Abstract
Background
Three non-synonymous single nucleotide polymorphisms (Q223R, K109R and K656N) of the leptin receptor gene (LEPR) have been tested for association with obesity-related outcomes in multiple studies, showing inconclusive results. We performed a systematic review and meta-analysis on the association of the three LEPR variants with BMI. In addition, we analysed 15 SNPs within the LEPR gene in the CoLaus study, assessing the interaction of the variants with sex.
Methodology/Principal Findings
We searched electronic databases, including population-based studies that investigated the association between LEPR variants Q223R, K109R and K656N and obesity- related phenotypes in healthy, unrelated subjects. We furthermore performed meta-analyses of the genotype and allele frequencies in case-control studies. Results were stratified by SNP and by potential effect modifiers. CoLaus data were analysed by logistic and linear regressions and tested for interaction with sex. The meta-analysis of published data did not show an overall association between any of the tested LEPR variants and overweight. However, the choice of a BMI cut-off value to distinguish cases from controls was crucial to explain heterogeneity in Q223R. Differences in allele frequencies across ethnic groups are compatible with natural selection of derived alleles in Q223R and K109R and of the ancient allele in K656N in Asians. In CoLaus, the rs10128072, rs3790438 and rs3790437 variants showed interaction with sex for their association with overweight, waist circumference and fat mass in linear regressions.
Conclusions
Our systematic review and analysis of primary data from the CoLaus study did not show an overall association between LEPR SNPs and overweight. Most studies were underpowered to detect small effect sizes. A potential effect modification by sex, population stratification, as well as the role of natural selection should be addressed in future genetic association studies.
Introduction
In the past few decades, the prevalence and incidence of obesity has rapidly increased globally and has reached epidemic proportions [1], [2]. Obesity is associated with many deleterious outcomes such as type 2 diabetes, hypercholesterolemia, hypertension or heart disease, and is directly related to increased mortality and reduced life expectancy [2]. With the completion of the human genome project and the first extensive genome-wide association studies, increasing numbers of risk alleles associated with obesity have been identified [3]–[5], some of them in genes not previously known to be associated with adiposity. However, to date, genome-wide association studies have only captured a small percentage of the genetic variance related to obesity or body mass index [6].
One commonly studied candidate gene for obesity, the leptin receptor gene (LEPR), is on a biologic pathway to obesity (leptin-insulin pathway) [7]. Leptin is produced in adipose tissue and in other organs. It is known to have pleiotropic actions, including regulation of several neuropeptides involved in appetite control [8] and thermogenesis [9]. Three non-synonymous single nucleotide polymorphisms of the LEPR gene (Q223R, K109R and K656N) have been tested for association with obesity-related outcomes in multiple studies, producing inconclusive results [e.g. 10], [11], [12], [13]. Two systematic reviews on these variants did not show an overall statistically significant association to obesity-related outcomes [14], [15]. However, many further studies on the association between LEPR variants and obesity have been published since the last review in 2005 [15], including studies on the interaction of these variants with sex or with other factors [e.g. 16]. We therefore performed a systematic review and meta-analysis on the association between the three LEPR variants Q223R, K109R and K656N and obesity-related outcomes.
As most studies do not report data stratified by sex or other possible effect modifiers, we additionally analysed 38 SNPs within the LEPR gene (among them K109R) in a cohort of more than 6000 Caucasian adults from the Swiss CoLaus study [17], (data unpublished to date). We assessed the association of the SNPs with a number of different obesity-related outcomes and for possible confounding variables, or interaction with sex. This approach allowed us to account for the need for more precise phenotyping of study subjects [18]. Furthermore, the issue of sample size in association studies is crucial [6]. Our approach provides data on a cohort with a larger sample size than the ones assessed in the systematic review.
Materials and Methods
Systematic review
We followed the guidelines PRISMA for the reporting of systematic reviews and meta-analyses [19]. The electronic databases Medline, Embase and ISI Web of Knowledge were searched (search date: 14 October 2009). The search strategy was carried out for all exposures and outcomes of interest. Search terms included LEPR, leptin receptor gene, Q223R, K109R, K656N, obesity, body mass index, BMI, weight, waist, waist-to-hip ratio, WHR, body fat, adiposity, overweight, fat mass, Quetelet index. Where possible, MeSH headings (or other standardized indexing terms) were used. The search was restricted to humans, but unrestricted for publication date or language (see supporting Table S1 for Medline search strategy. The search strategies for Embase and ISI were similar). We restricted the review to healthy people in order to separate potential associations of the SNPs with diabetes II, hypertension or with other diseases and only assess potential associations with overweight. This strategy was also followed by the previous review on the same topic by Paracchini and colleagues [15]. The reference lists of all included studies were examined to identify studies not found by the electronic databases search. The retrieved references were checked by title and abstract for inclusion or exclusion, according to the following criteria. Inclusion criteria: exposure: at least one of the LEPR SNPs Q223R, K109R or K656N; outcome: BMI, body fat percentage, weight, waist circumference, hip circumference, waist-to-hip ratio or other weight-related outcome; non related subjects, either sex, any ethnic group, any BMIs and all population based study designs. Exclusion criteria: studies not meeting the inclusion criteria; furthermore, studies including only non-healthy subjects (both study arms with e.g. diabetes, cancer or hypertension patients) and family-based studies. All included studies were retrieved as full text and reviewed again for inclusion and exclusion. The inclusion and exclusion process was performed at each level according to pre-established criteria by two independent reviewers (NB and NA), and consensus was reached by discussion.
Data was extracted from all the studies included as full text papers by the two independent reviewers on a standard data extraction sheet and entered into an electronic database (EpiData 3.1). Consensus was reached by discussion. Data extracted included reference details (author, year, journal), study design (case-control, cohort, comparative study), details of the population (sex, ethnicity, setting), sample size per comparison group, SNP studied, allele and genotype frequency per comparison group, obesity-related phenotypes tested (e.g. BMI, waist circumference), strength of association (odds ratios and confidence intervals), and potential confounders accounted for. If necessary data was not reported in the primary manuscript, the corresponding authors were contacted by email to request the missing data. Quality criteria were developed to assess the internal validity of the studies and the accuracy of reporting, using the guidelines for the assessment of cumulative evidence on genetic associations [20], the HuGe Review Handbook [21] and the extension of the STROBE statement STREGA (strengthening the reporting of genetic association studies) [22]. The quality of data was considered in the final interpretation of the findings.
Data analysis and meta-analysis
Data was analysed descriptively and statistically for each LEPR gene variant separately and stratified by ethnic group. Ethnic groups were defined according to Rosenberg et al [23]. For the analysis of association between LEPR variants and overweight, case-control studies, (where cases are obese people without other known disease and controls are healthy non-obese people), were included. For genotype and allele frequencies, single group studies, (consisting of cohorts, cross-sectional studies or healthy control arms of case-control studies), were also analysed. Reported statistical analyses on the association between a SNP and an obesity-related outcome that did not present results in a format convenient for a meta-analysis, (such as linear regression, ANOVA, and non-parametric analyses), were extracted and taken into account in the interpretation of the findings.
Derived allele frequencies of each SNP were summarized in Tables, stratified by ethnic group (including CoLaus data, see below). If a study reported data separately for subgroups (such as sex, country or cohort of origin), the subgroups were included in the analysis of genotype and allele frequencies. The heterogeneity between allele frequencies in different ethnic groups was assessed using Cochran's Q statistic. In case-control studies, associations between genotype, allele data and obesity were assessed by chi-square tests, general linear models and meta-analyses, according to different inheritance models (co-dominant, dominant and recessive), given the a priori absence of evidence on the allelic mode of action. If odds ratios and/or confidence intervals were missing, these were calculated. We used 0.006 (0.05/9, because three models were tested for 3 SNPs) as the cut-off p-value to declare an association as significant in case-control studies (Tables 1– 3).
Table 1. Analysis of genotypes for Q223R in case-control studies, according to different allelic modes of action.
| Reference | Co-dominant: OR (95% CI)* | Co-dominant: p-value* | Dominant: OR (95% CI)** | Dominant: Chi2 (p-value)** | Recessive: OR (95% CI)** | Recessive: Chi2 (p-value)** |
| Caucasians | ||||||
| Chagnon 1999 [61] | 0.89 (0.74–1.07) | 0.221 | 0.87 (0.52–1.45) | 0.34 (0.557) | 0.66 (0.36–1.19) | 2.19 (0.139) |
| Yiannakouris 2001 [10] | 1.45 (0.75–2.80) | 0.263 | 0.96 (0.38–2.44) | 0.01 (0.924) | 5.54 (1.13–27.27) | 7.40 (0.007) |
| Mattevi 2002 [31] | 1.62 (1.15–2.26) | 0.005 | 1.60 (0.99–2.59) | 4.05 (0.044) | 2.44 (1.16–5.12) | 6.92 (0.009) |
| Portolés 2006 [62] | 0.84 (0.68–1.04) | 0104 | 0.88 (0.65–1.18) | 0.84 (0.360) | 0.62 (0.38–1.03) | 3.96 (0.047) |
| De Krom 2007 [63] | 1.00 (0.73–1.37) | 0.991 | 0.77 (0.45–1.30) | 1.11 (0.293) | 1.33 (0.76–2.30) | 1.14 (0.285) |
| Mergen 2007 [32] | 1.43 (1.03–1.99) | 0.034 | 1.65 (1.06–2.56) | 5.45 (0.020) | 1.31 (0.60–2.83) | 0.55 (0.457) |
| Bienertova 2008 [64] | 0.88 (0.57–1.36) | 0.564 | 1.06 (0.50–2.23) | 0.03 (0.872) | 0.65 (0.30–1.44) | 1.33 (0.248) |
| Masuo 2008 [16] | 1.87 (1.10–3.16) | 0.021 | 2.10 (0.90–4.90) | 3.55 (0.060) | 2.89 (0.88–9.49) | 4.22 (0.040) |
| Overall | I2 = 69.8% | I2 p = 0.002 | 1.13 (0.87–1.45) | 0.368 | I2 = 66.5% | I2 p = 0.004 |
| Asians | ||||||
| Endo 2000 [65] | 0.89 (0.58–1.37) | 0.604 | 1.27 (0.20–8.25) | 0.10 (0.754) | 0.84 (0.49–1.42) | 0.51 (0.475) |
| Wang 2006 [30] | 1.32 (0.7–2.38) | 0.347 | 1.51 (0.75–3.04) | 1.57 (0.210) | 0.39 (0.01–13.37) | 0.61 (0.433) |
| Overall | 1.02 (0.71–1.47) | 0.924 | 1.48 (0.77–2.85) | 0.244 | 0.82 (0.49–1.38) | 0.459 |
| Mixed populations | ||||||
| Guízar-mendoza 2005 [66] | 0.70 (0.36–1.34) | 0.278 | 0.78 (0.33–1.84) | 0.40 (0.527) | 0.33 (0.04–2.74) | 1.86 (0.173) |
| Duarte 2007 [67] | 1.24 (0.89–1.71) | 0.202 | 1.65 (1.02–2.68) | 4.69 (0.030) | 0.86 (0.45–1.65) | 0.24 (0.628) |
| Overall | 1.00 (0.58–1.72) | 0.991 | 1.24 (0.60–2.54) | 0.564 | 0.79 (0.43–1.48) | 0.465 |
| Overall all populations | I2 = 56.7% | I2 p = 0.006 | 1.15 (0.93–1.43) | 0.185 | I2 = 52.0% | I2 p = 0.018 |
Odds ratios and 95% confidence intervals are given. Statistically significant results are shown in bold. Where the overall measure was significantly heterogeneous, the I2 value and its p-value are given instead of the overall measure.
*Values from generalized linear model, overall results from meta-analysis, random model.
**Values from meta-analysis, random model.
Table 2. Analysis of genotypes for K109R in case-control studies, according to different allelic modes of action.
| Reference | Co-dominant: OR (95% CI)* | Co-dominant: p-value* | Dominant: OR (95% CI)** | Dominant: Chi2 (p-value)** | Recessive: OR (95% CI)** | Recessive: Chi2 (p-value)** |
| Caucasians | ||||||
| Chagnon 1999 [61] | 0.82 (0.58–1.16) | 0.253 | 0.94 (0.59–1.51) | 0.07 (0.797) | 0.38 (0.14–1.04) | 4.70 (0.030) |
| Yiannakouris 2001 [10] | 1.49 (0.62–3.61) | 0.376 | 1.31 (0.45–3.83) | 0.32 (0.574) | Not calculable | 3.10 (0.079) |
| De Krom 2007 [63] | 1.13 (0.78–1.62) | 0.520 | 1.30 (0.77–2.18) | 1.10 (0.294) | 0.87 (0.34–2.23) | 0.11 (0.739) |
| Masuo 2008 [16] | 0.74 (0.37–1.48) | 0.391 | 0.81 (0.34–1.93) | 0.28 (0.599) | 0.22 (0.01–7.46) | 1.83 (0.177) |
| CoLaus men | 0.91 (0.77–1.08) | 0.298 | 0.91 (0.73–1.14) | 0.67 (0.413) | 0.81 (0.51–1.27) | 0.95 (0.329) |
| CoLaus women | 1.01 (0.86–1.18) | 0.918 | 1.01 (0.82–1.23) | 0.00 (0.959) | 1.04 (0.67–1.60) | 0.03 (0.872) |
| Overall | 0.96 (0.87–1.07) | 0.449 | 0.98 (0.86–1.12) | 0.782 | 0.84 (0.63–1.12) | 0.242 |
| Asians | ||||||
| Qu 2007 [68] | 1.04 (0.77–1.41) | 0.800 | 0.97 (0.34–2.76) | 0.00 (0.950) | 1.06 (0.73–1.53) | 0.10 (0.750) |
| Overall all populations | 0.97 (0.88–1.07) | 0.526 | 0.98 (0.86–1.12) | 0.779 | 0.90 (0.72–1.13) | 0.372 |
Odds ratios and 95% confidence intervals are given. Statistically significant results are shown in bold.
*Values from generalized linear model, overall results from meta-analysis, random model.
**Values from meta-analysis, random model.
Table 3. Analysis of genotypes for K656N in case-control studies, according to different allelic modes of action.
| Reference | Co-dominant: OR (95% CI)* | Co-dominant: p-value* | Dominant: OR (95% CI)** | Dominant: Chi2 (p-value)** | Recessive: OR (95% CI)** | Recessive: Chi2 (p-value)** |
| Caucasians | ||||||
| Chagnon 1999 [61] | 1.03 (0.72–1.48) | 0.879 | 1.08 (0.66–1.77) | 010 (0.752) | 0.90 (0.33–2.46) | 0.05 (0.820) |
| Yiannakouris 2001 [10] | 1.15 (0.60–2.24) | 0.670 | 1.25 (0.49–3.19) | 0.27 (0.600) | 1.03 (0.13–8.22) | 0.00 (0.977) |
| Masuo 2008 [16] | 1.71 (0.99–2.97) | 0.055 | 1.97 (0.86–4.50) | 3.12 (0.077) | 2.28 (0.60–8.72) | 2.04 (0.153) |
| Overall | 1.21 (0.89–1.63) | 0.226 | 1.26 (0.86–1.86) | 0.238 | 1.23 (0.58–2.59) | 0.596 |
| Asians | ||||||
| Qu 2007 [68] | 0.87 (0.52–1.46) | 0.587 | 0.91 (0.51–1.62) | 0.12 (0.731) | Not calculable | 1.42 (0.234) |
| Overall all populations | 1.12 (0.86–1.45) | 0.407 | 1.14 (0.83–1.57) | 0.421 |
For the meta-analyses, data was pooled using a random effects model, to calculate summary odds ratios with 95% confidence intervals, by SNP. The statistical evidence for heterogeneity between studies was assessed by I2 statistics [24]. Funnel plots of study precision were used to examine a possible small study bias, using Begg and Egger statistics [25].
CoLaus data
The CoLaus study (Cohorte Lausannoise) is a population-based study including 6'184 Caucasian adults aged 35–75 years from the city of Lausanne, Switzerland [17]. The study population consisted of 52.5% women. The mean age was 51.1 years (standard deviation of ±10.9). The following obesity-related phenotypic measurements were performed by trained nurses: body weight, body height, body fat percentage (by electrical bioimpedance using the Bodystat® 1500 analyzer [Isle of Man, British Isles]), waist and hip circumferences. Body weight and height were measured with participants standing without shoes in light indoor clothing. Body weight was measured in kilograms to the nearest 0.1 kg using a Seca® Scale (Hamburg, Germany), which was calibrated regularly. Height was measured to the nearest 5 mm using a Seca® height gauge (Hamburg, Germany). Waist circumference was measured twice with a nonstretchable tape over the unclothed abdomen at the mid-point between the lowest rib and the iliac crest. The mean of the two measurements was used for analyses. Furthermore, a number of additional potential confounders and effect modifiers were assessed by questionnaire or interview, including geographic origins [26], smoking status, alcohol consumption and menopausal status. Genotyping was performed using the Affymetrix 500K chip, 38 SNPs were located within the LEPR gene. In order to reduce the number of statistical tests performed, we chose 15 SNPs that tagged [27] all 38 SNPs (see complete list of covered SNPs on supporting Table S2), using the Haploview programme [28]. The 15 selected genotyped SNPs available at the LEPR locus tag 13, 26 and 33 SNPs from HapMap CEU release 22 using r2>0.8, >0.7 and >0.6, respectively, out of 208 SNPs (among which 188 SNPs have MAF >5%) available in this region. These tagging SNPs cover the entire gene region, including the 3′ UTR. We therefore consider that the set of SNPs we analyzed covers moderately well this locus. Relevant data on 5636 people was available for analysis in the present paper. Missing data was mostly due to missing genotype data.
We used logistic regressions to test the association of 15 tag LEPR SNPs (among them K109R) with the dichotomized body mass index (BMI) or waist circumference. Cut-off value for BMI was 25; cut-off value for waist circumference was 88 cm in women and 102 cm in men [29]. In addition, we performed linear regressions on body mass index, waist circumference, fat mass and leptin levels as continuous variables. Leptin level data was subjected to natural logarithmic transformation in order to better achieve normality of the residuals and homoscedasticity. We reported associations corrected for potential confounders like sex, age, height (for outcomes other than body mass index), alcohol consumption, smoking and geographic variation (expressed as principal components pc1 and pc2 from principal component analyses). These covariables showed an association with overweight-related phenotypes and with some of the SNPs studied in univariate regression analyses. We also assessed a potential interaction with sex. For analyses on the CoLaus data, we used 0.0033 (0.05/15) as the cut-off p-value to declare an association as significant. For interaction tests, we used 0.05 as the cut-off p-value to declare an interaction as significant. In the CoLaus analyses, we had more than 80% power to detect an additive association explaining 0.3% of trait variance for single SNP analysis and 80% power to detect an interaction explaining 0.14% of the variance.
All statistical analyses were conducted using STATA statistical package v 9.0 (Stata corp, College Station, TX, USA).
Results
Systematic review
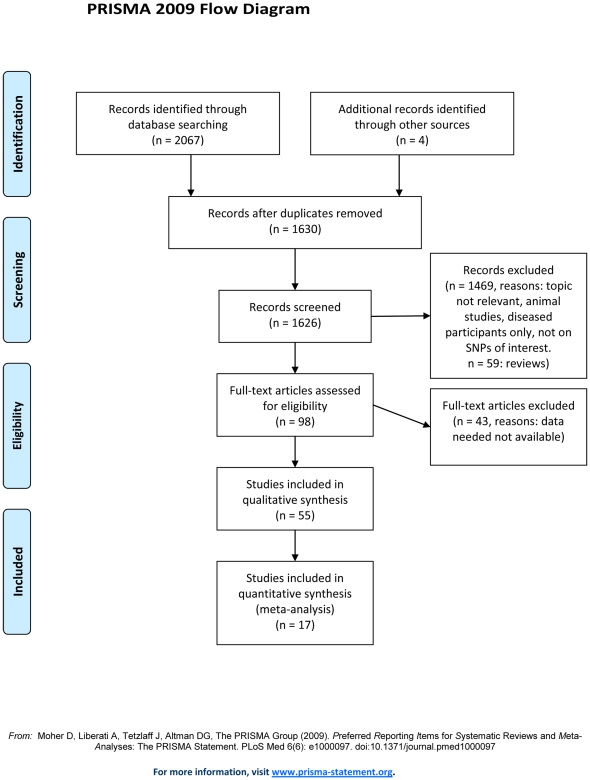
In total, 1630 papers were found through the search in electronic databases or by manual search. Fifty-five studies satisfied the inclusion and exclusion criteria and were obtainable as full text papers. Seventeen were case-control studies comparing obese with non-obese people and could be used for the meta-analyses. Thirty-eight studies were categorized as single group studies and contained data on genotype and allele frequencies (see the flow chart in Figure 1). supporting Table S3 presents participants' characteristics for case- control studies and supporting Table S4 for single group studies.
Figure 1. Flow diagram of studies included and excluded in the systematic review.
Numbers are given at each exclusion step.
Thirty studies were carried out on Caucasians, sixteen studies were carried out on Asians, four on people of African ancestry and six on populations of mixed ancestry, such as Brazilians or Mexicans. Most studies reported BMI as outcome, many studies reported several obesity-related outcomes, like body weight, body fat mass or waist-to-hip ratio. Most studies were carried out with participants of both sexes and with adults. Q223R was the most commonly studied SNP (52 studies), followed by K109R (20 studies) and K656N (20 studies).
Study quality
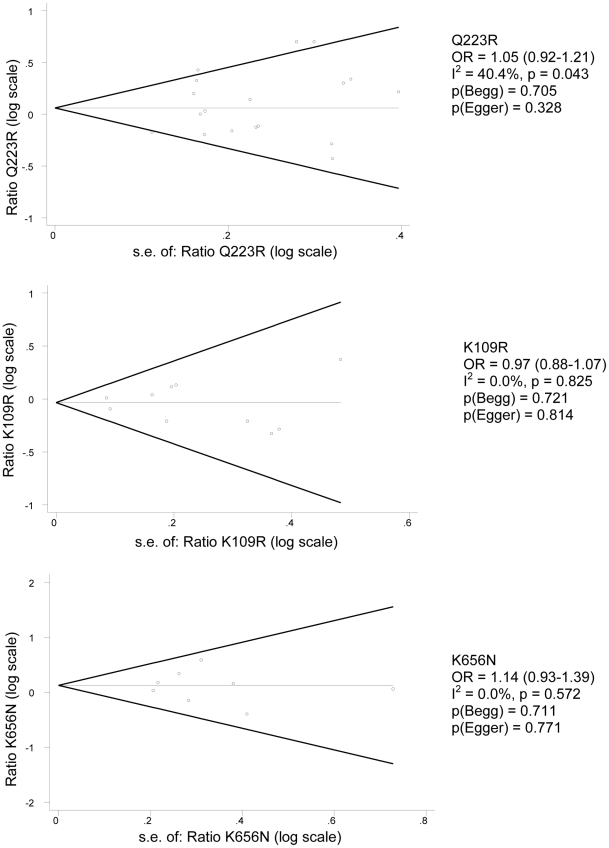
Overall, studies reported well on the participants' characteristics (summarized in supporting Tables S3 and S4) and on genotyping and analysis methods. Most studies did not report on missing data, and several call rates (the percentage of successfully genotyped individuals in the study population) were under 95%. Inclusion and exclusion criteria, as well as outcome assessments, were not always well described. Hardy-Weinberg equilibrium was mostly (but not always) reported as calculated. Genotypes were mostly in Hardy-Weinberg equilibrium. Potential confounders like sex and age were assessed in more than half of the studies. Surprisingly, the calculation or justification of the sample size was rarely described. The quality of study reporting is therefore of concern for the interpretation of the results. We assessed effects of small study bias by funnel plots (see Figure 2). There was no statistical evidence for publication bias or small study bias.
Figure 2. Begg's funnel plots of study precision.
For included studies on the association between Q223R, K109R and K656N and overweight, Begg's funnel plots were used to examine a possible small study bias. The natural logarithm of the odds ratio (OR) vs. its standard error and pseudo 95% confidence intervals are shown, together with Begg and Egger statistics.
Genotype and allele frequencies
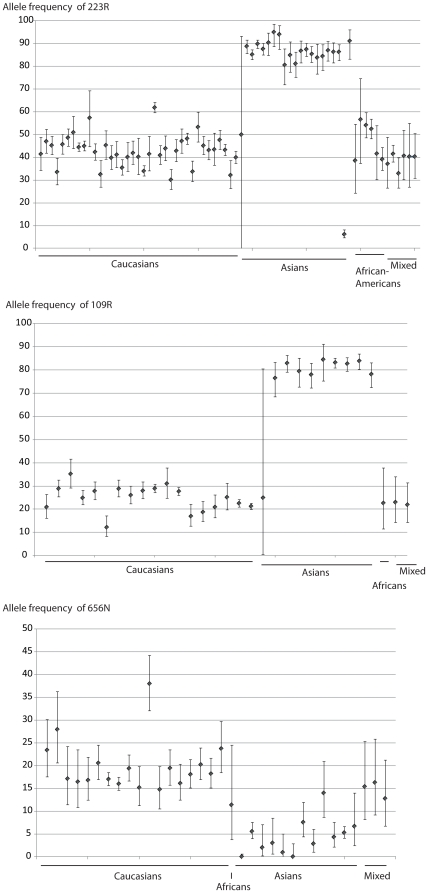
In this systematic review, A denotes the ancestral allele and D the derived allele (the derived alleles are 223R, 109R and 656N respectively). Reported genotype and reported or calculated allele D frequencies with according 95% confidence intervals of all included studies are reported in Figure 3 and supporting Tables S5, S6, S7, by SNP. For each ethnic group the Cochran's Q statistic and p-value are given to estimate heterogeneity in allele frequencies across studies. Allele frequencies differ strongly between Caucasians and Asians, with Asians showing much higher derived allele frequencies for 223R (80.56–95.00% compared to 30.18–56.67% in Caucasians) and 109R (76.47–84.44 compared to 12.29–35.25 in Caucasians). The Taiwanese aborigines [30] have lower allele frequency (6.07%) for 223R, than the other Asian populations. Asians have lower 656N allele frequencies than Caucasians (0.00–13.97 vs. 14.75–37.98, respectively).
Figure 3. Derived allele frequencies for Q223R, K109R and K656N, by ethnic group.
95% confidence intervals are shown for each derived frequency.
In Caucasians, allele frequencies are heterogeneous in all SNPs considered. For Q223R, there seems to be a north-south gradient in Caucasians, with the highest derived frequencies occurring in north Europe and the lowest derived frequencies found in Mediterranean countries.
Meta-analyses and further results
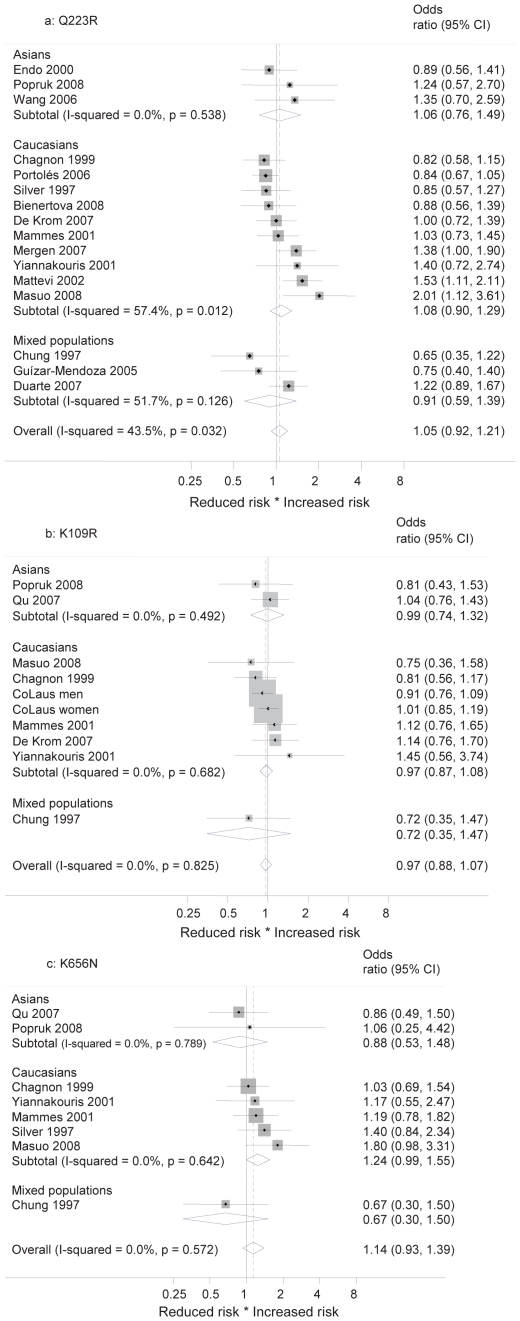
Odds ratios of allele frequencies of case-control studies were analyzed by random effects meta-analyses. Results by SNPs are shown in Figures 4a–4c. No meta-analysis showed overall significant results (overall OR for Q223R: 1.05 (95% CI 0.92–1.21), for K109R: 0.97 (0.88–1.07), for K656N: 1.14 (0.93–1.39)). Pooled results from meta-analyses of genotypes showed no significant results in any SNP for any of the three inheritance models (see Tables 1– 3).
Figure 4. Forest plot on the association between SNP alleles and overweight in case-control studies by ethnicity.
Figure 4a: Forest plot on the association between Q223R alleles and overweight in case-control studies by ethnicity. Overall association from random effects meta-analysis (odds ratio and 95% confidence intervals) and stratification by ethnic groups are shown, as well as heterogeneity by means of I2 value for overall measure and for subgroups. Figure 4b: Forest plot on the association between K109R alleles and overweight in case-control studies by ethnicity. Overall association from random effects meta-analysis (odds ratio and 95% confidence intervals) and stratification by ethnic groups are shown, as well as heterogeneity by means of I2 value for overall measure and for subgroups. Data from the CoLaus study are included for the Caucasian population, stratified by sex. Figure 4c: Forest plot on the association between K656N alleles and overweight in case-control studies by ethnicity. Overall association from random effects meta-analysis (odds ratio and 95% confidence intervals) and stratification by ethnic groups are shown, as well as heterogeneity by means of I2 value for overall measure and for subgroups.
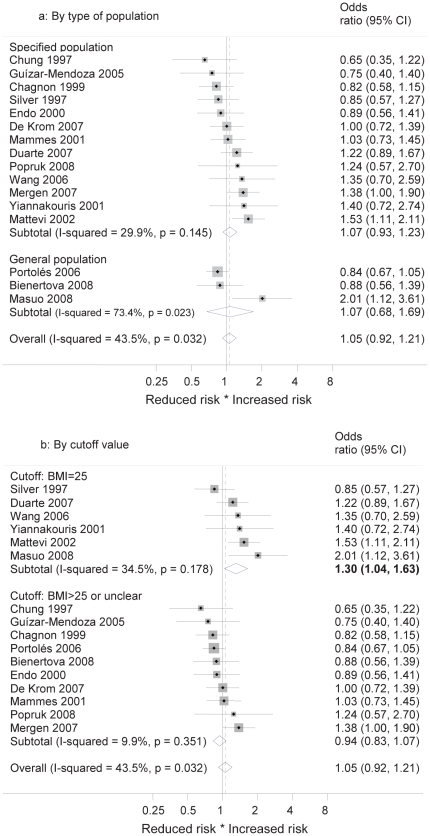
The meta-analysis of allele frequencies in Q223R showed significant heterogeneity (I2 = 43.5%, p = 0.032). Subgroup analyses of Q223R stratifying by ethnicity did not explain this heterogeneity and did not change any result to a significant association (Caucasians: OR 1.08 (0.90–1.29), Asians: OR 1.06 (0.76–1.49), mixed populations: OR 0.91 (0.59–1.39)), (see Figure 4a). The same is true for stratification by type of study population (see Figure 5a). However, if the meta-analysis of Q223R was stratified by BMI cutoff value, the heterogeneity became non-significant in each group (see Figure 5b). The overall OR for studies with a BMI cutoff value of 25 was significant with 1.30 (1.04–1.63). In studies using higher or unclear cutoff values the OR showed an opposite trend of 0.94 (0.83–1.07). Meta-analyses in K109R and K656N did not provide evidence for overall heterogeneity.
Figure 5. Forest plot on the association between Q223R alleles and overweight in case-control studies by potential effect modifier.
Figure 5a: Forest plot on the association between Q223R alleles and overweight in case-control studies by type of population. Overall association from random effects meta-analysis (odds ratio and 95% confidence intervals) and stratification by type of population are shown, as well as heterogeneity by means of I2 value for overall measure and for subgroups. Specified populations are all populations not declared as general populations in the original studies. Figure 5b: Forest plot on the association between Q223R alleles and overweight in case-control studies by BMI cut-off value. Overall association from random effects meta-analysis (odds ratio and 95% confidence intervals) and stratification by BMI cut-off value are shown, as well as heterogeneity by means of I2 value for overall measure and for subgroups. For definition of BMI cut-off values see main text.
From the single studies on Q223R, only three studies showed an association between Q223R and obesity [16], [31], [32], reporting an increased risk of overweight for the derived allele (OR 2.1 (1.12–3.61), 1.53 (1.11–2.11), and 1.38 (1.00–1.90) respectively). One study reported data on the association between Q223R alleles and waist circumference stratified by sex [31]: in men the OR was 2.1 (1.16–3.47), showing an increased risk of large waist circumference for the derived allele. In women the OR was 1.15 (0.74–1.79). In our meta-analysis of genotypes in case-control studies there was only one single statistically significant result for Q223R genotypes with overweight in the co-dominant model (OR 1.62 (1.15–2.26) [31]). In the reported results from ANOVAs or linear regressions for Q223R, eight studies reported an increased risk of overweight for the derived allele, five a protective effect and 18 studies did not show an association.
For K109R, one study reported an association with overweight in women (p = 0.011) [33]. From ANOVAs or linear regressions, one study reported an increased risk for the derived allele, while nine studies did not show significant results.
In the results from ANOVAs or linear regressions for K656N, four studies reported an increased risk of the derived allele, while five studies did not show significant results.
CoLaus data
Tables 4–5 show the results of the logistic regression analyses of 15 tag SNPs of the LEPR gene in CoLaus. For the association with overweight, only SNP rs9436746 showed an increased risk for the minor allele in the additive model (OR 1.13 (1.04–1.23)). No SNP showed an association with waist circumference and no SNP showed an interaction with sex. In the linear regressions (supporting Table S8), SNP rs10889553 showed an association with waist circumference in the additive model (beta 1.65 (SE 0.51), t-value 3.12, p = 0.001). In linear regressions three SNPs showed an interaction with sex (rs10128072, rs3790438 and rs3790437 (tag of K656N)) for the outcomes waist circumference and fat mass, and rs3790438 and rs3790437 showed an additional interaction with sex for the outcome BMI. If stratified by sex (supporting Table S9), all three SNPs show an increased risk for overweight-related outcomes for the minor alleles in men, and a decreased risk in women.
Table 4. Association from logistic regression models between LEPR variants and overweight in the CoLaus study.
| SNP | Allele minor/major | OR (95% CI)heterozygote | OR (95% CI)homozygote minor | P(chi2, 2df) | OR (95% CI)additive | P add(1df) | P interaction SNP*sex |
| rs10128072 | G/T | 1.02 (0.9–1.16) | 1.01 (0.71–1.44) | 0.96 | 1.01 (0.91–1.13) | 0.79 | 0.07 |
| rs7518849 | G/A | 1.01 (0.85–1.20) | 1.25 (0.43–3.69) | 0.91 | 1.02 (0.87–1.20) | 0.81 | 0.99 |
| rs970467 | T/C | 0.91 (0.79–1.05) | 0.72 (0.42–1.21) | 0.21 | 0.90 (0.79–1.02) | 0.09 | 0.58 |
| rs10889553 | T/C | 1.14 (0.94–1.38) | 2.72 (0.48–15.51) | 0.21 | 1.16 (0.96–1.40) | 0.12 | 0.21 |
| rs10889567* | C/T | 1.07 (0.94–1.22) | 1.17 (0.99–1.38) | 0.19 | 1.08 (0.99–1.17) | 0.07 | 0.23 |
| rs1137100** | G/A | 1.05 (0.92–1.19) | 1.14 (0.89–1.46) | 0.51 | 1.06 (0.96–1.16) | 0.26 | 0.86 |
| rs3790438 | A/T | 0.87 (0.76–0.99) | 0.94 (0.68–1.31) | 0.10 | 0.90 (0.81–1.01) | 0.07 | 0.08 |
| rs9436746 | A/C | 1.11 (0.98–1.26) | 1.29 (1.09–1.53) | 0.01 | 1.13 (1.04–1.23) | 0.003 | 0.25 |
| rs2025805 | A/G | 0.82 (0.71–0.93) | 0.87 (0.74–1.02) | 0.01 | 0.93 (0.86–1.00) | 0.06 | 0.35 |
| rs1805096 | T/C | 1.01 (0.90–1.14) | 0.88 (0.74–1.04) | 0.22 | 0.95 (0.88–1.03) | 0.25 | 0.51 |
| rs9436748 | T/G | 0.84 (0.74–0.97) | 0.94 (0.79–1.11) | 0.04 | 0.95 (0.88–1.04) | 0.25 | 0.14 |
| rs7531110 | G/T | 1.04 (0.93–1.18) | 1.18 (0.99–1.40) | 0.18 | 1.08 (0.99–1.17) | 0.08 | 0.16 |
| rs10158279 | C/A | 1.01 (0.89–1.16) | 1.13 (0.96–1.33) | 0.25 | 1.06 (0.98–1.15) | 0.15 | 0.41 |
| rs11585329 | A/C | 1.05 (0.93–1.20) | 0.88 (0.62–1.26) | 0.53 | 1.02 (0.91–1.13) | 0.78 | 0.57 |
| rs3790437*** | C/T | 0.92 (0.81–1.04) | 0.93 (0.70–1.25) | 0.37 | 0.94 (0.85–1.03) | 0.20 | 0.07 |
Results are odds ratios (95% confidence intervals) from logistic regression models (general model and additive model) including age, sex, alcohol consumption, smoking, and the first and second principal components, as covariates. In addition, the result of the interaction with sex is given. Statistically significant results are shown in bold.
*tag of Q223R.
**K109R.
***tag of K656N.
Table 5. Association from logistic regression models between LEPR variants and waist circumference in the CoLaus study.
| SNP | Allele minor/major | OR (95% CI)heterozygote | OR (95% CI)homozygote minor | P(chi2, 2df) | OR (95% CI)additive | P add(1df) | P interaction SNP*sex |
| rs10128072 | G/T | 1.11 (0.97–1.28) | 1.08 (0.74–1.58) | 0.31 | 1.09 (0.97–1.23) | 0.14 | 0.06 |
| rs7518849 | G/A | 1.04 (0.87–1.25) | 1.99 (0.70–5.66) | 0.40 | 1.08 (0.91–1.29) | 0.37 | 0.51 |
| rs970467 | T/C | 0.98 (0.84–1.14) | 0.53 (0.28–1.03) | 0.14 | 0.93 (0.81–1.07) | 0.32 | 0.56 |
| rs10889553 | T/C | 1.19 (0.97–1.45) | 1.94 (0.36–10.58) | 0.18 | 1.20 (0.99–1.46) | 0.06 | 0.08 |
| rs10889567* | C/T | 1.07 (0.93–1.23) | 1.02 (0.85–1.22) | 0.61 | 1.02 (0.93–1.11) | 0.65 | 0.30 |
| rs1137100** | G/A | 1.01 (0.88–1.16) | 0.93 (0.71–1.22) | 0.85 | 0.99 (0.90–1.10) | 0.91 | 0.25 |
| rs3790438 | A/T | 0.90 (0.78–1.04) | 0.90 (0.63–1.30) | 0.32 | 0.92 (0.82–1.03) | 0.16 | 0.16 |
| rs9436746 | A/C | 1.05 (0.92–1.20 | 1.19 (0.99–1.42 | 0.17 | 1.09 (1.00–1.18) | 0.06 | 0.29 |
| rs2025805 | A/G | 0.87 (0.76–1.09) | 0.92 (0.78–1.09) | 0.18 | 0.95 (0.87–1.03) | 0.22 | 0.22 |
| rs1805096 | T/C | 1.02 (0.90–1.16) | 0.84 (0.70–1.02) | 0.12 | 0.95 (0.87–1.03) | 0.21 | 0.53 |
| rs9436748 | T/G | 0.85 (0.74–0.98) | 0.95 (0.79–1.13) | 0.07 | 0.95 (0.87–1.04) | 0.24 | 0.34 |
| rs7531110 | G/T | 1.00 (0.88–1.14) | 1.03 (0.86–1.24) | 0.94 | 1.02 (0.93–1.11) | 0.71 | 0.06 |
| rs10158279 | C/A | 1.05 (0.91–1.21) | 1.04 (0.87–1.23) | 0.82 | 1.02 (0.94–1.11) | 0.63 | 0.17 |
| rs11585329 | A/C | 1.06 (0.92–1.21) | 0.96 (0.65–1.41) | 0.68 | 1.03 (0.92–1.16) | 0.59 | 0.58 |
| rs3790437*** | C/T | 0.91 (0.80–1.04) | 0.96 (0.70–1.31) | 0.37 | 0.94 (0.84–1.05) | 0.25 | 0.06 |
Results are odds ratios (95% confidence intervals) from logistic regression models (general model and additive model) including age, sex, height, alcohol consumption, smoking, and the first and second principal components, as covariates. In addition, the result of the interaction with sex is given. Statistically significant results are shown in bold.
*tag of Q223R.
**K109R.
***tag of K656N.
No SNP was associated in linear regressions with the outcome leptin levels. SNP rs7531110 showed an interaction with sex (supporting Tables S8 and S9).
Discussion
Overall results
In the present systematic review we analysed data on the association between three LEPR gene variants Q223R, K109R, K656N and overweight-related outcomes. In addition, we analysed primary data on the association of 15 LEPR tag SNPs with different overweight-related outcomes from a large, population based cross-sectional study. Overall, the meta-analysis of allele frequencies in obese cases and lean controls did not show an association between the three SNPs and overweight. In the present review, we also analysed genotype data, according to different genetic modes of action (co-dominant, dominant, recessive) and this did not reveal any clear pattern of association for any of the tested models. These results support previous findings [14], [15]. Studies published after our systematic review also confirm the unclear association between LEPR and overweight-related outcomes. Several studies reported no association between Q223R and overweight [34]–[36], while one study reported a protective effect of 223R for overweight in Pacific Islanders [37]. One study reported an increased risk of 109R for overweight in Asian children [38]. None of these studies would change our overall results. Most published studies are underpowered to detect small effect sizes. However, even in the large CoLaus study, we did not find evidence for an overall association between LEPR SNPs and overweight-related outcomes. Our search strategy is likely to have missed papers that included results on the association between the selected LEPR variants and obesity but have not mentioned LEPR in title or abstract. As these studies are likely to be negative, they would probably not change our overall conclusions.
Implications of findings on interactions and non-coding variants
Stratification by factors that were reported in the literature as significant effect modifiers, like ethnicity [39] or study population [40] (general population versus specified populations) did not change the results. Interestingly, a stratification by BMI cut-off value [41] reduced the heterogeneity for Q223R within each of the two subgroups to non-significant levels. In the stratum with a cut-off value of BMI = 25, the result showed an increased risk for overweight for the derived allele. Okorodudu and colleagues [41] showed that a cutoff value of BMI = 25 had a sensitivity of 0.50 (CI: 0.43–0.57) and a specificity of 0.90 (0.86–0.94) in their study sample to detect high adiposity. A cutoff value of BMI = 30 had a sensitivity of 0.42 (0.31–0.43) and a specificity of 0.97 (0.96–0.97).
The fact that stratification by ethnicity did not explain overall heterogeneity and did not show a significant difference in the association between SNPs and overweight in the different ethnic groups is surprising, especially if one considers the allele frequency differences across ethnic groups. The present review supports previous findings of much higher derived allele frequencies in Asians for the SNPs Q223R and K109R [15], [42]. These higher derived allele frequencies are compatible with evidence for a recent positive natural selection of LEPR in Asian populations [43]. Interestingly, the Taiwanese aborigine population [30] shows a different picture. The derived allele of Q223R does not only show a lower frequency like in the Caucasian population, but even an extremely low frequency, compatible with a selection of the ancestral allele. This result could be explained by the fact that Taiwanese aborigines have a different evolutionary history compared to the other Asian populations included in the present review (Taiwanese aborigines separated before the selection of the LEPR variants in continental Asia occurred) [44]–[46].
Considering major sex differences in leptin levels and fat distribution, a further potential effect modifier is sex [47], [48]. We could not perform stratification by sex in our meta-analysis as most studies did not report associations separately in men and women. One study reported stratified allelic results for the outcome waist circumference [31], showing an increased risk for large waist circumference for 223R in men but not in women. Another study reported an association of K109R with overweight in women [33]. In our linear regressions of the CoLaus data SNPs rs10128072, rs3790438 and rs3790437 (tag of K656N) showed an interaction with sex in their association with waist circumference and fat mass. Sex can therefore be considered as a potential effect modifier for associations between LEPR SNPs and overweight-related outcomes. Two studies on the association between Q223R and overweight published after our systematic review confirm this view. One study reports an increased risk for 223R for high BMI in Caucasian girls but not in boys [49]. The other study reports a protective effect of 223R in Caucasian men but not in women [50]. These results suggest that stratification by sex should be recommended for future association studies.The three SNPs of the CoLaus data showing a significant interaction with sex are all non-coding variants of the LEPR gene, as is often the case for genetic associations using high-throughput DNA chips. This may indicate that these variants tag functional variants located within coding regions or functional variants located within non-coding regions influencing gene expression or splicing sites (promoter, introns, etc). It is more and more recognized that non-coding variants may impact on disease [51]. A better knowledge of the exact mechanisms of gene regulation will be crucial to understand the general role of non-coding DNA in human phenotypic variation.
Evolutionary considerations
In our allele frequency data of Q223R we observed a north-south gradient in European Caucasians, with higher derived allele frequencies in the north and lower frequencies in the south. The same phenomenon for SNPs of other genes was reported in the Framingham Heart Study [52] and in a Europe wide analysis [26]. This phenomenon can be explained by the first settlement of Europe in Neolithic times from south to north [53]. It can lead to population stratification, a well known problem in large-scale association studies which can lead to false positive associations [39]. Furthermore, population stratification is a major issue in the interpretation of data from populations of known mixed ancestry, like Brazilians, Mexicans and certain US populations. For this reason, we considered such mixed populations separately in our systematic review and meta-analysis.
An alternative explanation for variation in allele frequency to the ancient migration hypothesis is positive natural selection. In fact, evidence for recent positive selection of variants in polymorphic genes was found in the major ethnic groups worldwide [43]. It was found that in selected genetic regions there is a significant over-representation of genetic association with complex diseases, a fact demonstrating that the understanding of recent genetic positive selection is important to comprehend the evolution of human disease [54]. Interestingly, the three LEPR SNPs Q223R, K109R and K656N show signals for positive selection in the Asian population [43]. This finding is compatible with the much higher derived allele frequencies for Q223R and K109R and the much lower derived allele frequency for K656N in Asians in our systematic review. It seems that the sequence changes were of advantage (or disadvantage) for Asian populations 6'000–8'000 years ago, a time that corresponds to the introduction of agriculture in Asia [55], [56]. It was therefore speculated that the LEPR gene could be considered a “thrifty” gene, leading to an accumulation of fat tissue in times of plenty, providing a reserve for times of hunger [57], [58]. As an alternative explanation for the positive selection of LEPR in Asian populations [59], Hancock et al. found associations of several LEPR variants (among them K109R) with climate variables suggesting a role of climate adaptations in the biological processes underlying cold adaptation and overweight. They suggest that variants like K109R might be deleterious in hot equatorial climates and advantageous in colder climates.
The influence of positive selection and the selective pressures operating in the past are issues of major importance that need further investigation. The human fat distribution, especially the subcutaneous fat, is unique among primates and among most land mammals. While primates have on average 5% body fat, a normal weight human male has approximately 10–15% body fat and a normal weight human female 20–25% [60]. Humans are born with a substantial layer of subcutaneous fat, showing the independence of this feature from diet at least in the first phase of life. The human subcutaneous fat and the susceptibility to obesity can therefore not only be explained by a disbalance between energy intake and expenditure, but also demands an evolutionary explanation.
Conclusions
In conclusion, our systematic review did not show an overall association between the LEPR SNPs Q223R, K109R and K656N and obesity-related outcomes, but Q223R showed a significant association with overweight in studies considering a BMI cut-off value of 25 to separate normal weight from overweight. In our analyses of primary data from the CoLaus study, rs9436746 was associated with overweight and rs10889553 with waist circumference. Our stratified analyses in CoLaus data suggest that sex could potentially modify the association of LEPR variants with obesity-related phenotypes, which is not surprising considering the major differences in both leptin levels and fat distribution between the sexes. Genetic association studies on obesity traits should consider sex as a potential effect modifier. Finally, the role of natural selection in allele frequency differences and the potential impact of selection on gene-phenotype associations also need further investigation.
Supporting Information
Search strategy for Medline, via platform OVID.
(DOC)
List of 38 SNPs within the LEPR gene covered by the Affymetrix 500K chip. The 15 tag SNPs are in bold.
(DOC)
Characteristics of case-control studies.
(DOC)
Characteristics of cohort or cross-sectional studies or control arms of case-control studies.
(DOC)
Genotype and derived allele frequencies (D) for Q223R, by ethnic group.
(DOC)
Genotype and derived allele frequencies (D) for K109R, by ethnic group.
(DOC)
Genotype and derived allele frequencies (D) for K656N, by ethnic group.
(DOC)
Association from linear regression models of LEPR variants with different outcomes.
(DOC)
Association from linear regression models of LEPR variants with different outcomes showing a significant interaction with sex, stratified by sex.
(DOC)
Acknowledgments
The authors are grateful to Dr O. Fletcher for comments on an early draft of the manuscript and to Erica Holt for English editing.
Footnotes
Competing Interests: The authors have the following competing interest: Vincent Mooser is an employee of GlaxoSmithKline, Philadelphia. There are no patents, products in development or marketed products to declare. This does not alter the authors′ adherence to all the PLoS ONE policies on sharing data and materials.
Funding: The CoLaus study was supported by GlaxoSmithKline, the Faculty of Biology and Medicine of Lausanne, and the Swiss National Science Foundation (33CSCO-122661, 3200BO-111361/2, 3100AO-116323/1). Dr. Bochud is supported by the Swiss School of Public Health Plus. The Faculty of Biology and Medicine of Lausanne, the Swiss National Science Foundation, and the Swiss School of Public Health Plus had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Vincent Mooser is an employee of GlaxoSmithKline, Philadelphia and helped perform the experiments; therefore, GlaxoSmithKline had a role in data collection.
References
- 1.Popkin BM. Global nutrition dynamics: the world is shifting rapidly toward a diet linked with noncommunicable diseases. Am J Clin Nutr. 2006;84:289–298. doi: 10.1093/ajcn/84.1.289. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. Obesity: Preventing and Managing the Global Epidemic. Geneva: WHO; 2000. [PubMed] [Google Scholar]
- 3.Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 4.Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li S, Loos RJF. Progress in the genetics of common obesity: Size matters. Curr Opin Lipidology. 2008;19:113–121. doi: 10.1097/MOL.0b013e3282f6a7f3. [DOI] [PubMed] [Google Scholar]
- 7.Yang W, Kelly T, He J. Genetic epidemiology of obesity. Epidemiol Rev. 2007;29:49–61. doi: 10.1093/epirev/mxm004. [DOI] [PubMed] [Google Scholar]
- 8.Jequier E. Leptin signaling, adiposity, and energy balance. [Review] [53 refs]. Ann New York Acad Sci. 2002;967:379–388. doi: 10.1111/j.1749-6632.2002.tb04293.x. [DOI] [PubMed] [Google Scholar]
- 9.Dulloo AG, Stock MJ, Solinas G, Boss O, Montani JP, et al. Leptin directly stimulates thermogenesis in skeletal muscle. FEBS Lett. 2002;515:109–113. doi: 10.1016/s0014-5793(02)02449-3. [DOI] [PubMed] [Google Scholar]
- 10.Yiannakouris N, Yannakoulia M, Melistas L, Chan JL, Klimis-Zacas D, et al. The Q223R polymorphism of the leptin receptor gene is significantly associated with obesity and predicts a small percentage of body weight and body composition variability. Journal of Clinical Endocrinology & Metabolism. 2001;86:4434–4439. doi: 10.1210/jcem.86.9.7842. [DOI] [PubMed] [Google Scholar]
- 11.Rosmond R, Chagnon YC, Holm G, Chagnon M, Perusse L, et al. Hypertension in obesity and the leptin receptor gene locus. Journal of Clinical Endocrinology & Metabolism. 2000;85:3126–3131. doi: 10.1210/jcem.85.9.6781. [DOI] [PubMed] [Google Scholar]
- 12.Silver K, Walston J, Chung WK, Yao F, Parikh VV, et al. The Gln223Arg and Lys656Asn polymorphisms in the human leptin receptor do not associate with traits related to obesity. Diabetes. 1997;46:1898–1900. doi: 10.2337/diab.46.11.1898. [DOI] [PubMed] [Google Scholar]
- 13.Gotoda T, Manning BS, Goldstone AP, Imrie H, Evans AL, et al. Leptin receptor gene variation and obesity: lack of association in a white British male population. Hum Mol Genet. 1997;6:869–876. doi: 10.1093/hmg/6.6.869. [DOI] [PubMed] [Google Scholar]
- 14.Heo M, Leibel RL, Fontaine KR, Boyer BB, Chung WK, et al. A meta-analytic investigation of linkage and association of common leptin receptor (LEPR) polymorphisms with body mass index and waist circumference. Int J Obes Relat Metab Disord. 2002;26:640–646. doi: 10.1038/sj.ijo.0801990. [DOI] [PubMed] [Google Scholar]
- 15.Paracchini V, Pedotti P, Taioli E. Genetics of leptin and obesity: a HuGE review. Am J Epidemiol. 2005;162:101–114. doi: 10.1093/aje/kwi174. [DOI] [PubMed] [Google Scholar]
- 16.Masuo K, Straznicky NE, Lambert GW, Katsuya T, Sugimoto K, et al. Leptin-receptor polymorphisms relate to obesity through blunted leptin-mediated sympathetic nerve activation in a Caucasian male population.[see comment]. Hypertension Research - Clinical & Experimental. 2008;31:1093–1100. doi: 10.1291/hypres.31.1093. [DOI] [PubMed] [Google Scholar]
- 17.Firmann M, Mayor V, Marques VP, Bochud M, Pecoud A, et al. The CoLaus study: a population-based study to investigate the epidemiology and genetic determinants of cardiovascular risk factors and metabolic syndrome. BMC Cardiovasc Disord. 2008;8:6. doi: 10.1186/1471-2261-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muller MJ, Bosy-Westphal A, Krawczak M. Genetic studies of common types of obesity: a critique of the current use of phenotypes. Obes Rev. 2010;11:612–618. doi: 10.1111/j.1467-789X.2010.00734.x. [DOI] [PubMed] [Google Scholar]
- 19.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. 10.1371/journal.pmed.1000097 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ioannidis JP, Boffetta P, Little J, O'Brien TR, Uitterlinden AG, et al. Assessment of cumulative evidence on genetic associations: interim guidelines. Int J Epidemiol. 2008;37:120–132. doi: 10.1093/ije/dym159. [DOI] [PubMed] [Google Scholar]
- 21.Little J, Higgins JPT. The HuGENet™ HuGE Review Handbook, version 1.0. 2006 February. Available: http://www.hugenet.ca/
- 22.Little J, Higgins JP, Ioannidis JP, Moher D, Gagnon F, et al. Strengthening the reporting of genetic association studies (STREGA): an extension of the strengthening the reporting of observational studies in epidemiology (STROBE) statement. J Clin Epidemiol. 2009;62:597–608. doi: 10.1016/j.jclinepi.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 23.Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, et al. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 24.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 25.Egger M, Davey-Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. British Medical Journal. 1997;315:634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novembre J, Johnson T, Bryc K, Kutalik Z, Boyko AR, et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, et al. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 28.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 29.Grundy SM, Cleeman JI, Daniels SR, Donato KA, Eckel RH, et al. Diagnosis and management of the metabolic syndrome: an American Heart Association/National Heart, Lung, and Blood Institute Scientific Statement. Circulation. 2005;112:2735–2752. doi: 10.1161/CIRCULATIONAHA.105.169404. CIRCULATIONAHA.105.169404 [pii];10.1161/CIRCULATIONAHA.105.169404 [doi] [DOI] [PubMed] [Google Scholar]
- 30.Wang TN, Huang MC, Chang WT, Ko AM, Tsai EM, et al. G-2548A polymorphism of the leptin gene is correlated with extreme obesity in Taiwanese aborigines. Obesity. 2006;14:183–187. doi: 10.1038/oby.2006.23. [DOI] [PubMed] [Google Scholar]
- 31.Mattevi VS, Zembrzuski VM, Hutz MH. Association analysis of genes involved in the leptin-signaling pathway with obesity in Brazil. International Journal of Obesity & Related Metabolic Disorders: Journal of the International Association for the Study of Obesity. 2002;26:1179–1185. doi: 10.1038/sj.ijo.0802067. [DOI] [PubMed] [Google Scholar]
- 32.Mergen H, Karaaslan C, Mergen M, Deniz OE, Ozata M. LEPR, ADBR3, IRS-1 and 5-HTT genes polymorphisms do not associate with obesity. Endocr J. 2007;54:89–94. doi: 10.1507/endocrj.k06-023. [DOI] [PubMed] [Google Scholar]
- 33.Mammes O, Aubert R, Betoulle D, Pean F, Herbeth B, et al. LEPR gene polymorphisms: associations with overweight, fat mass and response to diet in women. Eur J Clin Invest. 2001;31:398–404. doi: 10.1046/j.1365-2362.2001.00843.x. [DOI] [PubMed] [Google Scholar]
- 34.Constantin A, Costache G, Sima AV, Glavce CS, Vladica M, et al. Leptin G-2548A and leptin receptor Q223R gene polymorphisms are not associated with obesity in Romanian subjects. Biochem Biophys Res Commun. 2010;391:282–286. doi: 10.1016/j.bbrc.2009.11.050. [DOI] [PubMed] [Google Scholar]
- 35.Friedlander Y, Li G, Fornage M, Williams OD, Lewis CE, et al. Candidate molecular pathway genes related to appetite regulatory neural network, adipocyte homeostasis and obesity: results from the CARDIA Study. Ann Hum Genet. 2010;74:387–398. doi: 10.1111/j.1469-1809.2010.00596.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pyrzak B, Wisniewska A, Kucharska A, Wasik M, Demkow U. No association of LEPR Gln223Arg polymorphism with leptin, obesity or metabolic disturbances in children. Eur J Med Res. 2009;14(Suppl 4):201–204. doi: 10.1186/2047-783X-14-S4-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Furusawa T, Naka I, Yamauchi T, Natsuhara K, Kimura R, et al. The Q223R polymorphism in LEPR is associated with obesity in Pacific Islanders. Hum Genet. 2010;127:287–294. doi: 10.1007/s00439-009-0768-9. [DOI] [PubMed] [Google Scholar]
- 38.Okada T, Ohzeki T, Nakagawa Y, Sugihara S, Arisaka O. Impact of leptin and leptin-receptor gene polymorphisms on serum lipids in Japanese obese children. Acta Paediatr. 2010;99:1213–1217. doi: 10.1111/j.1651-2227.2010.01778.x. [DOI] [PubMed] [Google Scholar]
- 39.Marchini J, Cardon LR, Phillips MS, Donnelly P. The effects of human population structure on large genetic association studies. Nat Genet. 2004;36:512–517. doi: 10.1038/ng1337. [DOI] [PubMed] [Google Scholar]
- 40.Heid IM, Huth C, Loos RJ, Kronenberg F, Adamkova V, et al. Meta-analysis of the INSIG2 association with obesity including 74,345 individuals: does heterogeneity of estimates relate to study design? PLoS Genet. 2009;5:e1000694. doi: 10.1371/journal.pgen.1000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okorodudu DO, Jumean MF, Montori VM, Romero-Corral A, Somers VK, et al. Diagnostic performance of body mass index to identify obesity as defined by body adiposity: a systematic review and meta-analysis. Int J Obes (Lond) 2010;34:791–799. doi: 10.1038/ijo.2010.5. [DOI] [PubMed] [Google Scholar]
- 42.Kagawa Y, Dever GJ, Otto CT, Charupoonphol P, Supannatas S, et al. Single nucleotide polymorphism and lifestyle-related diseases in the Asia-Pacific region: comparative study in Okinawa, Palau and Thailand. Asia Pac J Public Health. 2003;15(Suppl):S10–S14. doi: 10.1177/101053950301500S04. [DOI] [PubMed] [Google Scholar]
- 43.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Melton T, Peterson R, Redd AJ, Saha N, Sofro ASM, et al. Polynesian Genetic affinities with southeast Asian populations as identified by mtDNA analysis. Am J Hum Genet. 1995;57:403–414. [PMC free article] [PubMed] [Google Scholar]
- 45.Chang J-G, Ko YC, Lee JCI, Chang S-J, Liu T-C, et al. Molecular analysis of mutations and polymorphisms of the Lewis secretor type α(1,2)-fucosyltransferase gene reveals that Taiwanese aborigines are of Austronesian derivation. J Hum Genet. 2002;47:60–65. doi: 10.1007/s100380200001. [DOI] [PubMed] [Google Scholar]
- 46.Capelli C, Wilson JF, Richards M, Stumpf MPH, Gratrix F, et al. A predominantly indigenous paternal heritage for the Austonesian-speaking peoples of insular Southeast Asia and Oceania. Am J Hum Genet. 2001;68:432–443. doi: 10.1086/318205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heid IM, Jackson AU, Randall JC, Winkler TW, Qi L, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42:949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Magi R, Lindgren CM, Morris AP. Meta-analysis of sex-specific genome-wide association studies. Genet Epidemiol. 2010;34:846–853. doi: 10.1002/gepi.20540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Riestra P, Garcia-Anguita A, Schoppen S, Lopez-Simon L, de OM, et al. Sex-specific association between leptin receptor polymorphisms and leptin levels and BMI in healthy adolescents. Acta Paediatr. 2010;99:1527–1530. doi: 10.1111/j.1651-2227.2010.01877.x. [DOI] [PubMed] [Google Scholar]
- 50.Ben AS, Kallel A, Sediri Y, Ftouhi B, Feki M, et al. LEPR p.Q223R Polymorphism influences plasma leptin levels and body mass index in Tunisian obese patients. Arch Med Res. 2009;40:186–190. doi: 10.1016/j.arcmed.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 51.Pastinen T. Genome-wide allele-specific analysis: insights into regulatory variation. Nat Rev Genet. 2010;11:533–538. doi: 10.1038/nrg2815. nrg2815 [pii];10.1038/nrg2815 [doi] [DOI] [PubMed] [Google Scholar]
- 52.Sebro R, Hoffman TJ, Lange C, Rogus JJ, Risch NJ. Testing for non-random mating: evidence for ancestry-related assortative mating in the Framingham heart study. Genet Epidemiol. 2010;34:674–679. doi: 10.1002/gepi.20528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Currat M, Excoffier L. The effect of the Neolithic expansion on European molecular diversity. Proc Biol Sci. 2005;272:679–688. doi: 10.1098/rspb.2004.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lappalainen T, Salmela E, Andersen PM, hlman-Wright K, Sistonen P, et al. Genomic landscape of positive natural selection in Northern European populations. Eur J Hum Genet. 2010;18:471–478. doi: 10.1038/ejhg.2009.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gepts P. Crop domestication as a long-term selection experiment. Plant Breeding Reviews. 2004;24:1–44. [Google Scholar]
- 56.Fuller DQ, Qin L, Zheng Y, Zhao Z, Chen X, et al. The domestication process and domesitcation rate in rice: spikelet bases from the lower Yangtze. Science. 2009;323:1607–1610. doi: 10.1126/science.1166605. [DOI] [PubMed] [Google Scholar]
- 57.Kagawa Y, Yanagisawa Y, Hasegawa K, Suzuki H, Yasuda K, et al. Single nucleotide polymorphisms of thrifty genes for energy metabolism: evolutionary origins and prospects for intervention to prevent obesity-related diseases. Biochem Biophys Res Commun. 2002;295:207–222. doi: 10.1016/s0006-291x(02)00680-0. [DOI] [PubMed] [Google Scholar]
- 58.Chakravarthy MV, Booth FW. Eating, exercise, and “thrifty” genotypes: connecting the dots toward an evolutionary understanding of modern chronic diseases. J Appl Physiol. 2004;96:3–10. doi: 10.1152/japplphysiol.00757.2003. [DOI] [PubMed] [Google Scholar]
- 59.Hancock AM, Witonsky DB, Gordon AS, Eshel G, Pritchard JK, et al. Adaptations to climate in candidate genes for common metabolic disorders. PLoS Genet. 2008;4:e32. doi: 10.1371/journal.pgen.0040032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pond CM. Fat and figures. New Scientist. 1987;114:62–66. [Google Scholar]
- 61.Chagnon YC, Chung WK, Perusse L, Chagnon M, Leibel RL, et al. Linkages and associations between the leptin receptor (LEPR) gene and human body composition in the Quebec Family Study. International Journal of Obesity & Related Metabolic Disorders: Journal of the International Association for the Study of Obesity. 1999;23:278–286. doi: 10.1038/sj.ijo.0800809. [DOI] [PubMed] [Google Scholar]
- 62.Portoles O, Sorli JV, Frances F, Coltell O, Gonzalez JI, et al. Effect of genetic variation in the leptin gene promoter and the leptin receptor gene on obesity risk in a population-based case-control study in Spain. Eur J Epidemiol. 2006;21:605–612. doi: 10.1007/s10654-006-9045-6. [DOI] [PubMed] [Google Scholar]
- 63.de Krom M, van der Schouw YT, Hendriks J, Ophoff RA, van Gils CH, et al. Common genetic variations in CCK, leptin, and leptin receptor genes are associated with specific human eating patterns. Diabetes. 2007;56:276–280. doi: 10.2337/db06-0473. [DOI] [PubMed] [Google Scholar]
- 64.Bienertova-Vasku J, Bienert P, Tomandl J, Forejt M, Vavrina M, et al. No association of defined variability in leptin, leptin receptor, adiponectin, proopiomelanocortin and ghrelin gene with food preferences in the Czech population. Nutr Neurosci. 2008;11:2–8. doi: 10.1179/147683008X301379. [DOI] [PubMed] [Google Scholar]
- 65.Endo K, Yanagi H, Hirano C, Hamaguchi H, Tsuchiya S, et al. Association of Trp64Arg polymorphism of the beta3-adrenergic receptor gene and no association of Gln223Arg polymorphism of the leptin receptor gene in Japanese schoolchildren with obesity. International Journal of Obesity & Related Metabolic Disorders: Journal of the International Association for the Study of Obesity. 2000;24:443–449. doi: 10.1038/sj.ijo.0801177. [DOI] [PubMed] [Google Scholar]
- 66.Guizar-Mendoza JM, Amador-Licona N, Flores-Martinez SE, Lopez-Cardona MG, Ahuatzin-Tremary R, et al. Association analysis of the Gln223Arg polymorphism in the human leptin receptor gene, and traits related to obesity in Mexican adolescents.[see comment]. J Hum Hypertens. 2005;19:341–346. doi: 10.1038/sj.jhh.1001824. [DOI] [PubMed] [Google Scholar]
- 67.Duarte SF, Francischetti EA, Genelhu VA, Cabello PH, Pimentel MM. Lepr p.Q223r, beta3-ar p.W64r and lep c.-2548G>A gene variants in obese brazilian subjects. Genetics & Molecular Research. 2007;6:1035–1043. [PubMed] [Google Scholar]
- 68.Qu Y, Yang Z, Jin F, Sun L, Zhang C, et al. Analysis of the relationship between three coding polymorphisms in LEPR gene and obesity in northern Chinese. Obes Res Clin Pract. 2007;1:261–266. doi: 10.1016/j.orcp.2007.10.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Search strategy for Medline, via platform OVID.
(DOC)
List of 38 SNPs within the LEPR gene covered by the Affymetrix 500K chip. The 15 tag SNPs are in bold.
(DOC)
Characteristics of case-control studies.
(DOC)
Characteristics of cohort or cross-sectional studies or control arms of case-control studies.
(DOC)
Genotype and derived allele frequencies (D) for Q223R, by ethnic group.
(DOC)
Genotype and derived allele frequencies (D) for K109R, by ethnic group.
(DOC)
Genotype and derived allele frequencies (D) for K656N, by ethnic group.
(DOC)
Association from linear regression models of LEPR variants with different outcomes.
(DOC)
Association from linear regression models of LEPR variants with different outcomes showing a significant interaction with sex, stratified by sex.
(DOC)