Abstract
Background
Cytomegalovirus (CMV) DNA viral load testing is routinely performed in centers that serve patients that are immunosuppressed from organ or hematopoietic stem cell transplantation. Clinical laboratories that offer this testing often face practical concerns about the storage of these specimens to ensure accurate measurement for patient care. The studies published that look at CMV DNA stability at 4°C have done so only up to 72 hours.
Objective
Our objective was to determine the stability of CMV DNA in whole blood and plasma for clinical viral load testing over a 14 day period.
Study Design
Twenty-one plasma samples that were CMV-positive and twenty whole blood samples (including eleven CMV-negative whole blood samples spiked with CMV-positive plasma) were stored at 4°C and underwent extraction and amplification at 3 time points: Day 0, Day 7, and Day 14.
Results
Log10 values were calculated and t-test was performed on the values comparing Day 0 to Day 14 for plasma and whole blood. There was no statistically significant difference between Day 0 and Day 14 for both specimen types, including the CMV-negative whole blood that was spiked with CMV-positive plasma.
Conclusions
CMV DNA in plasma and whole blood is stable for 14 days at a temperature of 4°C.
Background
Cytomegalovirus (CMV) viral load testing has become the standard of care for diagnosis of patients at risk for developing CMV disease after solid organ or hematopoietic stem cell transplant and for monitoring response to antiviral therapy. Clinical laboratories generally use EDTA anti-coagulated whole blood or EDTA plasma samples for CMV viral load testing. Studies that have assessed the stability of CMV DNA over short periods of time have shown mixed results. A study by Schafer et al. compared the stability of CMV DNA and pp65 antigen in peripheral blood leukocytes (PBL) and concluded that up to 72 hours delay in processing after sample collection did not decrease the amount of CMV DNA, though a decrease in pp65 antigen was observed 1. Likewise, Roberts et al. found CMV DNA to be stable at room temperature at 4°C over a 72 hour period 2. However Schafer et al. concluded an elevation of CMV viral load in plasma was due to WBC lysis in whole blood stored 24 hours before separation 3, while Boom et al. concluded that plasma separated within 48 hours of collection from EDTA whole blood did not affect CMV DNA level when stored 4°C 4. In a 2004 study Nesbitt et al. further demonstrated that a 24 hour delay in separation did not affect plasma viral load levels 5. None of these studies evaluated CMV DNA stability when storing whole blood or plasma for greater than 72 hours prior to testing.
Objective
The objective of this study was to assess the stability of CMV DNA in EDTA whole blood and plasma over a 14 day period.
Study Design
Residual EDTA plasma samples were stored after routine CMV viral load testing and residual corresponding EDTA whole blood samples were collected from the Core Laboratory. The specimens included nine CMV-positive EDTA anti-coagulated whole blood specimens, eleven CMV-negative EDTA whole blood specimens spiked with known CMV-positive plasma, and twenty-one CMV-positive EDTA plasma specimens. The whole blood specimens were stored at room temperature for a mean of 1.5 days prior to receipt in the testing laboratory. The plasma specimens were stored at 4°C for a mean of 3 days prior to receipt in the testing laboratory. Once the specimens arrived in the testing laboratory, DNA was extracted for the Day 0 time point. Specimens were then stored for Day 7 and Day 14 at 4°C prior to extracting for CMV viral load testing.
Due to the small number of whole blood specimens, eleven CMV-negative whole blood specimens were spiked with CMV positive plasma. For three of the spiked specimens, 100 µl of CMV-positive plasma was added to 900 µl of CMV-negative whole blood; this sample was further diluted to 1:100 in CMV-negative whole blood. One specimen was spiked as above and diluted 1:10, 1:100 and 1:1000 in CMV-negative whole blood and then tested as separate specimens. The final four specimens consisted of 500 µL of CMV-negative whole blood mixed with 500 µL of CMV-positive plasma. The spiked whole blood specimens were extracted on Day 0, Day 7 and Day 14. Each spiked specimen was treated as an individual sample for the purpose of this study.
Nucleic acid extraction was performed using the EZ1 virus Mini kit version 2.0 (Qiagen, Inc, Valencia, CA) following the manufacturer’s recommended protocol. An internal control (IC) (7.2 µL) was added to 200 µL of whole blood or plasma and eluted in 60 µL. CMV testing was performed using the artus CMV TM PCR kit (Qiagen, Inc, Valencia, CA) on the Qiagen Rotor Gene instrument. The amplification target is a 105 base pair region of the major immediate-early exon 4 gene of the CMV genome. For each amplification reaction, 20 µL of purified nucleic acid was added to 30 µL of a reaction mixture of master mix and magnesium (CMV RG PCR Kit, Qiagen, Inc). The thermocycler parameters were as follows: hold 10 min at 95°C followed by 15 seconds at 95°C, 30 seconds at 65°C and 20 seconds at 72°C for 45 cycles. A four point standard curve as well as a positive and a negative control were included on all runs. The laboratory- determined limit of detection (LOD) in this assay using whole blood and plasma was 1.76 log10 copies/mL and 1.46 log10 copies/mL, respectively. Statistics were performed using two tailed t-tests assuming equal variance.
Results
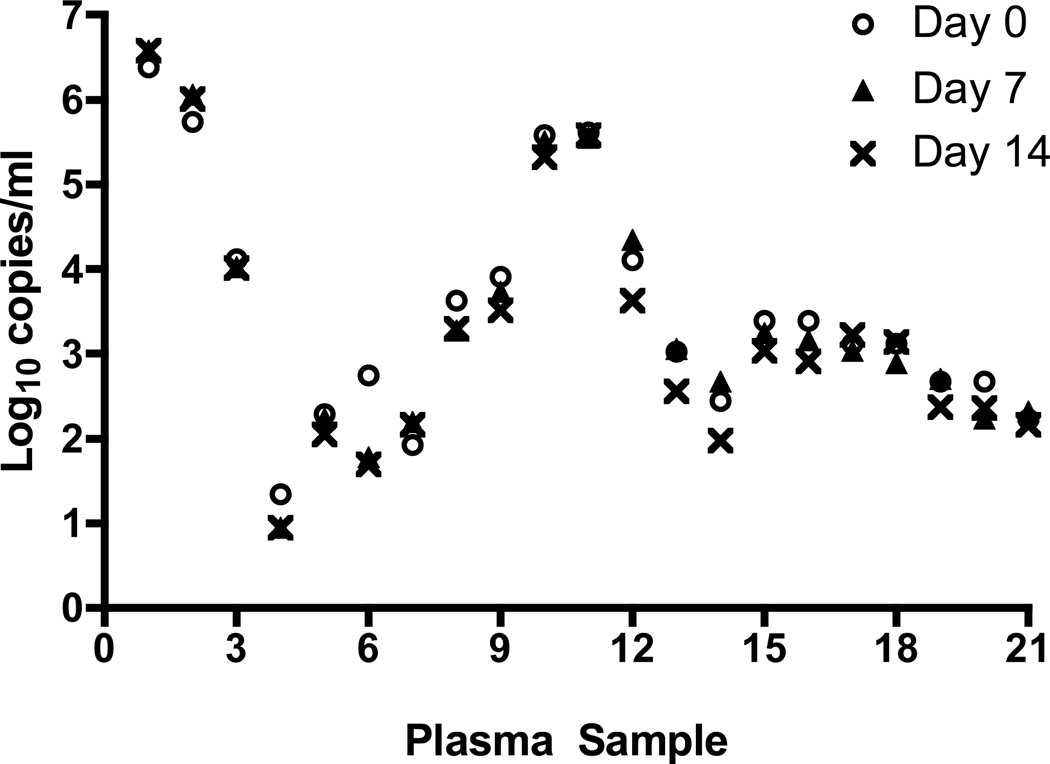
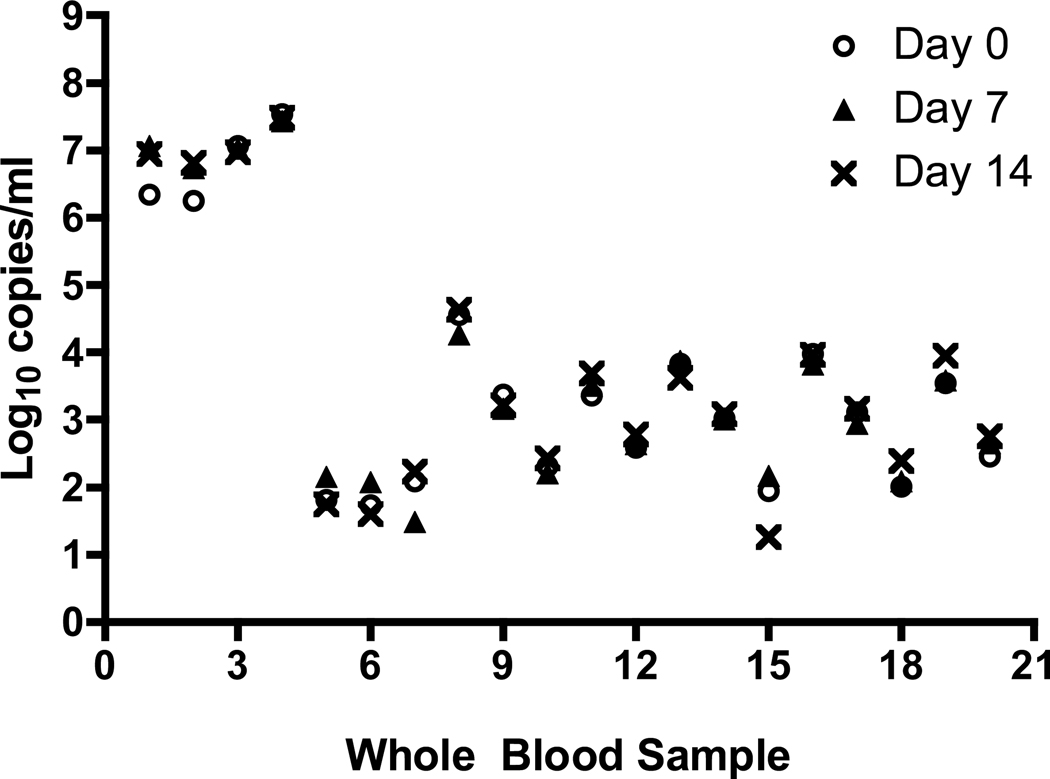
For all sample types the IC was successfully detected; there was no inhibition of amplification. The CMV viral load values for the 21 plasma specimens at Day 0, 7 and 14 are shown in Figure 1. The viral load values of the plasma specimens tested ranged from 1.1 log10 copies/mL to 6.5 log10 copies/mL. There was considerable overlap in the viral load values obtained for the three time points tested for most of the samples. The mean viral load over the 14 day period was stable and decreased from 3.50 log10 copies/mL (Day 0) to 3.26 log10 copies/mL on Day 14; this difference was not statistically different (p= 0.60) (Table 1). The CMV viral load values in the non-spiked whole blood specimens ranged from 1.80 log10 copies/mL to 7.48 log10 copies/mL, while the viral load values of the spiked whole blood specimens ranged from 1.79 log10 copies/ml to 4.49 log10 copies/mL. The CMV DNA concentration was stable over the 14 day period for both the non-spiked and spiked whole blood samples (Figure 2). The mean viral load of the spiked samples changed little over the 14 day period; 3.10 log10 copies/mL on Day 0 compared to 3.11 log10 copies/mL on Day 14 (p=0.97) (Table 1). For the non-spiked specimens, there was a slight increase in mean viral load on Day 14 (4.78 log10 copies/mL) compared to Day 0 (4.59 log10 copies/mL; p =0.88), which was not statistically significant.
Figure 1.
Stability of plasma samples. Individuals samples were de-identified using sample numbers and the log10 value of the sample at Day 0, 7 and 14 were plotted for each sample number.
Table 1.
Mean Log10 Results per Specimen Type
| Mean Viral Load (Standard Deviation) Log10 copies/ml |
|||
|---|---|---|---|
| Specimen Type | Day 0 | Day 7 | Day 14 |
| Plasma | 3.50 (1.30) | 3.41 (1.43) | 3.26 (1.46)* |
| Non-spiked Whole Blood | 4.59 (2.30) | 4.84 (2.27) | 4.78 (2.37)** |
| Spiked Whole Blood | 3.10 (0.80) | 3.06 (0.71) | 3.11 (0.86)*** |
p =0.60,
p=0.88,
p=0.97
Figure 2.
Stability of total whole blood samples. Individuals samples were de-identified using sample numbers and the log10 value of the sample at Day 0, 7 and 14 were plotted for each sample number.
The viral load values obtained from the initial clinical testing were available for the plasma samples; clinical testing was not done on the whole blood samples. This allowed comparison of the initial plasma clinical viral load with the plasma Time 0 viral load values, providing an opportunity to further assess the impact of storage at 4°C on the stability of CMV DNA. The mean difference for the initial clinical result and the Time 0 results was 0.22 log10 copies/ml. For 17 of the 21 specimens, the difference in the initial clinical and Time 0 value was ≤ 0.40 log10 copies/ml. Three of the four remaining specimens were within 0.55 log10 copies/ml, with one specimen showing a large decrease in viral load from 3.00 to 1.34 log10 copies/ml, although a greater inter-assay variability would be expected at this low viral load value. Of note the extraction method used for the clinical testing and the study were different, which could account for some of the differences in the viral load values.
Conclusion
These data show that CMV DNA is stable in whole blood and plasma over a 14 day period when stored at 4°C. Importantly, this finding was consistent for samples over a wide range of CMV DNA concentrations (1.1 to 7.48 log10 copies/ml), including specimens with a low viral load. There are several limitations of the study, including the use of spiked specimens, some of which were diluted with equal volumes of plasma, which may have impacted DNA degradation. However, there was not a difference in the trend of viral load values for the spiked and non-spiked whole blood specimens. Also, Day 0 of testing did not occur on the day the sample was collected. This was due to delays associated with clinical testing of plasma specimens and retrieval of corresponding whole blood specimens from the Core Laboratory. It is possible that there was a significant degradation of CMV DNA in the first few days after specimen collection, but it would be expected that the trend in degradation would have continued over the 14 days of testing and this was not observed. The comparison of the initial plasma clinical viral load to the plasma Time 0 viral load values, showed little decrease in CMV DNA levels (mean 0.22 log10 copies/ml) over this initial storage period (mean 3 days), further supporting that there is minimal degradation of CMV DNA when plasma specimens are stored at 4°C. The prolonged stability of CMV DNA observed in this study is important for 1) clinical laboratories that may perform batch testing a few times per week, 2) clinical laboratories that transport of specimens to reference laboratories for testing, and 3) blood banks in testing blood units stored for weeks before transfusion. However, laboratories should independently evaluate storage conditions for their individual needs.
Acknowledgments
Acknowledgements and COI
Funding: This work was supported in part by the Emory Center for AIDS Research (P30 AI050409).
Competing interests: Clinical Trial Research Grant support for AMC from Qiagen Ethical approval: This study was approved by the Emory Institutional Review Board (IRB# 247-2001)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Deborah Abdul-Ali, Emory University, 101 Woodruff Circle, Room 7007, Atlanta, GA 30322, 404-727-4483, FAX: 404-727-4382, dabdula@emory.edu.
Colleen S. Kraft, Emory University Hospital, 1364 Clifton Rd, F145C, Atlanta, GA 30322, 404-712-8889, FAX: 404-712-4632, colleen.kraft@emory.edu
Jessica Ingersoll, Emory University, 101 Woodruff Circle, Room 7007, Atlanta, GA 30322, 404-727-4483, FAX: 404-727-4382, jingers@emory.edu.
Mona Frempong, Emory University, 101 Woodruff Circle, Room 7007, Atlanta, GA 30322, 404-727-4483, FAX: 404-727-4382, mafremp@emory.edu.
Angela M. Caliendo, Emory University Hospital, H180, 1364 Clifton Rd, Atlanta, GA 30322, 404-712-5721, FAX: 404-727-3133, acalien@emory.edu
References
- 1.Schafer P, Tenschert W, Gutensohn K, Laufs R. Minimal effect of delayed sample processing on results of quantitative PCR for cytomegalovirus DNA in leukocytes compared to results of an antigenemia assay. J Clin Microbiol. 1997;35:741–744. doi: 10.1128/jcm.35.3.741-744.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roberts TC, Buller RS, Gaudreault-Keener M, et al. Effects of storage temperature and time on qualitative and quantitative detection of cytomegalovirus in blood specimens by shell vial culture and PCR. J Clin Microbiol. 1997;35:2224–2228. doi: 10.1128/jcm.35.9.2224-2228.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schafer P, Tenschert W, Schroter M, Gutensohn K, Laufs R. False-positive results of plasma PCR for cytomegalovirus DNA due to delayed sample preparation. J Clin Microbiol. 2000;38:3249–3253. doi: 10.1128/jcm.38.9.3249-3253.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boom R, Sol CJ, Schuurman T, et al. Human cytomegalovirus DNA in plasma and serum specimens of renal transplant recipients is highly fragmented. J Clin Microbiol. 2002;40:4105–4113. doi: 10.1128/JCM.40.11.4105-4113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nesbitt SE, Cook L, Jerome KR. Cytomegalovirus quantitation by real-time PCR is unaffected by delayed separation of plasma from whole blood. J Clin Microbiol. 2004;42:1296–1297. doi: 10.1128/JCM.42.3.1296-1297.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]