Summary
We have identified gene fusions of polyamine biosynthetic enzymes S-adenosylmethionine decarboxylase (AdoMetDC, speD) and aminopropyltransferase (speE) orthologues in diverse bacterial phyla. Both domains are functionally active and we demonstrate the novel de novo synthesis of the triamine spermidine from the diamine putrescine by fusion enzymes from β-proteobacterium Delftia acidovorans and δ-proteobacterium Syntrophus aciditrophicus, in a ΔspeDE gene deletion strain of Salmonella enterica sv. Typhimurium. Fusion proteins from marine α-proteobacterium Candidatus Pelagibacter ubique, actinobacterium Nocardia farcinica, chlorobi species Chloroherpeton thalassium, and β-proteobacterium Delftia acidovorans each produce a different profile of non-native polyamines including sym-norspermidine when expressed in Escherichia coli. The different aminopropyltransferase activities together with phylogenetic analysis confirm independent evolutionary origins for some fusions. Comparative genomic analysis strongly indicates that gene fusions arose by merger of adjacent open reading frames. Independent fusion events, and horizontal and vertical gene transfer contributed to the scattered phyletic distribution of the gene fusions. Surprisingly, expression of fusion genes in E. coli and S. Typhimurium revealed novel latent spermidine catabolic activity producing non-native 1,3-diaminopropane in these species. We have also identified fusions of polyamine biosynthetic enzymes agmatine deiminase and N-carbamoylputrescine amidohydrolase in archaea, and of S-adenosylmethionine decarboxylase and ornithine decarboxylase in the single-celled green alga Micromonas.
Keywords: evolution, gene fusion, polyamine, spermidine, S-adenosylmethionine decarboxylase, aminopropyltransferase
Introduction
Metabolic pathways evolve through diverse genetic and molecular mechanisms: gene fusion, duplication, extension, loss, and recruitment of genes from elsewhere in the same genome or from other genomes via horizontal and endosymbiotic gene transfer (Caetano-Anolles et al., 2009, Fondi et al., 2009). Although gene fusions are not infrequent, there are relatively few examples where fusion proteins have been biochemically characterised. However, gene fusion to produce multidomain proteins is a key mechanism in protein evolution (Yanai et al., 2002), and enzyme fusions are found in pathways such as histidine biosynthesis (Fani et al., 2007). Koonin and colleagues concluded that gene fusions arise through an intermediate stage of physical clustering of the component genes (Yanai et al., 2002) within operons. Fusion of two enzymes in a biosynthetic pathway may not necessarily produce a functional fusion protein due to structural constraints, and non-functional or dysfunctional fusion genes would be unlikely to persist in genomes. Enzymes in metabolic pathways can physically associate without fusion events, to enhance biosynthetic efficiency by forming complexes known as metabolons, which may involve metabolic channeling (Srere, 2000, Srere, 1987, Welch, 1977). A metabolon is a transient multienzyme complex involving metabolically sequential enzymes, and metabolons have been observed for tricarboxylic acid cylce enzymes in Pseudomonas aeruginosa (Mitchell, 1996), for branched-chain amino acid catabolism from mammalian mitochondia (Islam et al., 2007, Islam et al., 2010), bicarbonate transport (Alvarez et al., 2005) and purine biosynthesis (An et al., 2008, An et al., 2010). Evidence of a polyamine biosynthetic metabolon (spermidine and spermine synthase) was found in the model plant Arabidopsis thaliana (Panicot et al., 2002).
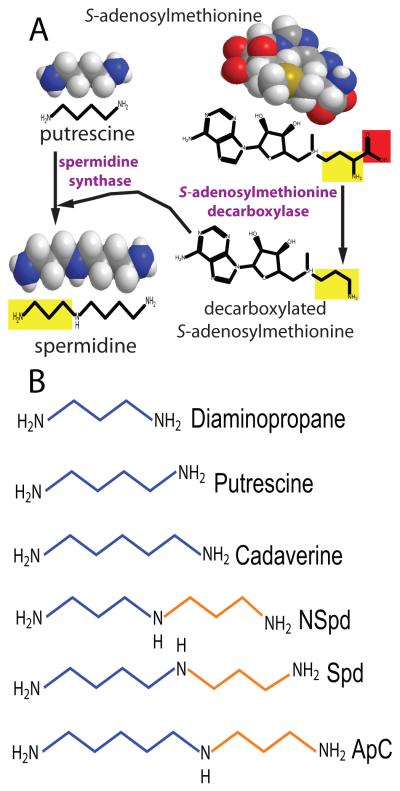
Polyamine biosynthesis is particularly interesting for studying the evolutionary mechanisms that form new pathways and generate biosynthetic diversity. Polyamines are small organic polycations that are found in all cells in all three domains of life and are essential for cell growth and proliferation in eukaryotes (Fig. 1). Biosynthesis of polyamines is modular, consisting of diverse modules for diamine, triamine and higher order polyamine formation. The diamine module converts an amino acid (glutamate, arginine, ornithine or lysine) directly or indirectly to a diamine. Triamines are formed from diamines by addition of an aminopropyl or aminobutyl group to form norspermidine (Lee et al., 2009, Cacciapuoti et al., 1986), spermidine (Tabor & Tabor, 1984), homospermidine (Shaw et al.), aminopropylcadaverine (Igarashi et al., 1986) or aminobutylcadaverine (Fujihara et al., 1995, Shaw et al., 2010). Longer linear or branched polyamine biosynthetic modules also exist (Knott et al., 2007, Knott, 2009). Different modules can make the same polyamine and different polyamines can be made using the same module (Lee et al., 2009, Shaw et al., 2010). In eukaryotes, archaea and many species of bacteria, the triamine spermidine is synthesized by transfer of an aminopropyl group to putrescine (Fig. 1). The aminopropyl group is transferred from decarboxylated S-adenosylmethionine (dcAdoMet) to putrescine by the aminopropyltransferase spermidine synthase (SpdSyn) (Wu et al., 2007), and the dcAdoMet is formed by S-adenosylmethionine decarboxylase (AdoMetDC) (Pegg, 2009). In some bacteria and archaea, these same enzymes can synthesize norspermidine from diaminopropane (Cacciapuoti et al., 1986) (Fig. 1). An alternative pathway for norspermidine biosynthesis based on asparate β-semialdehyde as source of the aminopropyl group is also found in some bacteria (Lee et al., 2009, Deng et al., 2010).
Fig. 1.
The decarboxylated S-adenosylmethionine pathway for polyamine biosynthesis. A, Formation of the triamine spermidine from the diamine putrescine by the sequential action of S- adenosylmethionine decarboxylase (AdoMetDC) and spermidine synthase (SpdSyn). The red box is the carboxyl group removed by AdoMetDC and the yellow box is the aminopropyl group transferred by SpdSyn from decarboxylated S-adenosylmethionine to putrescine to form spermidine. B, Structures of common diamine and triamine polyamines. Aminopropyl units transferred to the acceptor diamine by the aminopropyltransferase are shown in brown. NSpd, sym-norspermidine; Spd, spermidine; ApC, N1-aminopropylcadaverine.
AdoMetDC uses a pyruvoyl cofactor generated from an internal serine residue by autocatalytic self-cleavage of the proenzyme AdoMetDC protein to form an N-terminal β-subunit and a C-terminal α-subunit (Pegg, 2009). The pyruvoyl cofactor is formed at the N-terminus of the α-subunit. There are two classes of bacterial AdoMetDC: the Class 1a AdoMetDC is represented by the Escherichia coli enzyme and is Mg2+-activated (Toms et al., 2004, Bale & Ealick, Lu & Markham, 2007); the Class1b AdoMetDC is represented by the Thermotoga maritima enzyme and is not activated by Mg2+ and is a dimer (Toms et al., 2004). Eukaryotic AdoMetDC has evolved from the fusion of two bacterial Class 1 b-like AdoMetDC genes, fused in the same orientation and the autocatalytic cleavage site of the C-terminal AdoMetDC fusion component has been lost (Toms et al., 2004). The two bacterial AdoMetDC components of the eukaryotic AdoMetDC can be resolved only at the structural and not at the sequence level. Once AdoMet is decarboxylated by AdoMetDC, it is committed to polyamine biosynthesis (Pegg, 2009).
Aminopropyltransferases such as spermidine synthase are also found in eukaryotes, archaea and many bacteria (Pegg & Michael, 2010). The molecular mechanisms most prominent in the evolution of aminoproyltransferases are gene duplication followed by change of substrate specificity. All aminopropyltransferases bind dcAdoMet and transfer an aminopropyl group to their diamine or polyamine co-substrate. The product of AdoMetDC, dcAdoMet, is thus a co-substrate of aminopropyltransferases, which may use diaminopropane, putrescine, cadaverine, agmatine, norspermidine, spermidine, and homospermidine as co-substrates as well as longer chain and branched polyamines (Fuell et al., 2010). Additionally, with spermine synthase and thermospermine synthase, the aminopropyl group can be transferred to the N1- or N8-amino group of spermidine, respectively (Knott et al., 2007).
We noticed that gene fusions of AdoMetDC and an aminopropyltransferase are present in diverse bacterial genomes. Considering that the unfused SpdSyn of T. maritima is a tetramer (Wu et al., 2007), and its AdoMetDC is a dimer, we sought to determine if fusion proteins of these two enzymes types are functional, how they evolved and how extensive gene fusion is in polyamine metabolism. Here we show that bacterial AdoMetDC-aminopropyltransferase fusion proteins are enzymatically active, possessing AdoMetDC activity and functionally diverse aminopropyltransferase domains. Some of the fusion genes have evolved independently in different bacterial phyla although some have been acquired by horizontal and vertical gene transfer. We show that the fusion proteins from the β-proteobacterium Delftia acidovorans and δ-proteobacterium Syntrophus aciditrophicus are able to synthesize spermidine de novo from putrescine in cells lacking AdoMetDC and SpdSyn genes, demonstrating that the fusion proteins are functional covalent modules for polyamine biosynthesis.
Results
AdoMetDC-aminopropyltransferase gene fusions are present in diverse bacterial phyla
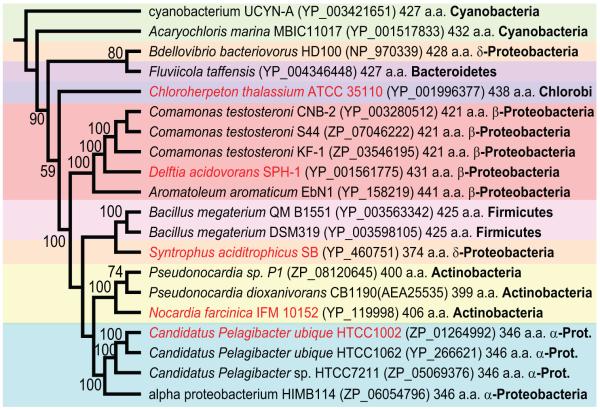
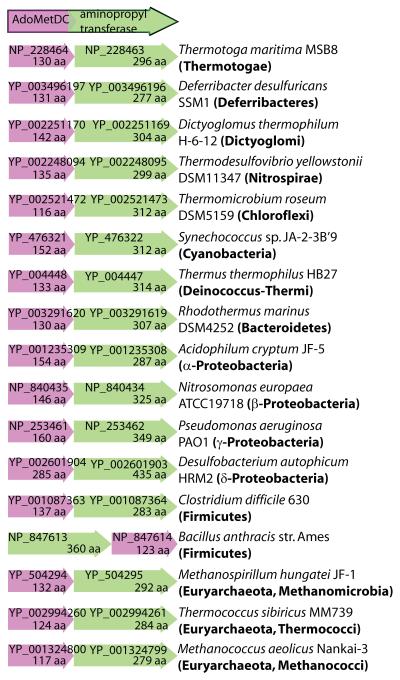
We identified gene fusions encoding ORFs with N-terminal AdoMetDC and C-terminal aminopropyltransferase domains in genomes from diverse bacterial phyla (Fig. 2). All AdoMetDC domains of the fusion proteins are of the Class 1b type (Bale & Ealick, 2010), similar to the AdoMetDC of T. maritima, and there are no fusions containing the Class 1a, E. coli-like AdoMetDC genes. AdoMetDC-aminopropyltransferase gene fusions are found in the α-, β- and δ-Proteobacteria, Cyanobacteria, Chlorobi, Bacteroidetes, Firmicutes and Actinobacteria and the encoded proteins range in size from 346 aa for the α-proteobacterium Ca. P. ubique protein to 441 aa for the β-proteobacterium Aromatoleum aromaticum EbN1 protein. The genome sequence of the cyanobacterium Acaryochloris marina MBIC11017 is annotated as having a discrete aminopropyltransferase ORF with no AdoMetDC ORF nearby. However, close inspection of the relevant genomic vicinity reveals that the aminopropyltransferase is longer at the N-terminus than annotated and the N-terminal extension is an AdoMetDC orthologous sequence, which, for whatever reason, was not annotated in the genome sequence. This AdoMetDC domain is the only AdoMetDC orthologous sequence in the A. marina genome. The complete amino acid sequence of the A. marina MBIC11017 AdoMetDC-aminopropyltransferase fusion ORF is shown aligned with the other fusion proteins in Fig. S1.
Fig. 2.
Unrooted Neighbour-Joining tree of AdoMetDC-aminopropyltransferase proteins. Sequences were aligned with ClustalW and the tree was made with PAUP* (Swofford, 2000). Numbers represent percentage support (above 50%) for nodes based on 1000 bootstrap replicates. Protein accession numbers in GenBank are displayed in parentheses. The alignment on which the Neigbour Joining tree is based is presented in Fig. S1. The full amino acid sequence of the Acaryochloris marina MBIC11017 AdoMetDC-aminopropyltransferase ORF is shown aligned with the other fusion proteins in Fig. S1. Accession number of the A. marina MBIC11017 AdoMetDC-aminopropyltransferase ORF refers only to the aminopropyltransferase domain. Fusion proteins analysed in the manuscript are indicated in red font.
The α-proteobacterium Ca. P. ubique is extremely abundant in the oceans (Rusch et al., 2007), and AdoMetDC-aminopropyltransferase gene fusion sequences are abundant in the marine metagenome. We conservatively found in the marine metagenome 124 fusion proteins clustering with the three Ca. P. ubique fusion protein sequences, one clustering with the fusion protein fromthe chlorobi species Chloroherpeton thalassium ATCC 35110 and two with the fusion protein from α-proteobacterium HIMB114 (Fig. S2). Sequence identity between the two most diverged marine metagenome fusion proteins that cluster with Ca. P. ubique sequences is only 61%, each protein sequence being exactly the same size with no gaps in the alignment (Fig. S3).
The gene fusion of AdoMetDC and an aminopropyltransferase from Ca. P. ubique is functional
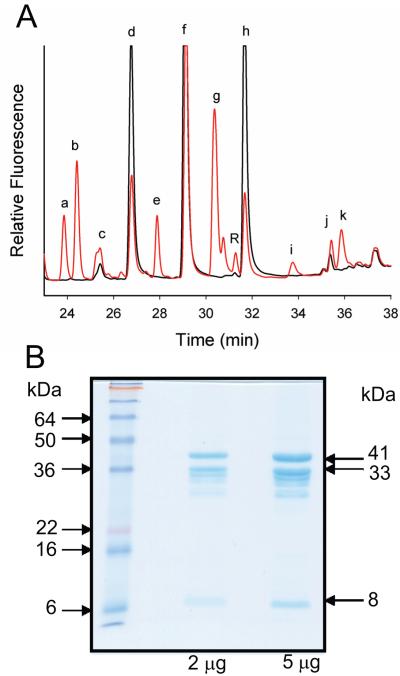
A gene fusion in the marine α-proteobacterium Can. P. ubique HTCC1002 encoding a 346 aa ORF (ZP_012649992) with N-terminal AdoMetDC and C-terminal aminopropyltransferase domains was synthesized with E. coli-optimised codons. An N-terminally T7-tagged Pelagibacter fusion ORF in pET21 was expressed in BL21 E. coli cells and induced at 30 °C for 24 h. Fig. 3A shows the polyamine profile detected by HPLC, of E. coli cells expressing the Pelagibacter fusion protein compared to cells expressing the empty pET21 vector. The host E. coli strain accumulates putrescine, cadaverine and spermidine. Several novel peaks appeared due to the expression of the Pelagibacter AdoMetDC-aminopropyltransferase fusion gene, the most prominent of which was norspermidine (peak g, Fig. 3A), a non-native polyamine in E. coli. Surprisingly, accumulation of non-native diaminopropane was observed, although this diamine could not have been a direct product of the fusion protein. Novel peaks of unknown identity appeared, including a prominent peak eluting between putrescine and cadaverine (peak e, Fig. 3A).
Fig. 3.
Expression of the Candidatus Pelagibacter ubique AdoMetDC-aminopropyltransferase fusion protein in E. coli. A, HPLC profile of Accu-Tag labelled polyamines detected by fluorescence in cellular extracts of BL21 E. coli expressing the Ca. P. ubique HTCC1002 AdoMetDC-aminopropyltransferase fusion ORF (ZP_012649992) in pET21b as an N-terminally T7-tagged protein. Black trace, empty pET21b plasmid; red trace, fusion protein in pET21b; a, b, e, i, k; unknown peaks; c, diaminopropane; d, putrescine; f, cadaverine; g, norspermidine; h, spermidine; j, internal standard (diaminoheptane). Identities based on comparison with authentic standards. B, Purified proteins obtained from E. coli BL21 cells expressing the Ca. P. ubique HTCC1002 AdoMetDC-aminopropyltransferase fusion ORF (ZP_012649992) in pQE80 as an N-terminal FLAG-tagged, C-terminal (6x)his-tagged protein and purified by nickel affinity column. The expected sizes of the unprocessed fusion protein (41 kDa) and the processed C-terminal α-(33 kDa) and N-terminal β-(8 kDa) polypeptides are indicated.
For the AdoMetDC domain of the Pelagibacter fusion protein to be enzymatically active, autocatalytic cleavage of the AdoMetDC proenzyme sequence must occur. To analyse the proteolytic processing of the Pelagibacter fusion protein, the fusion ORF was recloned with an 11 aa N-terminal FLAG-tag and an 8 aa C-terminal his-tag in pQE80 to allow greater protein recovery. The 41.3 kDa proenzyme-containing Pelagibacter fusion protein, if processed, should produce an approximately 8 kDa β-subunit consisting of the N-terminal part of the processed AdoMetDC domain and a 33.3 kDa α-domain consisting of the rest of the fusion protein. In Fig. 3B, prominent bands corresponding to the unprocessed fusion protein and the processed α- and β-subunits can be seen and approximately half of the fusion protein is unprocessed after expression in E. coli. The AdoMetDC specific activity of the purified Pelagibacter fusion protein, which was a mix of unprocessed and processed forms, was determined in the absence and presence of different polyamines (Table 1). AdoMetDC activity was stimulated to some extent by all polyamines tested but not by magnesium. Unusually, the aminopropyltransferase activity of the Pelagibacter fusion protein was promiscuous and clearly detected with a number of amine acceptors including putrescine, cadaverine, agmatine and spermidine (Table 2).
Table 1.
AdoMetDC activity of Ca. P. ubique fusion protein
| Additions to assay | nmol CO2/h/mg protein |
|---|---|
| No addition | 20.7 |
| 20 mM putrescine | 174.6 |
| 5 mM putrescine | 104.5 |
| 2.5 mM putrescine | 81.0 |
| 1.5 mM putrescine | 34.8 |
| 5 mM diaminopropane | 84.4 |
| 5 mM cadaverine | 155.0 |
| 5 mM spermidine | 263.8 |
| 5 mM agmatine | 101.4 |
| 10 mM MgCl2 | 13.2 |
Table 2.
Aminopropyltransferase activity of Ca. P. ubique fusion protein
| Aminopropyltransferase activity with 5 mM acceptor |
pmol MTA/h/mg protein |
|---|---|
| putrescine | 1387 |
| cadaverine | 1291 |
| spermidine | 1225 |
| agmatine | 1033 |
In 100 mM Na phosphate pH 7.5, 0.25 μM [35S]dcSAM (MTA, methylthioadenosine)
Functional diversity of fusion proteins from distant bacterial phyla
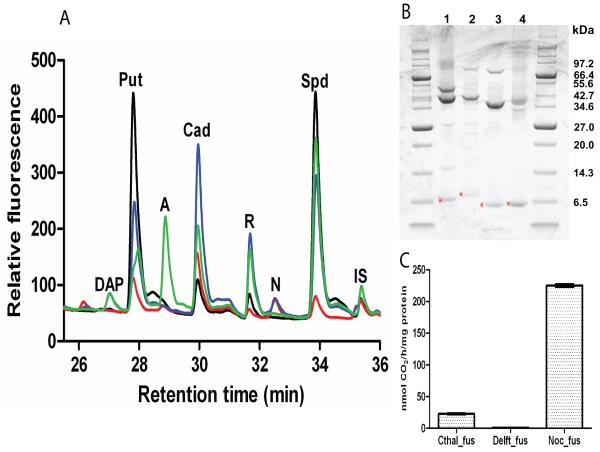
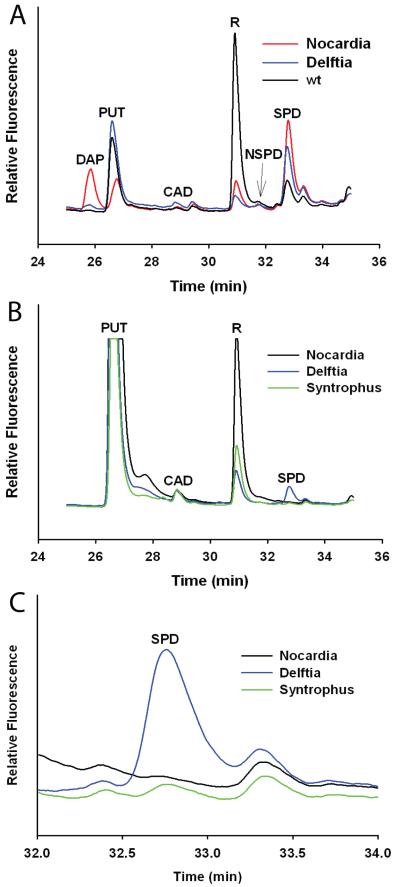
Genes encoding AdoMetDC-aminopropyltransferase fusion proteins from the chlorobi species C. thalassium ATCC 35110 (GenBank acc. no. YP_001996377), the actinobacterium Nocardia farcinica IFM 10152 (acc. no. YP_119998), the β-proteobacterium Delftia acidovorans SPH-1 (acc. no. YP_001561775) and the δ-proteobacterium Syntrophus aciditrophicus SB (acc. no. YP_460751) were synthesized with E. coli-optimised codons. Each fusion gene was cloned in pET21b and expressed in E. coli with an N-terminal T7 affinity tag. Extracts from E. coli cells expressing the induced fusion proteins were analysed by HPLC. Novel peaks appeared after expression of the fusion proteins (Fig. 4A): norspermidine accumulated with expression of the Chloroherpeton fusion protein, diaminopropane and norspermidine with the Delftia protein, diaminopropane and an unknown peak with the Nocardia protein but no novel peaks with the Syntrophus protein, although this probably reflects the insolubility of the Syntrophus protein, which was found almost entirely within the insoluble protein fraction. The abundant novel peak that accumulated on expression of the Nocardia fusion protein, eluting between the putrescine and cadaverine peaks, coincides with one of the Pelagibacter fusion protein peaks (peak e, Fig. 3A).
Fig. 4.
Expression of AdoMetDC-aminopropyltransferase fusion genes in E. coli. A, HPLC profile of Accu-Tag labelled polyamines detected by fluorescence in cellular extracts of E. coli BL21 expressing AdoMetDC-aminopropyltransferase fusion genes from Chloroherpeton thalassium ATCC 35110 (red), Delftia acidovorans SPH-1 (blue), Nocardia farcinica IFM 10152 (green), Syntrophus aciditrophicus SB (orange) and empty expression plasmid (black). DAP, diaminopropane; Put, putrescine; X, unknown; Cad, cadaverine; R, fluorescent label; NSpd, norspermidine; Spd, spermidine; IS, internal standard (diaminoheptane). B, T7 affinity tag purified proteins (4 μg per lane). The bands indicated by red asterisks are the putative processed N-terminal β-subunits of the fusion proteins and the calculated molecular masses are shown. 1, C. thalassium ATCC 35110 (predicted unprocessed fusion protein 49.9 kDa, processed α-subunit 43.0 kDa, β-subunit 7.0 kDa); 2, D. acidovorans SPH-1 (estimated unprocessed fusion protein 47.1 kDa, processed α-subunit 39.9 kDa, β-subunit 7.2 kDa); 3, N. farcinica IFM 10152 (unprocessed fusion protein 44.5 kDa, processed α-subunit 37.3 kDa, β-subunit 7.3 kDa); 4, S. aciditrophicus SB (unprocessed fusion protein 39.0 kDa, processed α-subunit 32.2 kDa, β-subunit 6.8 kDa). C, Specific AdoMetDC activities of the purified N-terminally T7-tagged fusion proteins. Cthal_fus, C. thalassium ATCC 35110; Delft_fus, D. acidovorans SPH-1; Noc_fus; N. farcinica IFM 10152.
Processing and activity of the AdoMetDC domains
The relative amount of unprocessed and processed fusion protein in E. coli extracts (Fig. 4B) was assessed for the Chloroherpeton, Delftia, Nocardia and Pelagibacter fusion proteins (the Pelagibacter protein was expressed from pET21b with an N-terminal T7-tag for comparison with the other fusion proteins). The Delftia and Nocardia fusion proteins were almost fully processed, the Chloroherpeton protein was more than 50% processed, and the Pelagibacter protein less than 50% processed. However, the Synthrophus enzyme was too insoluble for the analysis. Affinity tag-purified proteins for the Chloroherpeton, Delftia and Nocardia fusions were assayed for AdoMetDC activity (Fig. 4C). The Delftia fusion protein exhibited barely detectable activity; the Chloroherpeton fusion protein exhibited AdoMetDC specific activity similar to the unstimulated Pelagibacter activity described above, whereas the Nocardia fusion protein exhibited AdoMetDC activity similar to the Pelagibacter protein stimulated by the presence of 5 mM spermidine (Table 1).
De novo synthesis of spermidine from putrescine by the fusion genes
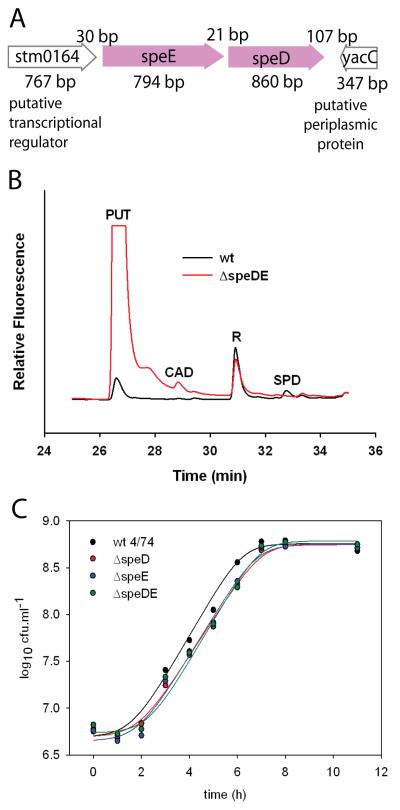
To assess whether AdoMetDC-aminopropyltransferase fusion proteins could synthesize spermidine, we deleted the AdoMetDC (speD) and SpdSyn (speE) genes of S. Typhimurium 4/74 using a homologous recombination approach and expressed them in this deletion strain. The speD and speE genes are immediately adjacent in the genome of S. Typhimurium 4/74 (Fig. 5A) and are probably co-transcribed. Deletion of the speD/speE genes abolished spermidine accumulation and caused a massive over-accumulation of putrescine in the ΔspeDE gene deletion strain (Fig. 5B). The ΔspeDE gene deletion strain and the individual AspeD and AspeE gene deletion strains grew approximately 40% less efficiently than the parental strain in defined, minimal liquid medium (Fig. 5C). Each AdoMetDC-aminopropyltransferase fusion gene was expressed in S. Typhimurium 4/74 and the derived ΔspeDE gene deletion strain, from a low copy number plasmid pWSK29 using a constitutive T7 promoter. The Pelagibacter fusion gene appeared to be completely inactive or non-expressed in either the parental or ΔspeDE gene deletion strain (results not shown). Expression of the Delftia fusion protein in the parental S. Typhimurium 4/74 strain resulted in a small accumulation of diaminopropane and norspermidine (Fig. 6A). In contrast, expression of the Nocardia fusion protein in the parental S. Typhimurium 4/74 strain resulted in an accumulation of more diaminopropane than putrescine, however, this did not result in norspermidine accumulation. Due to technical difficulties we were unable to express the Syntrophus fusion gene in the S. Typhimurium 4/74 parental strain. When expressed in the ΔspeDE gene deletion strain, neither the Delftia nor Nocardia fusion genes caused an accumulation of diaminopropane or norspermidine but there was clearly accumulation of spermidine with expression of the Delftia fusion gene (Figure 6B). Close inspection of the HPLC polyamine profile of the ΔspeDE gene deletion strain expressing the Syntrophus fusion gene revealed a small accumulation of spermidine only (Fig. 6C).
Fig. 5.
Deletion of the Salmonella enterica sv Typhimurium 4/74 speD and speE ORFs. A, Schematic representation of the organisation of the speD (AdoMetDC) and speE (spermidine synthase) ORFs in the genome of S. Typhimurium 4/74. B, HPLC profile of Accu-Tag labelled polyamines detected by fluorescence in late-log phase cellular extracts of the parental S. Typhimurium 4/74 strain (black) and the ΔspeDE gene deletion strain (red). PUT, putrescine; CAD, cadaverine; SPD, spermidine; R, fluorescent label. C, Growth of the parental S. Typhimurium 4/74 strain (wt 4/74) and the AspeDE, AspeD and AspeE gene deletion strains in polyamine-free liquid M9 defined minimal medium.
Fig. 6.
Expression of AdoMetDC-aminopropyltransferase fusion genes in Salmonella enterica sv Typhimurium 4/74. A, HPLC profile of Accu-Tag labelled polyamines detected by fluorescence in the S. Typhimurium 4/74 parental strain expressing AdoMetDC-aminopropyltransferase genes from N. farcinica IFM 10152 (red), D. acidovorans SPH-1 (blue) and the background host polyamines (black). B, Polyamine profile of the S. Typhimurium 4/74 ΔspeDE gene deletion strain expressing AdoMetDC-aminotransferase fusion genes from N. farcinica IFM 10152 (black), D. acidovorans SPH-1 (blue) and S. aciditrophicus SB (green). C, Detail from the polyamine profile of the S. Typhimurium 4/74 ΔspeDE gene deletion strain expressing AdoMetDC-aminotransferase fusion genes from N. farcinica IFM 10152 (black), D. acidovorans SPH-1 (blue) and S. aciditrophicus SB (green). Note the minor peak for spermidine in cells expressing the S. aciditrophicus SB (green) fusion protein. DAP, diaminopropane; PUT, putrescine; CAD, cadaverine; NSPD, norspermidine; SPD, spermidine; R, fluorescent label.
Origin of the AdoMetDC-aminopropyltransferase gene fusions
Of the bacterial genomes containing the AdoMetDC-aminopropyltransferase gene fusions (Fig. 2), none contain another unfused AdoMetDC paralogous gene except the two Bacillus megaterium strains (QM B1551 and DSM319). The AdoMetDC domain of the fusion protein from both strains of B. megaterium possesses an aberrant sequence at the putative proenzyme processing site (Fig. S1), with a threonine rather than a glutamate residue immediately preceeding the serine that generates the pyruvoyl cofactor. It is possible than the AdoMetDC domain of the B. megaterium fusion protein is therefore inactive, and the AdoMetDC function may be replaced by the paralogous unfused AdoMetDC gene, which possesses a normal proenzyme processing site.
Bacterial AdoMetDC-aminopropyltransferase gene fusions described above consist of an N-terminal class 1b AdoMetDC domain, similar to AdoMetDC of T. maritima, and a C-terminal aminopropyltransferase domain. The same arrangement, but of unfused Class 1b AdoMetDC and aminopropyltransferase unfused ORFs in the same order and orientation, is found in diverse bacterial and archaeal genomes (Fig. 7). An exception is Bacillus anthracis str. Ames, where the relative positions of the ORFs are reversed. Pairs of adjacent AdoMetDC and aminopropyltransferase ORFs are found for the Class 1a AdoMetDCs typified by the E. coli AdoMetDC. However, in the case of the class 1a AdoMetDC ORFs, the AdoMetDC ORFs are immediately downstream of the aminopropyltransferase ORFs (Fig. S4A). One possible mechanism for formation of the AdoMetDC-aminopropyltransferase fusions is point mutation of the AdoMetDC termination codon to allow translational readthrough into the downstream aminopropyltransferase ORF. This should produce a fusion protein if the two ORFs are in phase and there are no termination codons in the intervening linker region. Examples of where a simple nucleotide mutation of an AdoMetDC termination codon would result in an AdoMetDC-aminopropyltransferase fusion protein are shown in Fig. S5. For other pairs of unfused AdoMetDC and aminopropyltransferase ORFs, the downstream ORF is out of phase or there are several intervening termination codons. In such cases, short insertions or deletions would be required to form an AdoMetDC-aminopropyltransferase fusion protein.
Fig. 7.
Clustering of AdoMetDC and aminopropyltransferase genes in diverse bacterial and archaeal genomes. AdoMetDC (red) and aminopropyltransferase (green) ORFs cluster frequently in the same genomic arrangement found in the AdoMetDC-aminopropyltransferase fusion proteins. Protein accession numbers are displayed on the cartoon ORFs along with the amino acid size of the encoded protein.
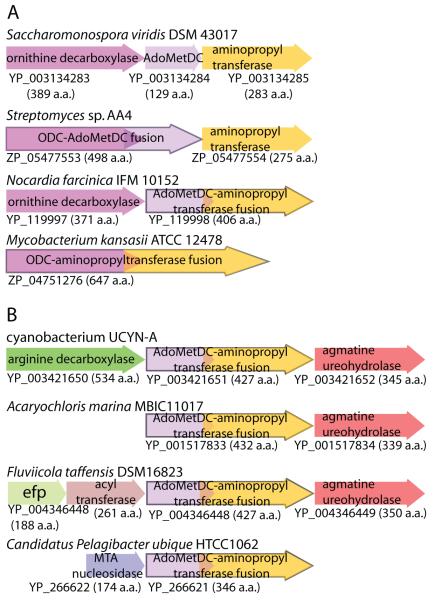
That fusion of physically adjacent ORFs is likely to be the mechanism by which fusion proteins arise is clearly illustrated by the actinobacterial Nocardia AdoMetDC-aminopropyltransferase fusion. A cluster of discrete, unfused ornithine decarboxylase, AdoMetDC and aminopropyltransferase ORFs in the same orientation is found in a number of actinobacterial genomes, eg. Saccharomonospora viridis DSM 43017 (Fig. 8A). In the genome of the actinobacterium Streptomyces sp. AA4, the ornithine decarboxylase ORF has fused with the immediately downstream AdoMetDC ORF to form a 498 aa ODC-AdoMetDC fusion protein while maintaining a separate downstream aminopropyltransferase ORF (Fig. 8A). In Nocardia farcinica IFM 10152, the upstream ODC ORF is maintained but the downstream AdoMetDC and aminopropyltransferase ORFs have fused to form the Nocardia AdoMetDC-aminopropyltransferase fusion protein described above. In the actinobacterium Mycobacterium kansasii ATCC 12478, the upstream ODC ORF has fused with the downstream aminopropyltransferase ORF to create a 647 aa ODC-aminopropyltransferase fusion protein but the intervening AdoMetDC ORF has been eliminated (Fig. 8A).
Fig. 8.
The formation of fusion proteins by coalescence of adjacent ORFs. A, Formation of the full panoply of possible fusion proteins from a polyamine biosynthetic gene cluster in actinobacterial genomes. Protein accession numbers are indicated below the corresponding ORFs. Ornithine decarboxylase (ODC, magenta); AdoMetDC (mauve), aminopropyltransferase (yellow). Fusion ORFs are outlined in black. B, Polyamine-related gene clusters containing other AdoMetDC-aminopropyltransferase fusion protein ORFs.
The mechanisms for formation of gene fusions proposed above suggests that fusions might arise independently in different bacterial lineages. Different polyamine products from the various AdoMetDC-aminopropyltransferase fusion proteins expressed in E. coli support distinct evolutionary origins. Phylogenetic comparison of the AdoMetDC domains of the fusion proteins indicates that the fusion proteins have arisen independently in the α- and β-Proteobacteria and in Actinobacteria and Firmicutes (Fig. S6). Horizontal gene transfer is probably responsible for the cluster of fusion proteins represented by the cyanobacteria UCYN-A and Acaryochloris marina MBIC11017, the Chlorobi species Chloroherpeton thalassium ATCC 35110, the Bacteroidetes species Fluviicola taffensis DSM16823 and the δ-proteobacterium Bdellovibrio bacteriovorus HD100 (Fig. S6). Adjacent pairs of unfused AdoMetDC and an aminopropyltransferase exhibiting a high degree of identity to the horizontally acquired fusion proteins described above are present in the fusobacterium Ilyobacter polytropus DSM2926 (YP_003967334 and YP_003967333) and in the Deferribacteres species Deferribacter desulfuricans SSM1 (YP_003496197 and YP_003496196). No other species within the Fusobacteria or Deferribacteres phyla contain adjacent AdoMetDC and aminopropyltransferase ORFs, suggesting that the I. polytropus DSM2926 and D. desulfuricans SSM1 AdoMetDC and aminopropyltransferase pairs were originally acquired as a fusion protein which then underwent scission to recreate discrete AdoMetDC and aminopropyltransferase ORFs. Thus there is strong evidence for both independent formation of the fusion proteins in different phyla, for horizontal (and vertical in the β-Proteobacteria) gene transfer of fusion protein genes, and for scisson of horizontally acquired fusion proteins to recreate individual AdoMetDC and aminopropyltransferase ORFs.
Some AdoMetDC-aminopropyltransferase fusion genes are found in polyamine-related gene clusters/operons. The cyanobacterium UCYN-A AdoMetDC-aminopropyltransferase fusion ORF is flanked upstream by an alanine racemase-fold arginine decarboxylase ORF and downstream by an agmatine ureohydrolase ORF (Fig. 8B). In the cyanobacterium A. marina MBIC11017, and in the Bacteroidetes species F. taffensis DSM16823, the AdoMetDC-aminopropyltransferase fusion ORF is immediately upstream of an agmatine ureaohydrolase ORF (Figure 8B). An ORF for methylthioadenosine nucleosidase, which metabolises methylthioadenosine, a co-product of aminopropyltransferase activity, is found immediately upstream of the AdoMetDC-aminopropyltransferase fusion ORF in the Ca. P. ubique genomes (Fig. 8B).
Additional polyamine biosynthetic gene fusions
Other polyamine-related gene fusions are present in different species. Gene fusions of agmatine deiminase and N-carbamoylputrescine amidohydrolase are found in four methanomicrobial species of the euryarchaeota (Fig. S4B). These two enzymes are responsible for the conversion of agmatine to putrescine via the intermediate metabolite N-carbamoylputrescine. The fusion protein has an N-terminal N-carbamoylputrescine amidohydrolase domain and a C-terminal agmatine deiminase domain. A similar arrangement of adjacent unfused N-carbamoylputrescine amidohydrolase and agmatine deiminase ORFs in found in diverse bacterial phyla although no bacterial genome appears to contain a fusion of the two ORFs (Fig. S4B). In eukaryotes, we found a gene fusion of a eukaryotic AdoMetDC and an ornithine decarboxylase in two species of single-celled prasinophyte green algae, Micromonas pusilla CCMP1545 (EEH58717, 1019 a.a.) and Micromonas sp. RCC299 (XP_002502359, 1039 a.a.). The AdoMetDC domain is N-terminal and the ODC domain C-terminal, and corresponding mRNA 5′ regions have a small upstream ORF similar to the upstream ORF in the mRNA 5′ ends of the related Ostreococcus AdoMetDC-encoding genes (Ivanov et al.). Although the Micromonas AdoMetDC-ODC gene fusions are arranged similarly to the Plasmodium AdoMetDC-ODC fusion genes (Muller et al., 2000), these fusion genes represent entirely independent eukaryotic gene fusion events because both domains of the Micromonas fusion protein exhibit greatest homology with the individual component genes from green algae and plants and present only distant similarity to the Plasmodium sequences. All gene fusions involved in polyamine biosythesis discovered to date are displayed in Fig. S7.
Discussion
Gene fusion is an important mechanism in the evolution of proteins and metabolic pathways. Polyamine biosynthesis is ideal for investigating the occurrence, evolution and function of gene fusions in metabolic pathways because polyamines are ubiquitous and the biosynthetic pathways are short. We wondered how pervasive are gene fusions in polyamine metabolism; how did the fusions evolve; are the fusion proteins functional; is the presence of fusions due to vertical descent, horizontal gene transfer or independent gene fusion events? An AdoMetDC-aminopropyltransferase fusion protein encoded by the genome of Ca. P. ubique HTCC102 was processed as expected within the AdoMetDC domain when expressed in E. coli. Purified recombinant Pelagibacter fusion protein exhibited AdoMetDC activity that was not stimulated by magnesium ions but was stimulated albeit weakly by diamines and spermidine. The aminopropyltransferase activity was highly promiscuous, recognising putrescine, cadaverine, spermidine and agmatine with similar efficiency as substrates. This substrate promiscuity explains the variety of products detected when the Pelagibacter gene fusion was expressed in E. coli.
Fusion protein activity reveals latent spermidine catabolic capacity in E. coli
Surprisingly, the E. coli cells expressing the Pelagibacter fusion gene accumulated diaminopropane and norspermidine, which are non-native polyamines in E. coli. Formation of norspermidine is explicable if diaminopropane is used as a substrate by the fusion protein. The accumulation of diaminopropane can only be explained by catabolism of spermidine. Spermidine dehydrogenase activities have been reported in the γ-proteobacteria Citrobacter freudii, Serratia marescens and Pseudomonas aeruginosa (Hisano et al., 1992, Tabor & Kellogg, 1970, Dasu et al., 2006). Each of these spermidine dehydrogenases cleaves spermidine to release diaminopropane and 4-aminobutyraldehyde. It is conceivable that the expression of the Pelagibacter AdoMetDC-aminopropyltransferase in E. coli produces an unprecedented surfeit of spermidine that results in catabolism of the excess spermidine by a latent spermidine dehydrogenase activity belonging to a promiscuous dehydrogenase. There is no orthologue of the cloned P. aeruginosa spermidine dehydrogenase spdH gene in the E. coli genome and no spermidine dehydrogenase activity has been documented for E. coli, so another dehydrogenase is likely to be responsible. Indeed, the P. aeruginosa spermidine dehydrogenase, which has a Km for spermidine of 36 μM, is not involved in spermidine catabolism in P. aeruginosa under normal physiological conditions due to low expression levels (Dasu et al., 2006). It is likely that the AdoMetDC-aminopropyltransferase fusion protein circumvents normal polyamine biosynthetic homeostatic mechanisms in E. coli and S. Typhimurium, causing excess spermidine accumulation and the novel formation of diaminopropane through spermidine catabolism. Norspermidine could then be synthesized by aminopropylation of the diaminopropane by the Pelagibacter fusion protein. It is also theoretically possible that accumulation of aminopropylcadaverine or aminopropylagmatine generated through the activity of the Pelagibacter fusion protein could result in catabolism of these compounds, in addition to spermidine, to release diaminopropane. When the Delftia fusion protein was expressed in the parental S. Typhimurium 4/74 strain, which normally accumulates spermidine, both diaminopropane and norspermidine were produced. In contrast, when the Delftia fusion protein was expressed in the ΔspeDE gene deletion mutant in which endogenously-produced spermidine is entirely absent, diaminopropane and norspermidine were not produced, however, spermidine was synthesized by the fusion protein through aminopropylation of putrescine. This result further supports the hypothesis that catabolism of excess spermidine, produced by the fusion protein in addition to the endogenous spermidine pool, through a latent promiscuous spermidine dehydrogenase activity, is responsible for the formation of diaminopropane.
Disparate origins of the AdoMetDC-aminopropyltransferase gene fusions
Accumulation of the non-native norspermidine in E. coli was also observed after expression of the Chloroherpeton and Delftia fusion genes. Diaminopropane in E. coli was detected after expression of the Nocardia fusion gene but there was no accumulation of norspermidine; the same result was obtained in the parental S. Typhimurium strain with the Nocardia fusion protein. This suggests that norspermidine is synthesized in E. coli by the Pelagibacter, Chloroherpeton and Delftia fusion proteins rather than by the endogenous E. coli SpdSyn and that the Nocardia fusion protein does not efficiently recognise diaminopropane as a substrate. The various fusion proteins synthesized different polyamine profiles in E. coli, indicating different substrate specificites of their respective aminopropyltransferase domains and suggesting that the fusion proteins evolved by independent gene fusion events involving distinct aminopropyltransferase genes. Further supporting independent gene fusion events, the AdoMetDC domain of the fusion proteins exhibits a highly scattered phyletic distribution (Fig. S6). There is a strong possibility of horizontal gene transfer being involved in the distribution of the Bdellovibrio (δ-Proteobacteria), Chloroherpeton (Chlorobi), cyanobacterium UCYN-A, Acaryochloris (Cyanobacteria) and Fluviicola (Bacteroidetes) fusion proteins, which are closely related at the amino acid sequence level. The sequence similarity for these fusions extends across both the AdoMetDC and aminpropyltransferase domains, and the horizontal gene transfer includes an adjacent putrescine-synthesising agmatine ureohydrolase gene in three of the species (Fig. 8) further supporting one gene fusion event and subsequent horizontal gene transfer. There also appears to be vertical inheritence for the five β-proteobacterial fusion genes.
The presence of a fused AdoMetDC-aminopropyltransferase gene in the genome of Ca. P. ubique is interesting in the context of the highly streamlined genome of this SAR11 clade marine species, which is amongst the smallest free-living bacterial genomes known (Giovannoni et al., 2005). The fusion protein is also the smallest encoded by the various AdoMetDC-aminopropyltransferase fusion genes. Ca. P. ubique is also extremely abundant at the ocean surface, the SAR11 clade comprising 12% of total marine prokaryotic biomass (Morris et al., 2002), and it exhibits a high level of intra-population sequence diversity, which can also be seen from the diversity of the AdoMetDC-aminopropyltransferase fusion protein sequences from the Global Ocean Sampling ocean metagenome indicated in supplementary Fig. S2. The nitrogen-fixing marine cyanobacterium UCYN-A also possess a highly streamlined genome (1.44 Mbp) with reduced metabolic capabilities (Tripp et al., 2010) and it too contains a fused AdoMetDC-aminopropyltransferase. However, the AdoMetDC-aminopropyltransferase genes fusions from these two pelagic species probably have independent origins. The cyanobacteria UCYN-A fusion protein is much more similar to that of the large-genomed (8.3 Mbp) marine cyanobacterium A. marina (Swingley et al., 2008) and it is possible that both the fusion protein and an adjacent agmatine ureohydrolase (Fig. 8b) have been horizontally transferred between the species as a gene cluster/operon. As discussed above, cyanobacterium UCYN-A and A. caryochloris fusion proteins are very similar to the fusion proteins from the marine chlorobi species C. thalassium and the river dwelling species B. bacteriovorus (δ-Proteobacteria) and F. fluviicola (Bacteroidetes). The acquisition by horizontal transfer of these related fusion genes may be a result of marine/aquatic interaction.
The mechanism of the gene fusion events between AdoMetDC and aminopropyltransferase genes is suggested by the prevalence of juxtaposed AdoMetDC and aminopropyltransferase gene pairs in diverse bacterial and archaeal phyla. In almost all cases, the class 1b AdoMetDC gene is found immediately upstream of the aminopropyltransferase gene and both genes are oriented in the same direction. Mutations allowing readthrough from the AdoMetDC ORF to the aminopropyltransferase ORF are the most probable cause of the fused protein. This putative mechanism is strongly supported by the different gene fusions found in some actinobacterial genomes where a cluster of unfused ODC/AdoMetDC/aminopropyltransferase ORFs has given rise to ODC-AdoMetDC, AdoMetDC-aminopropyltransferase, and ODC-aminopropyltransferase fusions. Intriguingly, although adjacent pairs of Class 1a AdoMetDC and aminopropyltransferase ORFs are widespread in γ-Proteobacteria, no fusions between the two ORFs are present. This may be due to the fact that the Class 1a AdoMetDC ORF is always downstream of the aminopropyltransferase ORF, and fusion might interfere with correct processing of the AdoMetDC proenzyme domain. The structural interference between protein domains may therefore be an important factor limiting gene fusion in general. There may be counter-selection of specific configurations of open reading frames in operons that, if fused, would produce a non-functional or dysfunction fusion protein due to steric or mechanistic interference.
Gene fusions are pervasive in polyamine metabolism
We also observed the presence of fusions of N-carbamoylputrescine amidohydrolase and agmatine deiminase in closely related methanomicrobial species of the Archaea. The NCPAH-AIH fusion protein is again probably a consequence of mutation and readthrough from juxtaposed NCPAH and AIH ORFs which are widepread in bacterial phyla as adjacent pairs. Gene fusions between polyamine biosynthetic enzymes can be found for other steps in polyamine biosynthesis. Previously we showed a functional gene fusion consisting of diaminobutyrate aminotransferase and diaminobutyrate decarboxylase domains is present in the marine γ-proteobacterium Vibrio vulnificus and the fusion gene produced diaminopropane when expressed in E. coli (Lee et al., 2009). This gene fusion is present throughout the Vibrionales but not elsewhere and so the diaminobutyrate aminotransferase-diaminobutyrate decarboxylase ORF fusion event may have occurred only once followed by vertical descent or horizontal gene transfer to closely related species. The E. coli acid-inducible ADC represents gene fusion between a biosynthetic ADC (typified by the Bacillus subtilis biosynthetic ADC) and a receiver domain response regulator, fused to the N-terminus of the ADC ORF. The fusion of the reponse regulator to a biosynthetic ADC has modified the behaviour of the enzyme so that it forms a decamer rather than a dimer, and it is now active only in acid conditions (Burrell et al., 2010). Human spermine synthase has evolved through the fusion of a bacterial Class 1 b-like AdoMetDC to the N-terminus of an aminopropyltransferase open reading frame (Wu et al., 2008). The AdoMetDC domain of spermine synthase is recognisable only through structural analysis and not through primary amino acid sequence; the AdoMetDC domain is not processed and does not possess AdoMetDC catalytic activity (Wu et al., 2008). Although not catalytically active, the AdoMetDC domain is essential for spermine synthase activity due to its role in dimer formation. Orthologues of the human spermine synthase are found throughout the metazoa and in the single-celled choanoflagellate Monosiga brevicollis, a sister clade to the metazoa. It is likely that the human spermine synthase was originally acquired by horizontal transfer of an AdoMetDC-aminopropyltransferase fusion gene from bacteria to a common ancestor of metazoa and M. brevicollis.
Gene fusions involving polyamine biosynthetic genes can be found in bacteria, archaea and eukaryotes. Three of the covalent biosynthetic modules described above, AdoMetDC/ aminopropyltransferase, agmatine deiminase/N-carbamoylputrescine amidohydrolase and diaminobutyrate aminotransferase/ diaminobutyrate decarboxylase are found only in prokaryotes. Limited phyletic distribution of the latter two fusions and their conserved sequences suggests a single origin for each of the two fusion genes. In contrast, the highly scattered phyletic distribution of the AdoMetDC-aminopropyltransferase fusion genes and their functional diversity indicates independent formation in diverse bacterial phyla. Most gene fusions in polyamine metabolism are between enzymes that are involved in the same biosynthetic module, i.e., the conversion of one intermediate to the next in sequence in the metabolic pathway. This suggests that an additional constraint on the formation of gene fusions in metabolic pathways is the requirement of component domains to participate in a shared biosynthetic step.
The phylogenetically broad but locally limited distribution of AdoMetDC-aminopropyltransferase gene fusions in bacteria suggests that fusion of these two enzymes is probably accompanied by eventual scission events. However, the abundance of the AdoMetDC-aminopropyltransferase fusions in the ocean metagenome (primarily Ca. P. ubique-related) indicates that there is positive selection to maintain the fusion in this environment. What are the possible consequences of the fusion of AdoMetDC and the aminopropyltransferase? The decarboxylation of AdoMet by AdoMetDC commits AdoMet to polyamine biosynthesis, thus the activity of AdoMetDC closely correlates with polyamine levels and is rate-limiting for polyamine biosynthesis (Pegg, 2009). Fusion of the aminopropyltransferase to AdoMetDC creates a problem because a side activity of AdoMetDC results eventually in irreversible transamidation of the AdoMetDC pyruvoyl cofactor, effectively killing the enzyme. The human AdoMetDC pyruvoyl group has been calculated to be transamidated after 15,000 turnover events (Bale & Ealick, 2010). Because of the transamidation, AdoMetDC has a relatively short activity half-life and so has to be synthesized at a relatively high rate. Fusion of the two enzymes will result in a rate of aminopropyltransferase synthesis that is likely to be more than required for the unfused enzyme. Nevertheless, fusion of these two enzymes could be beneficial independently of flux and biosynthetic cost considerations. The product of AdoMetDC, dcAdoMet is an inhibitor of methyltransferase reactions and is known to inhibit DNA methylation (Frostesjo et al., 1997). A recent study has also implicated dcAdoMet in the inhibition of histone H3 lysine 9 dimethylation, as well as general DNA hypomethylation (Yamamoto et al., 2010). Therefore, if the dcAdoMet product of AdoMetDC was channeled within the fusion protein to the aminopropyltransferase, the toxic impact of dcAdoMet on cellular transmethylation reactions would be greatly attenuated. Benefits of the AdoMetDC-aminopropyltransferase fusion might therefore be determined by the sensitivity of particular species to intereference of transmethylation reactions by dcAdoMet. The importance of detoxification to Ca. P. ubique is indicated by the fact that methylthioadenosine nucleosidase, which removes and salvages the co-product (methylthioadenosine, another inhibitor of methyltransferases) of the AdoMetDC-aminopropyltransferase fusion protein, is clustered immediately adjacent to the fusion protein in this species.
In conclusion, we have found that fusions of AdoMetDC and aminopropyltransferases in bacteria arise stochastically as a consequence of clustering of these ORFs in structurally favourable configurations, and mutation to allow translational readthrough from one ORF to the next. The phylogenetic distribution of the fusion proteins is determined by independent fusion events in diverse phyla, horizontal transfer between species in disparate phyla, limited vertical inheritance, and stochastic scission events, suggesting that the persistance of the fusion proteins in some species is dependent on physiological parameters specific to those species and their environments.
Experimental procedures
Protein expression and purification in E. coli
Coding sequences for the fusion proteins were ligated into pET21b (Novagen) for expression in E. coli. The expression host strain E. coli BL21 (DE3) was transformed with expression plasmids, and 1 ml of a 10 ml LB broth overnight culture was used to inoculate 100 ml of LB broth containing ampicillin. Recombinant protein expression was induced by incubating the cells with components of the Overnight Express™ Autoinduction System 1 (Novagen) at 25°C for 16 hours. T7-tagged proteins were purified using the T7-tag affinity purification kit (Novagen), following the manufacturer’s instructions for the column procedure. Fractions containing the target protein, as determined by SDS-PAGE, were pooled, buffer-exchanged, and concentrated. Fusion genes were synthesised de novo with codon usage optimised for expression in E. coli and Saccharomyces cerevisiae (by Genscript Corporation, Piscatway, NJ, USA).
HPLC analysis of cellular polyamines
Cells from 1 ml of induced E. coli cultures were harvested by centrifugation (10,000 g, 10 min), and washed twice in 1 ml phosphate-buffered saline. MOPS lysis buffer (100 mM MOPS, 50 mM NaCl, 20 mM MgCl2) was added at 5 μl per mg cell fresh weight, and cells were subjected to three cycles of freeze/thawing. TCA was added to a final concentration of 10%, and cells were incubated on ice for 5 min. After centrifugation (18,000 g, 5 min, 4 °C), 5 μl of supernatant were derivatized using the AccQ-Fluor reagent kit for labelling amino acids (Waters). For normalisation, 1,7-diaminoheptane was included as an internal standard. Labelled polyamines were separated by HPLC using a Luna 5μm C18 (2) 100A column (250 × 4.6 mm; Phenomenex) with fluorescence detection (excitation 248 nm, emission 398 nm). Solvent A was 70 mM acetic acid, 25 mM triethylamine, pH 4.82; solvent B was 80% acetonitrile, 20% H2O (v/v); solvent C was methanol, and the gradient was run for 65 min at a flow rate of 1.2 ml/min with the following concentrations: t = 0 min, 100% A; t = 1 min, 78% A, 22% B; t = 27 min, 55% A, 39% B, 6% C; t = 27.5 min, 53% A, 33% B, 14% C; t = 34 min, 20% A, 10% B, 70% C; t = 37 min, 100% B; t = 58 min, 100% A.
AdoMetDC activity analysis
Each purified T7-tagged recombinant fusion protein was present at 2 μg per assay. AdoMetDC activity was assayed by quantification of the released 14CO2 from S-adenosyl-L-[14C]methionine (GE Healthcare), as descibed previously (Michael et al., 1996). Background activity was determined from reactions with enzyme dilution buffer instead of protein.
Aminopropyltransferase activity assay
Activity was measured by following the production of [35S]MTA from [35S]dcAdoMet in 100 mM sodium phosphate buffer (pH 7.5) in the presence of the amine acceptor indicated as described (Michael et al., 1996).
Growth analysis of S. Typhimurium strains
To deplete cells of polyamines acquired from growth in rich medium (LB), cells picked from a single colony grown on solid LB agar plates were subcultured four times (1:100 dilution) in M9 minimal medium with selective antibiotic with the addition of 0.4% glucose. Polyamine-depleted gene deletion strains and the parental strain were then grown in 50 ml of M9 defined minimal growth medium in 250 ml erlenmayer flasks at 37°C and triplicate 1.0 ml samples were taken at hourly intervals and the OD600 nm was determined. For growth curves performed using colony-forming units (cfu) per ml measurements, samples were taken every hour and, in triplicate, diluted according to calculated approximations of cfu/ml measurements. These dilutions were used to make 10 μl spots on LB agar plates, which were incubated overnight at 30 °C to prevent overgrowth, facilitating counting of resultant colonies. All data produced were converted into cfu/ml values using reversal of dilution factors. These data were plotted on a log 10 scale over time to produce a growth curve.
Production of S. Typhimurium AspeDE gene deletion strain
The template plasmids pKD3 and pKD4 contain FRT (FLP-recombinase recognition target site)-flanked chloramphenicol or kanamycin resistance gene cassettes respectively (Datsenko & Wanner, 2000). These were used to generate a PCR product which contained the antibiotic resistance gene flanked by homologous regions on the targeted gene and the two FRT sites. The S. Typhimurium strain JH3006, which carries the Red helper plasmid pKD46, was grown at 30 °C to an OD600 nm of 0.6 and then made electrocompetent by washing three times in 10% glycerol. Electroporation was applied to 100 μl of cells mixed with 10 μl of PCR product, 1 ml of LB liquid growth medium was added and cells were incubated at 37 °C for 1 hour before being spread onto LB plates to select for kanamycin or chloroamphenical resistant colonies following incubation at 37 °C overnight. Verification of transformants was performed by PCR on single colonies. A verified colony for each mutant was then used to produce a P22int-4 HT λ-bacteriophage lysate; 50 μl of overnight culture for each mutant (in JH3006) was used to inoculate 5 ml of fresh LB and incubated at 37 °C until the OD600 nm reached ~0.15-0.2. To this, 5 μl of stock P22 phage was added and incubated overnight. Cells were lysed using chloroform and then incubated at 4 °C for two hours. This lysate was purified using a 0.22 μm syringe filter following centrifugation at 13,000 rpm for 15 minutes and stored at 4 °C under a layer of chloroform. Two hundred μl of a 5 ml overnight culture of recipient S. Typhimurium 4/74 (in LB at 37 °C) had 10 μl of phage lysate added and was incubated at 37 °C for 1 hour to allow expression of antibiotic resistance genes. Selective antibiotic LB plates were used to grow transductants overnight at 37 °C, negative controls were used to ensure there was no contamination of lysate and that there were no resistant recipient cells.
Construction of AdoMetDC-aminopropyltransferase expression plasmids for S. Typhimurium
The DNA encoding the different AdoMetDC-aminopropyltransferase fusion ORFs were ligated into the plasmid pWSK29 (Wang & Kushner, 1991). This low copy number plasmid has an origin of replication derived from pSC101 (Stoker et al., 1982) which allows 5-8 copies per cell in E. coli and Salmonella. Vector and insert were digested with the restriction enzymes BamHI and XhoI and the insert DNA was ligated into the vector. Plasmid pWSK29 was introduced into S. Typhimurium 4/74 cells by electroporation.
Transformation of S. Typhimurium with pWSK29-Electrocompetent Salmonella cells for both wild type S. Typhimurium 4/74, and the spermidine biosynthetic mutant AspeDE, were generated as follows: cells were grown at 37 °C to an OD600 nm of 0.6, washed three times in 10% glycerol, then snap frozen in liquid nitrogen and stored at −80 °C. Electroporation (3kW for 0.5 seconds) was applied to 100 μl of cells mixed with 10 μl of plasmid preparation, 1 ml of LB was added and cells were incubated at 37 °C for 1 hour before being spread onto LB plates to select for chloramphenical and ampicillin resistant colonies for AspeDE, and simply ampicillin resistantance for wild type Salmonella transformed with pWSK29.
Supplementary Material
Acknowledgements
AJM was supported by UT Southwestern Medical Center, a Biotechnological and Biological Sciences Research Council UK CSG grant and Institute Development Fellowship (BB/E024467/1). AEP was supported by grants CA-0181138 and GM-26290 from the National Institutes of Health. We thank Margaret A. Phillips for constructive criticism of the manuscript.
References
- Alvarez BV, Vilas GL, Casey JR. Metabolon disruption: a mechanism that regulates bicarbonate transport. EMBO J. 2005;24:2499–2511. doi: 10.1038/sj.emboj.7600736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S, Kumar R, Sheets ED, Benkovic SJ. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science. 2008;320:103–106. doi: 10.1126/science.1152241. [DOI] [PubMed] [Google Scholar]
- An S, Kyoung M, Allen JJ, Shokat KM, Benkovic SJ. Dynamic regulation of a metabolic multi-enzyme complex by protein kinase CK2. J Biol Chem. 2010;285:11093–11099. doi: 10.1074/jbc.M110.101139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale S, Ealick SE. Structural biology of S-adenosylmethionine decarboxylase. Amino Acids. 2010;38:451–460. doi: 10.1007/s00726-009-0404-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell M, Hanfrey CC, Murray EJ, Stanley-Wall NR, Michael AJ. Evolution and multiplicity of arginine decarboxylases in polyamine biosynthesis and essential role in Bacillus subtilis biofilm formation. J Biol Chem. 2010;285:39224–39238. doi: 10.1074/jbc.M110.163154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacciapuoti G, Porcelli M, Carteni-Farina M, Gambacorta A, Zappia V. Purification and characterization of propylamine transferase from Sulfolobus solfataricus, an extreme thermophilic archaebacterium. Eur J Biochem. 1986;161:263–271. doi: 10.1111/j.1432-1033.1986.tb10442.x. [DOI] [PubMed] [Google Scholar]
- Caetano-Anolles G, Yafremava LS, Gee H, Caetano-Anolles D, Kim HS, Mittenthal JE. The origin and evolution of modern metabolism. Int J Biochem Cell Biol. 2009;41:285–297. doi: 10.1016/j.biocel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Dasu VV, Nakada Y, Ohnishi-Kameyama M, Kimura K, Itoh Y. Characterization and a role of Pseudomonas aeruginosa spermidine dehydrogenase in polyamine catabolism. Microbiology. 2006;152:2265–2272. doi: 10.1099/mic.0.28920-0. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Lee J, Michael AJ, Tomchick DR, Goldsmith EJ, Phillips MA. Evolution of substrate specificity within a diverse family of beta/alpha-barrel-fold basic amino acid decarboxylases: X-ray structure determination of enzymes with specificity for L-arginine and carboxynorspermidine. J Biol Chem. 2010;285:25708–25719. doi: 10.1074/jbc.M110.121137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fani R, Brilli M, Fondi M, Lio P. The role of gene fusions in the evolution of metabolic pathways: the histidine biosynthesis case. BMC Evol Biol. 2007;7(Suppl 2):S4. doi: 10.1186/1471-2148-7-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondi M, Emiliani G, Fani R. Origin and evolution of operons and metabolic pathways. Res Microbiol. 2009;160:502–512. doi: 10.1016/j.resmic.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Frostesjo L, Holm I, Grahn B, Page AW, Bestor TH, Heby O. Interference with DNA methyltransferase activity and genome methylation during F9 teratocarcinoma stem cell differentiation induced by polyamine depletion. J Biol Chem. 1997;272:4359–4366. doi: 10.1074/jbc.272.7.4359. [DOI] [PubMed] [Google Scholar]
- Fuell C, Elliott KA, Hanfrey CC, Franceschetti M, Michael AJ. Polyamine biosynthetic diversity in plants and algae. Plant Physiol Biochem. 2010;48:513–520. doi: 10.1016/j.plaphy.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Fujihara S, Abe H, Yoneyama T. A new polyamine 4-aminobutylcadaverine. Occurrence and its biosynthesis in root nodules of adzuki bean plant Vigna angularis. J Biol Chem. 1995;270:9932–9938. doi: 10.1074/jbc.270.17.9932. [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D, Bibbs L, Eads J, Richardson TH, Noordewier M, Rappe MS, Short JM, Carrington JC, Mathur EJ. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- Hisano T, Murata K, Kimura A, Matsushita K, Toyama H, Adachi O. Characterization of membrane-bound spermidine dehydrogenase of Citrobacter freundii. Biosci Biotechnol Biochem. 1992;56:1916–1920. doi: 10.1271/bbb.56.1916. [DOI] [PubMed] [Google Scholar]
- Igarashi K, Kashiwagi K, Hamasaki H, Miura A, Kakegawa T, Hirose S, Matsuzaki S. Formation of a compensatory polyamine by Escherichia coli polyamine-requiring mutants during growth in the absence of polyamines. J Bacteriol. 1986;166:128–134. doi: 10.1128/jb.166.1.128-134.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MM, Nautiyal M, Wynn RM, Mobley JA, Chuang DT, Hutson SM. Branched-chain amino acid metabolon: interaction of glutamate dehydrogenase with the mitochondrial branched-chain aminotransferase (BCATm) J Biol Chem. 2010;285:265–276. doi: 10.1074/jbc.M109.048777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MM, Wallin R, Wynn RM, Conway M, Fujii H, Mobley JA, Chuang DT, Hutson SM. A novel branched-chain amino acid metabolon. Protein-protein interactions in a supramolecular complex. J Biol Chem. 2007;282:11893–11903. doi: 10.1074/jbc.M700198200. [DOI] [PubMed] [Google Scholar]
- Ivanov IP, Atkins JF, Michael AJ. A profusion of upstream open reading frame mechanisms in polyamine-responsive translational regulation. Nucleic Acids Res. 2010;38:353–359. doi: 10.1093/nar/gkp1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott JM. Biosynthesis of long-chain polyamines by crenarchaeal polyamine synthases from Hyperthermus butylicus and Pyrobaculum aerophilum. FEBS Lett. 2009;583:3519–3524. doi: 10.1016/j.febslet.2009.10.014. [DOI] [PubMed] [Google Scholar]
- Knott JM, Romer P, Sumper M. Putative spermine synthases from Thalassiosira pseudonana and Arabidopsis thaliana synthesize thermospermine rather than spermine. FEBS Lett. 2007;581:3081–3086. doi: 10.1016/j.febslet.2007.05.074. [DOI] [PubMed] [Google Scholar]
- Lee J, Sperandio V, Frantz DE, Longgood J, Camilli A, Phillips MA, Michael AJ. An alternative polyamine biosynthetic pathway is widespread in bacteria and essential for biofilm formation in Vibrio cholerae. J Biol Chem. 2009;284:9899–9907. doi: 10.1074/jbc.M900110200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu ZJ, Markham GD. Metal ion activation of S-adenosylmethionine decarboxylase reflects cation charge density. Biochemistry. 2007;46:8172–8180. doi: 10.1021/bi6025962. [DOI] [PubMed] [Google Scholar]
- Michael AJ, Furze JM, Rhodes MJ, Burtin D. Molecular cloning and functional identification of a plant ornithine decarboxylase cDNA. Biochem J. 1996;314(Pt 1):241–248. doi: 10.1042/bj3140241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell CG. Identification of a multienzyme complex of the tricarboxylic acid cycle enzymes containing citrate synthase isoenzymes from Pseudomonas aeruginosa. Biochem J. 1996;313(Pt 3):769–774. doi: 10.1042/bj3130769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RM, Rappe MS, Connon SA, Vergin KL, Siebold WA, Carlson CA, Giovannoni SJ. SAR11 clade dominates ocean surface bacterioplankton communities. Nature. 2002;420:806–810. doi: 10.1038/nature01240. [DOI] [PubMed] [Google Scholar]
- Muller S, Da’dara A, Luersen K, Wrenger C, Das Gupta R, Madhubala R, Walter RD. In the human malaria parasite Plasmodium falciparum, polyamines are synthesized by a bifunctional ornithine decarboxylase, S-adenosylmethionine decarboxylase. J Biol Chem. 2000;275:8097–8102. doi: 10.1074/jbc.275.11.8097. [DOI] [PubMed] [Google Scholar]
- Panicot M, Minguet EG, Ferrando A, Alcazar R, Blazquez MA, Carbonell J, Altabella T, Koncz C, Tiburcio AF. A polyamine metabolon involving aminopropyl transferase complexes in Arabidopsis. Plant Cell. 2002;14:2539–2551. doi: 10.1105/tpc.004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg AE. S-Adenosylmethionine decarboxylase. Essays Biochem. 2009;46:25–45. doi: 10.1042/bse0460003. [DOI] [PubMed] [Google Scholar]
- Pegg AE, Michael AJ. Spermine synthase. Cell Mol Life Sci. 2010;67:113–121. doi: 10.1007/s00018-009-0165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, Beeson K, Tran B, Smith H, Baden-Tillson H, Stewart C, Thorpe J, Freeman J, Andrews-Pfannkoch C, Venter JE, Li K, Kravitz S, Heidelberg JF, Utterback T, Rogers YH, Falcon LI, Souza V, Bonilla-Rosso G, Eguiarte LE, Karl DM, Sathyendranath S, Platt T, Bermingham E, Gallardo V, Tamayo-Castillo G, Ferrari MR, Strausberg RL, Nealson K, Friedman R, Frazier M, Venter JC. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw FL, Elliott KA, Kinch LN, Fuell C, Phillips MA, Michael AJ. Evolution and multifarious horizontal transfer of an alternative biosynthetic pathway for the alternative polyamine sym-homospermidine. J Biol Chem. 2010;285:14711–14723. doi: 10.1074/jbc.M110.107219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srere PA. Complexes of sequential metabolic enzymes. Annu Rev Biochem. 1987;56:89–124. doi: 10.1146/annurev.bi.56.070187.000513. [DOI] [PubMed] [Google Scholar]
- Srere PA. Macromolecular interactions: tracing the roots. Trends Biochem Sci. 2000;25:150–153. doi: 10.1016/s0968-0004(00)01550-4. [DOI] [PubMed] [Google Scholar]
- Stoker NG, Fairweather NF, Spratt BG. Versatile low-copy-number plasmid vectors for cloning in Escherichia coli. Gene. 1982;18:335–341. doi: 10.1016/0378-1119(82)90172-x. [DOI] [PubMed] [Google Scholar]
- Swingley WD, Chen M, Cheung PC, Conrad AL, Dejesa LC, Hao J, Honchak BM, Karbach LE, Kurdoglu A, Lahiri S, Mastrian SD, Miyashita H, Page L, Ramakrishna P, Satoh S, Sattley WM, Shimada Y, Taylor HL, Tomo T, Tsuchiya T, Wang ZT, Raymond J, Mimuro M, Blankenship RE, Touchman JW. Niche adaptation and genome expansion in the chlorophyll d-producing cyanobacterium Acaryochloris marina. Proc Natl Acad Sci U S A. 2008;105:2005–2010. doi: 10.1073/pnas.0709772105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (and Other Methods) Sinauer Associates; Sunderland, MA: 2000. [Google Scholar]
- Tabor CW, Kellogg PD. Identification of flavin adenine dinucleotide and heme in a homogeneous spermidine dehydrogenase from Serratia marcescens. J Biol Chem. 1970;245:5424–5433. [PubMed] [Google Scholar]
- Tabor CW, Tabor H. Polyamines. Annu Rev Biochem. 1984;53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- Toms AV, Kinsland C, McCloskey DE, Pegg AE, Ealick SE. Evolutionary links as revealed by the structure of Thermotoga maritima S-adenosylmethionine decarboxylase. J Biol Chem. 2004;279:33837–33846. doi: 10.1074/jbc.M403369200. [DOI] [PubMed] [Google Scholar]
- Tripp HJ, Bench SR, Turk KA, Foster RA, Desany BA, Niazi F, Affourtit JP, Zehr JP. Metabolic streamlining in an open-ocean nitrogen-fixing cyanobacterium. Nature. 2010;464:90–94. doi: 10.1038/nature08786. [DOI] [PubMed] [Google Scholar]
- Wang RF, Kushner SR. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- Welch GR. On the role of organized multienzyme systems in cellular metabolism: a general synthesis. Prog Biophys Mol Biol. 1977;32:103–191. [PubMed] [Google Scholar]
- Wu H, Min J, Ikeguchi Y, Zeng H, Dong A, Loppnau P, Pegg AE, Plotnikov AN. Structure and mechanism of spermidine synthases. Biochemistry. 2007;46:8331–8339. doi: 10.1021/bi602498k. [DOI] [PubMed] [Google Scholar]
- Wu H, Min J, Zeng H, McCloskey DE, Ikeguchi Y, Loppnau P, Michael AJ, Pegg AE, Plotnikov AN. Crystal structure of human spermine synthase: implications of substrate binding and catalytic mechanism. J Biol Chem. 2008;283:16135–16146. doi: 10.1074/jbc.M710323200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto D, Shima K, Matsuo K, Nishioka T, Chen CY, Hu GF, Sasaki A, Tsuji T. Ornithine decarboxylase antizyme induces hypomethylation of genome DNA and histone H3 lysine 9 dimethylation (H3K9me2) in human oral cancer cell line. PLoS One. 2010;5:e12554. doi: 10.1371/journal.pone.0012554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai I, Wolf YI, Koonin EV. Evolution of gene fusions: horizontal transfer versus independent events. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-5-research0024. research0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.