Abstract
Background
Nischarin (encoded by NISCH), an α5 integrin–binding protein, has been identified as a regulator of breast cancer cell invasion. We hypothesized that it might be a tumor suppressor and were interested in its regulation.
Methods
We examined nischarin expression in approximately 300 human breast cancer and normal tissues using quantitative polymerase chain reaction and immunohistochemistry. Loss of heterozygosity analysis was performed by examining three microsatellite markers located near the NISCH locus in normal and tumor tissues. We generated derivatives of MDA-MB-231 human metastatic breast cancer cells that overexpressed nischarin and measured tumor growth from these cells as xenografts in mice; metastasis by these cells after tail vein injection; and α5 integrin expression, Rac, and focal adhesion kinase (FAK) signaling using western blotting. We also generated clones of MCF-7 human breast cancer cells in which nischarin expression was silenced and measured tumor growth in mouse xenograft models (n = 5 for all mouse experiments). P values were from two-sided Student t tests in pairwise comparisons.
Results
Normal human breast tissue samples had statistically significantly higher expression of nischarin mRNA compared with tumor tissue samples (mean level in normal breast = 50.7 [arbitrary units], in breast tumor = 16.49 [arbitrary units], difference = 34.21, 95% confidence interval [CI] = 11.63 to 56.79, P = .003), and loss of heterozygosity was associated with loss of nischarin expression. MDA-MB-231 cells in which nischarin was overexpressed had statistically significantly reduced tumor growth and metastasis compared with parental MDA-MB-231 cells (mean volume at day 40, control vs nischarin-expressing tumors, 1977 vs 42.27 mm3, difference = 1935 mm3, 95% CI = 395 to 3475 mm3, P = .025). Moreover, MCF-7 tumor xenografts in which nischarin expression was silenced grew statistically significantly faster than parental cells (mean volume at day 63, tumors with scrambled short hairpin RNA [shRNA] vs with nischarin shRNA, 224 vs 1262 mm3, difference = 1038 mm3, 95% CI = 899.6 to 1176 mm3, P < .001). Overexpression of nischarin was associated with decreased α5 integrin expression, FAK phosphorylation, and Rac activation.
Conclusion
Nischarin may be a novel tumor suppressor that limits breast cancer progression by regulating α5 integrin expression and subsequently α5 integrin–, FAK-, and Rac-mediated signaling.
CONTEXT AND CAVEATS
Prior knowledge
Nischarin, a protein that binds the cytoplasmic tail of α5 integrin, has been shown to inhibit invasiveness of cells in culture. Because its gene maps to a chromosomal locus associated with several cancers, the authors investigated its role in cancer progression.
Study design
Expression of nischarin mRNA and protein was examined in breast cancer and normal tissue samples and in online databases, and loss of heterozygosity was tested in the clinical samples. Human breast cancer cell lines in which nischarin was overexpressed or silenced were used in vitro and as xenografts to examine its role in intracellular signaling, tumor growth, and metastasis.
Contribution
Normal breast tissue samples had higher levels of nischarin expression than breast cancers, and expression levels generally decreased with advancing cancer stage, often with loss of heterozygosity at the nischarin locus. Tumor growth and metastasis were reduced in human breast cancer cells in which nischarin was overexpressed, and increased when expression was silenced, compared with parental cell lines. Nischarin expression was associated with decreased α5 integrin expression and Rac and focal adhesion kinase activation.
Implication
Nischarin may inhibit the growth of cancer cells by limiting α5 integrin expression and cellular signaling pathways associated with invasiveness.
Limitations
Nischarin expression has not been studied in other types of cancer. The exact mechanisms whereby it affects α5 integrin and cell signaling levels have not been fully determined.
From the Editors
We previously identified a novel protein, nischarin, that selectively bound to the proximal transmembrane (IYILYKLGFFKR) region of the integrin α5 subunit cytoplasmic tail (1,2). Nischarin blocked Rac-induced cell migration and invasion in breast and colon epithelial cells, interacted with the p21 (cdc42/rac)-activated kinase 1 (PAK1) to block PAK activation, and influenced actin filament organization (1). Nischarin also blocked PAK-independent Rac signaling (3,4) and interacted with LIM kinase (LIMK) to inhibit LIMK activation and LIMK-driven cell invasion (5). A human ortholog of nischarin, IRAS, has been shown to bind to the adapter protein IRS4 to mediate translocation of α5 integrin from the cell membrane to endosomes (6).
Several studies, which included cytogenetic and homozygosity mapping, have indicated that distinct regions of human chromosome arm 3p are important for development of cancers including those of lung, breast, kidney, ovary, and cervix (7). Because nischarin was known to map at 3p21.1 (www.ncbi.nlm.nih.gov), we hypothesized that it may have an important role in cancer progression. In this study, we investigated the role of nischarin in breast cancer progression by overexpressing it or by silencing its expression in cultured cells. Furthermore, we examined the mechanism by which nischarin regulates breast cancer progression using various in vitro biochemical and in vivo mouse xenograft experiments.
Materials and Methods
Cell Culture
MDA-MB-231 and MCF-7 human breast cancer cell lines and COS-7–transformed African green monkey kidney cells were obtained in 2006 from American Type Culture Collection (Manassas, VA) and maintained in high-glucose Dulbecco’s Minimal Essential Medium (DMEM) with 2 mM L-glutamine, 110 mg/mL sodium pyruvate, and 10% fetal bovine serum (FBS; Gemini Bio-Products, Sacramento, CA) at 37°C in a humidified atmosphere of 5% CO2. T47D and BT474 cells were also obtained from American Type Culture Collection and maintained in RPMI medium with 10% FBS. Stable cell lines in which nischarin expression was increased or silenced were prepared as described elsewhere (see Supplementary Methods, available online). Experiments with T47D and BT474 cells were carried out soon after purchase. With regard to MCF7 and MDA-MB-231 cells, their morphologies were examined before conducting every crucial experiment, to confirm they were still the same lines.
Antibodies Used
The following antibodies were used for immunoblotting experiments: mouse monoclonal anti-nischarin (BD Biosciences, San Diego, CA; 1:1000 dilution for western blots; 1:300 for immunohistochemistry), mouse monoclonal anti–focal adhesion kinase (FAK) (BD Biosciences; 1:1000), rabbit polyclonal anti-α5 integrin (Millipore, Bedford, MA; 1:1000), anti-phosphoFAK (Invitrogen, Carlsbad, CA; 1:1000), mouse monoclonal anti-vinculin (catalog number V9264; Sigma, St Louis, MO; 1:1000), mouse monoclonal anti-cyclin D1 (catalog number 2926; Cell Signaling, Beverly, MA; 1:1000), mouse monoclonal anti-CDK4 (sc-70831; Santa Cruz Biotech, Santa Cruz, CA; 1:1000), anti-retinoblastoma (RB) protein (BD Biosciences; 1:1000), anti-phosphoRB (BD Biosciences; 1:1000), rabbit polyclonal anti-extracellular signal regulated kinase (ERK) (sc-93; Santa Cruz Biotech; 1:1000), mouse monoclonal anti-phosphoERK (sc-7383; Santa Cruz Biotech; 1:1000), mouse monoclonal anti-Rac1 (catalog number 610650; BD Biosciences; 1:1000), and anti-Ki67 (Novocastra Labs, Newcastle upon Tyne, UK; 1:50).
Tissues Used
Sixty human breast cancer surgical specimens, 60 noncancerous breast tissues, and 24 pairs of patient-matched breast cancer and nonmalignant breast tissue were obtained as frozen tissue sections from the Southern Division (Birmingham, AL), Eastern Division (Philadelphia, PA), Mid-Atlantic Division (Charlottesville, VA), Mid-Western Division (Columbus, OH), and Western Division (Nashville, TN) of the Cooperative Human Tissue Network. The patients ranged in age from 13 to 87 years. Sixty of the tumor samples represented six different histological types: there were 22 invasive ductal carcinomas (IDCs), four ductal carcinoma in situ (DCIS) samples, 17 mixtures of IDC and DCIS, five invasive lobular carcinomas (ILCs), six mixtures of ILC and lobular carcinoma in situ (LCIS), and a group of six miscellaneous samples. This group included two adenocarcinoma, one LCIS, one IDC + ILC mixture, one IDC + ILC + DCIS mixture, and one IDC + ILC + DCIS + LCIS mixture (see Figure 1, C).
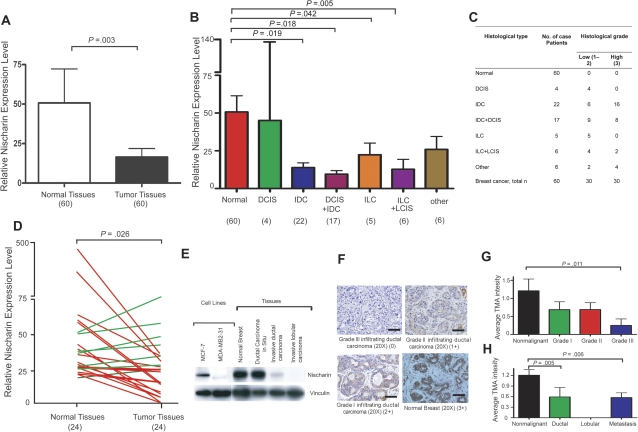
Figure 1.
Nischarin expression during breast cancer progression. A) Overall nischarin mRNA expression in normal vs breast cancer tissues. Expression was measured by quantitative polymerase chain reaction (qPCR) in 60 normal breast and 60 breast cancer tissues among which 24 samples of each type were paired specimens from the same patients. The experiment was repeated three times and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) expression was used for normalization. Paired t tests revealed that the expression of nischarin was higher in normal tissues relative to tumors. The means and 95% confidence intervals from two different experiments are shown (P = .003). B) Nischarin mRNA expression by tumor stage. qPCR data from the same 60 breast cancer and 60 normal breast tissue specimens shown in (A) were normalized to GAPDH expression and grouped by tumor stage. The numbers of samples in each group are shown in parentheses below the graphs. Expression of nischarin was statistically significantly higher in normal tissues than in invasive ductal carcinoma (IDC; P = .019), and in IDC + ductal carcinoma in situ (IDC + DCIS; P = .018). The means and 95% confidence intervals from two independent experiments are shown. C) Subcategories of breast cancer patients and their association to nischarin expression as demonstrated by quantitative reverse transcription-PCR assays. D) Relative expression of nischarin in the subset of 24 patient-matched breast cancers and adjacent noncancerous breast tissues. Nischarin was more highly expressed in normal tissues than in tumors (Wilcoxon matched pairs t test; P = .026). Red lines indicating decreased expression and green lines indicating increased expression from two independent experiments are shown. E) Nischarin protein levels in breast cancer cell lines and tissues. Protein lysates were prepared from MCF-7 and MDA-MB-231 cells and commercially obtained tissue lysates were analyzed by western blotting. Stripped blots were reprobed with an antibody to vinculin. A representative example of several blots is shown here. F) Immunohistochemical staining of nischarin in breast cancer tissues. At least three samples from each category were stained with a mouse monoclonal antibody to nischarin (brown), and the experiments were repeated three times. Representative examples of grade 1, 2, and 3 are shown: normal breast (score 3), grade 1 (score 2), grade 2 (score 1), and grade 3 (score 0) infiltrating ductal carcinoma (original magnification = 20×; scale bar = 500 μM). G) Mean anti-nischarin staining intensity among tissue microarray (TMA) samples, by tumor grade immunohistochemical staining scores for different breast cancer stages were analyzed by one-way analysis of variance, and Dunn post hoc test was applied to assess the possible differences between nonmalignant samples and breast cancers samples of different stages. There were 226 breast cancer and 10 normal breast tissue samples included. Nonmalignant samples had higher nischarin protein expression than grade 3 samples (P = .011). Means and 95% confidence intervals from two independent experiments are shown. H) Mean anti-nischarin staining intensity among TMA samples, by tumor type. The same samples were regrouped and analyzed as described in (F). Nonmalignant samples had statistically significantly higher levels of nischarin protein expression than invasive ductal samples (P = .005) and metastatic samples (P = .006). The means and 95% confidence intervals from two independent experiments are shown. All P values were from two-sided tests. ILC = invasive lobular carcinoma; LCIS = lobular carcinoma in situ.
RNA Isolation and Real-Time Quantitative Polymerase Chain Reaction (qPCR)
Total RNA was isolated from human breast tissues with an mRNeasy Kit (Qiagen, Valencia, CA). The quality of the RNA was determined using an Agilent 2100 bioanalyzer and an RNA Nano 6000 Lab chip kit (Agilent Technologies, Palo Alto, CA); the concentration was determined with a Nanodrop apparatus (Nanodrop Technologies, Wilmington, DE). Expression of nischarin (gene name: NISCH) was determined in triplicate using TaqMan real-time qPCR assays (Applied Biosystems, Foster City, CA). Reverse transcription was performed using 500 ng of total RNA in a 20-μL reaction volume. Real-time qPCR was performed using the standard TaqMan assay protocol and the ABI7900 Real-Time PCR Detection System (Applied Biosystems). Each 20-μL reaction included 2 μL of reverse transcription product, 10 μL of TaqMan Universal PCR Master Mix (Applied Biosystems), and 1 μL of 300 nM primer (primer sequences in Supplementary Methods, available online) and probe mix. The reactions were incubated in 96-well plates at 95°C for 10 minutes followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Expression of nischarin mRNA was measured using threshold cycle values (CT). The ΔCT was calculated by subtracting the CT of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA from the CT of nischarin mRNA.
Tissue Array Analyses and Immunohistochemistry
Tissue microarrays (CC08-01-005; CC08-00-001; CC08-21-002) were purchased from Cybrdi (Rockville, MD). Tissue microarrays were rehydrated and processed in 10 mM citrate buffer (pH 6.0) for four antigen retrieval cycles of 5 minutes each in a microwave oven (800 W). After blocking for 1 hour at room temperature with 0.1% Triton X-100 and 10% normal goat serum in phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.7 mM KH2PO4; pH 7.4), all tissue microarrays were incubated with primary anti-nischarin antibodies at a dilution of 1:300 in the same solution at room temperature overnight. The sections were further processed with 100 μL of biotinylated anti-mouse secondary antibodies and then with 100 μL of avidin–biotin–peroxidase complex (Vector Labs, Burlingame, CA) and finally visualized with 50 μL of 3-3′ diaminobenzidine according to the Vector Labs protocol. All slides were scored for average staining intensity by two pathologists blinded to patient information. Staining intensity was graded as follows: 0, no expression (0% of the total cell population is positive); 1, weak expression (1%–30% of the cells positive); 2, moderate expression (30%–60% of the cells positive); and 3, strong expression (60%–100% of the cells positive). Average staining intensity was compared with the pathological information provided by Cybrdi. Staining of control sections, in parallel, with a mouse IgG control antibody (Sigma; 1:300) was used to estimate background staining.
Loss of Heterozygosity Analysis
Genomic DNA was extracted using a blood and tissue DNA kit (Qiagen) from portions of most of the 24 matched pairs of breast cancer and normal breast tissue samples, and loss of heterozygosity analysis was performed on 18 of those specimens (for which sufficient DNA was available) using three polymorphic microsatellite markers that mapped across chromosome band 3p21.1 and contained the NISCH gene. Assays were performed by PCR amplification in 25 μL reaction volumes using 20 ng of genomic DNA, 300 nM fluorescent dye–labeled forward primer and unlabeled reverse primer (purchased from IDT technologies, Coralville, IA), and AmpliTaq gold polymerase (Applied Biosystems) and the following conditions: initial denaturation at 95°C for 2 minutes; 35 cycles at 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 60 seconds; final extension at 72°C for 10 minutes. Each 6-μL sample was loaded onto a 7.5% polyacrylamide gel, and allelic loss was determined upon ethidium bromide staining; the assay was repeated at least three times. Also, PCR products were visualized using an ABI genetic analyzer, and the data were analyzed using GeneScan software (Applied Biosystems, Foster City, CA). Microsatellite markers D3S3688, D35361, and D3S3026 were used in the reactions. The primers used in this study are listed elsewhere (Supplementary Methods and Supplementary Table 1, available online).
qPCR for Microdeletion Analysis of Genomic DNA from Breast Tissue
Applied Biosystems SYBR Green Mix was used for genomic qPCR. Primers were designed using Primer 3 software (http://frodo.wi.mit.edu/primer3/; see Supplementary Methods, available online, for sequences). Briefly, 20 ng genomic DNA was used in a 25-μL reaction with 300 nM primers. Reactions were performed in triplicate using an ABI 7900 HT real-time PCR thermocycler using the conditions: 95°C for 10 minutes, then 40 cycles at 95°C for 15 seconds, and 60°C for 1 minute. Standard curves for nischarin and GAPDH (control) primers were generated using 10-fold dilution series ranging from 0.1to 100 ng and the slope value was determined. The CT values for each primer pair were corrected or normalized using the CT value of the GAPDH products for the same sample. Corrected CT (KCT) values were determined using the absolute standard curve, comparative CT, and relative standard methods as described (8). If the difference (ΔKCT) in corrected CT (KCT) value in normal and tumor sample was less than −0.35, it was considered to indicate a microdeletion, and if the difference was −0.35 to +0.35, it was considered to indicate no deletion.
Anchorage-Independent Growth
To assess anchorage-independent growth of breast cancer cells in which nischarin expression was increased or silenced, we performed soft agar assays. In each well of six-well tissue culture plates, we plated 50 000 cells in 1 mL of 0.35% agarose in DMEM with 10% FBS (warmed to 40°C) on top of 1 mL of solidified 0.5% agarose in DMEM with 10% FBS. After the top layer of agarose solidified, 1.5 mL of DMEM with 10% FBS was added to each well, and the plates were incubated at 5% CO2 and 37°C. The medium was changed once every 3 days. After 3 weeks, colonies were stained with 0.1% crystal violet for 1 hour and destained with several washes of PBS. The number of colonies growing in soft agar that were greater than 100 microns in size were determined by examination under a simple Nikon microscope (Nikon Instruments, Melville, NY) and digital images were captured using Epson Perfection V700 Photo Scanner (Epson American, Long Beach, CA).
Tumor Growth Assays
A xenotransplantation assay was performed to understand the effect of nischarin on breast tumorigenesis using mouse models. Mouse experiments were done in accordance with protocols approved by the Institutional Animal Care and Use Committees of Rockefeller University and Louisiana State University Health Sciences Center (LSUHSC). We used 4- to 6-week-old female BALB/c nude mice in the xenograft studies (five mice per group). MDA-MB-231 cells overexpressing nischarin and their parental cells (as a control) or MCF-7 cells in which nischarin expression had been silenced and their parental cells (as a control) were incubated in trypsin, washed three times in PBS, and counted. Mice were anesthetized for 20 minutes with 50–100 mg/kg of ketamine–HCl and 5–10 mg/kg of xylazine–HCl, delivered intraperitoneally, and a small incision was made in the abdomen to visualize the mammary fat pads. We injected 1 × 106 cells, suspended in 100 μL of Matrigel diluted 1:1 in PBS, into each of the two fat pads per mouse. We also subcutaneously implanted a 60-day release tablet of 17β-estradiol (Innovative Research of America, Sarasota, FL) in each mouse when performing experiments with MCF-7 cells because these cells are estrogen-dependent. Tumor growth was measured twice per week using calipers, and the tumor volume was calculated using the formula π × length × width2/6 as previously described (9).
Experimental Metastasis Followed by Bioluminescent Imaging
MDA-MB-231 cells that stably expressed luciferase (referred to as MDA-MB-231-luc) were a generous gift from Dr Jerry W Shay, University of Texas Southwestern Medical Center at Dallas, TX. We transfected these cells to express nischarin (Supplemental Methods, available online). For lung metastasis experiments, 1 × 106 luciferase-expressing MDA-MB-231 cells that either did or did not overexpress nischarin were suspended in 100 μL of PBS and injected into the tail veins of 6-week-old BALB/c nude mice. On days 5, 10, 20, and 40 after injection of the cells, the mice were analyzed for metastatic disease by bioluminescence imaging using an IVIS-200 camera system for detection of luciferase expression (Xenogen, Hopkinton, MA) (10). Fifteen minutes before the in vivo imaging, the mice were anesthetized using the XGI-8 Gas Anesthesia System (Caliper Life Sciences, Hopkinton, MA) with 3% isofurane, and they were injected intraperitoneally with D-luciferin (150 mg/kg in PBS). Mice were kept inside the warm chamber of the Xenogen camera system with a continuous flow of 1.5% isofurane gas along with 100% oxygen to keep them anesthetized. For ex vivo lung imaging, 300 μL of luciferin (150 mg/kg in PBS) was injected intraperitoneally just before killing the mice by CO2 asphyxiation, and lungs were extracted and imaged using the IVIS-200 camera. Both experiments were performed using five or six mice per condition and were repeated three times. Also, intact lungs were extracted and subsequently fixed in 4% formalin (Sigma) and prepared for standard histological examination. The imaging results were analyzed using Living Image, version 3.0 (Xenogen software). A region of interest was manually selected over relevant regions of signal intensity, and the intensity was recorded as the maximum number of photons emitted within a region of interest. Luminescent intensity obtained from each mouse was plotted using Graph Pad Prism software version 5.0 (San Diego, CA).
Truncation Constructs of α5 Integrin
Full-length α5 integrin and truncation mutations that we made have been described previously (11). The α5 c-10 construct expressing α5 integrin had only 10 amino acids instead of 27 in the cytoplasmic region, whereas α5 c-1 had only one amino acid; the entire cytoplasmic domain of 27 C-terminal amino acids including KLGFFKR residues was deleted, except for lysine in the proximal transmembrane region. The latter construct is often referred to as a “tailless” mutant.
Luciferase Reporter Transfection and Dual Luciferase Assay
To measure activation of the α5 integrin promoter, 5 × 104 stably transfected MDA-MB-231 cells that overexpressed nischarin or 5 × 104 stably transfected MCF-7 cells in which nischarin expression had been silenced were transiently transfected with 400 ng of the pGL3 vector carrying either 923 bp or 26 bp of the integrin α5 promoter (12) (generous gift from Dr Jesse Raman, University of Louisville, KY), along with Renilla vector to normalize the expression. Cell lysates were prepared 48 hours after transfection, and reporter activity was measured with the Dual Luciferase Assay kit as recommended by manufacturer’s protocol (Promega, Madison, WI). Data represent the average of three independent experiments, and P values less than .05 were considered to be statistically significant.
Rac GTPase Assay
Rho/Rac/Cdc42 Activation Assay Combo Kit was used to perform Rac activation assay as described (Cell Biolabs, San Diego, CA). Briefly, MDA-MB-231 green fluorescence protein (GFP) and MDA-MB-231 GFP nischarin cells were washed two times in cold PBS and then lysed in buffer B (50 mmol/L Tris [pH 7.6], 150 mmol/L NaCl, 1% Triton X-100, 0.5 mmol/L MgCl2, supplemented with 1 mmol/L PMSF, 1 μg/mL aprotinin, and 1 μg/mL leupeptin). One milligram of total protein was then incubated with 30 μg glutathione S-transferase (GST)-PAK (PBD) beads for 30 minutes. The complexes were then washed, followed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and the amount of active-Rac was determined by western blotting using Rac1-specific antibodies. Total cellular lysates were also separated by SDS-PAGE, and western blot analysis with anti-Rac1 antibodies was done as a control for protein loading.
Lung Histology
Mice were killed, and lungs were inflated and fixed with 1.0 mL of Z-fix solution overnight followed by washing with PBS and dehydration in 70% ethanol. Tissue paraffin embedding, sectioning, and hematoxylin and eosin (H&E) staining were performed by Morphology and Imaging Core facility at LSUHSC.
Microarray Data Retrieval and Bioinformatics Analysis
Public-domain expression microarray data was obtained from web sites and further analyzed using web-based microarray analysis software (13–23).
Statistical Analyses
Statistically significant differences between samples were determined by paired two-tailed t tests. Statistical significance of nischarin expression differences between samples were also determined by paired two-tailed t tests. One-way analysis of variance was used to assess differences in nischarin expression between tumor types. Tukey multiple comparisons test was used for all possible pairwise comparisons.
In determinations of recurrence-free survival, nischarin expression levels from a microarray analysis of breast cancer patients were averaged, and patients were then stratified either into the low-nischarin expression group (below mean expression) or the high-nischarin group (above mean expression). Associated times to recurrence of breast cancer were then plotted using a Kaplan–Meier survival curve. For mouse experiments, we used two-sided student t tests to verify statistical significance.
Results
Nischarin Expression During Breast Cancer Progression
We investigated nischarin expression in human breast cancer cell lines and specimens from human breast cancer patients. Nischarin mRNA expression was lower in moderately invasive cells (MCF-7, T47D, BT474) than in nontumorigenic MCF-10A cells and was lowest in the highly invasive cancer cell line, MDA-MB-231, as assessed by reverse transcription-PCR (Supplementary Figure 1, available online). Using qPCR, we assessed nischarin mRNA expression in 60 human breast cancer specimens relative to 60 normal breast tissues (Figure 1, A and B). Nischarin mRNA was strongly expressed in normal tissues; however, in comparison, expression was lower in the breast cancer specimens (mean level in normal breast = 50.7 [arbitrary units], in breast tumor = 16.49 [arbitrary units], difference = 34.21, 95% confidence interval [CI] on difference = 11.63 to 56.79, P = .003; Figure 1, A). The 60 breast cancers were a mixture of different types, among them, in order of increasing aggressiveness, DCIS, LCIS, IDC, ILC, and various mixtures thereof. IDC samples had lower nischarin expression. Overall, all of the specimens with invasive components exhibited lower nischarin expression than the nonmalignant tissues, and the results were statistically significant (P = .019) (Figure 1, B and C).
To explore whether nischarin expression levels were associated with breast cancer stage, we used qPCR to quantify nischarin mRNA expression in a panel of 45 cDNAs prepared from breast cancer tissue at different stages. Nischarin expression was high in stage 0 specimens, whereas stage I–IV specimens had lower expression (Supplementary Figure 2, available online). To assess nischarin expression during breast cancer progression, we further quantified nischarin mRNA in a panel of 24 primary breast tumor samples with matched normal breast tissue from the same patient. Nischarin expression was statistically significantly higher in nonmalignant than in malignant specimens in 17 (71%) of 24 of these matched pairs (mean nischarin expression in normal breast = 61.76 arbitrary units, in breast tumor = 22.48 units, difference = 39.28, 95% CI on the difference = 8.66 to 88.15, P = .026 by Wilcoxon matched paired t test; Figure 1, D). These findings support the possibility that nischarin expression is lost during breast cancer progression.
To further examine whether nischarin expression was decreased in a broader spectrum of human breast cancers and further explore any associations with clinical and pathological data, we examined nischarin expression in three publicly available microarray datasets of human breast cancer specimens that we analyzed by the web-based microarray bioinformatics tool, ONCOMINE (13). Nischarin expression was decreased in breast cancers compared with levels in normal breast in the database (reference 14 dataset P < .001, reference 15 dataset P = .011, reference 16 dataset P = .014; Supplementary Figure 3,A, available online). We also used eight additional ONCOMINE datasets to examine the association of nischarin mRNA expression with breast carcinoma tumor grade (Supplementary Figure 3,B, available online). There were 258 grade 1, 536 grade 2, and 457 grade 3 samples in these datasets. Nischarin expression was statistically significantly lower in the high-grade tumors relative to the low-grade tumors (P < .05 for datasets 17–21 overall; reference datasets 17–21, P < .001; reference dataset 22, P = .002; reference dataset 23, P = .006; reference dataset 16, P = .007; Supplementary Figure 3, B, available online). Taken together, these data suggest that decreased nischarin expression is a common event in human breast cancer and that loss of expression is associated with higher tumor grade and with invasive disease.
We next examined nischarin protein expression in breast cancer cell lines and tumor tissues. Consistent with our nischarin mRNA expression data, western blot analysis detected little nischarin in highly invasive MDA-MB-231 cells or in invasive ductal and invasive lobular breast carcinoma tissues, whereas nischarin protein expression could be detected in MCF-7 cells, normal breast tissue, and in DCIS (Figure 1, E). To examine nischarin protein expression in a broader spectrum of human breast cancers, we performed immunohistochemical analysis on three commercially available tissue microarrays that collectively consisted of 226 breast cancer and 10 unmatched normal mammary tissue samples. Using an antibody that detects nischarin for which the specificity was independently verified (see Supplementary Figure 8, available online), these arrays were stained for nischarin and then scored for staining intensity by two pathologists. The intensity of staining was scored as the average staining intensity of the epithelial cells in each sample (0, no staining; 1, light staining; 2, medium staining; and 3, intense staining), as shown in representative examples (Figure 1, F). We used three different arrays and established that nischarin expression was highest in normal breast specimens, lower levels in grades 1 and 2 (which had approximately equal levels), and lowest for grade 3 (staining scores of nischarin expression in normal breast = 1.21 arbitrary units, in grade 3 breast tumor = 0.25 units, difference = 0.96, 95% CI = 0.329 to 1.639, P = .011; Figure 1, G). Furthermore, we compared mean staining intensity for nischarin in different types of breast cancer in all three microarrays (CC08-00-001, CC08-21-002, and CC08-01-005). Quantification of the mean nischarin staining intensity indicated that nischarin expression was lower in invasive carcinoma compared with normal breast tissue (Figure 1, H). These data are consistent with our nischarin mRNA level studies, supporting loss of nischarin expression in association with invasive disease and higher tumor grade.
Genomic Loss of the NISCH Locus in Human Breast Cancer
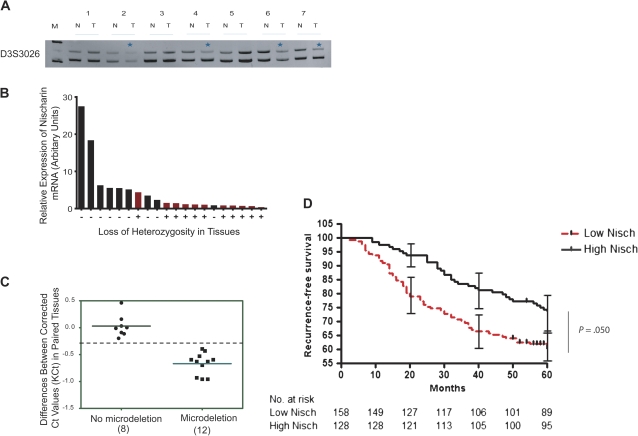
The nischarin gene, NISCH, maps to chromosomal locus 3p21, a cytogenetic region reported to exhibit loss of heterozygosity (LOH) in a variety of human cancers including breast cancer (24,25). To further investigate the mechanism for loss of nischarin expression during the breast cancer progression, we examined LOH at the NISCH locus using microsatellite markers in DNA samples extracted from 18 human breast cancers and patient-matched normal tissue counterparts. With three microsatellite markers, we established that 50% of tumor samples (nine of 18) exhibited LOH at the NISCH locus. Data are shown for the D3S3026 marker (Figure 2, A). Importantly, LOH at the NISCH locus was associated with decreased nischarin expression (Figure 2, B).
Figure 2.
Association of loss of the nischarin (NISCH) gene locus with loss of nischarin expression in breast cancers. A) Loss of heterozygosity (LOH) at the NISCH locus. Genomic DNAs were isolated from 20 matched normal and tumor samples and were amplified by polymerase chain reaction (PCR) using primers specific to microsatellite markers located near the NISCH locus; PCR products were separated on acrylamide gels to confirm the allelic loss in tumor samples. The experiment was repeated three times. Representative results show allelic loss in tumors 2, 3, 4, 6, and 7 (indicated by asterisks) after PCR products were separated on a 7.5% polyacrylamide gel. Data are shown for D3S3026 microsatellite marker, allele length: 209–226 (PCR product from two different alleles). B) Presence of nischarin mRNA compared with presence of LOH in breast cancer specimens. Nischarin mRNA expression was measured by quantitative PCR. Red bars represent the tissue samples with LOH and black bars represent the samples without apparent allelic loss. C) Microdeletion analysis of nischarin locus in matched breast tissue samples. Genomic DNA isolated from matched normal and tumor samples were amplified by PCR using primers located in introns 2, 3, and 6 of nischarin. The experiment was repeated three times. Eight paired tissues did not have microdeletions at the NISCH locus (ΔKCT values of 0 ± 0.35), whereas 12 paired tissues did have NISCH microdeletions (ΔKCT below −0.35) in tumor tissues. D) Nischarin message levels and recurrence-free survival in women with breast cancer. Nischarin expression levels from a microarray analysis of 286 breast cancer patients were averaged, and patients were then stratified either into the low-nischarin group (below mean expression, n = 158) or the high-nischarin group (above mean expression, n = 128). Associated times to recurrence of breast cancer were then plotted using a Kaplan–Meier survival curve (n = 286, P = .050 [two-sided log-rank test]). At month 40, recurrence-free survival among women with high nischarin (105 of 128) = 82%, among women with low nischarin (106 of 158) = 67.1%, difference = 14.9%, 95% CI = 13% to 16.8%, P = .046 (two-sided log-rank test). At 20, 40, and 60 months, 95% confidence intervals are shown. M = molecular weight marker; N = normal tissue; T = tumor tissue.
In addition, we performed microdeletion analysis on twenty matched pairs of breast tissue samples using qPCR with three pairs of primers (see “Methods” for details) designed to amplify within intron 2, 3, and 6 of the human nischarin gene. The data shown are for intron 6 only (Figure 2, C). The qPCR analysis further validated the loss of nischarin locus in 12 of 20 matched tumor samples. These results support the conclusion that nischarin expression is decreased in a large proportion of primary human breast cancers through LOH.
Because decreased nischarin expression is frequently observed in human breast cancers, we investigated whether nischarin expression could be a prognostic marker for breast cancer patients. We analyzed multiple expression datasets for which both nischarin expression and recurrence-free survival data were available (26). The patient population (n = 286) was stratified into two groups based on nischarin expression. The cut point was defined as the mean of the entire population, with high-expressing nischarin samples above the mean (n = 128 patients) and low-expressing nischarin samples below the mean (n = 158 patients). We found that patients with elevated nischarin expression showed statistically significantly increased recurrence-free survival compared with those with low expression levels of nischarin (hazard ratio of survival = 1.473, 95% CI = 1.003 to 2.148, P = .048) (Figure 2, D). Specifically, a twofold increase in nischarin expression conferred a 2.8-fold decrease in risk of tumor recurrence. These data suggest that decreased nischarin expression may predict decreased recurrence-free survival in breast cancer patients.
Effect of Nischarin on Cell Growth, Anchorage-Independent Growth, and Tumor Growth In Vivo
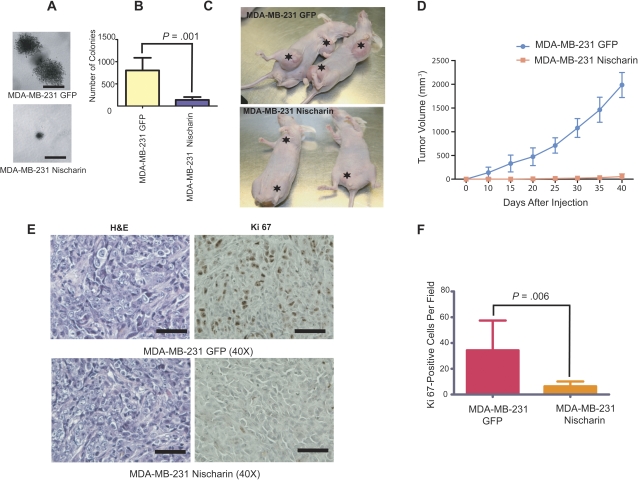
The loss of nischarin expression during breast cancer progression suggested that nischarin might normally function to prevent cancer progression. We previously demonstrated that nischarin decreases the migratory potential of breast cancer cells (1). Cell proliferation is an important determinant of tumor growth and metastasis. To examine whether nischarin could function as a putative tumor suppressor, we initially assessed the effects of nischarin expression on the growth of MDA-MB-231 cells. Whereas control MDA-MB-231 cells that carried the empty vector grew robustly, growth of nischarin-expressing cells was statistically significantly inhibited (at day 5, mean growth of MDA-MB-231 GFP cells = 1.000 absorbance units; mean growth of MDA-MB-231 GFP- and nischarin-expressing cells = 0.3267 absorbance units, difference = 0.6733, 95% CI = 0.6262 to 0.7206, P < .001; Supplementary Figure 4, available online), supporting an effect by nischarin on breast cancer cell growth. Because nischarin inhibited MDA-MB-231 cell growth in tissue culture dishes, we also investigated whether it could inhibit anchorage-independent growth. Vector-expressing MDA-MB-231 cells exhibited high potential to form colonies in soft agar; however, nischarin-expressing cells had a substantially reduced propensity for soft agar colony formation (Figure 3, A and B).
Figure 3.
Effect of restoring nischarin expression on anchorage-independent growth and tumor growth. A) Anchorage-independent growth of MDA-MB-231 cells overexpressing nischarin. Cells were stably transfected with a nischarin-expressing or green fluoresecence protein (GFP) control plasmid and assayed for growth in soft agar. Representative images of colonies formed from vector and nischarin-overexpressing cells are shown (scale bar = 100 μM). B) Quantification of soft agar colonies formed by nischarin-expressing MDA-MB-231 cells. Cells without nischarin overexpression (GFP control cells: MDA-MB-231 GFP) formed statistically significantly more colonies than nischarin-overexpressing MDA-MB-231 cells (two-sided paired t test; asterisks denote P = .001). The experiment was repeated three times. C) Effect of nischarin expression on growth of MDA-MB-231 cell xenografts. Nischarin- or GFP-expressing MDA-MB-231 cells were injected into the two breast fat pads per female nu/nu mouse (n = 5 per group). A representative photograph of the tumors produced by vector vs nischarin-expressing cells is shown. Tumors and the sites of injected tumor cells are shown with asterisks. D) Quantification of primary tumor growth from nischarin- or GFP-expressing MDA-MB-231 cells. Experiments were performed in triplicate. At day 40, mean volume of tumors from MDA-MB-231–GFP control cells = 1977 mm3, of tumors from MDA-MB-231–nischarin cells = 42.27 mm3, difference = 1935 mm3, 95% confidence interval = 395.0 to 3475 mm3, P = .003 (paired two-sided test). E) Cell proliferation in nischarin-expressing vs control tumors. Paraffin-embedded tissue sections of primary tumors from mice injected with MDA-MB-231 (Top panel) and MDA-MB-231-Nisch (bottom panel) cells were immunostained with anti-Ki67 antibody (brown stain; right) or hematoxylin and eosin (H&E; blue nuclei and red cytoplasm; left). Photomicrographs were taken at 40× magnification; scale bars = 100 μM. F) Quantification of cell proliferation in tumors from nischarin-expressing cells or control MDA-MB-231 cells. The number of Ki67-positive cells per field is shown, as an average of three of fields viewed. Error bars represent 95% confidence intervals, and the P value was calculated using a two-sided Student t test; P = .006.
Our in vitro studies suggested that decreased nischarin expression in MDA-MB-231 breast cancer cells may provide a growth advantage. To investigate this possibility, we stably expressed GFP–nischarin or GFP alone, as a control, in MDA-MB-231 cells using a lentiviral expression vector. Cells were injected into the mammary fat pads of BALB/c nu/nu mice and measured tumor volume once every 3 days for 40 days. Although tumors formed in both groups, tumors derived from cells expressing GFP alone consistently grew more rapidly and attained greater volumes than tumors derived from nischarin-expressing MDA-MB-231 cells. Overall, tumors from the control cells attained a mean calculated volume that was 10 times greater than that of tumors from nischarin-expressing cells (at day 40, mean volume of tumors from MDA-MB-231–GFP control cells = 1977 mm3, of tumors from MDA-MB-231–nischarin cells = 42.27 mm3, difference = 1935 mm3, 95% CI = 395.0 to 3475, P = .025; Figure 3, C and D). Western blot analysis of protein extracted from the tumors confirmed that tumors that carried GFP–nischarin plasmids continued to express nischarin (see Figure 5, E), suggesting that inhibition of tumor growth is due to nischarin expression. These results suggest that nischarin inhibits tumor growth and that it may act as a tumor suppressor in vivo. To investigate whether nischarin-expressing cells were less proliferative in vivo, we performed immunohistochemical staining for Ki67, a marker for proliferation. Nischarin-expressing tumors had statistically significantly less Ki67 staining than GFP-expressing controls (mean number of Ki67-positive MDA-MB-231 cells per field = 34.33, of Ki67-positive MDA-MB-231–nischarin cells = 6.333, difference = 28.00, 95% CI = 12.91 to 43.09, P = .006; Figure 3, E and F), which supports a role for nischarin in regulating tumor growth, in part via effects on cell proliferation.
Figure 5.
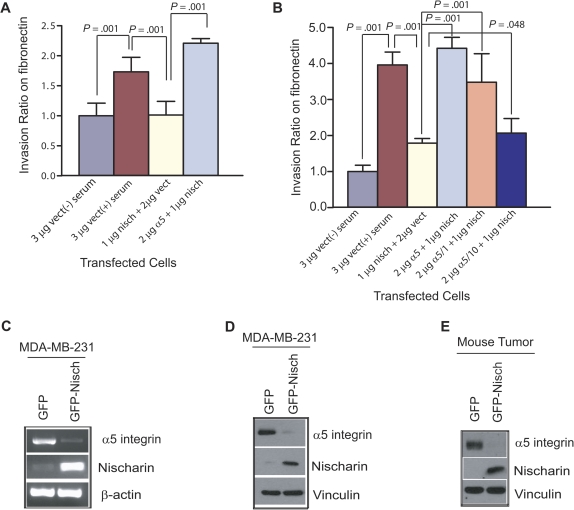
Effect of nischarin expression on α5 integrin expression and α5-mediated invasion. A) Effect of nischarin on invasiveness of MDA-MB-231 cells through fibronectin. MDA-MB-231 cells were transiently transfected with nischarin + vector, vector alone, or nischarin + α5 integrin. A plasmid encoding β-galactosidase was introduced in every transfection to visualize the cells. Invasiveness was tested by placing cells in serum-free medium on top of a fibronectin-coated filter with eight micron pores in a transwell dish and monitoring cell invasion toward serum-containing medium on the opposite side of the filter (except in one control sample, in which serum was omitted). Error bars represent 95% confidence intervals and for all three bracketed comparisons; P = .001 (two-sided Student t tests). Experiments were repeated three times. B) Role of α5 integrin in nischarin-mediated inhibition of MDA-MB-231 invasiveness on fibronectin. MDA-MB-231 cells were transfected with nischarin + α5 integrin, nischarin + vector, nischarin + α5 integrin c-1, or nischarin + α5 integrin c-10, and a serum-induced invasion assay was performed in triplicate. Error bars represent 95% confidence intervals, and P values were calculated using two-sided Student t tests. C) Effect of nischarin on α5 integrin mRNA expression. Nischarin, α5 integrin, and β-actin mRNA expression were examined by reverse transcription-polymerase chain reaction in MDA-MB-231 cells expressing green fluorescence protein (GFP) or nischarin. D) Effect of nischarin on α5 integrin protein expression. Nischarin, α5 integrin, and vinculin protein expression were examined by western blot analysis in MDA-MB-231 cells expressing GFP or nischarin. E) Effect of nischarin on α5 integrin protein expression in xenografted tumors. Nischarin, α5 integrin, and vinculin protein expression were examined by western blot analysis in MDA-MB-231 xenograft tumors expressing GFP or nischarin. nisch = nischarin; vect = vector.
Nischarin and Lung Metastasis
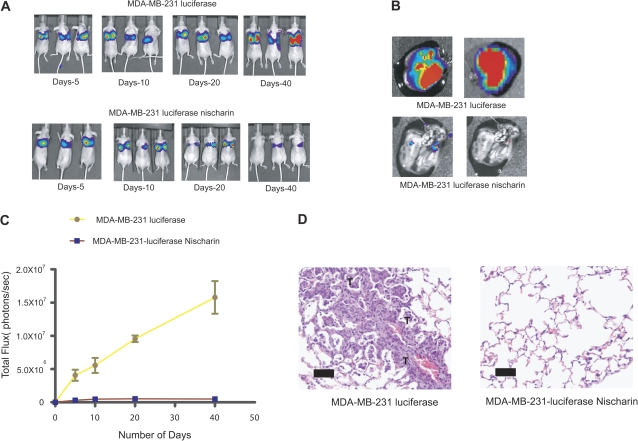
We further examined the function of nischarin in vivo using an experimental lung metastasis model. In this mouse model, tumor cells are injected directly into the blood stream, bypassing the invasion and intravasation steps of metastasis and seeding the lung. MDA-MB-231 cells (referred to as MDA-MB-231-luc) were genetically engineered to express the firefly luciferase gene. These cells were transduced with lentivirus particles that expressed nischarin or served as an empty vector control. Nischarin-expressing MDA-MB-231-luc or vector-expressing MDA-MB-231-luc cells were injected into the tail veins of nude mice. Two hours after injection, bioluminescent imaging was performed to establish whether the cells were homing to the lung, and subsequent bioluminescent imaging was performed every week for 8 weeks. In the mice injected with nischarin-expressing MDA-MB-231 cells, there was a statistically significantly lower bioluminescent signal than in mice injected with vector control MDA-MB-231 cells (at day 40, mean signal in lung for MDA-MB-231-luc = 17 110 000 photon per second; for MDA-MB-231-luc–nischarin = 513 300 photon per second, difference = 16 600 000 photon per second; 95% CI = 13 050 000 to 20 150 000 photon per second; P < .001; Figure 4, A). The bioluminescent signals from mice injected with nischarin-expressing MDA-MB-231 cells were six orders of magnitude less than those of controls injected with vector-expressing MDA-MB-231 cells (Figure 4, C). Ex vivo bioluminescent imaging of lungs from these mice after 8 weeks also revealed that nischarin suppressed lung metastasis (Figure 4, B).
Figure 4.
Nischarin overexpression and lung metastasis in a nude mouse model. A) Time course of lung metastasis in mice carrying nischarin-expressing or control MDA-MB-231 cells. One million MDA-MB-231–luc cells transfected with an empty luciferase vector (as a control) or with the same plasmid overexpressing nischarin were injected into the tail vein of nude mice (n = 5), and bioluminescent imaging was performed once a week. Here, bioluminescent images of three representative mice at 5, 10, 20, and 40 days after tail vein injection (post tail vein injection) are shown. B) Ex vivo bioluminescent imaging of lungs from mice injected with vector control–expressing cells or nischarin–expressing cells. Imaging was performed at 8 weeks after injection. C) Quantification of bioluminescence data. Photon intensity per second in whole mice was quantitated as in (A). Error bars represent 95% confidence intervals, and the P value was calculated at day 40 using a two-sided Student t test; P < .001 (n = 5 mice per group). D) Hematoxylin- and eosin-stained lung tissue sections from mice injected with MDA-MB-231 cells stably expressing nischarin or a vector control. The “T” indicates a tumor metastasis (Scale bar = 500 μM).
To further confirm the effect of nischarin on metastasis, we performed a histological analysis of lung tissues from the mice that were injected with breast cancer cells through the tail veins. Lung tissues were extracted and sectioned, and H&E staining revealed that very few metastases were formed in the lungs of mice injected with nischarin-expressing cells (Figure 4, D). By contrast, lungs from mice injected with vector control cells were heavily infiltrated by metastases. Further pathological examination of lung metastases in mice carrying nischarin-expressing MDA-MB-231-luc xenografts revealed that these metastases were small and isolated compared with the large locally invasive metastases observed in mice carrying control MDA-MB-231-luc xenografts (data not shown). These results suggest that nischarin is an important regulator of breast cancer cell invasion and metastasis in vivo.
Role of α5 Integrin in Nischarin-Mediated Inhibition of Cell Invasion
Cell invasion is an important event in tumor progression and both adhesion of tumor cells to the extracellular matrix and migration of tumor cells contribute to cancer cell invasion and metastasis (27). To define the mechanism whereby nischarin suppresses invasion, we examined invasion by nischarin-expressing vs vector-expressing MDA-MB-231 cells with in vitro assays in the presence of different types of extracellular matrices. Although nischarin overexpression did not affect basal or serum-stimulated invasion in transwell assays in the presence of polylysine (Supplementary Figure 5, A, available online) or collagen (Supplementary Figure 5, B, available online), nischarin expression did inhibit the invasiveness of MDA-MB-231 cells in the presence of fibronectin (Figure 5, A). These findings suggested that nischarin inhibits tumor cell invasion specifically in the context of fibronectin.
We previously established a specific protein-protein interaction between nischarin and α5 integrin (1): α5 integrin complexes with β1 integrin to form the classic fibronectin receptor, which plays a vital role in both cell migration and cell adhesion. Because nischarin specifically inhibited cancer cell invasion in the context of fibronectin, we investigated whether these effects were mediated through α5 integrin by expression of α5 integrin could abrogate nischarin-mediated inhibition of invasion. Although α5 integrin had no effect on the invasion through polylysine (Supplementary Figure 5, A, available online) or collagen (Supplementary Figure 5, B, available online) by MDA-MB-231 cells, exogenous α5 integrin did abrogate nischarin-mediated inhibition of invasion through fibronectin (Figure 5, A and B).
We previously demonstrated that the proximal transmembrane region (KLGFFKR) of α5 integrin is important for its interaction with nischarin (2). To examine whether nischarin inhibited cell invasiveness through α5 integrin, we introduced α5 truncation constructs into MDA-MB-231 cells before doing similar assays. One truncation mutant, α5 c-10, was missing 16 amino acids in the cytoplasmic domain but contained the nischarin binding region. The other, α5 c-1, lacked the entire cytoplasmic domain and did not include the nischarin binding region. Although α5 c-10 was partially able to recapitulate the effects of full-length α5 integrin, α5 c-1 was largely inactive (Figure 5, B). These data suggest that nischarin’s interaction with α5 integrin is critical for nischarin-mediated inhibition of invasion.
Effect of Nischarin on α5 Integrin Expression
We next investigated whether nischarin regulated α5 integrin expression. To examine this possibility, we performed reverse transcription-PCR on RNA from nischarin-expressing cells using α5 integrin–specific primers and western blot analysis using α5-specific antibodies. In MDA-MB-231 cells, nischarin expression was associated with potently decreased α5 integrin expression both at the mRNA level (Figure 5, C) and the protein level (Figure 5, D). These data suggested that a reduction of α5 integrin expression by nischarin could be one of the mechanisms by which nischarin functions as a tumor suppressor. To establish whether this mechanism was operating in vivo, we analyzed expression of nischarin and α5 integrin protein in MDA-MB-231 tumors formed in nude mice in the presence and absence of nischarin. Similar to our findings in vitro, nischarin expression was associated with potently decreased α5 integrin expression in vivo (Figure 5, E), supporting nischarin-induced loss of α5 integrin expression as one mechanism for the tumor-suppressing effects of nischarin.
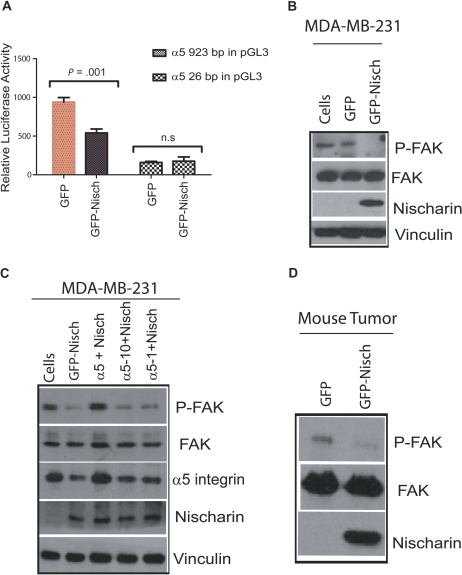
To examine whether nischarin expression modulates the transcriptional regulation of α5 integrin, we performed dual luciferase assays on stable clones of MDA-MB-231 cells that overexpressed nischarin and transiently expressed either the full-length (923 bp) α5 integrin promoter or a 26-bp segment of the α5 integrin promoter fused with the luciferase gene in a pGL3 vector (the 26-bp promoter construct served as a negative control). When nischarin was overexpressed, activity of the full-length α5 integrin promoter was statistically significantly decreased (P = .001; Figure 6, A). These data suggested that nischarin might transcriptionally regulate α5 integrin to decrease its expression.
Figure 6.
Nischarin and α5 integrin–mediated signaling. A) The α5 integrin promoter assay. In triplicate experiments, MDA-MB231-derived cell lines expressing green fluorescence protein (GFP)–nischarin or a control plasmid were transiently transfected with plasmids encoding firefly luciferase under the control of the α5 integrin promoter: either 500 ng of the full-length α5 integrin promoter (923 base pair, left two bars) or a truncated (25 base pair, right two bars) portion of the α5 integrin promoter. Transfections also included 50 ng of the Renilla luciferase vector as a control to normalize the experiments. Firefly luciferase activity, relative to Renilla luciferase activity, was measured in dual assays at 40 hours after transfection. Error bars represent 95% confidence intervals; asterisks indicate a P < .001 that was calculated using a two-sided Student t test. Experiments were performed in triplicate. B) Effect of nischarin on focal adhesion kinase (FAK) phosphorylation. Stable MDA-MB-231 cells, or derivatives that expressed GFP-nischarin or a control plasmid, were lysed and 30 μg of protein was used, and phosphorylation of FAK at tyrosine-397 (P-FAK), total FAK, and nischarin and vinculin protein expression were examined by western blot analysis. Thirty micrograms of protein were loaded in each lane. C) Effect of nischarin on FAK phosphorylation in presence of α5 integrin truncation constructs. MDA-MB-231 cells were transiently transfected as shown, and the lysates made from these cells were immunoblotted with FAK, phosphotyrosine 397-FAK (P-FAK), α5 integrin, nischarin, and vinculin antibodies. The rabbit polyclonal α5 integrin cytoplasmic domain–specific antibody (Millipore AB1928) detected a robust α5 integrin signal only in lysates from cells that were transfected with full-length α5 integrin. Experiments were performed in triplicate. D) Effect of nischarin on FAK phosphorylation in vivo. Phosphorylation of FAK at tyrosine-397 (P-FAK), total FAK, and nischarin protein expression were examined by western blot analysis in MDA-MB-231 xenograft tumors expressing GFP or nischarin. Experiments were performed in triplicate.
To further define the mechanism for nischarin’s tumor suppressor effects, we examined the effects of nischarin on α5-mediated signaling (28). FAK has been shown to promote cell migration and cell invasion, and it is overexpressed in human cancers. Because α5 integrin is known to stimulate FAK signaling (29), we examined whether nischarin affected FAK phosphorylation as a surrogate for FAK activation. Lysates prepared from MDA-MB-231 cells expressing nischarin and control cells were immunoblotted with an anti-phosphoFAK antibody. In the MDA-MB-231 model, nischarin expression was accompanied by strongly decreased FAK phosphorylation (Figure 6, B). To investigate whether these effects could be attributed to altered α5 integrin expression, we rescued the loss of α5 integrin expression in these cells by expression of exogenous α5 integrin or the α5 integrin cytoplasmic domain mutants α5 c-1 and α5 c-10. Although restoring α5 integrin expression reversed nischarin-mediated suppression of FAK phosphorylation, α5 c-1 and α5 c-10 were only partially able to do so (Figure 6, C). To establish whether this mechanism was operating in vivo, we analyzed the expression of nischarin and phosphorylated FAK in tumors formed in nude mice from MDA-MB-231 cells with and without nischarin expression. As in our in vitro studies, the expression of nischarin was accompanied by potently decreased FAK phosphorylation in vivo (Figure 6, D). Taken together, these results support a model in which nischarin functions as a tumor suppressor by decreasing α5 integrin expression and thereby decreasing FAK signaling.
Nischarin Silencing and α5 Integrin Expression
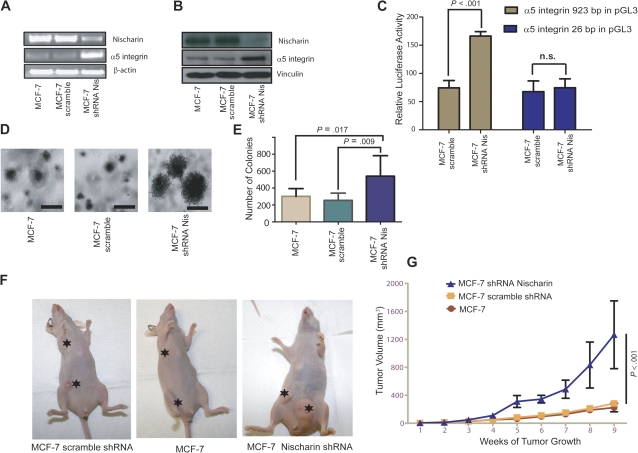
Because restoring expression of nischarin to MDA-MB-231 cells inhibited tumor growth and metastasis, we examined whether loss of nischarin expression could promote these processes in MCF-7 breast cancer cells, which are normally weakly tumorigenic and invasive. Expression of nischarin shRNA effectively reduced nischarin expression in MCF-7 cells at the mRNA (Figure 7, A) and the protein level (Figure 7, B). Whereas restoring nischarin expression in MDA-MB-231 cells reduced α5 integrin expression, reducing nischarin expression induced α5 integrin expression in MCF-7 cells (Figure 7, A and B, middle panels). Also, MCF-7 cells in which nischarin expression was silenced had statistically significantly increased α5 integrin promoter activity (Figure 7, C, P < .001). These data suggest that nischarin-mediated reduction of α5 integrin expression could be one mechanism by which nischarin functions as a tumor suppressor in breast cancer.
Figure 7.
Effects of nischarin silencing on α5 integrin expression and tumor growth. A) Expression of α5 integrin mRNA in cells with nischarin silencing. RNA was prepared from MCF-7 cells and from MCF-7 cells expressing short hairpin RNA (shRNA) to nischarin or scrambled shRNA. Nischarin, α5 integrin, and β-actin mRNA expression were examined using reverse transcription-polymerase chain reaction. B) Expression of α5 integrin protein. Lysates were prepared from MCF-7 cells and from MCF-7 cells expressing shRNA to nischarin or scrambled shRNA. Thirty micrograms of lysate proteins were loaded in each lane of a gel, and levels of nischarin, α5 integrin, and vinculin protein expression were examined by western blot analysis. C) The α5 integrin promoter assay. MCF-7 cell lines that stably expressed scrambled shRNA or shRNA to nischarin were transiently transfected with 400 ng of a plasmid that expressed firefly luciferase under the control of either the full-length α5 integrin promoter (923 bp) or a truncated (26 bp) α5 integrin promoter. They were also transfected with 50 ng of the Renilla luciferase vector as a control to normalize between experiments. Dual luciferase assays were conducted at 40 hours post-transfection. In the graph, 95% confidence intervals are shown and asterisks denote P = .001 from a two-sided Student t test. Experiments were performed in triplicate. D) Effect on anchorage-independent growth. MCF-7 cells or MCF-7 cells expressing scrambled shRNA or nischarin shRNA were suspended in culture medium containing 0.35% agarose, and the suspension was layered over 0.5% agarose. Representative images of the colonies formed from vector, scramble, and nischarin-silenced cells are shown (scale bar = 100 μM). E) Quantification of cell growth in the soft agar assay. Anchorage-independent growth is graphed for MCF-7 cells infected with lentiviruses expressing green fluorescence protein (GFP) alone, scrambled shRNA, or shRNA to nischarin. Error bars represent 95% confidence intervals; via two-sided Student t tests; experiments were performed in triplicate. F) Effect on tumor growth. To determine the effect of nischarin silencing on tumor growth in vivo, female nu/nu mice (n = 5 per group) were injected with 1 × 106 MCF-7-derived cells in each of two breast pads per mouse. The MCF-7 cells used expressed scrambled shRNA or nischarin shRNA or contained only the vector. Tumor volume was measured every 3 days. Tumors and their site of injections are shown with asterisks in representative mice. G) Quantitative analysis of tumor growth. Here, tumor volumes of MCF-7 cells with GFP alone, scrambled shRNA, or shRNA to nischarin (panel F) are quantified. The data are averaged from three independent experiments. Error bars represent 95% confidence intervals, and P values were calculated using two-sided Student t tests. n.s. = not significant; Nis = nischarin.
α5 Integrin Expression During Breast Cancer Progression
Because nischarin regulates α5 integrin expression, we examined the expression of α5 integrin mRNA in 54 human breast tumors relative to 54 normal tissues. By contrast to nischarin (shown in Figure 1), α5 integrin was weakly expressed in normal tissues and more strongly expressed in the breast cancer specimens (P = .002; Supplementary Figure 6, A, available online) including IDC and IDC + DCIS specimens (Supplementary Figure 6, B, available online). To further assess integrin α5 expression, we quantified ITGA5 mRNA in a panel of 24 primary breast tumor samples with matched normal breast tissue from the same patients. Average ITGA5 expression was higher in tumors compared with normal specimens in 22 (91%) of 24 of these matched pairs (P < .001; Supplementary Figure 6, C, available online). Furthermore, we compared the expression of nischarin mRNA and of ITGA5 mRNA in 54 tumor specimens (Supplementary Figure 6, D, available online). Our data clearly show that nischarin and ITGA5 are inversely related with regard to their expression levels.
Nischarin Silencing and Tumorigenic Potential
Because restoring nischarin expression inhibited the growth of MDA-MB-231 cells, we investigated the effect of silencing nischarin expression on anchorage-independent growth of MCF-7 cells. As expected, cells with nischarin shRNA had enhanced anchorage-independent growth relative to control shRNA–expressing cells (Figure 7, D and E), with an increase in both the size and the number of soft agar colonies. Consistent with these results, nischarin silencing was accompanied by increased proliferation of MCF-7 cells in culture (Supplementary Figure 7, A, available online). Silencing of nischarin expression also potently increased the invasiveness of MCF-7 cells through Matrigel (Supplementary Figure 7, B, available online). These data indicate that decreased expression of nischarin enhances in vitro tumorigenic functions.
We next examined whether reducing endogenous levels of nischarin would affect tumorigenesis in vivo. We injected MCF-7 cells that expressed GFP alone, control shRNA, or nischarin shRNA into the mammary fat pads of BALB/c nu/nu mice and measured tumor volume. Mice bearing MCF-7 cells that expressed nischarin shRNA developed statistically significantly larger tumors compared with either mice that bore cells with vector alone or scrambled control shRNA (at week 9, mean volume of nischarin shRNA–containing tumor = 1262 mm3, of vector-containing control tumor = 258 mm3, of scrambled shRNA–containing tumor = 224 mm3; difference, nischarin shRNA vs vector = 1003 mm3, 95% CI = 860.6 to 1146, P < .001; difference nischarin shRNA vs control scramble shRNA = 1038 mm3, 95% CI = 899.6 to 1176, P < .001; Figure 7, F and G). Taken together, these results support nischarin as a candidate tumor suppressor in breast cancer.
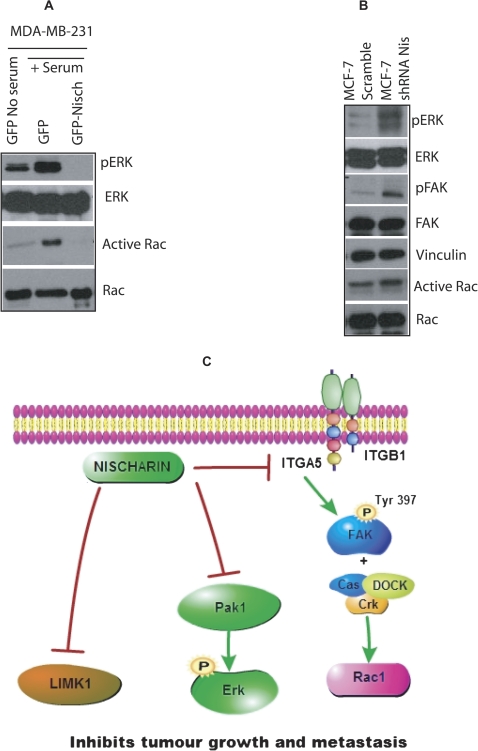
Model
Because it is known that α5 integrin activates Rac and Rac regulates PAK, we hypothesized that nischarin may regulate Rac activation. Nischarin-expressing MDA-MB-231 cells and GFP control cells were serum starved and stimulated with serum for 1 hour, and Rac GTPase assays were performed. In nischarin-expressing cells, Rac GTP loading was substantially reduced (Figure 8, A). These data suggest that nischarin regulates α5 integrin expression, which affects Rac-mediated signaling to regulate tumorigenesis. Moreover, PAK1 regulates ERK phosphorylation, and thus we examined whether nischarin regulates ERK phosphorylation. As predicted, restoring nischarin expression reduced ERK phosphorylation (Figure 8, A). By contrast, suppression of endogenous nischarin in MCF-7 xenograft tumors stimulated FAK and ERK phosphorylation as well as Rac GTP loading (Figure 8, B). These data suggest an important role for nischarin. Because we examined these signaling mechanisms in tumor xenografts from two distinct breast cancer models, using both overexpression and silencing of nischarin, our results strongly support a role for nischarin in regulating the FAK-, Rac-, and ERK-signaling cascades to regulate breast cancer progression (Figure 8, C).
Figure 8.
Effect of nischarin on the cell cycle and cell cycle regulators. A) Rac1 GTPase assay in MDA-MB-231 expressing green fluorescence protein (GFP) or nischarin. Nischarin and GFP stably transfected MDA-MB-231 cells were serum starved overnight and stimulated with serum for 1 hour. GTP bound Rac (active) was precipitated and blotted with anti-Rac1 antibody. B) Expression of the indicated proteins by western blots in MCF-7 scramble shRNA or MCF-7 shRNA nischarin xenograft tumors. Total cell lysates prepared from nischarin-silenced cells were immunoblotted with total FAK, ERK, and phospho-specific antibodies for ERK (tyrosine 204) and FAK (tyrosine 397) as described in the methods. C) Model of the mechanism of nischarin action during breast cancer progression. Nischarin, which is frequently expressed at lower levels in human breast cancers, regulates the expression of α5 integrin, which in turn modulates the FAK and Rac signaling to regulate breast cancer progression. Also, our previous data indicate that nischarin expression is associated with decreased PAK1 activation, and PAK1 is known to regulate tumor growth through ERK (4). Similarly, nischarin modulates LIM kinase (LIMK) activation, which is known to regulate tumor growth (5). Nis = nischarin.
Discussion
Based on our previous studies, which defined nischarin as a regulator of cell migration and invasion (3–5), we hypothesized that nischarin may function as a tumor suppressor in human breast cancer. Here, we demonstrate that nischarin expression is frequently decreased at the mRNA and protein level in human breast cancers. Loss of nischarin expression was associated with acquisition of an invasive phenotype, more advanced tumor grade, and decreased patient survival. LOH at the NISCH locus is one mechanism for loss of nischarin expression; 50% of breast cancer patients exhibited LOH, which was strongly associated with loss of nischarin expression. Moreover, a recent genome-wide study identified nischarin promoter methylation in 30% of breast cancers (30), which would also result in loss of nischarin expression in breast cancer. Functionally, restoration of nischarin expression to MDA-MB-231 cells, which lacked it, inhibited proliferation and soft agar colony formation in vitro and tumor growth in vivo, whereas directly silencing nischarin expression in MCF-7 cells, which had it, increased cell proliferation and soft agar colony formation in vitro and tumor growth in vivo. Mechanistically, restoring nischarin expression in MDA-MB-231 cells decreased α5 integrin expression and reduced FAK phosphorylation, leading to decreased ERK activation. In a reciprocal manner, shRNA-mediated silencing of nischarin expression in MCF-7 cells increased α5 integrin expression, FAK phosphorylation, and ERK phosphorylation. Furthermore, our data show an inverse relationship between α5 integrin and nischarin expression. Taken together, these results suggest that nischarin is a potential tumor suppressor in breast cancer and that it is likely to function by potently inhibiting α5 integrin, FAK, and Rac to regulate tumorigenesis.
Integrins have been shown to play an important role in cancer progression, especially by promoting tumor cell survival, tumor angiogenesis, and metastasis; thus, agents targeting integrins have great therapeutic potential (31). Our data agree with several recent studies showing that α5β1 integrin is important to induction of invasion of breast carcinoma cells. Also in several cell types, α5β1-mediated activation of ERK and FAK signaling has been demonstrated (32). Our data further support the conclusion that a nischarin-mediated decrease in α5 integrin expression affects ERK and FAK phosphorylation. A recent study (28) elegantly showed that FAK catalytic activity is required when α5β1 integrin stimulates Src activation through FAK phosphorylation. E-cadherin is known to function as a tumor suppressor that regulates epithelial mesenchymal transition (33). Furthermore, downregulation of E-cadherin upregulates α5 integrin protein expression through activation of the EGFR/FAK/ERK1 signaling pathway (34) suggesting an inverse correlation between E-cadherin and α5 integrin expression levels. Loss of E-cadherin has been reported to increase α5 integrin expression under conditions of hypoxia (35). FAK has been depicted to play an important role in transcriptional upregulation of mesenchymal markers and delocalization of membrane bound E-cadherin (36,37). E-cadherin levels, like nischarin levels, are sometimes substantially decreased in breast cancers, further suggesting that nischarin and E-cadherin reduce tumor growth through the modulation of α5-FAK signaling. Whereas α5β1 functions as a tumor promoter, an antagonist of this integrin was shown to block proliferation, adhesion, and anchorage-independent growth of human astrocytoma cells (38). Further support comes from a study demonstrating that α5 integrin expression is elevated in metastatic B16F10 melanoma cells (39). The exact mechanism by which nischarin regulates ITGA5 is currently unknown. One possibility is that the nischarin leucine zipper domains (1) could associate with other leucine zipper–containing proteins (40). For example, they might bind to AP1 complex proteins such as c-fos or c-jun, which are known to increase transcription of several genes involved in proliferation and have been shown to regulate the expression of ITGA5 (41,42). However, nischarin is primarily localized to the cytosol, and what, if anything, stimulates its translocation to the nucleus remains to be determined.
Although we have identified a tumor suppressor function for nischarin using qPCR expression analysis and in vitro and in vivo experiments with human breast cancer tissues and cell lines, there are several limitations for our study. First, nischarin also interacts with PAK and LIMK kinase, which have established roles in cancer migration and invasion (4,5), and how nischarin regulates those pathways in breast cancer is beyond the scope of this study. Second, our data strongly suggest that nischarin transcriptionally regulates α5 integrin, but the exact mechanism is not known. Third, it remains to be established whether the tumor suppressor function of nischarin is tissue-specific or a general phenomenon among various cancers. Furthermore, because FAK-Src signaling has been shown to play a critical role in epithelial mesenchymal transition and regulation of E-cadherin, it is important to examine whether nischarin plays a role in these events. Future studies with a mouse model in which nischarin expression has been eliminated will be a key step to identify the complex role played by nischarin in normal and cancer biology.
Based on our data, we propose a model in which nischarin reduces α5 integrin expression leading to reduction of FAK phosphorylation and Rac GTP loading, which in turn reduces tumor growth. In addition to effects on α5 integrin and FAK as shown above, nischarin also regulates PAK and LIMK signaling, all of which have defined roles in breast carcinogenesis (1,4,5). Whether nischarin mediates some of its tumor suppressor roles in breast cancer through regulation of these pathways is an area of current investigation.
Funding
This work was supported by grants from NIH (5RO1CA115706), Susan Komen Foundation (BCTR0600278), Louisiana Board of Regents (BOR-RD A-15), NIH COBRE (P20RR078766) and funds from the Louisiana Cancer Research Consortium as well as from the state of Louisiana to SKA.
S. Baranwal, Y. Wang contributed equally to this work.
The funding agencies had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the article, and the decision to submit the article for publication.
S. Baranwal and Y. Wang did several crucial experiments, R. Rathinam did several signaling experiments, J. Lee and G. C. Blobe did all bioinformatics analysis, L. Jin did α5 integrin qRT-PCR experiments, R. McGoey read immunohistochemistry slides, Y. Pylayeva and F. Giancotti helped with the initial mouse experiments, and S. K. Alahari designed the study, performed some mouse experiments, wrote the article, and submitted it to the Journal.
We are greatly indebted to Olli Kallioneimi and his colleagues from Finland for their initial help with bioinformatics approaches. We thank colleagues in the Alahari lab for their help with some experiments and for their valuable suggestions. We thank Valerie Weaver for accommodating us in her lab at the University of Pennsylvania after the Hurricane Katrina evacuation. We are grateful to M. B. Hatten for financial help and for arranging a visiting professorship in her lab for a period of 9 months. We thank Eve Govek and Traci Jackson of Hatten lab at Rockefeller for their help with animal experiments. In addition, we thank Rudy Juliano (UNC-CH) for critical reading of the article. Also, we thank Kong Chen (Jay Kolls lab, LSUHSC) for helping us with tail vein injections. Finally, we thank the DNA sequencing facility at the University of Pennsylvania for LOH analysis.
Supplementary Material
References
- 1.Alahari SK, Lee JW, Juliano RL. Nischarin, a novel protein that interacts with the integrin alpha5 subunit and inhibits cell migration. J Cell Biol. 2000;151(6):1141–1154. doi: 10.1083/jcb.151.6.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alahari SK, Nasrallah H. A membrane proximal region of the integrin alpha5 subunit is important for its interaction with nischarin. Biochem J. 2004;377(pt 2):449–457. doi: 10.1042/BJ20030411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alahari SK. Nischarin inhibits Rac induced migration and invasion of epithelial cells by affecting signaling cascades involving PAK. Exp Cell Res. 2003;288(2):415–424. doi: 10.1016/s0014-4827(03)00233-7. [DOI] [PubMed] [Google Scholar]
- 4.Alahari SK, Reddig PJ, Juliano RL. The integrin-binding protein Nischarin regulates cell migration by inhibiting PAK. EMBO J. 2004;23(14):2777–2788. doi: 10.1038/sj.emboj.7600291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding Y, Milosavljevic T, Alahari SK. Nischarin inhibits LIM kinase to regulate cofilin phosphorylation and cell invasion [published online ahead of print March 10, 2008] Mol Cell Biol. 2008;28(11):3742–3756. doi: 10.1128/MCB.01832-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lim KP, Hong W. Human Nischarin/imidazoline receptor antisera-selected protein is targeted to the endosomes by a combined action of a PX domain and a coiled-coil region. J Biol Chem. 2004;279(52):54770–54782. doi: 10.1074/jbc.M411315200. [DOI] [PubMed] [Google Scholar]
- 7.Kok K, Naylor SL, Buys CH. Deletions of the short arm of chromosome 3 in solid tumors and the search for suppressor genes. Adv Cancer Res. 1997;71:27–92. doi: 10.1016/s0065-230x(08)60096-2. [DOI] [PubMed] [Google Scholar]
- 8.Weksberg R, Hughes S, Moldovan L, Bassett AS, Chow EW, Squire JA. A method for accurate detection of genomic microdeletions using real-time quantitative PCR. BMC Genomics. 2005;6:180. doi: 10.1186/1471-2164-6-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tavazoie SF, Alarcon C, Oskarsson T, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451(7175):147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bos PD, Zhang XH, Nadal C, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459(7249):1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oh MA, Kang ES, Lee SA, et al. PKCdelta and cofilin activation affects peripheral actin reorganization and cell-cell contact in cells expressing integrin alpha5 but not its tailless mutant. J Cell Sci. 2007;120(pt 15):2717–2730. doi: 10.1242/jcs.003566. [DOI] [PubMed] [Google Scholar]
- 12.Boles BK, Ritzenthaler J, Birkenmeier T, Roman J. Phorbol ester-induced U-937 differentiation: effects on integrin alpha(5) gene transcription. Am J Physiol Lung Cell Mol Physiol. 2000;278(4):L703–L712. doi: 10.1152/ajplung.2000.278.4.L703. [DOI] [PubMed] [Google Scholar]
- 13.Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richardson AL, Wang ZC, De Nicolo A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9(2):121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 15.Turashvili G, Bouchal J, Baumforth K, et al. Novel markers for differentiation of lobular and ductal invasive breast carcinomas by laser microdissection and microarray analysis. BMC Cancer. 2007;7:55. doi: 10.1186/1471-2407-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivshina AV, George J, Senko O, et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66(21):10292–10301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- 18.Miller LD, Smeds J, George J, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A. 2005;102(38):13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van ’t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 20.Sotiriou C, Wirapati P, Loi S, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98(4):262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 21.Desmedt C, Piette F, Loi S, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007;13(11):3207–3214. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 22.Hess KR, Anderson K, Symmans WF, et al. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol. 2006;24(26):4236–4244. doi: 10.1200/JCO.2006.05.6861. [DOI] [PubMed] [Google Scholar]
- 23.Ma XJ, Wang Z, Ryan PD, et al. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5(6):607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 24.Martinez A, Walker RA, Shaw JA, Dearing SJ, Maher ER, Latif F. Chromosome 3p allele loss in early invasive breast cancer: detailed mapping and association with clinicopathological features. Mol Pathol. 2001;54(5):300–306. doi: 10.1136/mp.54.5.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Killary AM, Wolf ME, Giambernardi TA, Naylor SL. Definition of a tumor suppressor locus within human chromosome 3p21-p22. Proc Natl Acad Sci U S A. 1992;89(22):10877–10881. doi: 10.1073/pnas.89.22.10877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Klijn JG, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 27.Zlotnik A. Chemokines and cancer. Int J Cancer. 2006;119(9):2026–2029. doi: 10.1002/ijc.22024. [DOI] [PubMed] [Google Scholar]
- 28.Wu L, Bernard-Trifilo JA, Lim Y, et al. Distinct FAK-Src activation events promote alpha5beta1 and alpha4beta1 integrin-stimulated neuroblastoma cell motility. Oncogene. 2008;27(10):1439–1448. doi: 10.1038/sj.onc.1210770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Veevers-Lowe J, Ball SG, Shuttleworth A, Kielty CM. Mesenchymal stem cell migration is regulated by fibronectin through {alpha}5{beta}1-integrin-mediated activation of PDGFR-{beta} and potentiation of growth factor signals. J Cell Sci. 2011;124(pt 8):1288–1300. doi: 10.1242/jcs.076935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoque MO, Kim MS, Ostrow KL, et al. Genome-wide promoter analysis uncovers portions of the cancer methylome. Cancer Res. 2008;68(8):2661–2670. doi: 10.1158/0008-5472.CAN-07-5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moschos SJ, Drogowski LM, Reppert SL, Kirkwood JM. Integrins and cancer. Oncology (Williston Park). 2007;21(9 suppl 3):13–20. [PubMed] [Google Scholar]
- 32.Chen Q, Meng LH, Zhu CH, Lin LP, Lu H, Ding J. ADAM15 suppresses cell motility by driving integrin alpha5beta1 cell surface expression via Erk inactivation. Int J Biochem Cell Biol. 2008;40(10):2164–2173. doi: 10.1016/j.biocel.2008.02.021. [DOI] [PubMed] [Google Scholar]
- 33.Baranwal S, Alahari SK. Molecular mechanisms controlling E-cadherin expression in breast cancer. Biochem Biophys Res Commun. 2009;384(1):6–11. doi: 10.1016/j.bbrc.2009.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sawada K, Mitra AK, Radjabi AR, et al. Loss of E-cadherin promotes ovarian cancer metastasis via alpha 5-integrin, which is a therapeutic target. Cancer Res. 2008;68(7):2329–2339. doi: 10.1158/0008-5472.CAN-07-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arimoto-Ishida E, Sakata M, Sawada K, et al. Up-regulation of alpha5-integrin by E-cadherin loss in hypoxia and its key role in the migration of extravillous trophoblast cells during early implantation. Endocrinology. 2009;150(9):4306–4315. doi: 10.1210/en.2008-1662. [DOI] [PubMed] [Google Scholar]
- 36.Cicchini C, Laudadio I, Citarella F, et al. TGFbeta-induced EMT requires focal adhesion kinase (FAK) signaling. Exp Cell Res. 2008;314(1):143–152. doi: 10.1016/j.yexcr.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 37.Avizienyte E, Frame MC. Src and FAK signalling controls adhesion fate and the epithelial-to-mesenchymal transition. Curr Opin Cell Biol. 2005;17(5):542–547. doi: 10.1016/j.ceb.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 38.Maglott A, Bartik P, Cosgun S, et al. The small alpha5beta1 integrin antagonist, SJ749, reduces proliferation and clonogenicity of human astrocytoma cells. Cancer Res. 2006;66(12):6002–6007. doi: 10.1158/0008-5472.CAN-05-4105. [DOI] [PubMed] [Google Scholar]
- 39.Qian F, Zhang ZC, Wu XF, Li YP, Xu Q. Interaction between integrin alpha(5) and fibronectin is required for metastasis of B16F10 melanoma cells. Biochem Biophys Res Commun. 2005;333(4):1269–1275. doi: 10.1016/j.bbrc.2005.06.039. [DOI] [PubMed] [Google Scholar]
- 40.Venugopal R, Jaiswal AK. Nrf2 and Nrf1 in association with Jun proteins regulate antioxidant response element-mediated expression and coordinated induction of genes encoding detoxifying enzymes. Oncogene. 1998;17(24):3145–3156. doi: 10.1038/sj.onc.1202237. [DOI] [PubMed] [Google Scholar]
- 41.Gingras ME, Masson-Gadais B, Zaniolo K, et al. Differential binding of the transcription factors Sp1, AP-1, and NFI to the promoter of the human alpha5 integrin gene dictates its transcriptional activity. Invest Ophthalmol Vis Sci. 2009;50(1):57–67. doi: 10.1167/iovs.08-2059. [DOI] [PubMed] [Google Scholar]
- 42.Larouche K, Leclerc S, Salesse C, Guerin SL. Expression of the alpha 5 integrin subunit gene promoter is positively regulated by the extracellular matrix component fibronectin through the transcription factor Sp1 in corneal epithelial cells in vitro. J Biol Chem. 2000;275(50):39182–39192. doi: 10.1074/jbc.M002945200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.