Abstract
Many genomic alterations associated to human diseases localize in non-coding regulatory elements located far from the promoters they regulate, making the association of non-coding mutations or risk associated variants to target genes challenging. The range of action of a given set of enhancers is thought to be defined by insulator elements bound by CTCF. Here, we analyzed the genomic distribution of CTCF in various human, mouse and chicken cell types, demonstrating the existence of evolutionarily conserved CTCF-bound sites beyond mammals. These sites preferentially flank transcription factor-encoding genes, often associated to human diseases, and function as enhancer blockers in vivo, suggesting that they act as evolutionary invariant gene boundaries. We then applied this concept to predict and functionally demonstrate that the polymorphic variants associated to multiple sclerosis located within the EVI5 gene are actually impinging on the adjacent gene GFI1.
INTRODUCTION
While only a small proportion of the genome codes for proteins and regulatory RNAs, cis-regulatory elements (CREs), the DNA sequences controlling the expression of the coding segments, are located in the vast non-coding portion of the genome1. It is therefore not surprising that genome-wide association (GWA) studies are linking an increasing number of human diseases to non-coding DNA, most likely with regulatory function (reviewed in2,3). However, in these cases, the assignation of the candidate disease gene may not be straightforward: CREs can act at long distances, and their target gene may not be the one closest to the CRE (see, for example,4). Thereby, methods for predicting which gene, or genes are under regulation by particular non-coding genome segments should help in the identification of the candidate disease gene in cases where the lesion lies in non-coding regions.
Research from many laboratories has shown that the 11 zinc-finger nuclear factor CCCTC-binding protein (CTCF) contributes to the regulation of gene expression and higher order organization of the genome5. CTCF is evolutionarily conserved and widely distributed along the vertebrate and Drosophila genomes6–9. Although at present the primary function(s) of CTCF cannot be directly derived from its genomic distribution, some of the CTCF-bound sites are well known to function as regulatory boundaries, confining the range of actions of CREs to genes within those boundaries (reviewed in5,10). Different cofactors are able to interact with CTCF, including the SNF2-like chromodomain helicase CHD8 and, more recently, the DEAD-box RNA helicase p6811,12. CTCF also binds to the cohesin complex at a large number of genomic sites13–15. Indeed, at several loci, cohesin complex seems to regulate this insulator activity13–15. Constitutive CTCF-bound sites are more likely to serve this function, while more labile sites may be involved in tissue specific gene expression regulation. In fact, a proportion of CTCF sites have been shown to be constitutively occupied in several human cell types and even to be conserved between human and mice cell types7,16. This conservation might extend even further evolutionarily, since the development of the shared body plan of vertebrates is controlled by an also shared set of transcription factors and signaling molecules deployed in similar patterns17. However, genome-wide CTCF distribution has not yet been examined outside mammals. If CTCF-bound sites are found at syntenic positions in different vertebrates, these evolutionary conserved boundaries could be used to resolve ambiguous associations of target genes affected by mutation in non-coding regions in human diseases, as is the case of Multiple Sclerosis and the GFI1 and EVI5 genes.
Multiple Sclerosis (MS, [MIM 126200]) is the most common progressive and disabling neurological condition affecting young adults in the world today. The overall prevalence of MS ranges from 2 to 150 per 100,000 individuals. Pathogenetically, MS is considered an autoimmune disease leading to the demyelination of central nervous system axons18. From a genetic point of view, MS is considered a complex disorder resulting from a combination of genetic and non-genetic factors19. In addition to the human leukocyte antigen (HLA), which is recognized as the strongest locus for MS in most populations, other genetic factors involved in MS have remained elusive until the arrival of Genome-Wide Association Studies (GWAS) (The MSGene Database. http://www.msgene.org/.). To date, seven GWAS have been performed for MS; even though study design and results vary substantially between experiments, some new susceptibility genes have been identified and replicated using this approach20. However, even after convincing replications, the localization of the causal variant(s) of most of these loci remains to be determined. Several GWAS found a set of MS-associated polymorphisms belonging to the same linkage disequilibrium block located in a region containing the GFI1 (growth factor-independent 1), EVI5 (ecotropic viral integration site 5), RPL5 (ribosomal proteinL5) and FAM69 (family with sequence similarity 69)21,22,23. A fine mapping of this genomic region was performed pointing to polymorphisms located within the 17th intron of the EVI5 gene as the most probable causal variants of the association24. However, these findings did not clarify the functional role of this EVI5 risk region. Our analysis of the CTCF sites within this genetic block indicates that the 17th intron of the EVI5 gene likely belongs to the GFI1, and not the EVI5, regulatory domain. We further demonstrate that this intron indeed contains CREs that contact the GFI1, but not the EVI5, gene. We finally show that increased GFI1, but not EVI5, expression is associated by the MS risk haplotype. We therefore conclude that GFI1, and not EVI5, is the causal gene associated to MS.
RESULTS
CTCF occupies syntenic positions in vertebrate genomes
Recent studies have shown that an important fraction of CTCF sites in human cells are constitutive: that is, they are occupied by CTCF regardless of the cell type analyzed7. This led us to ask to what extent these constitutive sites are also bound in vivo by CTCF in equivalent syntenic positions (i.e. surrounded by the same orthologous genes) across vertebrate genomes. To investigate this, we collected available genome-wide CTCF ChIP-Seq data from human cells (CD4+, HeLa and Jurkat cells6,7) and produced CTCF ChIP-Seq data for mouse (Mus musculus) embryonic stem (ES) cells and embryonic fibroblasts (MEFs), as well as for chicken (Gallus gallus) red blood cells (RBC) isolated from embryos at 5 and 10 days of development. We derived potential CTCF-bound sites from the ChIP-Seq using previously described protocols6,7,25 (see Supplementary Fig. 1 for details). Within each species, we identified the set of sites common to all cell lines (species-specific constitutive CTCF sites), considering two sites as common between two cell types if they overlap in the genome by at least 50% of their length (see Methods for details). A large fraction of CTCF sites appear to be constitutive for the different cell types within each species (Fig. 1a).
Figure 1.
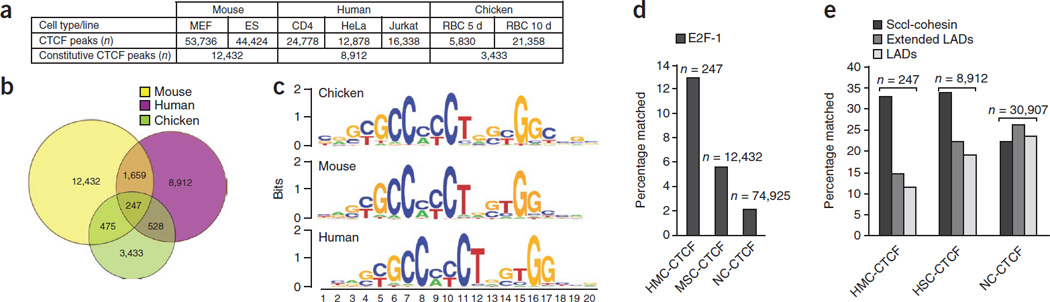
CTCF binding sites detection and conservation. (a) Number of CTCF peaks detected in each cell type/line investigated in each species. The bottom row shows the number of constitutive peaks within each species. (b) Venn diagram summarizing the inter-species conservation of CTCF sites. (c) Canonical CTCF motifs obtained by de novo motif discovery. (d) Genomic intersections (overlap ≥ 50%) of mouse CTCF sites with E2F-1 binding sites. CTCF sites were grouped according to their conservation status into: “HMC” (human/mouse/chicken conserved), “MSC” (mouse genome-specific) and “NC” (non-conserved). (e) Genomic intersections of human HMC, NC and HSC (human-specific) CTCF sites with LADs and SccI-Cohesin. LADs stands for Lamina-Associated Domains, while extended LADs corresponds to LADs extended for 10kb to each side.
We next wanted to identify the set of constitutive CTCF sites that are, in addition, evolutionary conserved in all investigated species. 74% and 5% of human constitutive CTCF sites lie within sequences conserved in mouse and chicken, respectively, displaying at least 50% identity in pairwise alignments. The global nucleotide coverage is 61.5% and 3.9% in mouse and chicken respectively, as inferred from multiple sequence alignments of 44 mammalian genomes26. However, we reasoned that a CTCF-bound site located at an equivalent position in two species (for example, between two paralogous genes) could play an equivalent function (i.e. be evolutionarily conserved), even if the sites were not residing in a conserved sequence. Therefore, relying exclusively on sequence conservation was not sufficient to identify these evolutionary conserved CTFC occupied sites. We therefore considered two CTCF sites as evolutionarily conserved if they were syntenic in two species, i.e., they separated the same evolutionary conserved regions (ECR,27), coding or non-coding, at orthologuous genome loci (see Methods and Supplementary Fig. 2). The outcome of our method is presented in Fig. 1b. We found 247 constitutive and syntenic (CONSYN)-CTCF sites among the three investigated genomes. Likely, this is an underestimation of the number of CONSYN sites because of the stringency in the definition of vicinity to conserved ECRs used, as well as the relatively low overall degree of sequence conservation between chicken and mammals. Still, these 247 CONSYN-CTCF sites correspond to 7% of the total constitutive sites in chicken, the species with the lowest number of identified CTCF-bound regions.
CONSYN sites co-localize with Cohesin and E2F-1, functioning as insulators
Next, we analyzed sequence features associated with this set of CTCF binding sites. We first found that the most over-represented motifs are three highly similar Position Weight Matrixes (PWMs) that matched the previously established CTCF consensus motif (Fig. 1c), consistent with the high conservation of the CTCF protein from mammals to birds28. Motif discovery on CONSYN sites (see Methods) identified a number of additional motifs over-represented in the CONSYN- CTCF set as compared to the species-specific constitutive ones (Table 1). This predicts the action of other nuclear factors cooperating with CTCF at these sites. Among the top ranking ones we found SAP-1a, E2F-1, HIC1 and AP-2. ChIP-Seq data available for E2F-1 in mouse29 confirms that E2F-1 is more frequently found in the vicinity of CONSYN CTCF sites than in the proximity of non-constitutive sites or species-specific constitutive ones (Fig. 1d). Using a very stringent set of simulated CONSYN sites as a random control (see Supplementary Methods), we have found the association of CONSYN CTCF sites with E2F-1 sites to be statistically significant (P-value < 0.001).
Table 1.
Transcription factor binding motifs over-represented in the CONSYN-CTCF set as compared to the species-specific constitutive ones.
| Rank | TRANSFAC ID |
Factor | Avg. rank |
P value (chicken) |
P value (human) |
P value (mouse) |
Domain |
|---|---|---|---|---|---|---|---|
| 1 | M01167 | SAP-1a | 9.3 | 9.62 × 10−5 | 4.72 × 10−7 | 2.77 × 10−5 | ETS |
| 2 | M00938 | E2F-1 | 22.0 | 0.00036 | 1.05 × 10−5 | 0.0028 | Forkhead |
| 3 | M01073 | HIC1 | 23.3 | 4.01 × 10−5 | 0.00035 | 0.0015 | CH+BTB/POZ |
| 4 | M00800 | AP-2 | 27.0 | 0.00012 | 0.00015 | 3.38 × 10−6 | bHSH |
| 5 | M00803 | E2F | 30.0 | 0.00026 | 6.38 × 10−5 | 0.0061 | Forkhead |
| 6 | M00469 | AP-2α | 36.3 | 0.0002 | 0.00055 | 0.0045 | bHSH |
| 7 | M00470 | AP-2γ | 36.7 | 0.0003 | 0.00029 | 0.0085 | bHSH |
| 8 | M00341 | GA BP | 43.7 | 0.0028 | 2.42 × 10−5 | 0.0075 | ETS |
| 9 | M01165 | Elk-1 | 44.7 | 0.00223 | 0.00147 | 0.0015 | ETS |
| 10 | M00333 | ZF5 | 46.3 | 0.00315 | 0.0013 | 0.00012 | CH+BTB/POZ |
| 11 | M01072 | HIC1 | 50.7 | 0.00253 | 0.0005 | 0.007 | CH+BTB/POZ |
CTCF sites have been proposed to serve four types of functions: (1) enhancer blocker, (2) barrier for the spreading of repressive heterochromatin and (3) genome organization (4) transcriptional enhancement5,10,30–32. It has been recently shown that in certain contexts cohesins act as mediators for the enhancer-blocking and/or three-dimensional genome organizing activities of CTCF15. On the other hand, CTCF is known to flank Lamina-Associated Domains (LADs), where it has been proposed to have a barrier function, preventing heterochromatin spreading into transcriptionally active chromosomal domains33. In order to determine whether CONSYN-CTCF sites could be linked preferentially to either of these functions, we correlated these sites to SccI-cohesin and LAD peaks33. We found that CONSYN sites overlap with cohesin-associated loci, while tend to avoid LADs, even when we extend the search to up to 10kb around each LAD site (Fig. 1e). Both, overlap with cohesin-associated loci and avoidance of LADs are statistically significant when compared the control set of simulated CONSYN sites (P-value < 0.001 on both cases.). These data are consistent with CONSYN-CTCF sites having an enhancer-blocking activity. To test this point, we assayed the insulator activity on a sample of six human CTCF sites, three conserved between human and mouse and three CONSYN sites, in two ways: through luciferase enhancer-blocking assays in human HEK 293 cells 34,35 and in vivo, using a recently described insulator assay in zebrafish36 (Fig. 2). All six sites showed consistent enhancer-blocking activity in the in vitro assays (>2 fold; Fig. 2a) and four of them, including the three CONSYN sites, reproducibly showed robust enhancer-blocking activity in vivo (Fig. 2b,c).
Figure 2.
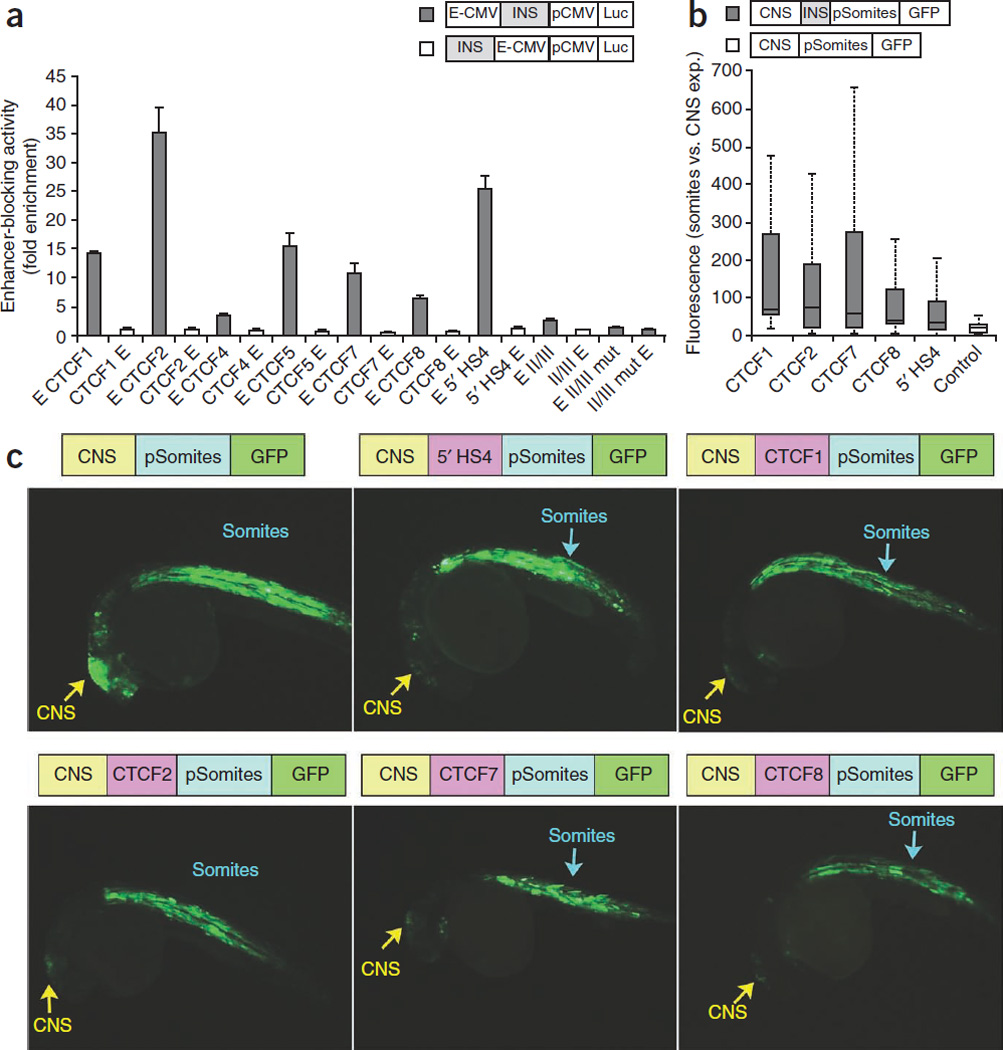
Functional validation of CTCF sites as insulators. (a) A set of three CTCF sites conserved between human and mouse (CTCF1, CTCF4 and CTCF5) and three CONSYN sites (CTCF2, CTCF7, CTCF8) were assessed for enhancer-blocking activity through a reported in vitro assay34. Data are presented as fold enhancer-blocking activity normalized by the value achieved by the reference pELuc vector +/− SD. The 5’HS4 chicken b-globin insulator (5’HS4) and the internal II/III element were used as positive controls. A mutated II/III element with an altered CTCF site was used as a negative control35. All CTCF sites tested showed a significant (>2 fold) enhancer-blocking activity. (b) Box plot representation of enhancer-blocking activity in vivo using a transgenic zebrafish assays. In this assay, the insulator is placed between a central nervous system (CNS)-enhancer and a promoter driving the expression of GFP to somites. Bars depict ratios between fluorescence in somites versus CNS. Only the four CTCF sites with significant insulator activity are depicted. (c) Images from zebrafish embryos after microinjection of each of the four CTCF sites shown in (b), along with positive (5’HS4) and negative (empty) controls. The construct used is shown above the image. Note the reduction of the activity of the midbrain enhancer (CNS; yellow arrow) relatively to the somite expression (blue arrow) when the insulators sites are placed in between.
CONSYN sites preferentially flank developmental and disease genes
All these facts led us to hypothesize that CONSYN-CTCF sites might be separating genes whose expression ought to be tightly regulated and their chromatin organized at the genomic level, at least from chicken to humans. To identify these genes, we assigned two neighbouring genes in each direction for each CONSYN site (lists are provided in Supplementary Table 1). Gene ontology term analysis identified an enrichment in transcription factors (TFs) involved in cell differentiation and embryonic development (Figure 3a and Supplementary Tables 2 and 3). Indeed, while TFs constitute ~ 5% of all genes in mouse and human genomes, ~ 12% and 10% of murine and human genes adjacent to CONSYN-CTCF sites are TFs. The difference is statistically significant compared with a set of random genomic sites (P-values < 0.01, Fig. 3b). We have analyzed recently published expression data for human and mouse37, and found that TFs flanking CONSYN sites do not particularly tend to have tissue specific significant expression patterns (Fig. 3b). However, we observed that adjacent genes separated by CTCF binding sites tend to have different patterns of expression as compared to all genes in the genome (Fig. 4a and Supplementary Fig.3), supporting a function of CTCF-bound sites as regulatory domain boundaries.
Figure 3.
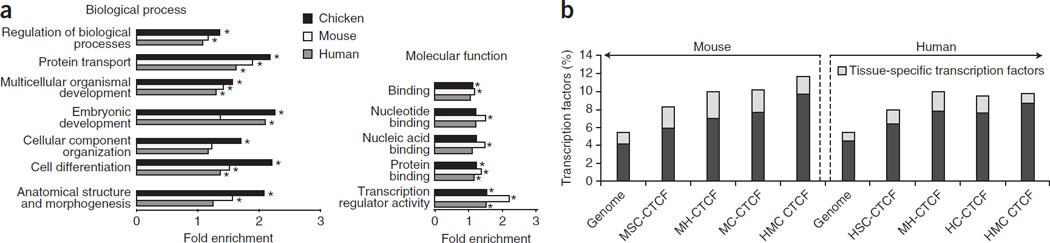
CONSYN-CTCF sites are preferentially flanking transcription factors involved in developmental processes. (a) Significantly enriched (p ≤ 0.05) Gene Ontology terms in genes associated to CONSYN-CTCF sites in chicken, mouse and human (black, white and gray bars, respectively). The bars marked with an asterisk correspond to highly significant p values (p ≤ 0.01) (b) Proportion of transcription factor-encoding genes associated to CONSYN CTCF sites in mouse and human genomes. The grey portion in each bar corresponds to the percentage of tissue specific expressed transcription factors. MSC (n=12,432) and HSC (n=8912) are constitutive sites in mouse and human cells, respectively. MH (n=1,659), MC (n=475), HC (n=528) and HMC (n=247) are mouse-human, mouse-chicken, human-chicken and human-mouse-chicken conserved sites, respectively.
Figure 4.
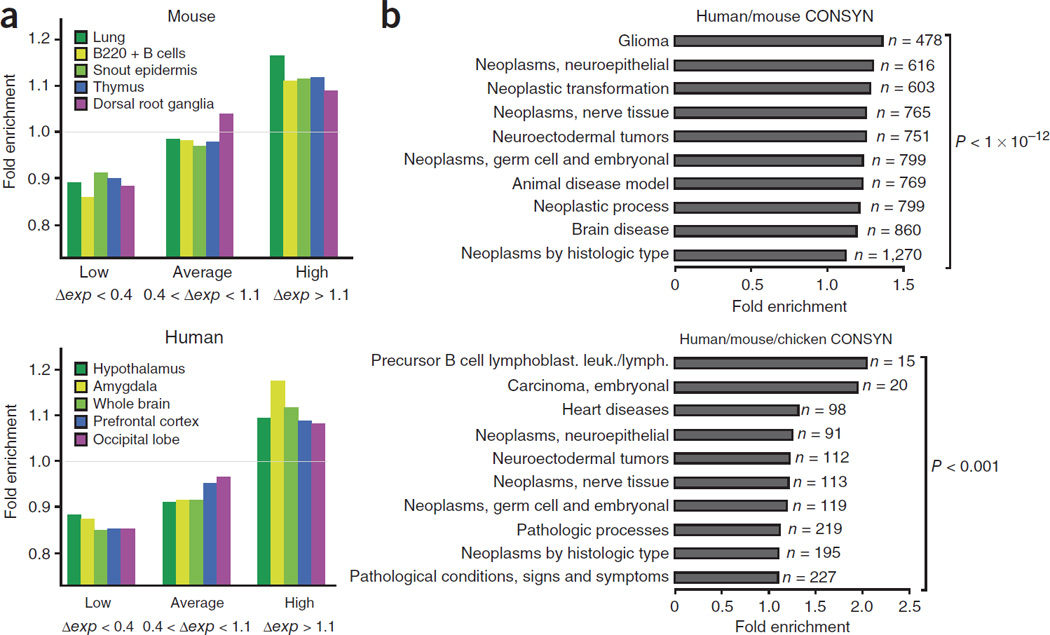
Genes separated by CTCF sites have differential expression patterns and are associated with human diseases. (a) The five tissues showing the most significant differences are shown for mouse and human CTCF sites. Genes separated by CTCF binding events exhibit greater difference in expression levels (Δexp) than expected. Genes separated by CTCF (as compared to random expectation) are enriched for pairs with high differences in expression and depleted for genes with low differences in expression, as compared to those with an average difference. (b) MESH analysis of genes flanking CONSYN-CTCF sites show high enrichment in disease-associated genes. Of the set of 2562 CTFC-bound genes conserved between human and mouse, genomatix identified 2273 genes, of which 1714 had a MeSH annotation. Of the set of 412 CTFC-bound genes that are conserved between human, mouse and chick genomatix identified 360 genes, of which 262 had a MeSH annotation. The top-ten overrepresented MeSH-disease terms in the input gene set (ranked by fold-enrichment) are listed together with their respective number of genes observed for each term. P-values for each set are also shown.
Altered regulation of genes is often associated with human diseases3,38. We therefore examined whether the set of genes flanking constitutive CTCF sites are enriched for diseases-associated genes. When subjected to MeSH analysis (http://www.nlm.nih.gov/mesh/genes), the human genes linked to human-mouse conserved CTCF sites and even to human-mouse-chicken (CONSYN) ones are significant associated with disease, including cardiovascular disease, neuroectodermal tumors and lymphomas (Fig. 4b).
CONSYN sites predict the association of GFI1 to MS susceptibility
A considerable number of the increasing available Genome Wide Association Studies (GWAS) indicate that many human diseases are caused by mutations in CREs. However, the identification of the target gene of each of these CREs is not trivial, since these may be located hundred of kilobases away from its target promoter, and even inside neighboring genes. Thus, often the gene truly implicated in the development of a particular disease cannot be directly identified. Since CONSYN-CTCF sites seem to define evolutionary conserved gene-regulatory boundaries and these boundaries are preferentially linked to genes encoding transcription factors whose malfunction is frequently associated with human diseases, we reasoned that these sites could aid to link mutations or polymorphisms in CREs associated to human diseases to their target “disease” gene.
As a proof of principle, we used the GFI1-EVI5 genomic region that has been associated with multiple sclerosis21. The most probable causal variants of the association to multiple sclerosis (MS) have been located in the last intron of the EVI5 gene24. Thus, one or several CREs within this intron may be affected in the risk haplotypes. Based on this evidence, EVI5 has been suggested as the potential target of these CREs22,23. However, examination of the human constitutive CTCF binding sites in the GFI1-EVI5 genomic locus indicates the presence of three sites separating the risk genomic area from the EVI5 promoter (Fig. 5a). Strong CTCF binding sites are also found separating this last EVI5 intron from its promoter in mouse and chicken genomes and in similar positions (Supplementary Fig. 4). Although these CTCF sites could not be identified as syntenic sites by our pipeline due to stringent criteria imposed, it is likely that they constitute functionally equivalent CTCF sites. The evolutionary conserved architecture of the GFI1-EVI5 genomic locus with CTCF-bound sites separating the last intron of EVI5 from its promoter in all vertebrates examined strongly suggests that potential CREs within this intron are preferentially acting on the neighbouring GFI1 gene, and not on EVI5. MS is a heterogeneous immunopathy likely caused by the joint participation of different peripheral blood cells in the central nervous system39. Interestingly, malfunction of GFI1, which encodes a zinc-finger transcription factor, causes abnormal or malignant haematopoiesis (reviewed in40), and therefore could play a role in an autoimmune disease such as MS. To evaluate whether CREs in the last intron of EVI5 act on either of the EVI5 or GFI1 promoters, we performed Chromatin Conformation Capture (3C) assays on control and PMA+ Io (phorbol-myristate-acetate plus ionomycin) activated human peripheral blood mononuclear cells (PBMCs). In these 3C assays we used two different anchor primers: one on the promoter region of each gene, and multiple primers spanning the whole genomic region of the last EVI5 intron (Fig. 5b). These primers allow detecting DNA interactions between different regions covering the whole intron with any of the two promoters. PCR product values for each primer pair were normalized against those obtained in control samples containing BAC clones spanning the tested genomic region (see Supplementary Methods). In non-activated PBMCs we found no significant interaction between any of the promoters and different regions of the intron (Fig. 5b, blue graph). The same was true in activated cells when the EVI5 promoter was surveyed (Fig. 5b, cyan graph). In contrast, the GFI1 promoter interacted with several regions of the intron, interaction that was stronger in the activated than in the control PBMCs (Fig. 5b, red and orange graphs, respectively).
Figure 5.
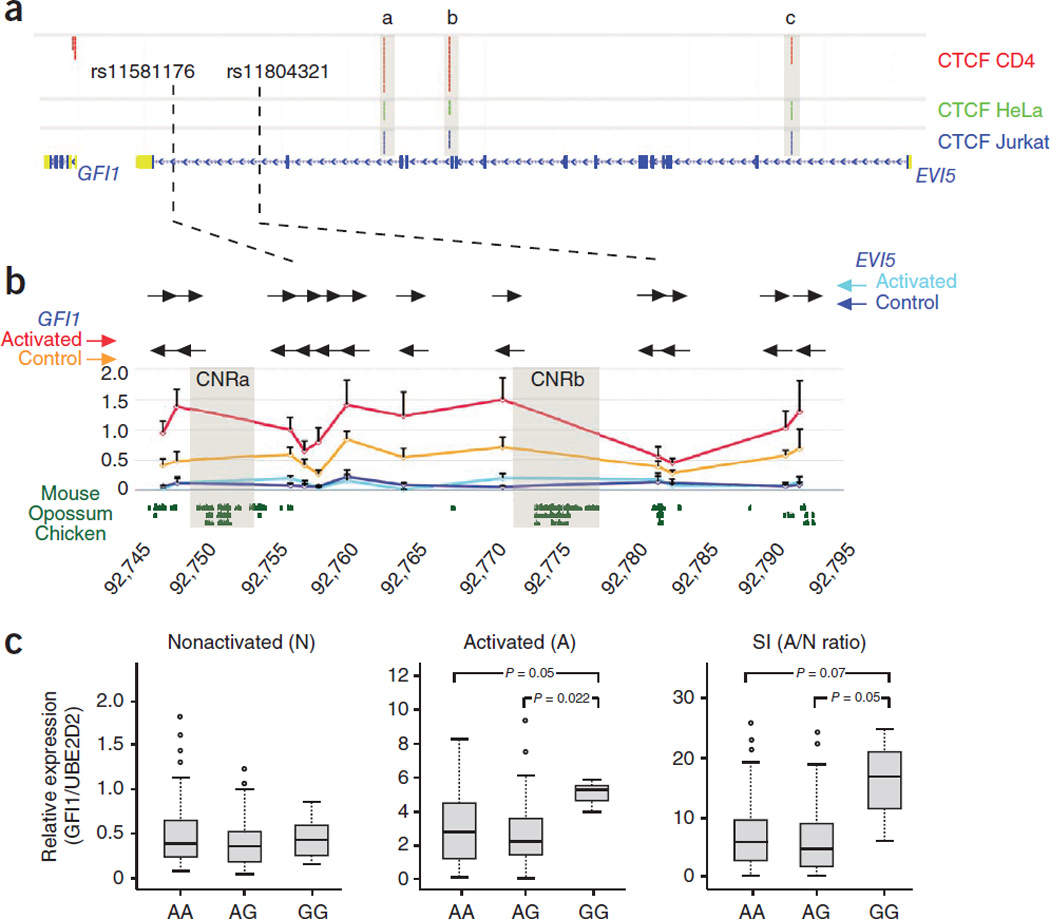
Constitutive CTCF sites help assign target genes for non-coding mutations associated with human diseases. (a) Distribution of CTCF bound sites in different human cell types along the GFI1-EVI5 genomic regions. Constitutive CTCF sites (grey boxes) separate the last intron of EVI5 (blue rectangle) from its promoter. Two SNPs associated with MS are marked by black dashed vertical lines. (b) Close view of this intron showing the graphical representation of the 3C results obtained from this intron and the two flanking promoters. The coordinates of the region are shown below in kilobases. The primers along the intron used are denoted with horizontal black arrows while the fixed primers at the promoters of EVI5 and GFI1 are shown with blue (control PBMCs)/cyan (activated PBMCs) and orange (control PBMCs)/red (activated PBMCs) horizontal arrows, respectively. Several intronic regions interact with GFI1 more strongly in the activated (red graph) than in control (orange graph) PBMCs. No interaction was observed between the different regions of the intron and EVI5 in neither control nor activated PBMCs The shadowed boxes mark the contacting region, conserved between human, mouse and chicken, and which were tested in functional assays in Fig. 6. (c) Association of GFI1 transcription with genotypes of the EVI5 rs11804321 polymorphism in PBMCs. Gene expression of GFI1 was assayed by real time PCR using as reference the UBcH5B gene (Relative expression, GFI1/UBcH5B). The expression obtained from non-activated PBMCs (N), activated PBMCs (A) and ratio A/N as stimulation index (SI) is represented in the panels. Statistical significance of GFI1 expression differences between genotypes was calculated using the nonparametric Mann-Whitney rank-sum test for a total of 108 PBMC independent samples, genotyped for rs11804321 with a distribution of 59 AA, 46 AG and 3 GG. Only significant (P≤ 0.05) or trend (P≤ 0.08) difference are indicated.
These results suggest that the EVI5 intron contains CREs that act on the promoter of GFI1, not on that of EVI5. Accordingly, GFI1 is robustly upregulated in activated PBMCs, while EVI5 is undetectable in both activated and non-activated blood cells (Supplementary Fig. 5). Strengthening this point further, a recent report identified a likely GFI1 haematopoietic stem cell-specific enhancer in this genomic area41.
An important prediction from these data is that it is GFI1, and not EVI5, the gene whose expression should be altered in risk haplotype carrying individuals. Indeed we found that this was the case. In PBMCs of the risk (G) allele within SNP rs11804321, the expression of GFI1 was increased, as compared to the levels found in samples carrying the protective (A) allele either in heterozygosity or in homozygozity (Fig. 5c). In contrast, no differences were found for the EVI5 expression levels (not shown). This correlates with a recent report showing that GFI1 expression levels are also increased in peripheral blood cells of individuals that will develop multiple sclerosis42, indicating that increased GFI1 is linked to higher risk of developing the disease. The regions from the EVI5 intron that interact with the GFI1 promoter in our 3C studies contain two evolutionary conserved non-coding sequence blocks (CNR-A and CNR-B; Fig. 5b), suggesting a possible regulatory function for them. To examine this possibility, we PCR-amplified these two regions and tested their potential enhancer or repressor activities in luciferase assays in THP-1 human acute monocytic leukemia cells. Both regions showed clear repression activity in these assays (Fig. 6a). Therefore, our results are compatible with a scenario in which an increased risk to develop multiple sclerosis is caused by a mutation in any of these two, or even other, repressors located in the last EVI5 intron, which would then lead to an abnormally high expression of GFI1.
Figure 6.
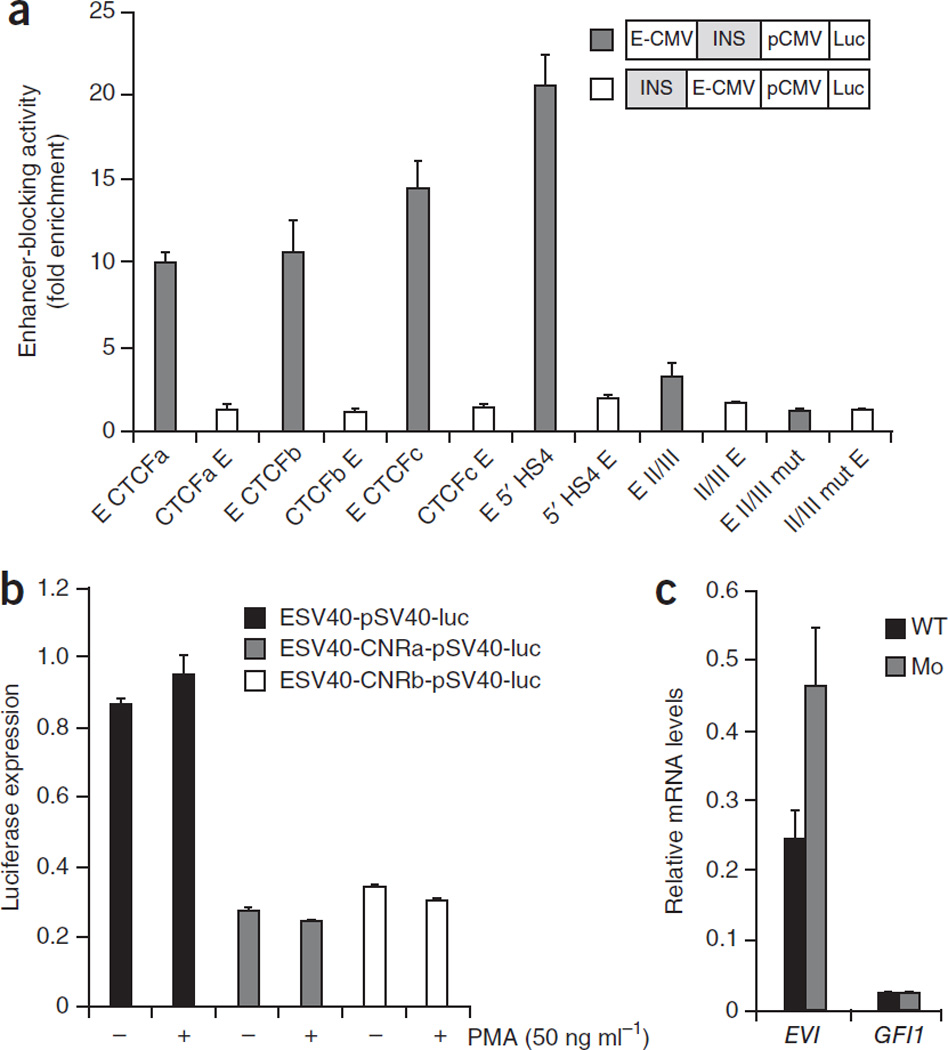
CTCF sites in the EVI5 gene act as insulators that prevent the interaction of GFI1-associated CREs with the EVI5 promoter. (a) Enhancer-blocking activity assays performed on three human CTCF-bound sites (a,b and c) shown in (Fig. 6) demonstrate that these sequences effectively work as insulators. (b) Luciferase assays performed in THP-1 cells indicate that the regions of the last EVI5 intron acting on the GFI1 promoter (CNRA and CNRB), behave as repressors in both control and activated THP-1 human acute monocytic leukemia cells. (c) evi5 and gfi1 mRNA levels in control (black bars) and CTCF-depleted (grey bar), 48 hours post fertilization zebrafish embryos, measured by quantitative RT-PCR. Error bars represent the SEM of three experiments.
Our starting prediction of a functional linkage between the risk haplotype and GFI1 was based on the location of a potential enhancer barrier separating the risk region and the EVI5 promoter. To test whether any of these three human CTCF-bound sites can function as insulators, we performed functional enhancer barrier cell culture assays with all three of them. Similar to the behavior displayed by other CTCF-bound sites we tested, all three regions clearly act as insulators in these experiments (Fig. 6b). These results strongly suggest that these CTCF sites are insulators separating GFI1 and EVI5 regulatory landscapes. If so, it would be expected that reducing CTCF function may affect this boundary, resulting in misregulation of any of these two genes. Since organization of GFI1 and EVI5 is syntenic in zebrafish, we tested this possibility by knocking-down CTCF function with a splicing-specific morpholino (MOsp1CTCF, see Methods and Supplementary Fig. 6 for details) in this organism. The MOsp1CTCF morpholino partially inhibits the correct removal of intron 2. The inclusion of intron 2 in the mRNA introduces several precocious stops codons that eliminate key domains of the CTCF protein (Supplementary Fig. 6). We then determined by qRT-PCR (quantitative real time PCR) the expression levels of both gfi1 and evi5 genes in control and morphant embryos. As shown in (Fig. 6c), in the CTCF morphant embryos the expression of evi5 is higher than in control individuals while that of gfi1 does not vary. These results indicate that reducing CTCF levels causes evi5 misregulation, which could be due to inappropriate contact with neighboring regulatory regions. Since the genomic organization of these two genes is conserved all along vertebrate evolution, we predict that a similar situation may also occur in humans.
DISCUSSION
Recent studies have shown that a large fraction of CTCF-bound sites in different human or mouse cell types are conserved within species7,16 defining what we denominate here constitutive CTCF-bound sites. Moreover, it has been also demonstrated that a number of these CTCF-bound sites lie within sequence stretches conserved between human and mouse genomes, and therefore are evolutionarily conserved in mammals16. However, this criterion is too restrictive. CTCF sites may serve similar insulator or enhancer blocking functions in two species if located at equivalent genomic position (i.e. syntenic), irrespectively of whether they are at conserved sequences or not. Therefore, here we have extended the set of conserved CTCF-bound sites to include those that are syntenic. With this approach, we identify at least two times more potentially equivalent CTCF-bound sites in mammalian genomes than just using sequence conservation, corresponding to 18% of the human constitutive sites. To further examine the existence of more deeply conserved CTCF syntenic sites in a non-mammalian genome, we determined by Chip-seq the genome-wide CTCF distribution in two chicken cell types. As in other species, we find that a large fraction of CTCF sites are constitutive occupied in the two different chicken cell types analyzed (59% of the sites form the cell type with less reads). Moreover, 7% of these chicken constitutive sites are placed at syntenic position in mice and humans, being most of them not conserved at the sequence level. We call these sites CONSYN (from constitutive (within each specie) and syntenic (between species)) CTCF sites. We therefore conclude that using synteny is a much more powerful way to identify equivalent positions occupied by a transcription factor in different species than using just sequence conservation.
Interestingly, our work demonstrates that these deeply evolutionary conserved CTCF-bound sites show enhancer-blocking activity and tend to flank developmental genes associated with human diseases. Therefore, our work identifies a set of gene boundaries that have remained constant, at least, from chicken to humans. This conservation may stem from the need of avoiding regulatory interference within and between these essential genes. Likely, disruption of these genes’ boundaries would impair development or cause disease. Therefore, we propose that evolutionarily conserved CTCF sites can serve as a general useful guide in assigning non-coding mutations to target genes, among them some associated with human diseases. Indeed, as a proof of principle, here we used this knowledge on conserved gene boundaries to identify a likely target gene affected by haplotypes associated to an increase risk of suffering multiple sclerosis located in the GFI1-EVI5 genomic region. Although, in previous works EVI5 was considered as the target gene likely involved in this disease22,23, we demonstrate that the last intron of this gene, which contains the multiple sclerosis risk haplotypes, is separate from its promoter by several syntenic CTCF-bound sites that can function as insulators. Indeed, these syntenic CTCF-bound sites suggest that the last EVI5 intron is within the GFI1 gene regulatory landscape. Therefore, CTCF potentially prevents the interaction of a number of GFI1 regulatory elements present in this EVI5 intron with its own promoter. Accordingly, evi5 expression is mis-regulated in zebrafish embryos with reduced CTCF function. We also find two repressor elements within this intron that are good candidate regions to be mutated in MS risk haplotypes. Accordingly, as expected by a malfunction of these repressors, we find that individuals that carry in homozygosity one of the MS risk SNPs have higher levels of expression of GFI1I, but not EVI5, in peripheral blood cells. Finally, in these cell type, and using 3C experiments, we further demonstrate that these repressors physically contact with the GFI1I, but not EVI5, promoter. Altogether, our results demonstrate that GFI1I, but not EVI5, is possibly the real gene associated with higher risk in developing multiple sclerosis, a prediction that was originally based on the distribution of syntenic CTCF sites in multiple vertebrates. Therefore, the location of these sites might inform on the associations between disease-linked SNPs at non-coding DNA and target genes by defining regulatory domains throughout the genome.
METHODS
Chromatin immunoprecipitation (ChiP)
Mouse CTCF ChIPs in ES cells and fibroblasts were performed as previously described43. The antibody used for immunoprecipitation was a rabbit polyclonal antibody against CTCF (07–729, Millipore). In order to evaluate the CTCF-ChIP quality, positive PCR controls were performed for the H19 Imprinting control region (Supplementary Fig. 7a). Chicken red blood cells (RBC) CTCF ChIP experiments were performed as previously described44,45. We evaluated the CTCF-ChIP quality with several positive and negative PCR controls (Supplementary Figure 7b).
Sequencing
Sequencing libraries were produced using the Illumina ChIP-Seq sample preparation kit, following the manufacturer’s instructions. Single read sequencing was performed on the Illumina Genome Analyzer platforms I and II, and images were analyzed using Illumina pipeline versions 1.3.2, and 1.4.
Short reads mapping and peak calling
Genomic coordinates of chicken and mouse ChIP-Seq sequence reads have been obtained using the GEM mapping software suite (http://gemlibrary.sourceforge.net). We used the quality files provided by the Illumina Genome Analyzer, and mapped them against the corresponding genome sequence (Galgal3, mm9 for chicken and mouse respectively). Using this same program, we determined the corresponding genome fractions to be used with the peak-calling program. For human data, since the quality files were not available, we used the provided mapping (Eland) on the hg18 genome assembly. We filtered out those reads not mapping uniquely to the reference genome sequence. Details are provided in Supplementary Fig.1.
Peak calling has been performed using SISSRs25, with the same parameters as those previously published for detection of the human CTCF binding sites6,7. The selected regions were then extended to 200 bp to each side, centered on the middle coordinate of the peak.
Evolutionary conservation
Sequence conservation analysis among species consisted in retrieving MAF blocks (from the UCSC multiple genome alignments) using one of the species as reference coordinates, and examining whether these blocks overlap a peak in the query species. The retrieval of the blocks has been performed using the ‘extract MAF blocks’ module from the Galaxy suite46,47.
We developed three methods (Supplementary Fig 2) to assess conservation based on anchoring peaks from one species (referred to as reference species) to peaks in another (query) species through sequence features. This allowed us to detect conserved peaks having no sequence conservation. Further details can be found in Supplementary Methods.
Motif analysis
Two types of analysis have been performed: de novo motif discovery and known motifs over-representation. To assess motif over-representation, we used Pscan48 with the Transfac49 Pro 2009.2 motif collection (892 matrices). Non- vertebrate motifs and low quality matrices (Q5 and Q6) have been removed from the collection. De novo motif discovery analysis was performed with MEME50 trying all possible motif widths from 6 to 20 bp, asking for 5 motifs to be found per run, and using two different distribution models: one occurrence of the motif per sequence (oops), and zero or one occurrence per sequence (zoops).
Gene Ontology analysis
Gene Ontology (GO) term enrichment analyses have been computed through the GOToolBox suite51. Term over-representation has been calculated using a hypergeometric-based test. P-values have been corrected for multiple testing using a Benjamini and Hochberg correction. We consider a p-value to be highly significant when lower than or equal to 0.01, and significant when lower than or equal to 0.05. We also computed the enrichment ratio for each over or under-represented term, dividing the frequency of this term in our gene set by its frequency in the whole genome. For clarity, we also mapped the over- and under-represented terms to the generic slim ontology provided by the GO consortium.
Identification of tissue differential expression of genes separated by CTCF peaks
First, we used the set of UCSC known genes52 to define non-overlapping gene clusters. Second, we associated each microarray probe in the Gene Expression Atlas 2 data provided by the Genomics Institute of the Novartis Research Foundation (GNF,53) with a given gene cluster. The GNF database (gnfAtlas2) contains two replicates each of 61 mouse tissues and 79 human tissues run over Affymetrix microarrays. The log2-ratios of the signals (expression score) of all probes associated with a given cluster were averaged. For each tissue, we then computed the absolute difference in the expression scores between pairs of adjacent gene clusters that are not isolated by CTCF binding events. This background distribution was subsequently compared to that corresponding to gene clusters isolated by CTCF binding events using the Wilcoxon-Mann-Whitney test. We corrected the resulting p-values for multiple testing with Bonferroni’s method using the number of tissues.
Enhancer-blocking assay
Enhancer-blocking assays were used to address the insulator activity of selected CTCF elements, using the pELuc backbone plasmid and human HEK 293 cells as reported34. For details see Supplementary Methods.
Repressor Luciferase assays
CNRA and CNRB were PCR amplified, cloned in TOPO T/A vector and transferred using the Gateway system to the destiny vector pGL3 control, which contains the SV40 enhancer and the SV40 minimal promoter. The constructs were transfected to exponentially growing THP1 cells. Triplicated of transfected cell cultures were treated or non-treated with PMA+Io for 4 h and then harvested. Luciferase activity was evaluated using Dual-Luciferase®. For more details see Supplementary Methods.
In vivo insulator activity in zebrafish and morpholino injections
The insulator activity of selected CTCF elements was analyzed in vivo by microinjection in one-cell zebrafish embryos as reported36. About 10 to 40 individual zebrafish were analyzed and quantified for each condition. Each set of experimental constructs was always injected and analyzed with their corresponding set of controls. The LaserPix (Bio-Rad) image analysis software was used for quantification.
MOsp1 was designed to bind to the acceptor-splicing site between intron 2 and exon 3 (5’- AGCAAATATCACACACTCACCTTTC-3’). A total of 15 ng of MOsp1 morpholino was injected into one cell-stage embryos. More details can be found in Supplementary Methods.
Chromosome conformation capture assay (3C)
3C assay was performed in control and PMA+Io-activated human PBMCs cells as previously described54. See Supplementary Methods for details.
Supplementary Material
ACKNOWLEDGEMENTS
Grant support: grants BFU2007-60042/BMC, BFU2010-14839, Petri PET2007_0158, CONSOLIDER CSD2007-00008 (Spanish MICINN) and Proyecto de Excelencia CVI-3488 (Junta de Andalucía)(JLG-S); BFU2009-07044 (Spanish MICINN) and Proyecto de Excelencia CVI 2658 (Junta de Andalucía)(FC); FIS PI081636 (Instituto de Salud Carlos III)(FM); PN-SAF2009-11491 (Spanish MICINN) and Proyecto de Excelencia P07-CVI-02551 (Junta de Andalucía)(AA); BFU2008-00838, CONSOLIDER CSD2007-00008 (Spanish MICINN), Regional Government of Madrid (CAM S-SAL-0190-2006) and the Pro-CNIC Foundation (MM); BFU2006-12185 and BIO2009-12697 (Spanish MICINN)(LM); Dirección General de Asuntos del Personal Académico-Universidad Nacional Autónoma de México (IN209403, IN214407 and IN203811) and Consejo Nacional de Ciencia y Tecnología, México (CONACyT: 42653-Q, 58767 and 128464)(FR-T); Intramural Research Program of the National Center for Biotechnology Information (NIH)(IO). BIO2006-03380, CONSOLIDER CSD2007-00050 (Spanish MICINN), and RETICS RD07/0067/0012 (Spanish MSC)(RG). LM thanks Almudena Fernández for technical assistance, Laura Barrios for statistical analysis. FR-T thanks Georgina Guerrero Avendaño for excellent technical assistance.
Footnotes
AUTHOR CONTRIBUTIONS
JLGS and FC conceived the study, designed the experiments, interpreted results and wrote the manuscript. DM devised bioinformatics methods, performed data analysis and wrote the paper. CP, MS, MAB, conducted the mouse ChIP experiments. CVQ, MFM and FRT performed the chicken ChIP experiments. EdCM, EM and LM performed the insulator assays. AFM conducted the 3C experiments. FM, AA and NF provided the PBMCs from blood cells and performed the qRT-PCR, CNRA/CNRB activity on a luciferase reported system, quantification of GFI1 relative expression of 108 PBMC samples, genotyping of the EVI5 rs11804321 and statistical analysis OD performed the high-throughput sequencing. OB, LT, IO and PSP performed data analysis. MM and RG collaborated in the experimental design, discussion of results and in writing the manuscript.
REFERENCES
- 1.Elgar G, Vavouri T. Tuning in to the signals: noncoding sequence conservation in vertebrate genomes. Trends Genet. 2008;24:344–352. doi: 10.1016/j.tig.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 363:166–176. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- 3.Epstein DJ. Cis-regulatory mutations in human disease. Brief Funct Genomic Proteomic. 2009;8:310–316. doi: 10.1093/bfgp/elp021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ragvin A, et al. Long-range gene regulation links genomic type 2 diabetes and obesity risk regions to HHEX, SOX4, and IRX3. Proc Natl Acad Sci U S A. 2010;107:775–780. doi: 10.1073/pnas.0911591107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 7.Cuddapah S, et al. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bushey AM, Ramos E, Corces VG. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev. 2009;23:1338–1350. doi: 10.1101/gad.1798209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Negre N, et al. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 2010;6:e1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohlsson R, Bartkuhn M, Renkawitz R. CTCF shapes chromatin by multiple mechanisms: the impact of 20 years of CTCF research on understanding the workings of chromatin. Chromosoma. 2010;119:351–360. doi: 10.1007/s00412-010-0262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishihara K, Oshimura M, Nakao M. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol Cell. 2006;23:733–742. doi: 10.1016/j.molcel.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 12.Yao H, et al. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 2010;24:2543–2555. doi: 10.1101/gad.1967810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parelho V, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 14.Rubio ED, et al. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci U S A. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wendt KS, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 16.Mikkelsen TS, et al. Comparative epigenomic analysis of murine and human adipogenesis. Cell. 143:156–169. doi: 10.1016/j.cell.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shubin N, Tabin C, Carroll S. Deep homology and the origins of evolutionary novelty. Nature. 2009;457:818–823. doi: 10.1038/nature07891. [DOI] [PubMed] [Google Scholar]
- 18.Oksenberg JR, Baranzini SE, Sawcer S, Hauser SL. The genetics of multiple sclerosis: SNPs to pathways to pathogenesis. Nat Rev Genet. 2008;9:516–526. doi: 10.1038/nrg2395. [DOI] [PubMed] [Google Scholar]
- 19.Handel AE, Handunnetthi L, Giovannoni G, Ebers GC, Ramagopalan SV. Genetic and environmental factors and the distribution of multiple sclerosis in Europe. Eur J Neurol. 2010;17:1210–1214. doi: 10.1111/j.1468-1331.2010.03003.x. [DOI] [PubMed] [Google Scholar]
- 20.Hoffjan S, Akkad DA. The genetics of multiple sclerosis: an update 2010. Mol Cell Probes. 2010;24:237–243. doi: 10.1016/j.mcp.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 21.Hafler DA, et al. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 22.Hoppenbrouwers IA, et al. EVI5 is a risk gene for multiple sclerosis. Genes Immun. 2008;9:334–337. doi: 10.1038/gene.2008.22. [DOI] [PubMed] [Google Scholar]
- 23.(ANZgene), A.a.N.Z.M.S.G.C. Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nat Genet. 2009;41:824–828. doi: 10.1038/ng.396. [DOI] [PubMed] [Google Scholar]
- 24.Alcina A, et al. Tag-SNP analysis of the GFI1-EVI5-RPL5-FAM69 risk locus for multiple sclerosis. Eur J Hum Genet. 2010;18:827–831. doi: 10.1038/ejhg.2009.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhead B, et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2009;38:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004;32:W280–W286. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Filippova GN, et al. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol. 1996;16:2802–2813. doi: 10.1128/mcb.16.6.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 30.Pikaart M, Recillas-Targa F, Felsenfeld G. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev. 1998;12:2852–2862. doi: 10.1101/gad.12.18.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Recillas-Targa F, et al. Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities. Proc Natl Acad Sci U S A. 2002;14:6883–6888. doi: 10.1073/pnas.102179399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wallace JA, Felsenfeld G. We gather together: insulators and genome organization. Curr Opin Genet Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 34.Lunyak VV, et al. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science. 2007;317:248–251. doi: 10.1126/science.1140871. [DOI] [PubMed] [Google Scholar]
- 35.Recillas-Targa F, Bell AC, Felsenfeld G. Positional enhancer-blocking activity of the chicken beta-globin insulator in transiently transfected cells. Proc Natl Acad Sci U S A. 1999;96:14354–14359. doi: 10.1073/pnas.96.25.14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bessa J, et al. Zebrafish enhancer detection (ZED) vector: A new tool to facilitate transgenesis and the functional analysis of cis-regulatory regions in zebrafish. Dev Dyn. 2009;238:2409–2417. doi: 10.1002/dvdy.22051. [DOI] [PubMed] [Google Scholar]
- 37.Ravasi T, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleinjan DA, van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hemmer B, Cepok S, Zhou D, Sommer N. Multiple sclerosis -- a coordinated immune attack across the blood brain barrier. Curr Neurovasc Res. 2004;1:141–150. doi: 10.2174/1567202043480152. [DOI] [PubMed] [Google Scholar]
- 40.Phelan JD, Shroyer NF, Cook T, Gebelein B, Grimes HL. Gfi1-cells and circuits: unraveling transcriptional networks of development and disease. Curr Opin Hematol. 17:300–307. doi: 10.1097/MOH.0b013e32833a06f8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson NK, et al. Gfi1 expression is controlled by five distinct regulatory regions spread over 100 kilobases, with Scl/Tal1, Gata2, PU.1, Erg, Meis1, and Runx1 acting as upstream regulators in early hematopoietic cells. Mol Cell Biol. 2010;30:3853–3863. doi: 10.1128/MCB.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Achiron A, et al. Microarray analysis identifies altered regulation of nuclear receptor family members in the pre-disease state of multiple sclerosis. Neurobiol Dis. 38:201–209. doi: 10.1016/j.nbd.2009.12.029. [DOI] [PubMed] [Google Scholar]
- 43.Gonzalez S, et al. Oncogenic activity of Cdc6 through repression of the INK4/ARF locus. Nature. 2006;440:702–706. doi: 10.1038/nature04585. [DOI] [PubMed] [Google Scholar]
- 44.Escamilla-Del-Arenal M, Recillas-Targa F. GATA-1 modulates the chromatin structure and activity of the chicken alpha-globin 3' enhancer. Mol Cell Biol. 2008;28:575–586. doi: 10.1128/MCB.00943-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rincon-Arano H, Guerrero G, Valdes-Quezada C, Recillas-Targa F. Chicken alpha-globin switching depends on autonomous silencing of the embryonic pi globin gene by epigenetics mechanisms. J Cell Biochem. 2009;108:675–687. doi: 10.1002/jcb.22304. [DOI] [PubMed] [Google Scholar]
- 46.Blankenberg D, et al. Galaxy: a web-based genome analysis tool for experimentalists, Chapter 19. Curr Protoc Mol Biol. 2010;Unit 19:10 1–10 21. doi: 10.1002/0471142727.mb1910s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blankenberg D, et al. A framework for collaborative analysis of ENCODE data: making large-scale analyses biologist-friendly. Genome Res. 2007;17:960–964. doi: 10.1101/gr.5578007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zambelli F, Pesole G, Pavesi G. Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res. 2009;37:W247–W252. doi: 10.1093/nar/gkp464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matys V, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bailey TL, Elkan C. The value of prior knowledge in discovering motifs with MEME. Proc Int Conf Intell Syst Mol Biol. 1995;3:21–29. [PubMed] [Google Scholar]
- 51.Martin D, et al. GOToolBox: functional analysis of gene datasets based on Gene Ontology. Genome Biol. 2004;5:R101. doi: 10.1186/gb-2004-5-12-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hsu F, et al. The UCSC Known Genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 53.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


