, since they were detected at week 2 (in most cases, the whole viral population in the animal had that mutation) and persisted until week 12 (not shown). These data suggest that variants in the inoculum harboring polymorphisms at particular nucleotide sites are preferentially transmitted.
DISCUSSION
HIV-1 preclinical trials performed with rhesus macaques play a critical role in assessing potential interventions against infection. The use of a high-dose challenge setting ensures that all control animals become infected after a single exposure; however, it is questionable whether they reflect natural HIV infection, considering the relatively low frequency of HIV-1 acquisition by heterosexual transmission (14). It is thus arguable that the protective properties of vaccine candidates or prophylactic measures could be underestimated when evaluated under such stringent conditions. The introduction of repeated low-dose challenges allows a more realistic transmission scenario, although it also brings higher costs and more-complex data interpretation. In light of the need to improve preclinical nonhuman primate models, we provided evidence here supporting the use of phylogenetic tools to define dose settings that overcome some of the drawbacks of the commonly used challenge approaches.
We showed that all the animals involved in this study became infected after a single intrarectal exposure despite receiving wide dose ranges of the SHIVsf164p4 challenge stock. Moreover, all animals displayed comparable infection kinetics, reaching peak viremia at similar time points. This demonstrated that the use of lower challenge doses did not compromise the infectious rate of the study, at least for macaques infected with SHIV via the intrarectal route, in agreement with previous findings for macaques infected with SIV by the same route (28). However, our results contrast with the findings for macaques infected with repeated low doses of SHIV via the vaginal route, where substantially lower values for viral RNA copies/ml were found at peak viremia (54). Inherent properties of the virus stocks and the route of infection could account for the differences observed between the two studies. However, data from the present study suggest that the use of intermediate doses (groups 2 and 3) could generate more-comparable study groups and avoid the appearance of transient viremia often observed in repeated low-dose challenge studies. The appearance of transient viremia complicates data interpretation, since it is open to individual interpretation whether this should be considered a sign of infection or whether animals should be rechallenged.
We also found a correlation between the number of variants transmitted and the infectious dose administered. We were able to show clearly that animals that received the highest challenge dose were infected with multiple variants, while single transmissions occurred in animals receiving the lowest dose. Interestingly, we observed that in an animal infected with an intermediate dose (group 2), infection was established by a few variants, while in the other animal in this group, infection was established by multiple variants. Therefore, infection with intermediate doses leads to a transmission scenario that more closely reflects natural HIV transmission (22). However, it should be noted that the enumeration of the founder variants is a minimum estimate, since it is feasible that more variants were transmitted but were missed due to restricted sampling, lower fitness, and compartmentalization within the inoculation site, among other reasons. Indeed, more transmission events were detected when sequences from week 12 postchallenge were analyzed. The correlation between the virus challenge dose and the number of variants transmitted is in agreement with the findings of Liu et al. (28) for macaques infected intrarectally with a single injection of descending doses of SIV. However, our results contrast with the findings of Keele et al. (23), in which no clear correlation was found between the challenge dose and the number of variants transmitted in macaques infected intrarectally with repeated low doses of SIV. The major difference in the latter study is that animals of the lower-dosage group were challenged repeatedly until positive viremia was detected. It is possible that the challenges that passed undetected resulted in effective transmission with a delayed onset of detectable viremia. Another explanation could be that these undetected transmissions could have induced local inflammatory changes that would predispose the animal to the transmission of multiple variants upon subsequent exposures, as observed for HIV-infected individuals in whom higher numbers of variants are detected when other concomitant infections are present (15). This idea is further supported by the finding that exposure to HIV impairs the mucosal epithelial barrier, allowing viral and bacterial translocation, which is thought to be mediated by enhanced inflammatory cytokine production (31). Therefore, as demonstrated here and in the study of Liu et al. (28), the use of a single intermediate dose administered via the intrarectal route may provide more-predictable transmission dynamics and outcomes that would better resemble those of HIV-1 infection. On the other hand, a different picture is presented by macaques vaginally infected with SIV, where an inverse correlation between the number of challenges and the number of variants transmitted was found (50). Given the study variables, all the conclusions and extrapolations from different studies should be taken with caution.
The use of SGA followed by direct sequencing allowed us to determine, for the first time, whether the predictions of the mathematical model (22) developed for the identification of early HIV diversification events applied to SHIV-infected macaques. We found that the Hamming distances of animals receiving low challenge doses followed a Poisson distribution and a star-like phylogeny, which were indicative of infections initiated by the transmission of a single variant. Moreover, the model estimates of the time to the MRCA for these animals closely reflected the known time of infection (Table 3). On the other hand, the Hamming distances of animals receiving higher challenge doses did not conform to a Poisson distribution or a star-like phylogeny, and the estimated times to the MRCA were greatly overestimated, indicative of infections initiated by the transmission of multiple variants. By the use of the recently developed algorithm SeqTrack, an algorithm previously applied to analyzing the dynamics of transmission of influenza A virus between hosts, we were able to formally enumerate and identify the variants that were transmitted from the inoculum to the animals. We identified nonuniform transmissibility of variants in the inoculum, although we could not identify any individual amino acid substitutions that correlated with transmissibility. In the absence of a simple way to independently validate the transmissibility of particular variants, the use of simulated data may be a means of validating the performance of SeqTrack in estimating transmissibility. Importantly, the number of transmitted variants estimated using SeqTrack coincided with the predictions of the mathematical model of Keele et al. (22), supporting the use of SeqTrack for the inference of transmission events in HIV. Moreover, we found no evidence of immune selection in any of the animals at week 2 postchallenge. We also observed an extreme genetic bottleneck occurring upon transmission, which was evidenced by the transmission of single variants and a shortening in the branch lengths of the phylogenies reconstructed with sequences from animals with respect to the inoculum phylogeny. Taken together, these results demonstrate that early virus diversification events in macaques infected with SHIV via the mucosal route emulate key features of HIV transmission (1, 6, 7, 13, 15, 21, 45, 46, 56) and thus provide an alternative model to SIV-infected macaques for the unraveling of the mechanisms fueling mucosal transmission.
The virus challenge inoculum revealed a heterogeneous composition characterized by the presence of viral variants that were mostly distinct from each other. Variants that were successfully transmitted represented 12.8% or less of the whole viral population present in the inoculum, in agreement with the findings for discordant couples infected with HIV, where generally a small fraction of the variants found in the donor are transmitted to the recipient (15). Several distinct variants of the inoculum were transmitted, demonstrating that a wide range of variants is capable of establishing infection for this particular stock, an observation in agreement with previous findings for SIV-infected macaques (23, 28, 50). However, we observed that certain variants in the inoculum were preferentially transmitted, since they were identified as the ancestral sequence in more than one animal. With the aid of a recently developed Bayesian technique (29), we were able to identify nucleotide sites of interest that may potentially correlate with viral fitness. The study of these sites could unravel viral determinants of transmission, and they could serve as targets for vaccination.
Here we studied, for the first time, the early diversification events undergone by the HIV-1 envelope gene in the context of the chimeric virus SHIVsf162p4. This analysis provided evidence supporting a correlation between the inoculum dose administered and the number of viral variants transmitted. This information could be used to inform the design of preclinical intervention experiments with nonhuman primates. The dynamics of viral genetic transmission has been overlooked both in the single high-dose and the repeated-low-dose mucosal challenge settings, and thus, we propose the use of molecular evolution tools to supplement and perfect these models. We believe that the study of early diversification events occurring upon transmission is critical to an accurate understanding of the animal model of interest and that this information can contribute to a better interpretation of the outcome of preclinical trials. We are aware that the conclusions from this study may be restricted to SHIV-infected macaques, which is the animal model of necessity for HIV-1 antibody-based vaccine development, and the challenge inoculum used here. It would thus be important to properly design in vivo dosage experiments to dissect the impact of dosage for other virus challenge inocula, such as SIV. This would be of particular interest for repeated low-dose challenge settings, where the exposure dynamics and the viral transmission dynamics through the mucosal surfaces would then better mimic natural HIV transmission. We appreciate the fact that the use of SGA followed by direct sequencing is costly and that not every facility is equipped to carry out molecular evolution analysis. However, we propose as a simpler alternative the amplification of the envelope gene by bulk PCR, followed by direct sequencing. Then the analysis of the number of ambiguous bases would give an indication of the diversity of the sample that would translate into an approximation of the number of variants transmitted.
ACKNOWLEDGMENTS
M.V. is a Wellcome Intermediate Clinical Fellow. Evolutionary analysis was supported by a Wellcome Intermediate Clinical Fellowship, animals by NIH grant 1P01AI06628, and neutralization through the Bill and Melinda Gates Foundation's CAVD. S.D.W.F. is supported in part by a Royal Society Wolfson Merit Award.
We thank Pablo R. Murcia and Fabian Schmidt for intellectual and technical support, respectively.
Footnotes
Published ahead of print on 27 July 2011.
REFERENCES
- 1. Abrahams M. R., et al. 2009. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-Poisson distribution of transmitted variants. J. Virol. 83:3556–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson R. E., et al. 1990. Use of β2 microglobulin level and CD4 lymphocyte count to predict development of AIDS in persons with human immunodeficiency virus infection. Arch. Intern. Med. 150:73–77 [PubMed] [Google Scholar]
- 3. Baba T. W., et al. 2000. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat. Med. 6:200–206 [DOI] [PubMed] [Google Scholar]
- 4. Bogers W. M., et al. 2008. Systemic neutralizing antibodies induced by long interval mucosally primed systemically boosted immunization correlate with protection from mucosal SHIV challenge. Virology 382:217–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. DeGruttola V., Seage G. R., III, Mayer K. H., Horsburgh C. R., Jr 1989. Infectiousness of HIV between male homosexual partners. J. Clin. Epidemiol. 42:849–856 [DOI] [PubMed] [Google Scholar]
- 6. Derdeyn C. A., et al. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303:2019–2022 [DOI] [PubMed] [Google Scholar]
- 7. Edwards C. T., et al. 2006. Population genetic estimation of the loss of genetic diversity during horizontal transmission of HIV-1. BMC Evol. Biol. 6:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ellenberger D., et al. 2006. HIV-1 DNA/MVA vaccination reduces the per exposure probability of infection during repeated mucosal SHIV challenges. Virology 352:216–225 [DOI] [PubMed] [Google Scholar]
- 9. Emini E. A., et al. 1992. Prevention of HIV-1 infection in chimpanzees by gp120 V3 domain-specific monoclonal antibody. Nature 355:728–730 [DOI] [PubMed] [Google Scholar]
- 9a. European Council 24 November 1986. Council Directive on the approximation of laws, regulations and administrative provisions of the Member States regarding the protection of animals used for experimental and other scientific purposes (86/609/EEC). http://ec.europa.eu/food/fs/aw/aw_legislation/scientific/86-609-eec_en.pdf [PubMed]
- 9b. European Council 18 March 1986. European Convention for the Protection of Vertebrate Animals used for Experimental and Other Scientific Purposes (ETS no. 123), including the revised Appendix A. http://conventions.coe.int/treaty/en/treaties/html/123.htm
- 10. Felber B. K., et al. 2009. Monospecific expansion of SIVmac251 during acute infection masks multiple transmitted virus variants revealed during the chronic phase. Retrovirology 6(Suppl. 3):O38 [Google Scholar]
- 11. Galvin S. R., Cohen M. S. 2004. The role of sexually transmitted diseases in HIV transmission. Nat. Rev. Microbiol. 2:33–42 [DOI] [PubMed] [Google Scholar]
- 12. Giorgi E. E., et al. 2010. Estimating time since infection in early homogeneous HIV-1 samples using a Poisson model. BMC Bioinform. 11:532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gottlieb G. S., et al. 2008. HIV-1 variation before seroconversion in men who have sex with men: analysis of acute/early HIV infection in the multicenter AIDS cohort study. J. Infect. Dis. 197:1011–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gray R. H., et al. 2001. Probability of HIV-1 transmission per coital act in monogamous, heterosexual, HIV-1-discordant couples in Rakai, Uganda. Lancet 357:1149–1153 [DOI] [PubMed] [Google Scholar]
- 15. Haaland R. E., et al. 2009. Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype A and C HIV-1. PLoS Pathog. 5:e1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harouse J. M., Gettie A., Tan R. C., Blanchard J., Cheng-Mayer C. 1999. Distinct pathogenic sequela in rhesus macaques infected with CCR5 or CXCR4 utilizing SHIVs. Science 284:816–819 [DOI] [PubMed] [Google Scholar]
- 17. Heeney J. L., et al. 1998. β-Chemokines and neutralizing antibody titers correlate with sterilizing immunity generated in HIV-1 vaccinated macaques. Proc. Natl. Acad. Sci. U. S. A. 95:10803–10808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jewell N. P. 1990. Some statistical issues in studies of the epidemiology of AIDS. Stat. Med. 9:1387–1416 [DOI] [PubMed] [Google Scholar]
- 19. Jewell N. P., Shiboski S. C. 1990. Statistical analysis of HIV infectivity based on partner studies. Biometrics 46:1133–1150 [PubMed] [Google Scholar]
- 20. Jombart T., Eggo R. M., Dodd P. J., Balloux F. 2011. Reconstructing disease outbreaks from genetic data: a graph approach. Heredity 106:383–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kearney M., et al. 2009. Human immunodeficiency virus type 1 population genetics and adaptation in newly infected individuals. J. Virol. 83:2715–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Keele B. F., et al. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105:7552–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Keele B. F., et al. 2009. Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J. Exp. Med. 206:1117–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim C. N., et al. 2006. Repetitive exposures with simian/human immunodeficiency viruses: strategy to study HIV pre-clinical interventions in non-human primates. J. Med. Primatol. 35:210–216 [DOI] [PubMed] [Google Scholar]
- 25. Kosakovsky Pond S. L., Posada D., Gravenor M. B., Woelk C. H., Frost S. D. 2006. Automated phylogenetic detection of recombination using a genetic algorithm. Mol. Biol. Evol. 23:1891–1901 [DOI] [PubMed] [Google Scholar]
- 26. Kraft Z., et al. 2007. Macaques infected with a CCR5-tropic simian/human immunodeficiency virus (SHIV) develop broadly reactive anti-HIV neutralizing antibodies. J. Virol. 81:6402–6411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li M., et al. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108–10125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu J., et al. 2010. Low-dose mucosal simian immunodeficiency virus infection restricts early replication kinetics and transmitted virus variants in rhesus monkeys. J. Virol. 84:10406–10412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McKinley T. J., Murcia P. R., Gog J. R., Varela M., Wood J. L. 2011. A Bayesian approach to analyse genetic variation within RNA viral populations. PLoS Comput. Biol. 7:e1002027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mooij P., et al. 1998. A clinically relevant HIV-1 subunit vaccine protects rhesus macaques from in vivo passaged simian-human immunodeficiency virus infection. AIDS (London, England) 12:F15–F22 [DOI] [PubMed] [Google Scholar]
- 31. Nazli A., et al. 2010. Exposure to HIV-1 directly impairs mucosal epithelial barrier integrity allowing microbial translocation. PLoS Pathog. 6:e1000852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Neuhaus J. M., Jewell N. P. 1990. The effect of retrospective sampling on binary regression models for clustered data. Biometrics 46:977–990 [PubMed] [Google Scholar]
- 33. Padian N. S., Shiboski S. C., Jewell N. P. 1990. The effect of number of exposures on the risk of heterosexual HIV transmission. J. Infect. Dis. 161:883–887 [DOI] [PubMed] [Google Scholar]
- 34. Parren P. W., et al. 2001. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J. Virol. 75:8340–8347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pilcher C. D., Eron J. J., Jr., Galvin S., Gay C., Cohen M. S. 2004. Acute HIV revisited: new opportunities for treatment and prevention. J. Clin. Invest. 113:937–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pilcher C. D., et al. 2004. Frequent detection of acute primary HIV infection in men in Malawi. AIDS (London, England) 18:517–524 [DOI] [PubMed] [Google Scholar]
- 37. Pilcher C. D., et al. 2004. Brief but efficient: acute HIV infection and the sexual transmission of HIV. J. Infect. Dis. 189:1785–1792 [DOI] [PubMed] [Google Scholar]
- 38. Pond S. L., Frost S. D. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics (Oxford, England) 21:2531–2533 [DOI] [PubMed] [Google Scholar]
- 39. Posada D., Crandall K. A. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics (Oxford, England) 14:817–818 [DOI] [PubMed] [Google Scholar]
- 40. Quinn T. C., et al. 2000. Viral load and heterosexual transmission of human immunodeficiency virus type 1. Rakai Project Study Group. N. Engl. J. Med. 342:921–929 [DOI] [PubMed] [Google Scholar]
- 41. R Development Core Team 2011. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org [Google Scholar]
- 42. Regoes R. R., Longini I. M., Feinberg M. B., Staprans S. I. 2005. Preclinical assessment of HIV vaccines and microbicides by repeated low-dose virus challenges. PLoS Med. 2:e249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rerks-Ngarm S., et al. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361:2209–2220 [DOI] [PubMed] [Google Scholar]
- 44. Rybarczyk B. J., et al. 2004. Correlation between env V1/V2 region diversification and neutralizing antibodies during primary infection by simian immunodeficiency virus sm in rhesus macaques. J. Virol. 78:3561–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Salazar-Gonzalez J. F., et al. 2008. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J. Virol. 82:3952–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Salazar-Gonzalez J. F., et al. 2009. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J. Exp. Med. 206:1273–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shankarappa R., et al. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489–10502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shedlock D. J., Silvestri G., Weiner D. B. 2009. Monkeying around with HIV vaccines: using rhesus macaques to define ‘gatekeepers’ for clinical trials. Nat. Rev. 9:717–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shibata R., et al. 1999. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat. Med. 5:204–210 [DOI] [PubMed] [Google Scholar]
- 50. Stone M., et al. 2010. A limited number of simian immunodeficiency virus (SIV) env variants are transmitted to rhesus macaques vaginally inoculated with SIVmac251. J. Virol. 84:7083–7095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Subbarao S., et al. 2006. Chemoprophylaxis with tenofovir disoproxil fumarate provided partial protection against infection with simian human immunodeficiency virus in macaques given multiple virus challenges. J. Infect. Dis. 194:904–911 [DOI] [PubMed] [Google Scholar]
- 52. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 53. ten Haaft P., et al. 2004. Readily acquired secondary infections of human and simian immunodeficiency viruses following single intravenous exposure in non-human primates. J. Gen. Virol. 85:3735–3745 [DOI] [PubMed] [Google Scholar]
- 54. Tsai L., et al. 2007. Efficient repeated low-dose intravaginal infection with X4 and R5 SHIVs in rhesus macaque: implications for HIV-1 transmission in humans. Virology 362:207–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Watkins D. I., Burton D. R., Kallas E. G., Moore J. P., Koff W. C. 2008. Nonhuman primate models and the failure of the Merck HIV-1 vaccine in humans. Nat. Med. 14:617–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wolinsky S. M., et al. 1992. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science 255:1134–1137 [DOI] [PubMed] [Google Scholar]
- 57. Yeh W. W., et al. 2010. Autologous neutralizing antibodies to the transmitted/founder viruses emerge late after simian immunodeficiency virus SIVmac251 infection of rhesus monkeys. J. Virol. 84:6018–6032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang M., et al. 2004. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 14:1229–1246 [DOI] [PubMed] [Google Scholar]