Abstract
Nature possesses an unlimited number and source of biologically-relevant natural glycans, many of which are too complicated to synthesize in the laboratory. To capitalize on the naturally-occurring plethora of glycans, we have developed a method to fluorescently tag the isolated free glycans, which maintains the closed-ring structure. After purification of the labeled glycans, they can be printed on a glass surface to create a natural glycan microarray, available for interrogation with potential glycan-binding proteins. The derivatization of these natural glycans has vastly expanded the number of glycans for functional studies.
Keywords: Fluorescence, reductive amination, glycan microarray, conjugation
Introduction
Glycan microarrays have become an important hypothesis-generating platform in studies of glycan-protein interactions important in signal transduction, cellular adhesion, and host-pathogen interaction (Alvarez and Blixt, 2006; de Paz and Seeberger, 2006; Feizi and Chai, 2004; Horlacher and Seeberger, 2008; Paulson et al., 2006; Stevens et al., 2006). With hundreds of defined glycans presented and available simultaneously on the same microarray for interrogation, the specific structures or structural motifs recognized by glycan binding proteins (GBPs) or microorganisms can be rapidly discovered. Understanding these specificities and motifs provides important clues to glycan function. For example, the realization that certain galectins had high affinity for specific human blood groups, led to the discovery of their function as innate immune proteins (Stowell et al., 2010). However, the power of glycan microarrays to detect specificities and motifs of GBPs is directly related to the availability of diverse structures, which have been generated largely by chemical and chemo/enzymatic synthesis (Blixt et al., 2004) The current version of the Glycan Microarray that is publicly available through the NIGMS-funded Consortium for Functional Glycomics (CFG) (http://www.functionalglycomics.org/) is comprised of over 500 defined glycans; however, the human glycome is estimated to possess in excess of 7,000 glycan determinants (Cummings, 2009). Expanding the defined array is fraught with the difficulties associated with the chemical synthesis of large and complex oligosaccharides, and although an approachable task, other methods to enhance the diversity of these platforms are being explored.
An obvious source of glycans for this expansion is nature itself, which offers an inexhaustible supply of complex glycan structures, and since they are natural products made by complex genetic/biochemical pathways, they may be presumed to be biologically relevant. Mining these structures from nature, however, presents the biochemist with significant challenges, and to this end, we have developed chemical methods enabling us to acquire naturally occurring glycans on a microscale. Since these glycans are generally invisible to non-destructive detection methods and often available in only nanomole quantities, it is necessary to introduce linkers at their reducing ends that are fluorescent to provide sensitive detection, and chemically functionalized to permit immobilization on array surfaces (Song et al., 2009; Song et al., 2009; Xia et al., 2005) The resulting derivatives can then be quantified and printed in the same way as synthetic glycans with a primary amino group. The bifunctional fluorescent labeling of free glycans, as a complementary method to synthetic approaches, has shown great potential in increasing the size of the glycan library and facilitating the study of GBPs.
In addition to their use for expanding the defined glycans on glycan microarrays, the fluorescent glycan derivatives that also possess an alkyl primary amine function can be used for linking defined glycans to any type of derivatized surface for use in exploring glycan recognition. For example, fluorescent glycan derivatives can be coupled to biotin using commercially available NHS–biotin and the resulting reagent can be used for coupling glycan to any surface coated with streptavidin. Alternatively the glycans can be covalently coupled to epoxide or NHS-derivatized surfaces or to carboxylated microspheres.
Basic Protocol
Preparation of Closed ring Glycan-AEAB from free reducing glycans
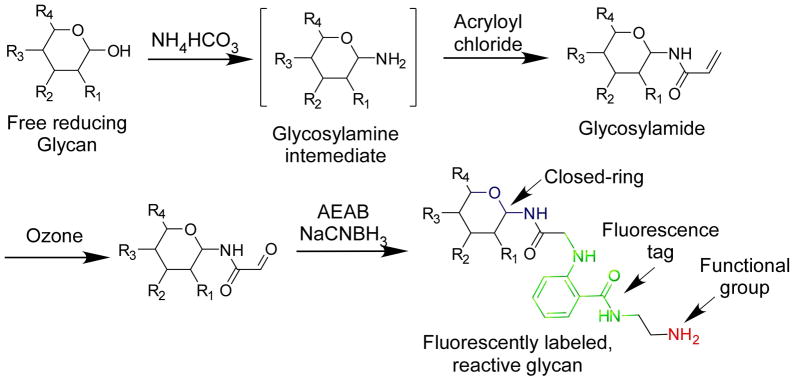
Even with efficient extraction and isolation methods, naturally occurring glycans are often only available in sub-milligram quantities. Many free reducing glycans are commercially available. Thus, to utilize such quantities of glycans in studies on their interactions with GBPs, the glycan amounts need to be accurately quantified and immobilized to solid phases, such as microarray chips. Bifunctional fluorescent tags such as 2,6-diaminopyridine (DAP) and 2-amino(N-aminoethyl) benzamide (AEAB) can be coupled to reducing glycans through reductive amination, and the resulting glycan conjugates are fluorescent and possess a primary amino group (Song et al., 2009; Xia et al., 2005). While the fluorescence greatly facilitates its separation and quantification, the amino group is used to immobilize the conjugates to solid surfaces for further functional study with GBPs. However, the direct reductive amination of free reducing glycans destroys the ring structure at the reducing end, which is desired in many situations. We therefore developed a microscale process combining the glycosylamide formation, ozonolysis, and AEAB reductive amination (summarized in Fig. 1), to prepare a glycan derivative retaining its ring structure (Song et al., 2009).
Fig. 1.
The closed-ring fluorescent conjugation of reducing glycans.
Materials
Solutions and reagents:
Free reducing glycan (0.05–1 mg lyophilized) (Commercially available from several suppliers such as Sigma-Aldrich, V-labs, Carbosynth etc.)
Filtered MilliQ Water
Ammonium bicarbonate
Acetonitrile (Fisher Scientific, HPLC grade)
Sodium bicarbonate
Acryloyl chloride
Trifluoroacetic acid (TFA) (Fisher Scientific, HPLC grade)
Methanol
Ethanol
Methyl sulfide
Ozone
Nitrogen
Dimethyl sulfoxide (DMSO) (Fisher Scientific, ACS grade)
Acetic acid (Fisher Scientific, ACS grade)
2-(N-aminoethyl)amino benzamide (AEAB) (Song et al., 2009)
Sodium cyanoborohydride
- All chemicals are analytical grade and used without further purification.
Special equipment:
Carbograph SPE columns (Alltech, 150mg, 300 mg and 1g)
Speed-vac
MALDI-TOF mass spectrometer
HPLC
Porous graphitized carbon (PGC) analytical HPLC column (Thermo Scientific)
Prepare AEAB conjugates
-
Dissolve free reducing glycan (0.05 mg – 1 mg) in water (50 μL) in a 1.5 mL screw cap tube. Ammonium bicarbonate (100 mg) is added. Incubate the mixture at 55°C for 1.5 hours and cool to room temperature.
Note: Normal polypropylene centrifuge tubes can be used as the reaction vessel throughout the protocol. Glassware is not necessary.
Note: For the carbograph desalting, the glycan size needs to be bigger than trisaccharide to ensure the absorption on carbograph.
Condition a small-size carbograph SPE column (150 mg, Alltech) using 50% acetonitrile with 10 mM ammonium bicarbonate (1 column volume, ~ 3 mL) followed by 10 mM aqueous ammonium bicarbonate (3 column volumes).
-
Use 1 mL 10 mM ammonium bicarbonate to transfer the glycan sample to the preconditioned carbograph column. Wash the column with 10 mM ammonium bicarbonate (2 column volumes) to remove salts. Elute the carbograph with 50% acetonitrile containing 10 mM ammonium bicarbonate (2 column volumes).
Note: For larger amounts of glycans, the reaction can be easily scaled up. For example, for 10 mg free reducing glycan, 1 mL water and 2 g ammonium bicarbonate are used. The reaction should be incubated in a 15 mL conical tube and a 1 g carbograph should be used in the desalting procedures.
Evaporate the glycosylamine eluate in 50% acetonitrile containing 10 mM ammonium bicarbonate in a Speed-vac for 2 hours to remove acetonitrile, and remove residual water and ammonium bicarbonate by freeze drying in a conical tube.
To the lyophilized powder in a 15 mL conical tube (sitting on ice), add 50 mg sodium bicarbonate and 0.5 mL ice-cold saturated sodium bicarbonate solution. Add acryloyl chloride (20 μL) and immediately cap the mixture and agitate by vortexing for 5 minutes. Slightly open the cap to release the pressure and close again. Then shake the mixture at room temperature for another 60 minutes.
-
(Optional) For a mixture of glycans, for example, N-glycans released from a glycoprotein by PNGase F, use a sodium borohydride reduction. For x mg glycan starting material, directly add x mg sodium borohydride (dissolved in water) under cooling on ice after the acryloylation reaction. Shake the reaction with frequent cooling for 10 minutes. Add acetic acid (2x μL) under cooling and incubate for 10 minutes.
Note: Due to the instability of the glycosylamine, a percentage of reducing glycan often occurs in the mixture after acryloylation. This portion of reducing glycan is also conjugated with AEAB to form the corresponding open-ring conjugate. While this is not a problem for a conjugation started with a single glycan, the purification by HPLC of a glycan mixture is somewhat more complicated. Thus, it is important to introduce the step of sodium borohydride reduction before the AEAB conjugation to solve this problem. This minor fraction of reduced glycans is not reactive with AEAB or DAP.
Condition a small-size carbograph SPE column (150 mg, Alltech) using 50% acetonitrile with 0.1% trifluoroacetic acid (TFA) (1 column volume, ~ 3 mL) followed by water (3 column volumes). Dissolve the acryloylated glycan from step 5 (or optional step 6) in 3 mL water and apply to the carbograph column. Wash the column with water (6 column volumes) and elute the glycosylamide with 50% acetonitrile and 0.1% TFA (2 column volumes). Evaporate the eluted solution by Speed-vac for 2 hours to remove acetonitrile, and remove water by freeze drying.
Dissolve the lyophilized material in 1 mL methanol and chill to −78°C (dry ice – ethanol). Bubble ozone through for ~1 minute until the blue color remains. Then turn off the ozone generator, bubble nitrogen through the solution for ~1 minute until the blue color disappears. Add 50 μL methyl sulfide. Let the mixture warm up to room temperature and stand for 1 hour.
-
Dry the solution under a stream of nitrogen. To the dried residue, add AEAB solution (0.35 M in DMSO/acetic acid = 7/3 (v/v), 10 μL – 50 μL) and equal-volume of sodium cyanoborohydride solution (1 M in DMSO/acetic acid = 7/3 (v/v)). A precipitate forms briefly after mixing. Heat the suspension at 65°C for 2 hours.
Note: Prepare the AEAB solution by adding 1 mL DMSO/acetic acid = 7/3 (v/v) to 88 mg AEAB hydrochloride and shake at room temperature for 10 minutes. After centrifugation, collect the supernatant and use it for conjugation. Prepare a solution of sodium cyanoborohydride by adding 1 mL DMSO/acetic acid = 7/3 (v/v) to 64 mg sodium cyanoborohydride.
-
Purify glycan-AEAB conjugates or the closed-ring conjugates from the reaction mixture by addition of 10 volumes acetonitrile, which causes the conjugates to precipitate. Transfer the mixture to a 1.5 mL Eppendorf tube and chill to −20°C. Collect the precipitated glycan conjugates after the tube is centrifuged at 10,000xg for 3 minutes. Remove the supernatant and discard. Dissolve the pellet in 100 μL water and perform MALDI-TOF analysis and HPLC purification.
Note: If closed-ring conjugates are not required, direct conjugation of AEAB starting with lyophilized or dry, free reducing glycans can be easily conducted by simply following steps 9 and 10. The HPLC purification procedure of open-ring and closed-ring AEAB conjugates is essentially identical, with the latter showing relatively longer retention times.
Purification of the derivatives
A Hypercarb (porous graphitized carbon) analytical HPLC column coupled with a Javelin guard cartridge is used to purify both closed-ring and open-ring glycan AEAB derivatives. The column has sufficient capacity and resolving power to purify conjugates at a scale of 1 μg to 1 mg.
-
The HPLC program for the purification of the conjugates:
-
Solvents:
Acetonitrile (A)
1% Trifluoroacetic acid in water (B)
Water (C)
Flow rate: 1 mL/min.
-
Linear gradient:
0 min: 15% A, 10% B and 75% C;
30 min, 45% A, 10% B and 45% C;
30.1 min: 15% A, 10% B and 75% C;
40 min: stop.
Fluorescence detection: 330 nm (Ex)/420 nm (Em).
UV detection: 330 nm.
-
-
The derivatives are usually found at retention time 10–30 minutes. Collect peaks, evaporate in a Speed-vac for 2 hours, and lyophilize to remove water. Based on its fluorescence and UV absorption, reconstitute each glycan or glycan fraction to 200 μM in water. This solution of closed ring AEAB derivative is suitable for printing in the appropriate buffer and may be stored at −20°C. The printing of microarray, however, is out of the scope of this chapter and can be found elsewhere in this series. (Smith et al., 2010).
Note: The quantification and reconstitution of the glycan-AEAB conjugates are based on standard curves generated from accurately weighed standard lactose-AEAB conjugate. Lactose- AEAB from 10 pmol to 1 nmol can be detected by fluorescence linearly and lactose-AEAB from 500 nmol can be detected by UV absorbance at 330nm linearly.
Alternate/Support Protocols
Prepare AEAB conjugates using commercial chemicals
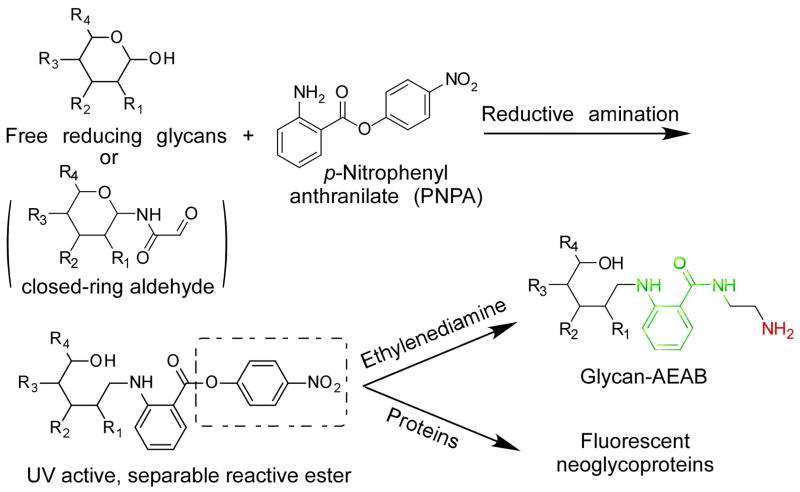
The microscale preparation of open-ring and closed-ring AEAB conjugates from free reducing glycans provides a quick and potentially high throughput way to obtain natural glycan derivatives that can be immobilized on microarrays for study of interactions with GBPs. The AEAB conjugates have a primary amino group as an active nucleophile. To expand the scope and application of glycan derivatives, we also developed another bifunctional tag, p-nitrophenyl anthranilate (PNPA) (Luyai et al., 2009). The preparation of glycan conjugates with reductive amination, as described above, can be readily applied to the conjugation of PNPA. However, the PNPA derivatives have a p-nitrophenyl ester as an active leaving group, which can react easily with nucleophiles. Here we describe a protocol for PNPA conjugation and the conversion of PNPA derivatives to AEAB conjugates (Fig. 2).
Fig. 2.
The preparation of AEAB conjugates from PNPA conjugation.
Materials
Solution and Reagents:
DMSO
Acetic acid
p-Nitrophenyl anthranilate (Fisher Scientific, 98%)
Sodium cyanoborohydride (Sigma-Aldrich, 95%)
Ethylenediamine
Acetonitrile
Water
Trifluoroacetic acid
Special equipment:
HPLC
C18 analytical column
MALDI-TOF
Prepare PNPA conjugates
-
To free reducing glycan (0.05 – 1 mg), add a solution of PNPA (0.35 M in DMSO/acetic acid = 7/3 (v/v), 10 μL – 50 μL) and an equal-volume of sodium cyanoborohydride solution (1 M in DMSO/acetic acid = 7/3 (v/v)). Heat the mixture at 65°C for 2 hours.
Note: PNPA solution is prepared by adding 1 mL DMSO/acetic acid = 7/3 (v/v) to 90 mg PNPA. While the protocol is for reducing glycans, it can also be used for the ozone-treated closed ring glycan derivatives.
Precipitate the conjugates by addition of 10 volumes of acetonitrile. Transfer the mixture to a 1.5 mL Eppendorf tube and cool down at −20°C. Centrifuge the tube at 10,000xg for 3 minutes. Remove the supernatant and discard. Dissolve the pellet in 100 μL water and it is suitably ready for MALDI-TOF analysis, HPLC purification, or direct conjugation to proteins.
To convert PNPA conjugates to AEAB conjugates, add 25 μL ethylenediamine solution in DMSO (dissolve 100 μL ethylenediamine in 1 mL DMSO) to the pellet until the pellet is dissolved. The reaction is completed immediately during this process. Add 75 μL 10% acetic acid in water. The mixture is ready for HPLC purification.
Purification of the derivatives
AEAB conjugates transformed from PNPA conjugates can be purified using Hypercarb-HPLC as described above. The PNPA conjugates can also be purified on a C18-HPLC column prior to the reaction with ethylenediamine, due to the greater hydrophobicity of the PNPA tag. A normal C18 analytical column (4.6mm x 250mm) has sufficient capacity and resolving power to purify conjugates at a scale of 1 μg to 1 mg.
-
The HPLC program for the purification of the PNPA conjugates:
-
Solvents:
Acetonitrile (A)
1% Trifluoroacetic acid in water (B)
Water (C)
Flow rate: 1 mL/min.
-
Linear gradient:
0 min: 1% A, 10% B and 89% C;
30 min, 90% A, 10% B and 0% C;
30.1 min: 1% A, 10% B and 89% C;
40 min: stop.
UV detection: 330 nm.
-
The PNPA derivative is usually found at retention time 10–20 minutes. Collect peaks, evaporate in a Speed-vac for 2 hours, and remove the residual water by freeze drying. Quantify the conjugates based on UV absorption in comparison with a standard glycan-PNPA derivative such as lactose-PNPA of known concentration.
Commentary
Background information
Due to the lack of suitable chromophores for spectrometric detection of carbohydrates, their analysis had historically relied on chemical colorimetric methods such as phenol-sulfuric acid assay (Dubois et al., 1951), which are destructive and relatively insensitive. The derivatization of free reducing glycans with fluorescent small molecules by reductive amination has greatly facilitated the HPLC profiling of glycans (Bigge et al., 1995). Other than the alteration at the reducing end, these reaction products retain the structures of the derivatized glycans, but without a functional group these conjugates are of limited practical value in small scale other than use in structural analyses by mass spectrometry. On the other hand, generating conjugates that have, in addition to fluorescent properties, a functional group that does not severely alter chromatographic properties, provides a derivative that can be quantified with high sensitivity and coupled to derivatized surfaces. Such an approach has an obvious application for derivatizing larger, biologically-relevant glycans from natural glycans to construct libraries to extend the diversity of defined, synthetic glycan microarrays. For this reason we have investigated a variety of methods for preparing bifunctional fluorescent labeling reagents (Song et al., 2009; Song et al., 2009; Song et al., 2009; Xia et al., 2005). Here we describe the utility of AEAB, which imparts fluorescence properties, and also has two functional amino groups. Due to the different reactivities of the alkyl amine and the aryl amine of AEAB, the resulting glycan-AEAB conjugate can be prepared as a pure primary alkyl amine, suitable for immobilization both epoxy- and NHS-coated glass slides. Since some protein-carbohydrate interactions may require a natural, acetyl or ring structure, we developed a more sophisticated approach by introducing the glycosylamine formation to generate the natural ring structure at the reducing end. Several high yielding chemical reactions, including glycosylamine formation, amidation, ozonolysis, and reductive amination have been elegantly combined to provide a microscale preparation of fluorescent and immobilizable glycan conjugates.
Critical Parameters and Troubleshooting
Commonly used polypropylene tubes including microcentrifuge tubes can be used throughout the protocol with no need for glassware. AEAB is prepared as the HCl salts. It reacts with free aldehydes through the aryl amine function making the more reactive alkyl amine available for subsequent reactions or covalent coupling to activated surfaces. It is important to remove all free AEAB from the glycan-AEAB derivative to ensure its accurate quantification and efficient coupling the glycan derivative to an activated surface. For defined glycan arrays, it is imperative that the glycan-AEAB conjugate is comprised of a single, defined glycan or be at least >95% free of contaminating structures. This can be accomplished by starting the process with a purified, structurally defined glycan or by derivatizing a mixture of glycans and separating the glycan-AEAB by chromatographic procedures to derive the principal components. Purification of either of the AEAB derivatives is accomplished by 2-dimensional HPLC separations as described above, and this is greatly facilitated by their fluorescent properties. It is recommended that a normal phase separation be used for the first dimension and the PGC be used as the second dimension. PGC is a convenient step in the second dimension because elution is with acetonitrile/water without a non-volatile counter ion so that the resulting peaks may be collected and dried to obtain salt-free fractions for MALDI analysis or for direct coupling or other reactions.
The retention of glycan structures on carbograph SPE column used for desalting varies greatly among different glycans, with larger and charged glycans generally being absorbed much stronger. Therefore, this procedure is not suitable for preparation of mono- and disaccharide conjugates. Furthermore, highly charged, especially highly sulfated, oligosaccharides such as heparin oligosaccharides are not compatible with either carbograph SPE or PGC-HPLC, presumably due to extremely strong retention. The capacity of carbograph SPE columns is approximately 100 mg carbon/1 mg glycan.
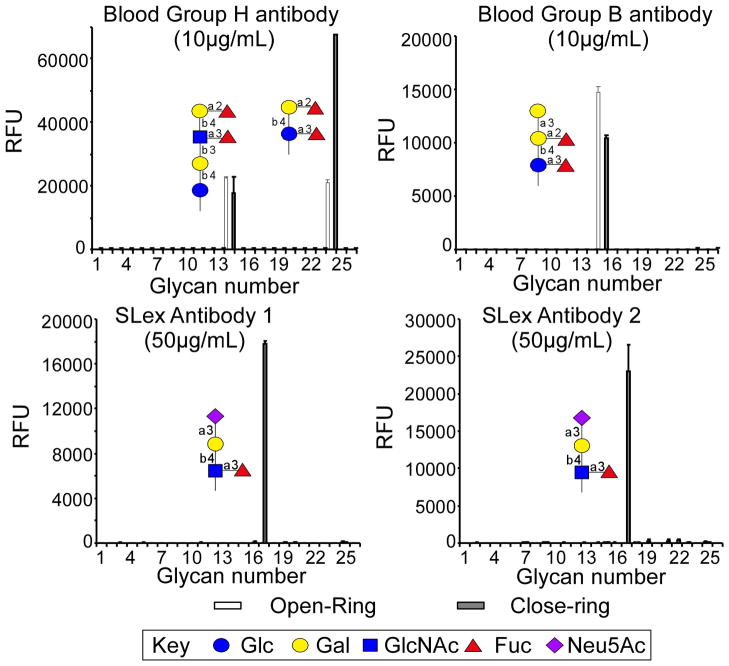
Anticipated Results
The purified “closed-ring” and “open-ring” conjugates can be treated as any other synthetic material with a primary alkyl amine group. Their reactivity toward both epoxy- and NHS-derivatized surfaces is high. Although the printing, assay and data analysis of glycan microarray is not within the scope of this Unit, Fig. 3 shows several histograms that demonstrate the comparison of 4 antibodies binding to a glycan microarray printed with 26 different open-ring Glycan-AEAB derivatives and 26 corresponding closed-ring Glycan-AEAB derivatives (Song et al., 2009). The structures of the bound glycans are indicated using symbols in each panel. In the case of the human anti-H and anti-B blood group antibodies, the binding is apparently directed more toward the non-reducing end of the oligosaccharide as binding is observed with both the natural closed-ring and open-ring structure. However in the case of the two anti-sialyl-Lewis x (SLex) antibodies, the open-ring GlcNAc of the AEAB derivative destroys the glycan epitope. Thus, if the binding motif of a GBP is at the non-reducing end of the glycan, which is generally the case, both types of conjugates show similar binding. However, if the binding motif is close to or includes the reducing end, the open-ring AEAB conjugates may not be appropriate for binding assays.
Fig. 3.
Comparison of 4 different antibodies binding to open-ring and closed-ring AEAB conjugates. The arrays were printed using a piezo printer (Perkin Elmer) with open- and closed-ring AEAB derivatives at 300 μM on NHS-derivatized slides. Antibodies were applied to the glycan array at the indicated concentrations above and detected with appropriate fluorescently labeled secondary antibodies (Song et al., 2009). The X-axis represents different glycans and the Y-axis represents the relative fluorescent unit (RFU) detected on the microarray.
Time Considerations
With the available reagents, the procedure for preparing 1 to 20 glycans can be completed in 3–4 days. This includes scheduling the freeze drying overnight between steps. The procedure can be shortened significantly if the drying steps are concluded more quickly. In addition, multiple parallel syntheses can be carried out at the same time, which will greatly increase the efficiency. Testing the glycan array with GBPs, although not covered in this unit, takes 3–4 hours depending on the number of samples and number of steps to detect the GBPs (Smith et al., 2010).
Literature Cited
- Alvarez RA, Blixt O. Identification of ligand specificities for glycan-binding proteins using glycan arrays. Methods Enzymol. 2006;415:292–310. doi: 10.1016/S0076-6879(06)15018-1. [DOI] [PubMed] [Google Scholar]
- Bigge JC, Patel TP, Bruce JA, Goulding PN, Charles SM, Parekh RB. Nonselective and efficient fluorescent labeling of glycans using 2-amino benzamide and anthranilic acid. Anal Biochem. 1995;230:229–38. doi: 10.1006/abio.1995.1468. [DOI] [PubMed] [Google Scholar]
- Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, Razi N, Stevens DJ, Skehel JJ, van Die I, Burton DR, Wilson IA, Cummings R, Bovin N, Wong CH, Paulson JC. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci U S A. 2004;101:17033–8. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings RD. The repertoire of glycan determinants in the human glycome. Mol Biosyst. 2009;5:1087–104. doi: 10.1039/b907931a. [DOI] [PubMed] [Google Scholar]
- de Paz JL, Seeberger PH. Recent advances in carbohydrate microarrays. QSAR & Combinatorial Science. 2006;25:1027–1032. [Google Scholar]
- Dubois M, Gilles K, Hamilton JK, Rebers PA, Smith F. A colorimetric method for the determination of sugars. Nature. 1951;168:167. doi: 10.1038/168167a0. [DOI] [PubMed] [Google Scholar]
- Feizi T, Chai W. Oligosaccharide microarrays to decipher the glyco code. Nat Rev Mol Cell Biol. 2004;5:582–8. doi: 10.1038/nrm1428. [DOI] [PubMed] [Google Scholar]
- Horlacher T, Seeberger PH. Carbohydrate arrays as tools for research and diagnostics. Chem Soc Rev. 2008;37:1414–22. doi: 10.1039/b708016f. [DOI] [PubMed] [Google Scholar]
- Luyai A, Lasanajak Y, Smith DF, Cummings RD, Song X. Facile Preparation of Fluorescent Neoglycoproteins Using p-Nitrophenyl Anthranilate as a Heterobifunctional Linker. Bioconjug Chem. 2009 doi: 10.1021/bc900189h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson JC, Blixt O, Collins BE. Sweet spots in functional glycomics. Nat Chem Biol. 2006;2:238–48. doi: 10.1038/nchembio785. [DOI] [PubMed] [Google Scholar]
- Smith DF, Song X, Cummings RD. Use of glycan microarrays to explore specificity of glycan-binding proteins. Methods Enzymol. 2010;480:417–44. doi: 10.1016/S0076-6879(10)80033-3. [DOI] [PubMed] [Google Scholar]
- Song X, Lasanajak Y, Rivera-Marrero C, Luyai A, Willard M, Smith DF, Cummings RD. Generation of a natural glycan microarray using 9-fluorenylmethyl chloroformate (FmocCl) as a cleavable fluorescent tag. Anal Biochem. 2009;395:151–60. doi: 10.1016/j.ab.2009.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Lasanajak Y, Xia B, Smith DF, Cummings RD. Fluorescent glycosylamides produced by microscale derivatization of free glycans for natural glycan microarrays. ACS Chem Biol. 2009;4:741–50. doi: 10.1021/cb900067h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Xia B, Stowell SR, Lasanajak Y, Smith DF, Cummings RD. Novel fluorescent glycan microarray strategy reveals ligands for galectins. Chem Biol. 2009;16:36–47. doi: 10.1016/j.chembiol.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J, Blixt O, Paulson JC, Wilson IA. Glycan microarray technologies: tools to survey host specificity of influenza viruses. Nat Rev Microbiol. 2006;4:857–64. doi: 10.1038/nrmicro1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowell SR, Arthur CM, Dias-Baruffi M, Rodrigues LC, Gourdine JP, Heimburg-Molinaro J, Ju T, Molinaro RJ, Rivera-Marrero C, Xia B, Smith DF, Cummings RD. Innate immune lectins kill bacteria expressing blood group antigen. Nat Med. 2010;16:295–301. doi: 10.1038/nm.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B, Kawar ZS, Ju T, Alvarez RA, Sachdev GP, Cummings RD. Versatile fluorescent derivatization of glycans for glycomic analysis. Nat Methods. 2005;2:845–50. doi: 10.1038/nmeth808. [DOI] [PubMed] [Google Scholar]