Abstract
Telomeres protect the normal ends of chromosomes from being recognized as deleterious DNA double-strand breaks. Recent studies have uncovered an apparent paradox: although DNA repair is prevented, several proteins involved in DNA damage processing and checkpoint responses are recruited to telomeres in every cell cycle and are required for end protection1. It is currently not understood how telomeres prevent DNA damage responses from causing permanent cell cycle arrest. Here we show that fission yeast (Schizosaccharomyces pombe) cells lacking Taz1, an orthologue of human TRF1 and TRF2 (ref. 2), recruit DNA repair proteins (Rad22RAD52 and Rhp51RAD51, where the superscript indicates the human orthologue) and checkpoint sensors (RPA, Rad9, Rad26ATRIP and Cut5/Rad4TOPBP1) to telomeres. Despite this, telomeres fail to accumulate the checkpoint mediator Crb253BP1 and, consequently, do not activate Chk1-dependent cell cycle arrest. Artificially recruiting Crb253BP1 to taz1Δ telomeres results in a full checkpoint response and cell cycle arrest. Stable association of Crb253BP1 to DNA double-strand breaks requires two independent histone modifications: H4 dimethylation at lysine 20 (H4K20me2) and H2A carboxy-terminal phosphorylation (γH2A)3–5. Whereas γH2A can be readily detected, telomeres lack H4K20me2, in contrast to internal chromosome locations. Blocking checkpoint signal transduction at telomeres requires Pot1 and Ccq1, and loss of either Pot1 or Ccq1 from telomeres leads to Crb253BP1 foci formation, Chk1 activation and cell cycle arrest. Thus, telomeres constitute a chromatin-privileged region of the chromosomes that lack essential epigenetic markers for DNA damage response amplification and cell cycle arrest. Because the protein kinases ATM and ATR must associate with telomeres in each S phase to recruit telomerase6, exclusion of Crb253BP1 has a critical role in preventing telomeres from triggering cell cycle arrest.
Taz1 is required for proper telomere DNA replication and prevents end-joining reactions at chromosome ends that occur in the G1 phase of the cell cycle7–9. As fission yeast spend most of the cell cycle in the S and G2 phases, chromosome-end fusions and loss of viability are undetectable in taz1Δ mutants. Despite this, taz1Δ chromosome ends undergo frequent rearrangements10. To determine the nature of these rearrangements, we used a combination of live-cell imaging and chromatin immunoprecipitation (ChIP) to monitor the recruitment of various DNA repair and checkpoint proteins to telomeres in taz1Δ cells.
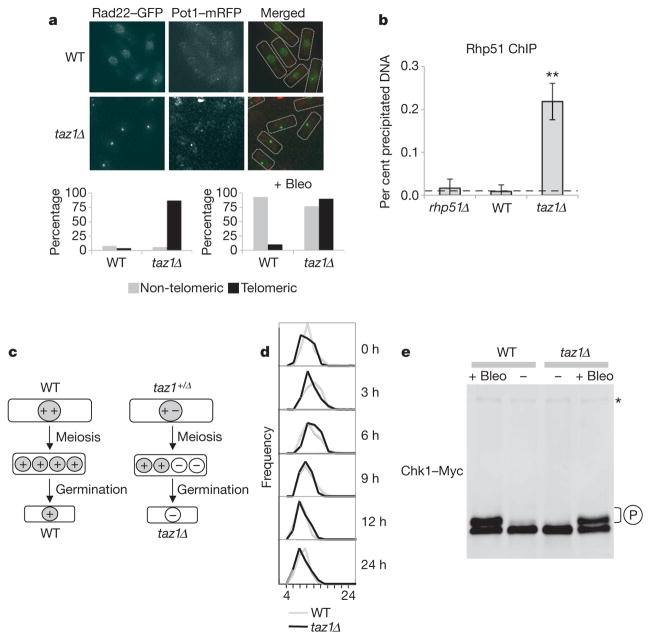
We endogenously tagged the homologous recombination protein Rad22RAD52 with green fluorescent protein (GFP) and, using Pot1 tagged with monomeric red fluorescent protein (mRFP) as a marker for telomeres (Supplementary Fig. 1), we studied its co-localization to chromosome ends (Fig. 1a). Few wild-type cells have Rad22–GFP foci at non-telomeric sites but these become prevalent when exposed to the radiomimetic drug bleomycin. In contrast, taz1Δ cells had Rad22–GFP foci at telomeres throughout the cell cycle (Fig. 1a). Using ChIP, we also found that taz1Δ cells strongly accumulate another homologous recombination repair protein, Rhp51RAD51, at telomeres (Fig. 1b). Thus, DNA repair is ongoing at taz1Δ chromosome ends.
Figure 1. taz1Δ telomeres undergo the DNA damage response without eliciting a checkpoint-dependent cell cycle arrest.
a, Live analysis of Rad22RAD52–GFP and its co-localization with Pot1–mRFP used as a telomeric marker. WT, wild type. Bleo, bleomycin. b, Dot-blot ChIP quantification of Rhp51RAD51 bound to telomeres for the indicated strains. rhp51Δ cells were used as a control. n ≥ 3; ** P < 0.01 based on a two-tailed Student’s t-test to control sample. Error bars represent mean ± standard deviation (s.d.). c, Strategy to generate taz1Δ mutants de novo. d, Histograms depict the distribution of cell sizes (μm) after germination over the course of the experiment. e, Immunoblot analysis of Chk1–Myc in wild-type and taz1Δ cells. The asterisk depicts an antibody cross-reactive band used as a loading control. The DNA damaging agent bleomycin (Bleo; 5 mU ml−1) was added to cultures as a positive control. P indicates hyperphosphorylation.
DNA damage initiates signal transduction pathways, known as checkpoints, which culminate in cell cycle arrest. Telomeres protect chromosome ends from DNA repair and do not induce cell cycle arrest via checkpoint pathways. Unexpectedly, taz1Δ telomeres undergo the DNA damage response and yet these cells did not show any cell cycle delay. We were also able to rule out adaptation phenomena by analysing newly generated taz1Δ mutants; they were also unable to elicit a checkpoint response (Fig. 1c, d).
Next we examined the phosphorylation status of the Chk1 protein kinase, a well-characterized checkpoint target responsible for inhibiting Cdk1 and preventing entry into mitosis in response to DNA damage. Consistent with a lack of checkpoint activation in taz1Δ cells, phosphorylated Chk1–Myc was absent in taz1Δ cell extracts (Fig. 1e). As expected, exposure to bleomycin caused strong Chk1 phosphorylation in both wild-type and taz1Δ cells (Fig. 1e). We performed similar experiments to investigate the phosphorylation status of Cds1, which is involved in DNA-replication checkpoints, and failed to detect any activation of this branch of the checkpoint pathway unless hydroxyurea was added (Supplementary Fig. 2). Thus, even though taz1Δ cells are checkpoint proficient, they are unable to arrest the cell cycle in response to telomeres undergoing the DNA damage response.
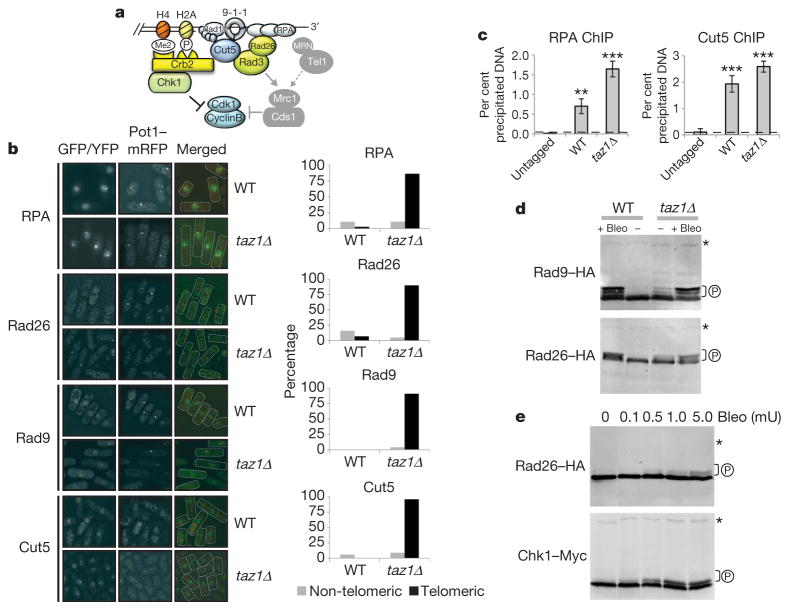
The absence of cell cycle delay prompted us to examine the checkpoint pathway to identify at which step it was disrupted. Telomeres in taz1Δ cells are long and accumulate single-stranded DNA throughout the cell cycle11. We started probing the checkpoint pathway (Fig. 2a) at its upstream point by looking for the accumulation of RPA at telomeres by co-localization of Rad11–GFP, the largest subunit of RPA, with Pot1–mRFP. Pot1 protects telomeres from triggering cell cycle arrest via the ATR checkpoint pathway12–14. It has been proposed that Pot1 accomplishes this function directly by outcompeting RPA from telomere ends, as both molecules bind to telomeric single-stranded DNA13. Surprisingly, Pot1–mRFP co-localized with Rad11–GFP in taz1Δ cells (Fig. 2b), indicating that Pot1 does not abrogate checkpoint signalling by exclusion of RPA from dysfunctional telomeres. We quantitatively confirmed increased recruitment of RPA to telomeres in taz1Δ compared with wild-type cells using ChIP (Fig. 2c). Because telomere-associated RPA greatly increases during S phase15, we conclude that we are able to measure protein interaction with telomeres in S-phase cells using an asynchronously growing population.
Figure 2. taz1Δ telomeres initiate a DNA damage checkpoint response.
a, Checkpoint pathways in fission yeast. DNA-damage checkpoint (left) and replication checkpoint (shaded on the right). b, Co-localization of Rad11RPA–GFP, Rad26ATRIP–GFP, Rad9–YFP, Cut5TOPBP1–GFP with Pot1–mRFP (used as a telomere marker) (left) and quantification of cells showing telomeric and non-telomeric foci (right). c, Telomeric dot-blot ChIP quantification of Rad11RPA–Flag and Cut5TOPBP1–Myc. Untagged strains were used as controls. n ≥ 3; ** P < 0.01 and *** P < 0.001 based on a two-tailed Student’s t-test to control sample. Error bars represent mean ± s.d. d, Immunoblot analysis of Rad9–HA and Rad26ATRIP–HA. Bleomycin (5 mU ml−1) was added to cultures as a positive control. e, Immunoblot analysis of Rad26ATRIP–HA and Chk1–Myc in wild-type cells treated with increasing concentrations of bleomycin. Asterisks point to a cross-reactive band that serves as a loading control. P indicates hyperphosphorylation.
Next we analysed Rad26ATRIP, Rad9 (a component of the 9-1-1 complex) and Cut5/Rad4TOPBP1. In contrast to wild type, the majority of taz1Δ cells had telomeric foci for all three components (Fig. 2b), indicating that taz1Δ dysfunctional telomeres show increased recruitment for components of the DNA-damage checkpoint pathway. ChIP analysis revealed that Cut5TOPBP1 is also strongly recruited to wild-type telomeres (Fig. 2c), consistent with previous observations that telomeres normally engage DNA damage response components15,16. Rad3ATR kinase was active in growing taz1Δ cells, because hyperphosphorylation of its known substrates, Rad26ATRIP and Rad9, was detected in untreated taz1Δ extracts as in control cells exposed to bleomycin (Fig. 2d). Further analysis revealed that phosphorylation levels of haemagglutinin (HA) tagged Rad26ATRIP observed in taz1Δ cells were otherwise sufficient to trigger cell cycle arrest. Exposure to increasing doses of bleomycin revealed that, at levels of DNA damage where phosphorylation of Rad26ATRIP–HA becomes detectable (0.5–1.0 mU bleomycin; Fig. 2e), Chk1–Myc is clearly hyperphosphorylated. Thus, Rad3ATR activity equivalent to that observed in taz1Δ cells is sufficient to activate a full checkpoint response if DNA damage occurs at non-telomeric regions.
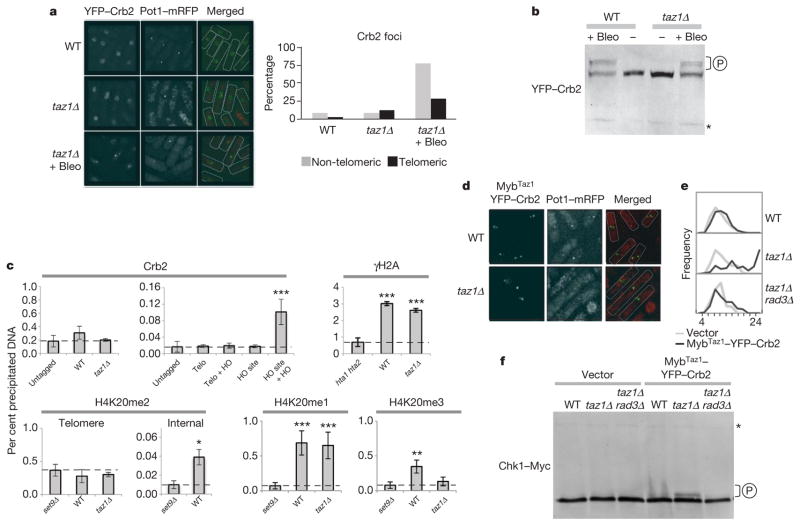
Upon Rad3ATR activation and Cut5TOPBP1 recruitment, Crb253BP1 is brought to sites of DNA damage and, on phosphorylation by Rad3ATR, induces activation of the Chk1 kinase and cell cycle arrest17. Unlike other upstream checkpoint proteins, yellow fluorescent protein (YFP)-tagged Crb2 protein did not co-localize with Pot1–mRFP in most taz1Δ cells (Fig. 3a). In accordance with this, Crb2 was not phosphorylated in taz1Δ cells (Fig. 3b), even though treatment with bleomycin strongly induces Crb2 phosphorylation. We were also unable to detect telomere-bound tandem affinity purification (TAP)-tagged Crb2 in either wild-type or taz1Δ cells using ChIP, whereas controls showed clear recruitment of TAP–Crb2 to an HO-endonuclease-induced DNA double-strand break (Fig. 3c). Therefore, unlike other DNA damage response components, Crb2 is unable to stably bind to telomeres in either wild-type or taz1Δ cells, indicating that the checkpoint signalling pathway is blocked at the step between Cut5TOPBP1 and Crb253BP1.
Figure 3. Dysfunctional taz1Δ telomeres avert cell cycle arrest by preventing recruitment of Crb253BP1.
a, Visualization and quantification of YFP–Crb2 foci in the indicated strains. b, Immunoblot analysis of YFP–Crb2. Bleomycin (5 mU ml−1) was added to cultures where indicated (+). c, Telomeric dot-blot ChIP quantification for Crb253BP1–TAP, γH2A, H4K20me1, H4K20me2 and H4K20me3. Positive controls: quantitative polymerase chain reaction (qPCR) ChIP was performed for TAP–Crb253BP1 using primers adjacent to an internal double-strand break generated by HO cleavage (HO site) and to wild-type telomeres (Telo) and for H4K20me2 using primers for the ade6+ locus (Internal). For negative controls we used H2A phosphorylation mutants hta1-S129A hta2-S128A (hta1 hta2) for γH2A and set9Δ for all forms of H4K20 methylation. n ≥ 3; * P < 0.05, ** P < 0.01 and *** P < 0.001 based on a two-tailed Student’s t-test to controls. Error bars represent mean ± s.d. d, Localization of MybTaz1–YFP–Crb2 expressed from a low expression vector (pREP81). e, Distribution of sizes (μm) in cells carrying either MybTaz1–YFP–Crb2 or an empty plasmid control. f, Immunoblot analysis of Chk1–Myc. P indicates hyperphosphorylation. Asterisk indicates a cross-reactive band that serves as a loading control.
We next investigated what prevents the stable association of Crb253BP1 with telomeres. Apart from Cut5, Crb2 requires two histone modifications for stable association to double-strand breaks: C-terminal phosphorylation of γH2A (γH2AX in higher eukaryotes) and H4K20me2 (refs 3–5). These modifications show different responses to DNA damage: whereas γH2A/X is locally phosphorylated by ATM or ATR at DNA double-strand breaks, H4K20me2 levels do not change on DNA damage18. Instead, H4K20me2 seems to be ubiquitously distributed even though it participates in the DNA damage response and is essential for sustained checkpoint activation19. Consistent with the observed recruitment of Rad3ATR–Rad26ATRIP to telomeres, we readily detected γH2A by ChIP at both wild-type and taz1Δ telomeres (γH2A; Fig. 3c). In contrast, H4K20me2 was undetectable at telomeres in either wild-type or taz1Δ cells (H4K20me2 telomere; Fig. 3c and Supplementary Fig. 3). The absence of H4K20me2 could not be explained by exclusion of the methyltransferase Set9/Kmt5 from chromosome ends. Telomeres show Set9-dependent mono- and trimethylated forms of H4K20 even though taz1Δ telomeres lacked the latter (H4K20me1 and H4K20me3; Fig. 3c). Similarly, Clr4 methyltransferase, responsible for H3K9 methylation, is not required to maintain the pattern of H4K20 methylation, despite a marked reduction in H4K20me3 (Supplementary Fig. 3). Thus, although H3K9me3 inversely correlates with H4K20me2 at fission yeast telomeres, these are likely to be independently regulated.
Because checkpoint signalling seems to be achieved through the exclusion of Crb2, we reasoned that artificial recruitment of Crb2 to taz1Δ telomeres might be sufficient to induce cell cycle arrest. Taz1 binds to telomeres via its C-terminal Myb domain (MybTaz1); MybTaz1 has been previously used to tether Rap1 directly to taz1Δ telomeres20,21. Likewise, we ectopically expressed a MybTaz1–YFP–Crb2 chimaeric protein in fission yeast. On induction of MybTaz1–YFP–Crb2, taz1Δ cells became extremely elongated (Fig. 3d, e), denoting a full checkpoint response confirmed by Chk1 phosphorylation (Fig. 3f). None of these phenomena was visible in wild-type cells expressing the chimaeric protein (Fig. 3e, f). Also consistent with a checkpoint response, recruitment of MybTaz1–YFP–Crb2 to telomeres in taz1Δ rad3Δ cells resulted in neither cell elongation nor Chk1–Myc phosphorylation (Fig. 3e, f). These data demonstrate that Crb2 recruitment to telomeres in taz1Δ cells is sufficient to restore the full checkpoint pathway.
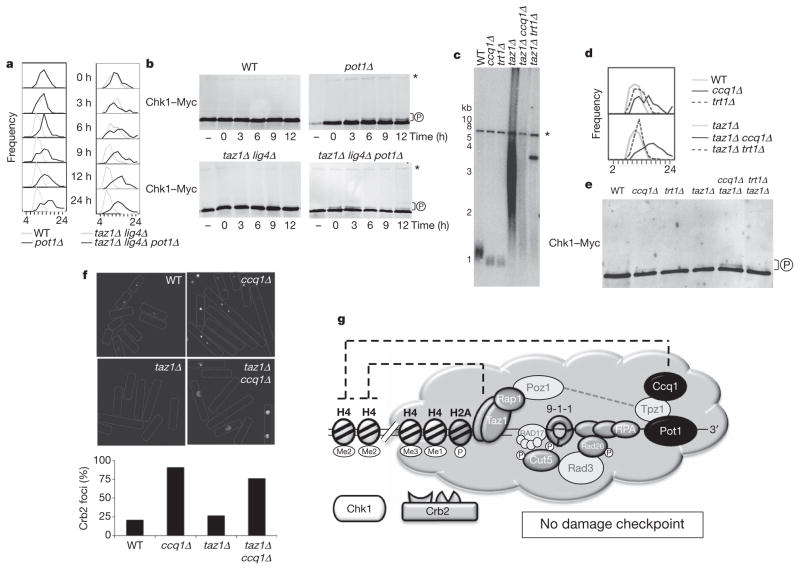
Next we investigated what marks telomeres different from the rest of the genome. Pot1 and its interacting protein Ccq1 are likely candidates as they bind telomeres independently of Taz1 and prevent checkpoints at telomeres12–14,22. Indeed, in contrast to wild type, both germinating pot1Δ and taz1Δ lig4Δ pot1Δ cells derived from heterozygous diploids became elongated and presented Chk1–Myc phosphorylation (Fig. 4a, b). Cell elongation was dependent on Rad3ATR, indicating an activation of this checkpoint pathway (Supplementary Fig. 4). Therefore, Pot1 is required to prevent telomeres from eliciting Rad3ATR-dependent checkpoints not only in wild-type but also in taz1Δ cells. Ccq1 is required for telomerase recruitment and inhibition of Rad3ATR checkpoints at fission yeast telomeres22,23. Loss of Ccq1 results in progressive telomere shortening and checkpoint activation before complete telomere erosion. Pre-senescent ccq1Δ and trt1Δ single mutants have comparable short telomeres that initiate checkpoints on telomere erosion (Fig. 4c–e). Likewise, ccq1Δ taz1Δ and trt1Δ taz1Δ double mutants undergo ALT-like telomere elongation, preserving long and heterogenous telomeres (Fig. 4c). However, in contrast to trt1Δ taz1Δ double mutants, ccq1Δ taz1Δ mutants had a strong immediate checkpoint response, becoming extremely elongated with strong Chk1 phosphorylation (Fig. 4d, e). As expected, quantification of plasmid-borne YFP–Crb2 revealed that both ccq1Δ and ccq1Δ taz1Δ cells had YFP–Crb2 foci in the majority of observed cells (Fig. 4f). Moreover, ccq1Δ telomere deprotection partially requires H4K20 methylation. Preventing H4K20 methylation at telomeres by removing Set9 from either ccq1Δ or ccq1Δ taz1Δ cells reduces checkpoint activation and YFP–Crb2 foci formation (Supplementary Fig. 5). Thus, absence of the Pot1–Ccq1 complex results in prompt Crb2 foci formation, Chk1–Myc phosphorylation and cell cycle arrest, indicating that Crb2 telomere exclusion constitutes a barrier imposed by normal telomeres to a full checkpoint response. However, loss of Ccq1 in wild-type cells results in a weaker and delayed checkpoint response than in taz1Δ mutants, establishing that, in addition to Pot1–Ccq1, the Taz1 complex participates in checkpoint inhibition at normal telomeres (Fig. 4d, e).
Figure 4. Pot1 and Ccq1 prevent Crb253BP1-dependent checkpoints at telomeres.
a, Distribution of cell sizes over time (μm) after germination of spores with the indicated genotype. b, Immunoblot analysis of Chk1–Myc. Extracts derived from diploid cells are indicated with a dash. P indicates hyperphosphorylation. Asterisks indicate a cross-reactive band used as a loading control. c, Telomere-length analysis by Southern blotting using a telomere probe. d, Distribution of cell sizes (μm) in the absence of either Ccq1 or Trt1 in wild-type and taz1Δ cells. e, Immunoblot analysis of Chk1–Myc. f, Visualization and quantification of plasmid borne YFP–Crb253BP1 foci. g, Model for checkpoint inhibition at fission yeast telomeres. Pot1 and Ccq1, together with the Taz1 complex, define a chromatin-privileged region that excludes H4K20me2 and prevents stable Cbr253BP1 association. As a result, the full checkpoint response is severed at telomeres and cell cycle arrest is averted.
Our work provides new insight into how chromosome ends are protected from DNA damage checkpoints (Fig. 4g). Instead of preventing the detection of DNA damage, fission yeast telomeres constitute a chromatin-privileged region on the chromosome that blocks transduction of an active checkpoint signal. This is achieved by preventing stable association of Crb2 with telomeres, a checkpoint adaptor protein required for the delivery and activation of Chk1. Studies in budding yeast using a short telomere seed adjacent to a HO break postulated an anticheckpoint activity at telomeres24. Following studies showed that checkpoint repression in budding yeast is primarily regulated by the inhibition of Mec1ATR recruitment to telomeres25–27. Fission yeast telomeres initiate a checkpoint response that fails to phosphorylate Crb2 and Chk1. However, rather than preventing recruitment and activation of Rad3ATR, exclusion of Crb253BP1 from telomeres represents the critical step in the inhibition of the checkpoint pathway.
The inability of telomeres to stably recruit Crb253BP1 is probably due to the lack of H4K20me2 epigenetic marks at chromosome ends. Studies in both mammalian cells and in fission yeast demonstrate that H4K20me2 is the relevant histone modification for Crb2 binding and is required for the stable recruitment of Crb2 to sites of DNA damage3,4,19,28. Although H4K20me2 is the most abundant form of H4K20me18, how it is regulated or localized is not understood. Our finding that telomeres have low levels of H4K20me2 provides a rationale for its ubiquitous nature throughout the genome. Conceivably, H4K20me2 marks regions of the genome required for chromosome contiguity, and cell division should not be attempted unless the DNA damage response is disengaged. In contrast, the absence of H4K20me2 marks regions where DNA perceived as damaged, such as chromosome ends, would not interfere with genome stability thus precluding a full checkpoint response.
Normal telomeres undergoing DNA replication temporarily lose DNA protection. Studies in both yeast model systems and mammalian cells show that during S phase, concomitant with the replication fork passage, chromosome ends engage ATR and ATM15,16,29,30, which are redundantly required for telomerase recruitment6. However, the periodic activation of ATM and ATR at telomeres does not result in either Chk1 or Chk2 phosphorylation or in cell cycle delay. As dividing cells use cell-cycle-staged telomere exposure to process chromosome ends and engage telomerase, it is likely that these mechanisms have evolved to prevent cell cycle arrest in response to normal replicating telomeres.
METHODS SUMMARY
All experiments were performed using the Schizosaccharomyces pombe strains listed in Supplementary Information. Checkpoint studies were performed using a combination of microscopy, ChIP and western blotting. Telomere length was analysed by Southern blotting using telomere probes. For details of the methods used, see Methods.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Material
Acknowledgments
We thank J. Cooper, K. Tomita and the rest of the Telomere Biology Laboratory (Cancer Research UK, London) for support at the start of this project. We thank J. Cooper, T. Wolkow, P. Russell and A. M. Carr for strains and plasmids. We thank W. Kaufman for her initial effort in the establishment of HO strains. We are grateful to S. Grewal for sharing unpublished results and to D. Lydall for insights on the quantitative analysis of checkpoint activation. We thank K. Labib, L. Jansen, K. Xavier, R. Martinho and S. Lopes for critically reading the manuscript. T.C. and C.C.R. are supported by Fundação para a Ciência e a Tecnologia (FCT) postdoctoral fellowships. T.M.N. was supported by the Sidney Kimmel Scholar Program and his laboratory is supported by NIH grant GM078253. This work was supported by the FCT (PTDC/BIA-BCM/67261/2006) and the Association for International Cancer Research (06-396).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions T.C. helped with the design and executed most experiments. L.K. performed ChIP experiments. C.C.R. performed western and Southern blotting experiments. V.B. performed live cell analysis. B.A.M. established the ChIP and HO assays. T.M.N. contributed to the design of the ChIP and HO assays. All authors contributed with strains and data analysis. M.G.F. conceived the study, performed live cell analysis and wrote the paper.
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.de Lange T. How telomeres solve the end-protection problem. Science. 2009;326:948–952. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li B, Oestreich S, de Lange T. Identification of human Rap1: implications for telomere evolution. Cell. 2000;101:471–483. doi: 10.1016/s0092-8674(00)80858-2. [DOI] [PubMed] [Google Scholar]
- 3.Du LL, Nakamura TM, Russell P. Histone modification-dependent and -independent pathways for recruitment of checkpoint protein Crb2 to double-strand breaks. Genes Dev. 2006;20:1583–1596. doi: 10.1101/gad.1422606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greeson NT, Sengupta R, Arida AR, Jenuwein T, Sanders SL. Di-methyl H4 lysine 20 targets the checkpoint protein Crb2 to sites of DNA damage. J Biol Chem. 2008;283:33168–33174. doi: 10.1074/jbc.M806857200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakamura TM, Du LL, Redon C, Russell P. Histone H2A phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol Cell Biol. 2004;24:6215–6230. doi: 10.1128/MCB.24.14.6215-6230.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moser BA, Subramanian L, Khair L, Chang YT, Nakamura TM. Fission yeast Tel1ATM and Rad3ATR promote telomere protection and telomerase recruitment. PLoS Genet. 2009;5:e1000622. doi: 10.1371/journal.pgen.1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferreira MG, Cooper JP. The fission yeast Taz1 protein protects chromosomes from Ku-dependent end-to-end fusions. Mol Cell. 2001;7:55–63. doi: 10.1016/s1097-2765(01)00154-x. [DOI] [PubMed] [Google Scholar]
- 8.Ferreira MG, Cooper JP. Two modes of DNA double-strand break repair are reciprocally regulated through the fission yeast cell cycle. Genes Dev. 2004;18:2249–2254. doi: 10.1101/gad.315804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller KM, Rog O, Cooper JP. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 2006;440:824–828. doi: 10.1038/nature04638. [DOI] [PubMed] [Google Scholar]
- 10.Rog O, Miller KM, Ferreira MG, Cooper JP. Sumoylation of RecQ helicase controls the fate of dysfunctional telomeres. Mol Cell. 2009;33:559–569. doi: 10.1016/j.molcel.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 11.Tomita K, et al. Fission yeast Dna2 is required for generation of the telomeric single-strand overhang. Mol Cell Biol. 2004;24:9557–9567. doi: 10.1128/MCB.24.21.9557-9567.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Churikov D, Price CM. Pot1 and cell cycle progression cooperate in telomere length regulation. Nature Struct Mol Biol. 2007;15:79–84. doi: 10.1038/nsmb1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 14.Guo X, et al. Dysfunctional telomeres activate an ATM-ATR-dependent DNA damage response to suppress tumorigenesis. EMBO J. 2007;26:4709–4719. doi: 10.1038/sj.emboj.7601893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moser BA, et al. Differential arrival of leading and lagging strand DNA polymerases at fission yeast telomeres. EMBO J. 2009;28:810–820. doi: 10.1038/emboj.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verdun RE, Crabbe L, Haggblom C, Karlseder J. Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol Cell. 2005;20:551–561. doi: 10.1016/j.molcel.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 17.Mochida S, et al. Regulation of checkpoint kinases through dynamic interaction with Crb2. EMBO J. 2004;23:418–428. doi: 10.1038/sj.emboj.7600018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanders SL, et al. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119:603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Jia S. Degrees make all the difference: the multifunctionality of histone H4 lysine 20 methylation. Epigenetics. 2009;4:273–276. doi: 10.4161/epi.4.5.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chikashige Y, Hiraoka Y. Telomere binding of the Rap1 protein is required for meiosis in fission yeast. Curr Biol. 2001;11:1618–1623. doi: 10.1016/s0960-9822(01)00457-2. [DOI] [PubMed] [Google Scholar]
- 21.Miller KM, Ferreira MG, Cooper JP. Taz1, Rap1 and Rif1 act both interdependently and independently to maintain telomeres. EMBO J. 2005;24:3128–3135. doi: 10.1038/sj.emboj.7600779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomita K, Cooper JP. Fission yeast Ccq1 is telomerase recruiter and local checkpoint controller. Genes Dev. 2008;22:3461–3474. doi: 10.1101/gad.498608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyoshi T, Kanoh J, Saito M, Ishikawa F. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science. 2008;320:1341–1344. doi: 10.1126/science.1154819. [DOI] [PubMed] [Google Scholar]
- 24.Michelson RJ, Rosenstein S, Weinert T. A telomeric repeat sequence adjacent to a DNA double-stranded break produces an anticheckpoint. Genes Dev. 2005;19:2546–2559. doi: 10.1101/gad.1293805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdallah P, et al. A two-step model for senescence triggered by a single critically short telomere. Nature Cell Biol. 2009;11:988–993. doi: 10.1038/ncb1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirano Y, Sugimoto K. Cdc13 telomere capping decreases Mec1 association but does not affect Tel1 association with DNA ends. Mol Biol Cell. 2007;18:2026–2036. doi: 10.1091/mbc.E06-12-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Negrini S, Ribaud V, Bianchi A, Shore D. DNA breaks are masked by multiple Rap1 binding in yeast: implications for telomere capping and telomerase regulation. Genes Dev. 2007;21:292–302. doi: 10.1101/gad.400907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Botuyan MV, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goudsouzian LK, Tuzon CT, Zakian VA. Scerevisiae Tel1p and Mre11p are required for normal levels of Est1p and Est2p telomere association. Mol Cell. 2006;24:603–610. doi: 10.1016/j.molcel.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 30.Verdun RE, Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127:709–720. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.