Abstract
Chloracne is commonly observed in people exposed to dioxins, yet the mechanism of toxicity is not well understood. The pathology of chloracne is characterized by hyperkeratinization of the interfollicular squamous epithelium, hyperproliferation and hyperkeratinization of hair follicle cells as well as a metaplastic response of the ductular sebum secreting sebaceous glands. In vitro studies using normal human epidermal keratinocytes to model interfollicular human epidermis demonstrate a 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD)-mediated acceleration of differentiation and increase in gene expression of several prodifferentiation genes, including filaggrin (FLG). Here, we demonstrated that the TCDD-activated aryl hydrocarbon receptor (AHR) bound a small fragment of DNA upstream of the transcriptional start sites of the FLG gene, containing one of two candidate xenobiotic response elements (XREs). Reporter assays using the promoter region of FLG containing the two putative XREs indicated that the increase in this messenger RNA (mRNA) was due to TCDD-mediated enhanced transcription, which was lost when both XREs were mutated. As FLG is part of the human epidermal differentiation complex (EDC) found on chromosome 1, we measured mRNAs from an additional 18 EDC genes for their regulation by TCDD. Of these genes, 14 were increased by TCDD. Immunoblot assays demonstrated that the proteins of FLG as well as that of another prodifferentiation gene, small proline rich protein 2, were increased by TCDD. In utero exposure to TCDD accelerated the formation of the epidermal barrier in the developing mouse fetus by approximately 1 day. These results indicate that the epidermal permeability barrier is a functional target of the TCDD-activated AHR.
Keywords: AHR, gene regulation, TCDD, keratinocyte, epidermal barrier, filaggrin
Normal human skin epidermis is composed of highly organized stratified epithelium. Each of the morphologically distinct strata, the basal, spinous, granular, and cornified, have unique functional characteristics. The proliferative basal cell layer of keratinocytes is found attached to the basement membrane and gives rise to the polyhedron-shaped cells of the spinous layer. Keratinocytes continue to migrate outward as they change to the more differentiated phenotype of the granular layer, which is visually characterized by cells with keratohyalin- and profilaggrin-filled granules and lipid-filled lamellar bodies. The outermost cornified layer is composed of terminally differentiated anucleated corneocytes held together by tight and adherens junctions as well as corneodesmosomes. Anchored to the corneodesmosomes, intracellular structural proteins such as intermediate filaments provide cellular strength. Together with extruded extracellular lipids, the stratum corneum and the nucleated cells of the epidermis provide protection from water loss, exposure to infectious or chemical agents, and loss of heat. Eventually, corneocytes are sloughed from the epidermis as they are replaced with new ones (Proksch et al., 2008).
The synchronized processing of numerous proteins and lipids is critical to establishing and maintaining an epidermal barrier. Disruption of this barrier is now understood to be important in the pathogenesis of multiple skin diseases including contact dermatitis, ichthyosis vulgaris, psoriasis, and atopic dermatitis (Proksch et al., 2008). Numerous structural proteins have been shown to be important to the formation of the epidermal barrier, many of which are present in the epidermal differentiation complex (EDC) locus, spanning 1.6 Mb on human 1q21 (Mischke et al., 1996; Volz et al., 1993). Loss-of-function mutations in one of these proteins, filaggrin (FLG), have been shown to underlie the skin disease icthyosis vulgaris (Smith et al., 2006) and have been associated with atopic dermatitis (Palmer et al., 2006; Smith et al., 2006). In addition, atopic dermatitis and psoriasis have shown genetic linkage to the EDC, which contains nearly 60 genes as possible candidates for disease causation (de Cid et al., 2009; Palmer et al., 2006; Zhang et al., 2009).
Previously, we and others have demonstrated that the environmental contaminant, 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD, dioxin), alters keratinocyte terminal differentiation in vitro (Greenlee et al., 1985; Loertscher et al., 2001; Sutter et al., 2009). TCDD increases the quantity of cornified envelopes in monolayer cultures of normal human keratinocytes; organotypic cultures of keratinocytes treated with TCDD develop a cornified layer earlier than controls as well as a thicker keratinized layer and more compact spinous and granular layers (Loertscher et al., 2001). Increases in the expression of genes important to keratinocyte differentiation have been reported in these two in vitro models. In monolayers of normal keratinocytes, we and others have reported TCDD-mediated increases in FLG messenger RNA (mRNA) (Sutter et al., 2009) and protein (Ray and Swanson, 2003), involucrin (IVL) protein (Ray and Swanson, 2003), as well as the expression of additional mRNAs of genes involved in terminal differentiation, including sphingolipid C4-hydroxylase/delta 4-desaturase (DEGS2), UDP-glucose ceramide glucosyltransferase (UGCG) (Sutter et al., 2009), interleukin-1 beta (IL-1B), and plasminogen activator inhibitor-2 (Sutter et al., 1991). In organotypic cultures, TCDD causes increased and aberrant expression of FLG, IVL, and transglutaminase (TGM) proteins (Loertscher et al., 2001).
In utero exposure models demonstrate that skin from fetuses exposed to TCDD at gestational day 13 have an increase in FLG and loricrin (LOR) proteins at gestational day 16 (Loertscher et al., 2002). While the authors did not see any morphological differences between the skin of the fetuses from the control and TCDD-treated mice, functional assays were not performed. We hypothesized that the formation of an epidermal barrier in fetuses following in utero exposure to TCDD would be accelerated.
The mechanism of regulation of genes involved in epidermal differentiation by TCDD has not been explored. A decrease in FLG, DEGS2, and UGCG mRNA in aryl hydrocarbon receptor (AHR) nuclear translocator (ARNT) null keratinocytes indicates that ARNT is possibly directly regulating the transcription of these genes (Geng et al., 2006). As the AHR is one of the known dimerization partners of ARNT and is activated by TCDD, we investigated whether the AHR, activated by TCDD, can increase the transcription of FLG. As FLG is located within the EDC, we selected 18 other genes found in the EDC to determine whether their expression was increased by TCDD. The selection of genes to study was based on their association with differentiated normal skin (Smiley et al., 2005) or with the more differentiated suprabasal cell layer (Radoja et al., 2006). One of these genes, small proline-rich(SPRR)2A, is decreased in the ARNT null keratinocytes, similar to FLG (Geng et al., 2006), indicating that it may be regulated by TCDD as well.
Activation of the epidermal growth factor receptor (EGFR) pathway in normal human epidermal keratinocytes (NHEKs) inhibits TCDD-mediated differentiation and inhibits TCDD-mediated increases in the mRNAs of FLG, DEGS2, and UGCG (Sutter et al., 2009), each of which is associated with epidermal differentiation. EGFR activation decreases AHR transcription of cytochrome P450 (CYP) 1A1 by inhibiting the binding of the coactivator, p300, to the AHR transcriptional complex (Sutter et al., 2009). To explore further the interaction of the AHR and EGFR pathways, we investigated the effect of EGF on the AHR transcriptional regulation of FLG. In addition, we studied effects of EGF on TCDD-mediated increases in the genes of the EDC.
The formation of an epidermal barrier is essential for life. Without a fully functioning barrier, losses of water and heat, as well as increased exposure to infectious and chemical agents, have serious detrimental consequences to survival. Agents that increase epidermal differentiation, such as glucocorticoids (Aszterbaum et al., 1993), estrogen (Hanley et al., 1996), and activators of peroxisome proliferator-activated receptor (PPAR)α (Hanley et al., 1997), PPARδ (Jiang et al., 2010), and liver X receptor (Hanley et al., 1999), accelerate the formation of the epidermal barrier in utero. As activation of the AHR caused increases in numerous genes located within the EDC and in utero TCDD exposure increased fetal FLG and LOR protein expression in the skin (Loertscher et al., 2002), we sought to determine whether activation of the AHR by TCDD would alter the development of the cornified layer and the formation of the epidermal barrier in utero.
MATERIALS AND METHODS
Chemicals.
Dimethyl sulfoxide (DMSO) and alpha-napthoflavone (ANF) were purchased from Sigma (St Louis, MO).
Keratinocyte cell culture.
Neonatal foreskin NHEKs, purchased from Lonza (Walkersville, MD), were grown in keratinocyte serum-free media ([KSFM] Invitrogen, Carlsbad, CA). Confluent fifth passage NHEKs were pretreated in basal medium with or without EGF for 24 h. Treatments (with or without EGF and with or without TCDD) were carried out for 24 h unless otherwise indicated (Sutter et al., 1994, 2009).
Quantitative real-time PCR.
Total RNA was isolated using RNA Stat-60 (Tel-Test, Friendswood, TX). Quantitative real-time PCR was carried out in an iCycler (Bio-Rad) with M-MLV RT (Invitrogen), iQ SYBR Green Supermix (Bio-Rad), and the primers listed in Table 1 (Integrated DNA Technologies, Coralville, IA), as described in (Sutter et al., 2009). Cyclophilin (PPIA) was used as the reference for sample normalization. Levels of mRNA (n = 3 ± SD) were plotted relative to DMSO control sample, given a value of one.
TABLE 1.
Primer Pairs Used in Quantitative Real-Time PCR
| Gene | Forward primer (5′–3′) | Reverse complement primer (5′–3′) |
| S100A11 | TTGACCGCATGATGAAGAAACTGG | GATGGGTGGGCTGTGGAGATGATG |
| RPTN | GAGGCCCCAAACCAGAGAAC | AGGGAGATACAGCAGAGGATAGAG |
| HRNR | CCAGCACCAAGAGGAACAAGAAGA | GCCGCGGCCTGAAGACTGATG |
| FLG | GACACCCCGGATCCTCTCACC | AGCTGCCATGTCTCCAAACTAAAC |
| FLG2 | GTGGGCATGAATCAAACTCTACTC | ACTTCGCCTCCCACTGTCTCCT |
| LCE3E | CGAGGGCGGCTGCTTCC | GCCCCCGCCTTGCTGAC |
| LCE3A | CCAGCCTCCGCCCAAGTG | TCAGCAGCAGCCCGCAGAA |
| LCE2B | TGCGTGTGACCAGGGTTGACTAA | TTTGGGGTACACTTGGGAGGACA |
| LCE2A | AGCCCCGATTGTTGTGAGTGTG | AGAAAAAGGGGCAGGAGAAGCATC |
| LCE1C | TGTCCCCCAAAGTGTCCCCCTAAG | CAGCAGCCACCTCCCCCAGAAC |
| IVL | GGCCACCCAAACATAAATAACCAC | CACCTAGCGGACCCGAAATAAGT |
| SPRR1A | CTGGCCACTGGATACTGAACACC | GCACCCGAGCAACAAGAAGA |
| SPRR2A | CCCCACCCTGCCAGTCAAAGTA | GCAGTATGGCAGCCTCAGAAAAGA |
| SPRR2B | CCCACCCTGCCAGCCAAAGTA | CATGCCCAGGTGAAAGACAGACA |
| LOR | GGCCACCCAAACATAAATAACCAC | CACCTAGCGGACCCGAAATAAGT |
| S100A9 | GTGGCTCCTCGGCTTTGACAG | GATCTTTTCGCACCAGCTCTTTGA |
| S100A12 | CAAGGCCTGGATGCTAAT | AAGGCTGGGTTTTGGTGA |
| S100A7 | AATTACCTCGCCGATGTCTTTGA | ATGGCTCTGCTTGTGGTAGTCTGT |
| PPIA | GCAGAGGGTTAAGGCGCAGACTAC | TAAGGTGGGCAGAGAAGGGGTTTT |
Western blots.
Protein was isolated by boiling cells in lysis buffer (2% SDS, 1% beta-mercaptoethanol, Tris-HCl [pH 6.8]) for 10 min. Samples were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membrane, and incubated with FLG (Leica, Buffalo Grove, IL) or SPRR2 (Enzo, Plymouth Meeting, PA) antibodies and species appropriate horseradish peroxidase-conjugated secondary antibodies (Jackson Immunoresearch Laboratories, West Grove, PA). Signals were visualized by chemiluminescence after incubation with Supersignal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL) (Sutter et al., 2009) and quantified by densitometry using ImageJ (National Institutes of Health).
Chromatin immunoprecipitation (ChIP) assays.
ChIP assays were performed as in (Beischlag et al., 2004) except that DNA was purified using QIAquick DNA Purification Kit (Qiagen). The AHR antibody and normal rabbit IgG were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Primers used for PCR were 5′-CACCAGTGCTATTGAGAGG-3′ and 5′-CATATTGGTCCACTACTGTTC-3′, which amplified the sequence from −1565 to −1368, relative to the FLG transcriptional start site (TSS) at +1. The conditions for PCR using HotStart Taq DNA Polymerase with Q-Solution (Qiagen) included an initial 15 min 95°C incubation followed by cycling between 95°C for 30 s, 49°C for 30 s, and 72°C for 30 s and then a single incubation at 72°C for 2 min. PCR products were analyzed on 1.8% agarose gels. To analyze input DNA, DNA-protein crosslinks were reversed by adding NaCl to 0.45mM and heating to 95°C for 15 min. RNase A was added and samples were incubated at 37°C for 30 min, followed by purification using QIAquick DNA Purification Kit (Qiagen). PCR was preformed and analyzed using the above primers and conditions.
Luciferase reporter constructs.
To generate pGL4.23-1A1 (−815/−1216), a 402-bp region of genomic DNA containing two minimal and three canonical xenobiotic response elements (XREs) was amplified using the following primer sequences: 5′-TATGGTACCGCAGAAGCCACACGCAGACC-3′, containing a Kpn I site, and 5′-TATGAGCTCGGGGGCCACGGAAAGACTC-3′, containing a Sac I site. For the creation of pGL4.23-FLG (−1346/−2058), a 712-bp region containing two canonical XREs was amplified using the following primers: 5′-TATGGTACCCTTGGGTCGCTTATCAGTAT-3′, containing a Kpn I site, and 5′-TATGAGCTCAAATCTTTGCGTGCTATCC-3′, containing a Sac I site. PCR amplified fragments were digested with Kpn I and Sac I and ligated into Kpn I- and Sac I-digested pGL4.23 (Luc2/minP) vector (Promega, Madison, WI). Mutagenesis of the two XRE regions of FLG was conducted using the Quick Change II Site Directed Mutagenesis Kit (Agilent Stratagene, Santa Clara, CA). Briefly, the synthetic oligonucleotide primers, each complementary to opposite strands of the insert of interest, and containing the desired mutation are extended during temperature cycling by PfuUltra HF DNA polymerase, without primer displacement. The following primers were used for mutagenesis: M1: 5′-CCTGATGATTAAAATTAATACACTCAAAATCTTAGCACATTCC-3′ and 5′-GGAATGTGCTAAGATTTTGAGTGTATTAATTTTAATCATCAGG-3′; M2: 5′-GGTAGTGGAGGATAGCACTCAAAGATTTGAGCTCGCTA-3′ and 5′-TAGCGAGCTCAAATCTTTGAGTGCTATCCTCCACTACC-3′. The extension of the oligonucleotide primers generated the mutated M1 and/or M2 XRE sequence. All constructs were verified by DNA sequence analysis.
Luciferase activity.
NHEKs were cultured in 12-well plates and cotransfected with 0.1 μg of Renilla luciferase DNA pGL4.74 (Promega) and 0.9 μg of the following firefly luciferase plasmids: pGL4.23 (Promega), pGL4.23-1A1, pGL4.23-FLG, pGL4.23-FLG M1, pGL4.23-FLG M2, or pGL4.23-FLG M1 and M2. Transient transfections of NHEKs were performed using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions. Briefly, 100 μl of basal KSFM (Invitrogen) containing DNA and 2.5 μl of Lipofectamine 2000 were added to the cells for 6 h, after which the media was changed to complete media for 18 h. Pretreatment was carried out for 6 h (± EGF, 10 ng/ml), followed by treatment with vehicle control (0.1% DMSO) or TCDD (10nM) for 24 h, ± EGF. Dual luciferase assays (Renilla and firefly) were performed according to the manufacturer's instructions (Promega) using a TD 20/20 Luminometer (Turner Biosystems, Sunnyvale, CA). Firefly luciferase activity was normalized to Renilla luciferase.
XRE sequence analysis.
Gene sequences, 5 kb upstream to 2 kb downstream of the TSSs, were obtained from NCBI Human Genome Build 37.2 and analyzed for the presence of the XRE core consensus sequence, 5′-GCGTG-3′, using Lasergene (DNASTAR, Madison, WI). A specific 5-bp sequence is expected to be found approximately every 500 bp by chance alone. In order to increase the likelihood that the core XRE consensus identifies an actual XRE, a position weight matrix (Quandt et al., 1995) was calculated using an additional 7 bps upstream and 7 bps downstream from the 5 bp core consensus site of authenticated XREs. From this position weight matrix, a cut off matrix similarity score was determined to be 0.8473, the lowest score of the authenticated XREs (Dere et al., 2011). This approach considers mismatches at less conserved positions, by giving each nucleotide a score based on its frequency in authenticated XREs. We calculated matrix similarity scores for each XRE (5 bp core XRE consensus site plus 7 upstream and 7 downstream bps) using the described position weight matrix (Dere et al., 2011) and the matrix similarity equation found in Quandt et al. (1995).
Skin permeability assay.
Time-mated presumed pregnant C57BL/J6 mice (Jackson Laboratories) were fed Harlan Teklad No 216C Certified Global 16% Protein Rodent Diet (Harlan Teklad, Madison, WI) ad libitum. Mice were treated with TCDD (10 μg/kg) or vehicle control in corn oil on embryonic day (E)12 or E13 by gavage and sacrificed on E15 and E16, respectively (n = 3/group). The skin permeability assay was carried out according to published methods using 5-bromo-4-chloro-3-indolyl-beta-D-galactopyranoside (X-gal) as the substrate (Hardman et al., 1998). Fetuses were immersed in X-gal reaction mix for 18 h before photographs were taken. Digital color photographs were converted to 8-bit gray scale for densitometric analysis using ImageJ.
Statistical analyses.
Statistical analyses were performed using Student’s t-test with Sidak-Bonferroni correction (p < 0.0253).
RESULTS
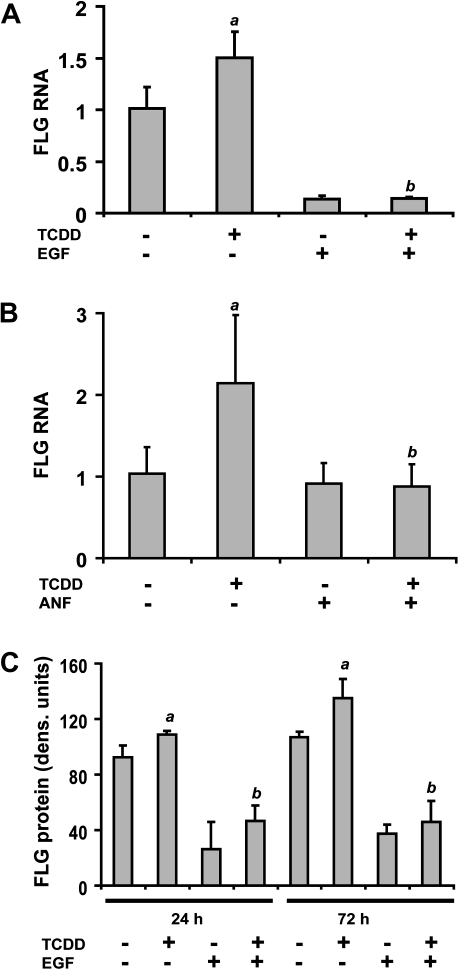
Exposure of confluent NHEKs to TCDD (10nM, 24 h) caused a significant increase in the FLG RNA. TCDD-dependent FLG RNA increases were repressed by activation of the EGFR (Fig. 1A and Sutter et al., 2009). Immunoblots demonstrated that the TCDD-mediated increase in FLG RNA resulted in an increase in FLG protein. Statistically significant increases were detected after 24 and 72 h of TCDD treatment (10nM). TCDD-mediated increases in FLG mRNA were decreased to control levels when NHEKs were cotreated with the AHR antagonist, ANF (Fig. 1B). Both the control levels of FLG protein as well as the TCDD-mediated increased levels were decreased by the presence of EGF, correlating with the RNA results in Figure 1A (Fig. 1C).
FIG. 1.
Increases in FLG RNA and protein levels by TCDD. (A) NHEKs were grown to confluence before EGF (10 ng/ml) was added as indicated for 24 h before treatment. RNA was isolated after treatment with either control vehicle (0.1% DMSO) or TCDD (10nM) for 24 h. Real-time PCR was used to determine the relative mRNA expression of FLG (y-axis). Levels of mRNA (mean [n = 3], ± SD) are expressed relative to DMSO control, given a value of one. The a indicates a significant difference from control samples (i.e., no EGF and no TCDD), p < 0.0253; the b indicates a significant difference from samples treated only with TCDD, p < 0.0253. (B) NHEKs were grown to confluence before basal media was added for 24 h. RNA was isolated after pretreatment with either control (0.1% DMSO) or ANF (1μM) for 1 h followed by the indicated treatments for 24 h. The concentration of TCDD was 10nM and the final concentration of DMSO was 0.2%. (C) NHEKs were treated as indicated for 24 or 72 h in the presence of 1mM calcium chloride. Using densitometry, the signals generated by immunoblots incubated with FLG antibody were quantified (mean [n = 3], ± SD). The a indicates a significant difference from control samples at the corresponding time point (i.e., no EGF and no TCDD), p < 0.0253; the b indicates a significant difference from samples treated only with TCDD, p < 0.0253.
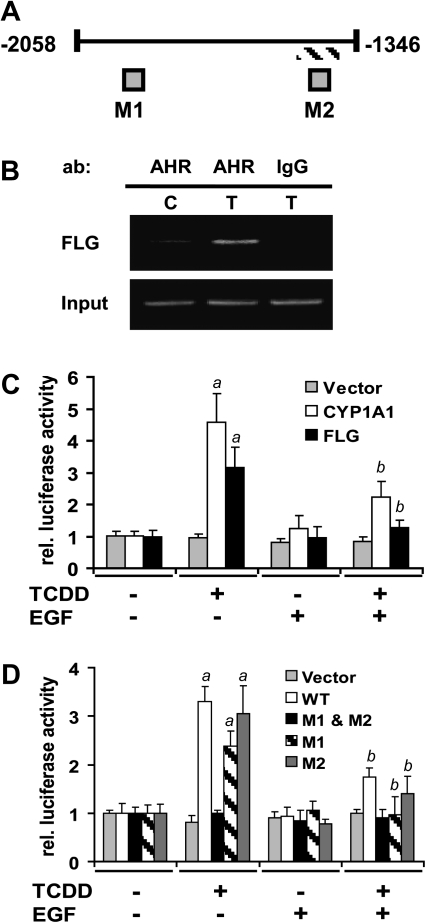
Analysis of the human FLG gene (−5000 to +2000 kb) revealed two XRE consensus sites (5′-CACGC-3′), one at −2011 to −2007 and the other at −1402 to −1398, relative to the transcription start site at +1 (Fig. 2A). ChIP assays demonstrated that after treatment with TCDD, there was an increase in the amount of AHR protein bound to the XRE consensus site found at −1402 to −1398 (Fig. 2B). The XRE consensus site, located at −2011 to −2007, was not analyzed by ChIP. To determine whether FLG was transcriptionally regulated by the AHR, luciferase reporter assays were carried out in NHEKs treated with TCDD with or without EGF. The wild-type FLG reporter construct contains the FLG promoter with both of the XRE consensus sites. The TCDD-mediated increase in relative transcriptional activity of the wild-type FLG promoter (3.2-fold) was similar to that of CYP1A1 (4.6-fold), a gene well characterized for its regulation by the AHR (Fig. 2C).
FIG. 2.
Transcriptional regulation of FLG by the AHR. (A) Scheme of the FLG promoter region used in the studies presented in panel (B–D). Fragment −1346 through −2058 is relative to TSS at +1. Gray boxes indicate the locations of two consensus XREs (CACGC). Hatched line indicates the fragment amplified during ChIP analysis using an AHR antibody in (B). M1 and M2 (CACTC) indicate the designations of the two mutant XREs used in (D). (B) NHEKs treated with either 0.1% DMSO (C) or 10nM TCDD (T) were subjected to ChIP assay using the AHR antibody or normal IgG for immunoprecipitation and FLG primers to generate a DNA fragment. (C) and (D) NHEKs transfected with the indicated luciferase reporter constructs were treated as indicated. Levels of luciferase (mean [n = 5], ± SD) are expressed in units relative to the control (no EGF and no TCDD) for each construct, given a value of one. The a indicates a significant difference from control samples (i.e., no EGF and no TCDD), p < 0.0253; the b indicates a significant difference from samples treated only with TCDD, p < 0.0253.
To provide further evidence that FLG is directly regulated by the AHR, single and double XRE mutants were generated by site-directed mutagenesis, where only the G in the consensus sequence is changed to a T (5′-CACTC-3′), in either site (M1 or M2) or both sites (M1 and M2). Mutation of this nucleotide was chosen based on its effectiveness in inhibiting binding of the AHR (Shen and Whitlock, 1992; Yao and Denison, 1992). Similar to results of Figure 2C, there was a transcriptional increase following treatment with TCDD of 3.2-fold in the NHEKs transfected with the wild-type FLG XRE. When both XREs were mutated, the levels of transcription after TCDD treatment did not increase above the DMSO control levels providing strong evidence that FLG is directly regulated by the AHR. To determine the importance of each of the two different consensus sites, individual sites were mutated and tested for transcriptional activation. When the XRE located at −2011 to −2007 (M1) was mutated, there was a slight decrease in transcriptional activity indicating that the XRE found at −1402 to −1398 was able to sustain transcriptional activity although at a slightly lower level than when both sites were functional. When the XRE located at −1402 to −1398 (M2) was mutated, no transcriptional activity was lost indicating that the XRE located at −2011 to −2007 was sufficient to sustain transcriptional activity. Therefore, although there was a slight decrease in transcription when only the XRE located at −1402 to −1398 was present, either site was capable of increasing transcription in response to treatment by TCDD.
Activation of the EGFR was previously shown to decrease the transcription rate of CYP1A1 and CYP1B1, both AHR-regulated genes (Sutter et al., 2009). This was replicated in Figure 2B, where EGF decreased the AHR-mediated transcriptional activity of CYP1A1 by 2.1-fold. To broaden the role of EGFR in regulating AHR-mediated transcription, we tested the effects of EGF on AHR-driven FLG transcription. As shown in Figures 2C and 2D, EGF is able to decrease the AHR-mediated transcription of FLG by 2.5- and 1.9-fold, respectively; illuminating the possibility that activation of the EGFR pathway in NHEKs may oppose the transcription of all genes regulated by the AHR.
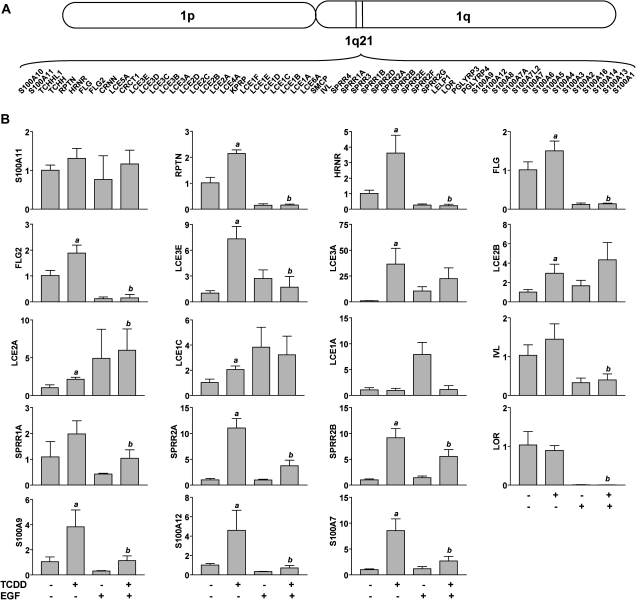
As TCDD is known to accelerate keratinocyte differentiation and FLG is part of the EDC located on chromosome one, 18 other genes in this complex were examined for their regulation by TCDD (Fig. 3). TCDD exposure significantly increased mRNA expression of 14 genes besides FLG (p < 0.0253). These included repetin (RPTN) (2.1-fold), hornerin (HRNR) (3.6-fold), FLG (1.5-fold), FLG2 (1.9-fold), late cornified envelope (LCE)3E (7.3-fold), LCE3A (36.6-fold), LCE2B (3.0-fold), LCE2A (2.2-fold), LCE1C (2.0-fold), SPRR1A (2.0-fold), SPRR2A (11.0-fold), SPRR2B (9.2-fold), S100A9 (3.8-fold), S100A12 (4.6-fold), and S100A7 (8.5-fold). Compared with TCDD alone, TCDD plus EGF significantly decreased the following RNA levels, RPTN (4.3-fold), HRNR (16.1-fold), FLG (10.7-fold), FLG2 (20.0-fold), LCE3E (4.3-fold), LCE3A (1.6-fold), SPRR1A (1.9-fold), SPRR2A (2.9-fold), SPRR2B (1.6-fold), S100A9 (3.4-fold), S100A12 (6.4-fold), and S100A7 (3.2-fold).
FIG. 3.
Regulation of genes in EDC on chromosome 1 by TCDD. (A) Scheme of human chromosome 1, indicating 1q21, and the genes associated with the EDC. (B) NHEKs were grown to confluence and treated as indicated. Quantitative real-time PCR was used to determine the relative mRNA expression of each indicated gene (y-axis). Levels of mRNA (mean [n = 3], ± SD) are expressed in units relative to the DMSO control, given a value of one. The a indicates that the value from treatment with TCDD is significantly different from the DMSO control, p < 0.0253; the b indicates that the value from cotreatment with TCDD and EGF is significantly different from TCDD alone, p < 0.0253.
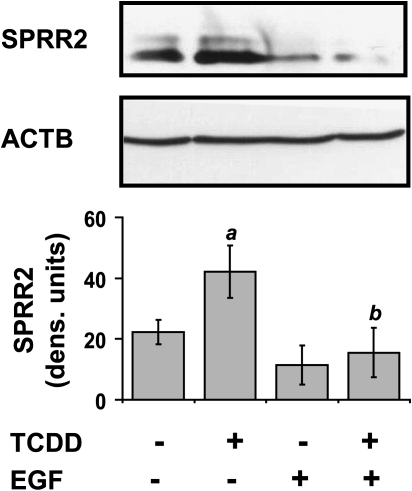
Immunoblot assays using a SPRR2 antibody, which detects both SPRR2A and SPRR2B proteins demonstrated that the SPRR2 proteins levels corresponded to the mRNA levels, as SPRR2 protein level was increased (2-fold) by TCDD alone and this increase was repressed by EGF (3-fold) (Fig. 4). SPRR2 was chosen for protein analysis due to the fact that SPRR2A and SPRR2B mRNAs were highly increased by TCDD and there was an antibody readily available to the protein.
FIG. 4.
Repression of TCDD-mediated increases in SPRR2 protein by EGF. Upper panel, representative immunoblot of SPRR2 detected in protein isolated from NHEKs after the indicated treatment. Expression of β-actin (ACTB) was used as the loading control. Lower panel, levels of SPRR2 protein are expressed in densitometric units after normalizing with β-actin (mean [n = 3] ± SD). The a indicates that the value from treatment with TCDD is significantly different from the DMSO control, p < 0.0253; the b indicates that the value from co-treatment with TCDD and EGF is significantly different from TCDD alone, p < 0.0253.
Analysis of DNA sequence 5 kb upstream and 2 kb downstream of the TSSs resulted in detection of the core XRE (5′-GCGTG-3′) in each of the genes selected to study (Table 2). Based on a previous analysis of confirmed XRE sequences, a matrix similarity score of 0.8473 was proposed as the threshold value of putative 19 bp XREs (7 bp upstream and 7 bp downstream of 5 bp consensus) (Dere et al., 2011). Of the 14 genes with mRNA expression significantly increased by TCDD, only HRNR, FLG, FLG2, and LCE3E had at least one XRE with a matrix similarity score ≥0.8473.
TABLE 2.
XRE Sequence Analysis
| Gene | Increased by TCDDa | Increases by TCDDdecreased by EGFa | XRE relative to TSSb | MSSc |
| S100A11 | + | − | −4210 | 0.7773 |
| −3053 | 0.7881 | |||
| −872 | 0.8121 | |||
| −524 | 0.6862 | |||
| −513 | 0.7904 | |||
| +20 | 0.7755 | |||
| +49 | 0.7837 | |||
| +1040 | 0.7514 | |||
| RPTN | + | + | −4676 | 0.7226 |
| −2397 | 0.7476 | |||
| HRNR | + | + | −4415 | 0.8432 |
| −4411 | 0.8745d | |||
| −4407 | 0.8600d | |||
| −4403 | 0.8409 | |||
| −617 | 0.7448 | |||
| +1089 | 0.8077 | |||
| FLG | + | + | −2018 | 0.8105 |
| −1409 | 0.8747d | |||
| FLG2 | + | + | −4115 | 0.7828 |
| −3277 | 0.8595d | |||
| +1908 | 0.7567 | |||
| +1964 | 0.7887 | |||
| LCE3E | + | + | −4853 | 0.8298 |
| −1828 | 0.8630d | |||
| LCE3A | + | − | −276 | 0.8303 |
| LCE2B | + | − | +15 | 0.8260 |
| +387 | 0.7413 | |||
| LCE2A | + | − | −2229 | 0.7532 |
| −1752 | 0.7550 | |||
| LCE1C | + | − | −4039 | 0.7727 |
| −487 | 0.7942 | |||
| +524 | 0.8307 | |||
| +1716 | 0.8103 | |||
| +1758 | 0.7313 | |||
| LCE1A | − | − | −2026 | 0.7381 |
| −474 | 0.8465 | |||
| +430 | 0.7743 | |||
| +1742 | 0.7759 | |||
| +1781 | 0.7580 | |||
| IVL | − | − | −3210 | 0.7623 |
| SPRR1A | + | + | −2776 | 0.8193 |
| −2213 | 0.8125 | |||
| −1486 | 0.7997 | |||
| +1775 | 0.7579 | |||
| SPRR2A | + | + | −4623 | 0.7845 |
| −2260 | 0.7761 | |||
| +827 | 0.7886 | |||
| SPRR2B | + | − | −3605 | 0.7777 |
| −2369 | 0.7877 | |||
| +819 | 0.7886 | |||
| LOR | − | − | +934 | 0.7532 |
| +1287 | 0.8179 | |||
| S100A9 | + | + | −3383 | 0.7623 |
| S100A12 | + | + | −4900 | 0.7668 |
| −4768 | 0.8023 | |||
| −4711 | 0.7329 | |||
| −4600 | 0.7660 | |||
| −3503 | 0.7627 | |||
| −3457 | 0.7918 | |||
| −2033 | 0.8057 | |||
| −1231 | 0.8423 | |||
| +263 | 0.7623 | |||
| +388 | 0.7427 | |||
| +444 | 0.8023 | |||
| S100A7 | + | + | −3875 | 0.7801 |
| −2662 | 0.8126 | |||
| −1351 | 0.7769 | |||
| +1074 | 0.7412 |
RNA levels shown in Figure 3
Location of the 5′ nucleotide of the 19 analyzed (5′-GCGTG-3′ plus seven flanking bases) relative to the TSS.
Matrix similarity score using reported position weight matrix and conservation indices (Dere et al., 2011).
≥ threshold value (0.8437) determined in Dere et al. (2011).
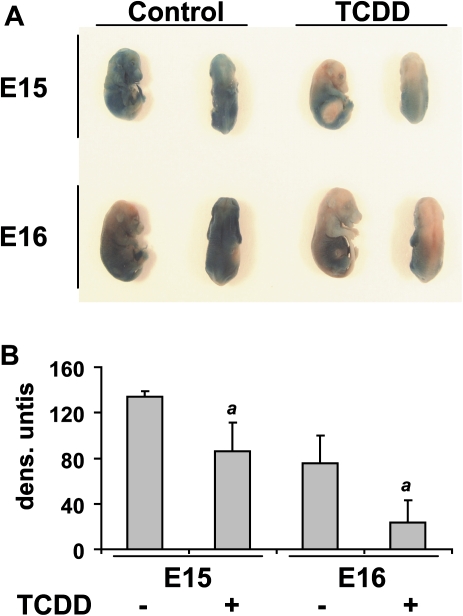
To investigate the effect of TCDD-mediated activation of the AHR on skin development in utero, pregnant mice were treated with TCDD (10 μg/kg) on E12 or E13 by gavage and sacrificed on E15 and E16, respectively. Fetuses were subjected to an epidermal permeability assay. After submerging fetuses in the reaction mix containing X-gal, a substrate for endogenous β-galactosidase, the color of the fetus is assessed. A blue color indicates that the barrier has not formed, as the X-gal is able to enter the fetuses, whereas a pink color indicates that the barrier has formed, as the X-gal is not able to enter the fetuses. As shown in Figure 5A and quantified in Figure 5B, TCDD exposure caused a significant acceleration in epidermal barrier formation in fetuses from mice treated on either E12 or E13 and sacrificed on E15 and E16, respectively. TCDD accelerated the barrier formation by one day, as the TCDD-treated E15 mice have a similar barrier to the control E16 fetuses.
FIG. 5.
Acceleration of in utero epidermal barrier formation in C57BL/J6 mice by TCDD. (A) Representative photograph taken following X-gal skin permeability assay of fetuses from time-mated mice treated by gavage with either vehicle control or TCDD (10 μg/kg) in corn oil at E12 or 13 and sacrificed on E15 or 16, respectively. (B) Quantification of skin permeability in fetuses by densitometry. The a indicates that the value from treatment with TCDD is significantly different from the DMSO control of the corresponding gestational day, p < 0.0253.
DISCUSSION
Whether increased FLG expression is associated with chloracne has not been studied. However, following exposure of organotypic cultures of human keratinocytes to TCDD, increased aberrant expression of FLG associates with accelerated differentiation, premature cornification, hyperkeratinization, and compacted spinous and granular layers (Loertscher et al., 2001). The most well-characterized function of FLG in the differentiating keratinocyte is to aggregate keratin into filaments, which stabilize a network of structural proteins. When degraded, FLG provides a source of free hygroscopic amino acids needed to produce pyrrolidone carboxylic acid and trans-urocanic acid, important components of the natural moisturizing factor produced in the cornified layer of skin (Sandilands et al., 2009). Premature over expression of FLG in vitro causes a collapse of the keratin and vimentin intermediate filaments networks, a disruption of the nuclear envelope, a loss of cell-cell adhesion, and an increase in cell cycle arrest (Dale et al., 1997; Presland et al., 2001), each a feature of terminal differentiation. Transgenic mice prematurely expressing human FLG demonstrate accelerated barrier recovery following an acute barrier disruption (Presland et al., 2004). These in vitro and in vivo effects of over expressed FLG in keratinocyte differentiation support a potential role for FLG in TCDD-mediated acceleration of differentiation and barrier formation reported previously and here, respectively. Whether premature expression of FLG is associated with or contributes to the deleterious skin effects found in chloracne is an interesting, yet unanswered, question.
FLG joins an increasingly diverse list of genes transcriptionally regulated by the AHR. Some of these genes include the metabolizing enzymes CYP1A1, CYP1A2, and CYP1B1 as well as more recently discovered AHR-regulated genes such as ABCG2 (Tompkins et al., 2010), AHRR (Baba et al., 2001), HES1 (Thomsen et al., 2004), and Slug (Ikuta and Kawajiri, 2006). Computational sequence analysis of 19 of the genes found in the EDC demonstrated that the position weight matrix described in Dere et al. (2011) would have predicted the functional XRE found at −1409 of FLG, as it had a matrix similarity score of 0.8747 greater than the threshold cut off of 0.8473. However, the matrix similarity score of the XRE of FLG found at −2018 (0.8105) was below the threshold, despite evidence that it was transcriptionally responsive to treatment with TCDD. This suggests that the position weight matrix for XRE similarity may be improved with additional experimental analysis, such as reported here for FLG. HRNR, FLG2, and LCE3E are the other mRNAs most likely to be transcriptionally regulated by the AHR as their matrix similarity scores were above the published threshold and EGF inhibited their TCDD-mediated mRNA increases. Increases in some of the other mRNAs could possibly be regulated transcriptionally by AHR despite having matrix similarity scores below the threshold cut off, as was the case with the FLG XRE found at −2018. Biological experimentation will provide necessary evidence for transcriptional regulation, which will improve the predictability of matrix similarity scores.
While FLG plays an important role in keratinocyte differentiation, the action of TCDD in the skin likely involves the concerted effects of multiple genes. Increased expression of FLG, as well as that of many of the other genes of the EDC studied here, is associated with human skin diseases such as psoriasis and/or atopic dermatitis, indicating that the increased expression of these genes by TCDD may cause deleterious effects. Premature expression of FLG in the spinous layer of cells instead of the granular or lower cornified layer is reported in skin samples from patients with psoriasis (Guttman-Yassky et al., 2009). Psoriatic samples have increased expression of IVL, S100A2, S100A7, S100A8, S100A9, S100A11, S100A12, S100A14, SPRR1A, SPRR1B, SPRR2A, SPRR2D, SPRR3, LCE3A, and LCE3E (Bergboer et al., 2011; Bowcock et al., 2001; Guttman-Yassky et al., 2009). S100A2, S100A7, S100A9, and S100A12 are also elevated in samples of atopic dermatitis (Glaser et al., 2009; Guttman-Yassky et al., 2009). SPRRs, which are calcium binding proteins cross-linked in the cornified envelope, are often increased under stress producing conditions such as inflammation or tissue remodeling (Demetris et al., 2008) and protect against reactive oxygen species during wound healing (Vermeij and Backendorf, 2010). Secreted S100A7 protein leads to increases in immune responses, which likely contributes to the increased inflammation seen in psoriasis and atopic dermatitis (Wolf et al., 2010). We studied the expression of nine of the genes associated with psoriasis and/or atopic dermatitis, of which seven, FLG, S100A7, S100A9, S100A12, SPRR2A, LCE3A, and LCE3E, were increased by TCDD, whereas two, S100A11 and IVL, were not. Taken together, a number of genes that TCDD increased are associated with the diseases psoriasis and/or atopic dermatitis, which are characterized by abnormal skin phenotypes, increased inflammation, and a compromised epidermal barrier. Increases in these genes of the EDC by TCDD may contribute to the hyperkeratosis, a thickening of the cornified and granular epidermal layers, observed in the interfollicular skin of humans with chloracne as well as to the atopic dermatitis and inflammation observed in the skin of mice expressing constitutively active AHR (Tauchi et al., 2005).
The regulation of epidermal barrier formation in utero is complex. In mice, a barrier initiates dorsally at gestational day 16, spreads ventrally, and is completed by gestational day 18 (Hardman et al., 1998). Several transcription factors are essential in establishing a prenatal epidermal barrier in mice including distal-less homeobox 3 (Morasso et al., 1996), Kruppel-like factor 4 (Segre et al., 1999), GATA-binding protein 3 (GATA3) (de Guzman Strong et al., 2006), and ARNT (Geng et al., 2006; Takagi et al., 2003). Our results demonstrating that TCDD accelerated mouse epidermal barrier formation by approximately 1 day correspond well to the report of detectable FLG protein expression on gestational day 16, approximately 1 day before expression is normally detected, following a similar in utero exposure to TCDD beginning on gestational day 13 (Loertscher et al., 2002). AHR null mice were used to investigate the role of the AHR in regulating the in utero increase in FLG expression by TCDD. Unexpectedly, in the absence of the AHR, FLG protein expression in the vehicle-treated mice was even more pronounced than in the wild-type mice treated with TCDD (Loertscher et al., 2002). Thus, these results of Loertscher et al. (2002) indicate that loss-of-function studies of the AHR using AHR null mice may not be able to validate the AHR dependency of the gain-of-function actions of TCDD in the developing epidermis, even though the effects of TCDD are generally believed to be mediated by the AHR. While the longer term deleterious effects of in utero accelerated epidermal barrier formation by TCDD are unknown, our studies demonstrate that TCDD can dramatically alter the epidermal permeability function of the skin during mouse development.
Although we show here that activation of the AHR in utero is able to accelerate epidermal barrier formation, we know that the AHR is not essential to epidermal barrier formation as AHR knock-out mice survive, albeit with dermatological lesions characterized by epidermal hyperplasia, hyperkeratosis, fibrosis, and anagenic hair follicles (Fernandez-Salguero et al., 1997). This is in contrast to skin-specific ARNT null mice, which die shortly after birth due to excessive loss of water because of a dysfunctional epidermal barrier. Therefore, with respect to barrier formation, the AHR is not an essential dimerization partner of ARNT, yet activation of the AHR modulates the expression of a number of genes in the EDC.
It is clear now that the AHR transcriptionally regulates a number of genes besides its well-characterized metabolizing and detoxifying enzyme targets. However, there is still little understanding of how the transcriptional regulation of any of these target genes manifests in toxicity in the skin or in other target organs. Because increases in many of the EDC genes are associated with abnormal epidermal barrier formation, our findings warrant further investigation into the transcriptional regulation of these genes and the resulting aberrant epidermal protein expression which may be contributing to the etiology of chloracne or other skin conditions related to exposure to exogenous AHR ligands.
FUNDING
National Institute of Environmental Health Sciences at the, National Institutes of Health (R01 ES017014) and the W. Harry Feinstone Center for Genomic Research at the University of Memphis.
Acknowledgments
We thank Edward R. Frizell of Huntingdon Life Sciences (Somerset, NJ) for aiding us in animal studies, and the Molecular Resource Core for sequence analysis (University of Tennessee, Memphis, TN), as well as Gaylene Stevens, Quynh T. Tran, Shirlean Goodwin, Lawrence H. Kennedy, and Sandra Leon Carrion for helpful discussions.
References
- Aszterbaum M, Feingold KR, Menon GK, Williams ML. Glucocorticoids accelerate fetal maturation of the epidermal permeability barrier in the rat. J. Clin. Invest. 1993;91:2703–2708. doi: 10.1172/JCI116509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Mimura J, Gradin K, Kuroiwa A, Watanabe T, Matsuda Y, Inazawa J, Sogawa K, Fujii-Kuriyama Y. Structure and expression of the Ah receptor repressor gene. J. Biol. Chem. 2001;276:33101–33110. doi: 10.1074/jbc.M011497200. [DOI] [PubMed] [Google Scholar]
- Beischlag TV, Taylor RT, Rose DW, Yoon D, Chen Y, Lee WH, Rosenfeld MG, Hankinson O. Recruitment of thyroid hormone receptor/retinoblastoma-interacting protein 230 by the aryl hydrocarbon receptor nuclear translocator is required for the transcriptional response to both dioxin and hypoxia. J. Biol. Chem. 2004;279:54620–54628. doi: 10.1074/jbc.M410456200. [DOI] [PubMed] [Google Scholar]
- Bergboer JG, Tjabringa GS, Kamsteeg M, van Vlijmen-Willems IM, Rodijk-Olthuis D, Jansen PA, Thuret JY, Narita M, Ishida-Yamamoto A, Zeeuwen PL, et al. Psoriasis risk genes of the late cornified envelope-3 group are distinctly expressed compared with genes of other LCE groups. Am. J. Pathol. 2011;178:1470–1477. doi: 10.1016/j.ajpath.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowcock AM, Shannon W, Du F, Duncan J, Cao K, Aftergut K, Catier J, Fernandez-Vina MA, Menter A. Insights into psoriasis and other inflammatory diseases from large-scale gene expression studies. Hum. Mol. Genet. 2001;10:1793–1805. doi: 10.1093/hmg/10.17.1793. [DOI] [PubMed] [Google Scholar]
- Dale BA, Presland RB, Lewis SP, Underwood RA, Fleckman P. Transient expression of epidermal filaggrin in cultured cells causes collapse of intermediate filament networks with alteration of cell shape and nuclear integrity. J. Invest. Dermatol. 1997;108:179–187. doi: 10.1111/1523-1747.ep12334205. [DOI] [PubMed] [Google Scholar]
- de Cid R, Riveira-Munoz E, Zeeuwen PL, Robarge J, Liao W, Dannhauser EN, Giardina E, Stuart PE, Nair R, Helms C, et al. Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat. Genet. 2009;41:211–215. doi: 10.1038/ng.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Guzman Strong C, Wertz PW, Wang C, Yang F, Meltzer PS, Andl T, Millar SE, Ho IC, Pai SY, Segre JA. Lipid defect underlies selective skin barrier impairment of an epidermal-specific deletion of Gata-3. J. Cell Biol. 2006;175:661–670. doi: 10.1083/jcb.200605057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demetris AJ, Specht S, Nozaki I, Lunz JG, III, Stolz DB, Murase N, Wu T. Small proline-rich proteins (SPRR) function as SH3 domain ligands, increase resistance to injury and are associated with epithelial-mesenchymal transition (EMT) in cholangiocytes. J. Hepatol. 2008;48:276–288. doi: 10.1016/j.jhep.2007.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dere E, Forgacs AL, Zacharewski TR, Burgoon LD. Genome-wide computational analysis of dioxin response element location and distribution in the human, mouse, and rat genomes. Chem. Res. Toxicol. 2011;24:494–504. doi: 10.1021/tx100328r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Salguero PM, Ward JM, Sundberg JP, Gonzalez FJ. Lesions of aryl-hydrocarbon receptor-deficient mice. Vet. Pathol. 1997;34:605–614. doi: 10.1177/030098589703400609. [DOI] [PubMed] [Google Scholar]
- Geng S, Mezentsev A, Kalachikov S, Raith K, Roop DR, Panteleyev AA. Targeted ablation of Arnt in mouse epidermis results in profound defects in desquamation and epidermal barrier function. J. Cell Sci. 2006;119:4901–4912. doi: 10.1242/jcs.03282. [DOI] [PubMed] [Google Scholar]
- Glaser R, Meyer-Hoffert U, Harder J, Cordes J, Wittersheim M, Kobliakova J, Folster-Holst R, Proksch E, Schroder JM, Schwarz T. The antimicrobial protein psoriasin (S100A7) is upregulated in atopic dermatitis and after experimental skin barrier disruption. J. Invest. Dermatol. 2009;129:641–649. doi: 10.1038/jid.2008.268. [DOI] [PubMed] [Google Scholar]
- Greenlee WF, Dold KM, Osborne R. Actions of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) on human epidermal keratinocytes in culture. In Vitro Cell. Dev. Biol. 1985;21:509–512. doi: 10.1007/BF02620843. [DOI] [PubMed] [Google Scholar]
- Guttman-Yassky E, Suarez-Farinas M, Chiricozzi A, Nograles KE, Shemer A, Fuentes-Duculan J, Cardinale I, Lin P, Bergman R, Bowcock AM, et al. Broad defects in epidermal cornification in atopic dermatitis identified through genomic analysis. J. Allergy Clin. Immunol. 2009;124:1235–1244.e58. doi: 10.1016/j.jaci.2009.09.031. [DOI] [PubMed] [Google Scholar]
- Hanley K, Jiang Y, Crumrine D, Bass NM, Appel R, Elias PM, Williams ML, Feingold KR. Activators of the nuclear hormone receptors PPARalpha and FXR accelerate the development of the fetal epidermal permeability barrier. J. Clin. Invest. 1997;100:705–712. doi: 10.1172/JCI119583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley K, Komuves LG, Bass NM, He SS, Jiang Y, Crumrine D, Appel R, Friedman M, Bettencourt J, Min K, et al. Fetal epidermal differentiation and barrier development in vivo is accelerated by nuclear hormone receptor activators. J. Invest. Dermatol. 1999;113:788–795. doi: 10.1046/j.1523-1747.1999.00743.x. [DOI] [PubMed] [Google Scholar]
- Hanley K, Rassner U, Jiang Y, Vansomphone D, Crumrine D, Komuves L, Elias PM, Feingold KR, Williams ML. Hormonal basis for the gender difference in epidermal barrier formation in the fetal rat. Acceleration by estrogen and delay by testosterone. J. Clin. Invest. 1996;97:2576–2584. doi: 10.1172/JCI118706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardman MJ, Sisi P, Banbury DN, Byrne C. Patterned acquisition of skin barrier function during development. Development. 1998;125:1541–1552. doi: 10.1242/dev.125.8.1541. [DOI] [PubMed] [Google Scholar]
- Ikuta T, Kawajiri K. Zinc finger transcription factor Slug is a novel target gene of aryl hydrocarbon receptor. Exp. Cell. Res. 2006;312:3585–3594. doi: 10.1016/j.yexcr.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Jiang YJ, Barish G, Lu B, Evans RM, Crumrine D, Schmuth M, Elias PM, Feingold KR. PPARdelta activation promotes stratum corneum formation and epidermal permeability barrier development during late gestation. J. Invest. Dermatol. 2010;130:511–519. doi: 10.1038/jid.2009.245. [DOI] [PubMed] [Google Scholar]
- Loertscher JA, Lin TM, Peterson RE, Allen-Hoffmann BL. In utero exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin causes accelerated terminal differentiation in fetal mouse skin. Toxicol. Sci. 2002;68:465–472. doi: 10.1093/toxsci/68.2.465. [DOI] [PubMed] [Google Scholar]
- Loertscher JA, Sattler CA, Allen-Hoffmann BL. 2,3,7,8-Tetrachlorodibenzo-p-dioxin alters the differentiation pattern of human keratinocytes in organotypic culture. Toxicol. Appl. Pharmacol. 2001;175:121–129. doi: 10.1006/taap.2001.9202. [DOI] [PubMed] [Google Scholar]
- Mischke D, Korge BP, Marenholz I, Volz A, Ziegler A. Genes encoding structural proteins of epidermal cornification and S100 calcium-binding proteins form a gene complex (“epidermal differentiation complex”) on human chromosome 1q21. J. Invest. Dermatol. 1996;106:989–992. doi: 10.1111/1523-1747.ep12338501. [DOI] [PubMed] [Google Scholar]
- Morasso MI, Markova NG, Sargent TD. Regulation of epidermal differentiation by a Distal-less homeodomain gene. J. Cell Biol. 1996;135:1879–1887. doi: 10.1083/jcb.135.6.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CN, Irvine AD, Terron-Kwiatkowski A, Zhao Y, Liao H, Lee SP, Goudie DR, Sandilands A, Campbell LE, Smith FJ, et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat. Genet. 2006;38:441–446. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- Presland RB, Coulombe PA, Eckert RL, Mao-Qiang M, Feingold KR, Elias PM. Barrier function in transgenic mice overexpressing K16, involucrin, and filaggrin in the suprabasal epidermis. J. Invest. Dermatol. 2004;123:603–606. doi: 10.1111/j.0022-202X.2004.23226.x. [DOI] [PubMed] [Google Scholar]
- Presland RB, Kuechle MK, Lewis SP, Fleckman P, Dale BA. Regulated expression of human filaggrin in keratinocytes results in cytoskeletal disruption, loss of cell-cell adhesion, and cell cycle arrest. Exp. Cell. Res. 2001;270:199–213. doi: 10.1006/excr.2001.5348. [DOI] [PubMed] [Google Scholar]
- Proksch E, Brandner JM, Jensen JM. The skin: an indispensable barrier. Exp. Dermatol. 2008;17:1063–1072. doi: 10.1111/j.1600-0625.2008.00786.x. [DOI] [PubMed] [Google Scholar]
- Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radoja N, Gazel A, Banno T, Yano S, Blumenberg M. Transcriptional profiling of epidermal differentiation. Physiol. Genomics. 2006;27:65–78. doi: 10.1152/physiolgenomics.00031.2006. [DOI] [PubMed] [Google Scholar]
- Ray SS, Swanson HI. Alteration of keratinocyte differentiation and senescence by the tumor promoter dioxin. Toxicol. Appl. Pharmacol. 2003;192:131–145. doi: 10.1016/s0041-008x(03)00277-1. [DOI] [PubMed] [Google Scholar]
- Sandilands A, Sutherland C, Irvine AD, McLean WH. Filaggrin in the frontline: role in skin barrier function and disease. J. Cell. Sci. 2009;122:1285–1294. doi: 10.1242/jcs.033969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat. Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- Shen ES, Whitlock JP., Jr Protein-DNA interactions at a dioxin-responsive enhancer. Mutational analysis of the DNA-binding site for the liganded Ah receptor. J. Biol. Chem. 1992;267:6815–6819. [PubMed] [Google Scholar]
- Smiley AK, Klingenberg JM, Aronow BJ, Boyce ST, Kitzmiller WJ, Supp DM. Microarray analysis of gene expression in cultured skin substitutes compared with native human skin. J. Invest. Dermatol. 2005;125:1286–1301. doi: 10.1111/j.0022-202X.2005.23971.x. [DOI] [PubMed] [Google Scholar]
- Smith FJ, Irvine AD, Terron-Kwiatkowski A, Sandilands A, Campbell LE, Zhao Y, Liao H, Evans AT, Goudie DR, Lewis-Jones S, et al. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat. Genet. 2006;38:337–342. doi: 10.1038/ng1743. [DOI] [PubMed] [Google Scholar]
- Sutter CH, Yin H, Li Y, Mammen JS, Bodreddigari S, Stevens G, Cole JA, Sutter TR. EGF receptor signaling blocks aryl hydrocarbon receptor-mediated transcription and cell differentiation in human epidermal keratinocytes. Proc. Natl. Acad. Sci. U.S.A. 2009;106:4266–4271. doi: 10.1073/pnas.0900874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter TR, Guzman K, Dold KM, Greenlee WF. Targets for dioxin: genes for plasminogen activator inhibitor-2 and interleukin-1 beta. Science. 1991;254:415–418. doi: 10.1126/science.1925598. [DOI] [PubMed] [Google Scholar]
- Sutter TR, Tang YM, Hayes CL, Wo YY, Jabs EW, Li X, Yin H, Cody CW, Greenlee WF. Complete cDNA sequence of a human dioxin-inducible mRNA identifies a new gene subfamily of cytochrome P450 that maps to chromosome 2. J. Biol. Chem. 1994;269:13092–13099. [PubMed] [Google Scholar]
- Takagi S, Tojo H, Tomita S, Sano S, Itami S, Hara M, Inoue S, Horie K, Kondoh G, Hosokawa K, et al. Alteration of the 4-sphingenine scaffolds of ceramides in keratinocyte-specific Arnt-deficient mice affects skin barrier function. J. Clin. Invest. 2003;112:1372–1382. doi: 10.1172/JCI18513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauchi M, Hida A, Negishi T, Katsuoka F, Noda S, Mimura J, Hosoya T, Yanaka A, Aburatani H, Fujii-Kuriyama Y, et al. Constitutive expression of aryl hydrocarbon receptor in keratinocytes causes inflammatory skin lesions. Mol. Cell. Biol. 2005;25:9360–9368. doi: 10.1128/MCB.25.21.9360-9368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen JS, Kietz S, Strom A, Gustafsson JA. HES-1, a novel target gene for the aryl hydrocarbon receptor. Mol. Pharmacol. 2004;65:165–171. doi: 10.1124/mol.65.1.165. [DOI] [PubMed] [Google Scholar]
- Tompkins LM, Li H, Li L, Lynch C, Xie Y, Nakanishi T, Ross DD, Wang H. A novel xenobiotic responsive element regulated by aryl hydrocarbon receptor is involved in the induction of BCRP/ABCG2 in LS174T cells. Biochem. Pharmacol. 2010;80:1754–1761. doi: 10.1016/j.bcp.2010.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeij WP, Backendorf C. Skin cornification proteins provide global link between ROS detoxification and cell migration during wound healing. PLoS One. 2010;5:e11957. doi: 10.1371/journal.pone.0011957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz A, Korge BP, Compton JG, Ziegler A, Steinert PM, Mischke D. Physical mapping of a functional cluster of epidermal differentiation genes on chromosome 1q21. Genomics. 1993;18:92–99. doi: 10.1006/geno.1993.1430. [DOI] [PubMed] [Google Scholar]
- Wolf R, Mascia F, Dharamsi A, Howard OM, Cataisson C, Bliskovski V, Winston J, Feigenbaum L, Lichti U, Ruzicka T, et al. Gene from a psoriasis susceptibility locus primes the skin for inflammation. Sci. Transl. Med. 2010;2:61ra90. doi: 10.1126/scitranslmed.3001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao EF, Denison MS. DNA sequence determinants for binding of transformed Ah receptor to a dioxin-responsive enhancer. Biochemistry. 1992;31:5060–5067. doi: 10.1021/bi00136a019. [DOI] [PubMed] [Google Scholar]
- Zhang XJ, Huang W, Yang S, Sun LD, Zhang FY, Zhu QX, Zhang FR, Zhang C, Du WH, Pu XM, et al. Psoriasis genome-wide association study identifies susceptibility variants within LCE gene cluster at 1q21. Nat. Genet. 2009;41:205–210. doi: 10.1038/ng.310. [DOI] [PubMed] [Google Scholar]