Abstract
Lrig1 is the founding member of the “Lrig” family and has been implicated in the negative regulation of several oncogenic receptor tyrosine kinases including ErbB2. Lrig1 is expressed at low levels in several cancer types but is over-expressed in some prostate and colorectal tumors. Given this heterogeneity, whether Lrig1 functions to suppress or promote tumor growth remains a critical question. Previously, we found that Lrig1 was poorly expressed in ErbB2-positive breast cancer, suggesting that Lrig1 has a growth inhibitory role in this tumor type. However, breast cancer is a complex disease, with ErbB2-positive tumors accounting for just 25% of all breast cancers. To gain a better understanding of the role of Lrig1 in breast cancer, we examined its expression in estrogen receptor alpha (ERα)-positive disease which accounts for the majority of breast cancers. We find that Lrig1 is expressed at significantly higher levels in ERα-positive disease as compared to ERα-negative disease. Our study provides a molecular rationale for Lrig1 enrichment in ERα-positive disease by demonstrating that Lrig1 is a target of ERα. Estrogen stimulates Lrig1 accumulation and disruption of this induction enhances estrogen-dependent tumor cell growth, suggesting that Lrig1 functions as an estrogen regulated growth suppressor. Additionally, we find that Lrig1 expression correlates with prolonged relapse-free survival in ERα-positive breast cancer, identifying Lrig1 as a new prognostic marker in this setting. Finally, we demonstrate that ErbB2 activation antagonizes ERα-driven Lrig1 expression, providing a mechanistic explanation for Lrig1 loss in ErbB2-positive breast cancer. This work provides strong evidence for a growth inhibitory role for Lrig1 in breast cancer.
Keywords: Lrig1, negative regulator, estrogen receptor alpha, breast cancer
Introduction
Lrig1 is a member of the Lrig family of transmembrane leucine-rich repeat proteins and has been found to negatively regulate several oncogenic receptor tyrosine kinases (RTKs), including all members of the ErbB family (1–3), the Met (4) and Ret receptors (5). Lrig1 functions by promoting receptor degradation (1, 2) although the precise mechanisms by which Lrig1 engages the degradation machinery are not yet understood.
Lrig1 has growth suppressive properties (3, 6) and was proposed to be a tumor suppressor nearly ten years ago (7). Lrig1 null mice are known to develop epidermal hyperplasia (8, 9) but the tumor susceptibility of these mice remains uncharacterized. Lrig1 plays a critical role in maintaining epidermal stem cell quiescence (9, 10) and consequently, is down-regulated in poorly differentiated squamous cell carcinomas (11). Lrig1 expression is also decreased in other tumor types, including renal cell carcinoma (12), cervical cancer (13) and breast cancer (3). However, in colorectal and prostate cancer, Lrig1 over-expression has been reported (14, 15). Interestingly, Lrig1 correlated with poor prognosis in a Swedish cohort of prostate cancer patients who were followed by watchful waiting but with good prognosis in an American cohort who were treated with radical prostatectomy (15), raising the question as to whether and under what circumstances Lrig1 functions as a growth suppressor.
Lrig1 is down-regulated in ErbB2-positive breast cancer, an aggressive subtype of the disease with poor patient prognosis (16). Lrig1 loss in this setting is functionally significant since restoration of Lrig1 to ErbB2-overexpressing breast cancer cells decreases ErbB2 expression and limits tumor cell proliferation. Conversely, knock-down of residual Lrig1 in these cells enhances ErbB2 expression and augments tumor cell growth (3). Interestingly, ErbB2 activation was found to reduce Lrig1 expression, suggesting that ErbB2 takes an “active role” in its own overexpression through the marginalization of negative regulators (3). Currently, the mechanisms which govern Lrig1 expression and how ErbB2 activation intersects with these pathways remains completely unexplored.
Breast cancer is a heterogeneous disease, classified on the basis of clinical marker expression (ERα, Her2) and more recently, gene expression profiles. Given the variation in Lrig1 expression in human cancer, we sought to expand our analysis of Lrig1 beyond ErbB2-positive breast cancer (which represents just 25% of all breast cancers). In this study, we find that Lrig1 is enriched in ER-α positive breast cancer and mechanistically, we demonstrate that Lrig1 is a direct transcriptional target of ER-α. Lrig1 silencing experiments indicate that Lrig1 acts to restrict estrogen-driven tumor cell growth, strongly suggesting that Lrig1 functions as an estrogen-regulated growth suppressor. In support of this, ERα-positive breast cancer patients with high Lrig1 expression show significantly longer relapse-free survival, identifying Lrig1 as a new prognostic marker. In addition, our study reveals one mechanism by which ErbB2 suppresses Lrig1. ErbB2 activation antagonizes ER-α regulation of Lrig1, acting in a dominant manner to limit Lrig1 expression. Collectively, these data indicate that Lrig1 is a key growth suppressor in ERα–positive breast cancer and that Lrig1 suppression negatively impacts patient prognosis.
Materials and Methods
Reagents and Cell Culture
MCF7, ZR75-1 and T47D cells were purchased from the American Type Culture Collection and used at low passage. Growth rate and cell morphology of all cell lines were monitored on a continual basis. β-estradiol, Tamoxifen and Fulvestrant was purchased from Sigma-Aldrich. EGF was purchased from BD Biosciences. Neuregulin-1β (Nrg1β) was produced and purified as previously described (17).
Cell assays
T47D and ZR75-1 cells were plated into 24-well plates (Nunc, Rochester, NY, USA) at a density of 4.0 × 104 cells per well and 3.0 × 104 cells per well, respectively. After 72 hours of hormone starvation, cells were treated with vehicle control (VC) or 10 nM E2 and allowed to proliferate for another 48 hours at 37°C, changing the media every 24 hours. During the last 2 hours of growth, 500 ul of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Sigma, St. Louis, MO, USA) was added to the medium at a concentration of 0.5 mg/ml to measure activity. Crystals formed from the MTT were dissolved in acidic isopropanol and the absorption was measured at 570nm with a baseline subtraction at 655 nm. Plates were read within 30 minutes of adding the isopropanol. At least five points were averaged for each condition, and the experiment was repeated at least three times with a representative experiment selected.
RNAi
T47D, ZR75-1 or MCF7 cells were plated at a density of 1 × 105 cells per well in 24-well culture plates. Cells were transfected 24h after plating with 100nM siRNA targeting Lrig1 or FOXA1 (OnTarget plus SMARTpool, Dharmacon, Lafayette, CO, USA) or with non-targeting pool (OnTarget plus non-targeting pool, Dharmacon) using DharmaFect 1 (Dharmacon). Medium was replaced after 24h, and cells were treated as indicated. ON-TARGET plus SMARTpool siRNA_human FOXA1 (NM_004496) product # L-010319, ON-TARGET plus SMARTpool siRNA_human LRIG1 product # L-013940-00-0020, ON-TARGET plus non-targeting pool product # D-001810-10-20.
RNA isolation and Taqman real-time PCR
RNA was purified using a commercial kit (Qiagen; RNAeasy Kit). RNA was converted to cDNA using the High Capacity cDNA Reverse Transcription Synthesis Kit (Applied Biosystems). Analyses were carried out using TaqMan Gene Expression primers (Applied Biosystems) and probes that were labeled with FAM and Two Step RT qPCR Master Mix (EuroGentec, Freemont, CA, USA). Further details are listed in supplementary methods.
Chromatin Immunoprecipitation
ChIP was carried out following the method previously described (18). Please see supplementary methods for additional information. The four ER-alpha binding sites (Enh-1 to Enh-4) were submitted to the ORegAnno database (www.oreganno.org) and were assigned accession numbers OREG0052356 to OREG0052359 (19).
Analysis of Breast Cancer Microarray Data Sets for correlation between Lrig1 and ER status
Data was collected from publicly available microarray data sets from Oncomine (www.oncomine.org). ERα phenotypic data and LRIG1 gene expression profiles from two independent studies were obtained (20, 21). The log scale expression level of LRIG1 was determined and plotted according to the estrogen receptor status for each sample and the significance was determined using a t-test.
Analysis of Breast Cancer Microarray Data Sets for correlation between Lrig1 and relapse-free survival
Nine publicly available microarray studies were chosen and combined for analysis (21–29). The following criteria were employed to select usable samples: 1) study size greater than 100 samples 2) ER+ (as determined by clinical information or by array if unavailable) 3) Lymph node negative 4) HER2-negative (determined by array) 5) no systemic therapy or endocrine-therapy-only (e.g., tamoxifen) 6) outcome available: relapse free survival, distant metastasis free survival or disease free survival 7) Event time must be greater than 0 years. Data pre-processing: Duplicates were removed if they had the same GSM number, were indicated as having the same sample or patient identifier, or displayed a perfect correlation (>0.99) with another sample in correlation analysis. Cel files were downloaded from GEO and processed in R/Bioconductor using the ‘affy’ and ‘gcrma’ libraries. All samples were normalized together using GCRMA and mapped to Entrez gene symbols using the standard affy CDF. ESR1 expression status was determined using probe “ 205225_at” which was found to be the most useful probe by visual inspection (it also had by far the greatest variance) (30). Similarly, 4 probes were chosen from the ERBB2 amplicon for the genes ERBB2, GRB7, STARD3, PGAP3. Other genes in the amplicon (e.g., NEUROD2, TCAP, PNMT, IKZF3) either did not have probes or had very low variance. Expression values for the 4 probes in the ERBB2 amplicon were combined using a ranksum approach. ESR1- and ERBB2+ cutoff values were then chosen by mixed model clustering of their expression values. A total of 858 samples passed all filtering steps. Of these, 371 had insufficient follow-up for 10 year analysis but were included in the dataset for use in survival analysis. All filtered data were re-normalized together as above. Mapping was again performed with standard CDF files but also with custom CDFs (31). The probeset for Lrig1 with highest coefficient of variation was chosen for analysis (211596_s_at).
Statistics
Association between LRIG1 expression and 5year/10year relapse status was determined by Mann-Whitney U-test (MU). Patients were also divided into groups based on Lrig1 expression levels. Two grouping systems were used: (1) tertiles; (2) cutoffs determined by mixed model (MM) clustering. Kaplan-Meier survival analysis was then performed for relapse-free-survival (RFS) with these expression groups as a factor. Significant survival differences between the groups were determined by log rank (Mantel-Cox) test (with linear trend for factor levels). Events beyond 10 years were censored.
Results
Lrig1 is enriched in ER-positive breast cancer
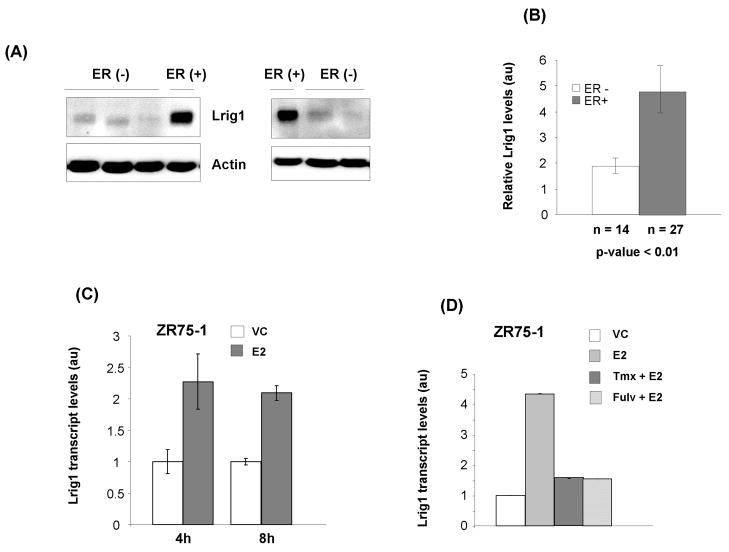
Prior work from our lab revealed that Lrig1 is down-regulated in ErbB2-positive breast cancer (3). To explore Lrig1 expression in other types of breast cancer, we conducted a survey of publicly available gene expression studies at the Oncomine database (www.oncomine.org). This survey revealed that Lrig1 transcript is enriched in ERα-positive breast tumors when compared to ERα-negative breast tumors. This correlation was observed in multiple independent studies (n > 8) and in Figure 1, two such studies are shown, the Chin study (20) and the Ivshina study (21) (p < 0.0001 for both). This correlation was also observed in a panel of 51 human breast cancer cell lines (Supplementary Figure 1, p < 0.001) (32). We next examined whether Lrig1 protein is enriched in ERα-positive human breast tumor specimens. 41 specimens with known ER status were surveyed by western blotting and densitometric analysis. A representative Lrig1 immunoblot is shown in Figure 2A with quantification of Lrig1 in the 41 specimens shown in Figure 2B. Additional blots are shown in Supplementary Figures 2 and 3. In agreement with the gene expression studies, Lrig1 protein was more abundant overall in ERα-positive tumors compared to ERα-negative tumors (p < 0.01).
Figure 1.
Lrig1 expression is associated with ERα status in human breast cancer. LRIG1 gene expression profiles and ERα phenotypic data of 363 breast carcinomas were obtained from two publicly available breast cancer microarray data sets. Correlation of Lrig1expression levels with ERα status of breast cancer specimens. Lrig1 is significantly overexpressed in the ER-positive tumors versus the ER-negative tumors using the t-test (p=0.0001). Left panel, Chin study. Right panel, Ivshina study.
Figure 2.
Lrig1 is elevated in ER-positive tumors. (A) Western blot analysis of lysates from ER-negative and ER-positive tumors. Tissue lysates were blotted for Lrig1 and actin (loading control). Representative samples shown. (B) Densitometric analysis of ER-negative (n = 14) and ER-positive (n = 27) tumors. (C) qPCR analysis of Lrig1 transcript in ZR75-1 breast cancer cells. Hormone starved cells were treated with either vehicle control (VC) or E2 for 4 or 8 hours. (D) qPCR analysis of Lrig1 transcript in ZR75-1 breast cancer cells. Hormone starved cells were treated for 72 hours with either vehicle control (VC), 10 nM E2, 10 nM E2 plus 1 μM tamoxifen or 10 nM E2 plus 100 nM Fulvestrant. Columns, representative experiment performed in triplicate from at least three independent experiments bars, standard deviation.
Lrig1 is induced in breast cancer cells by E2 stimulation
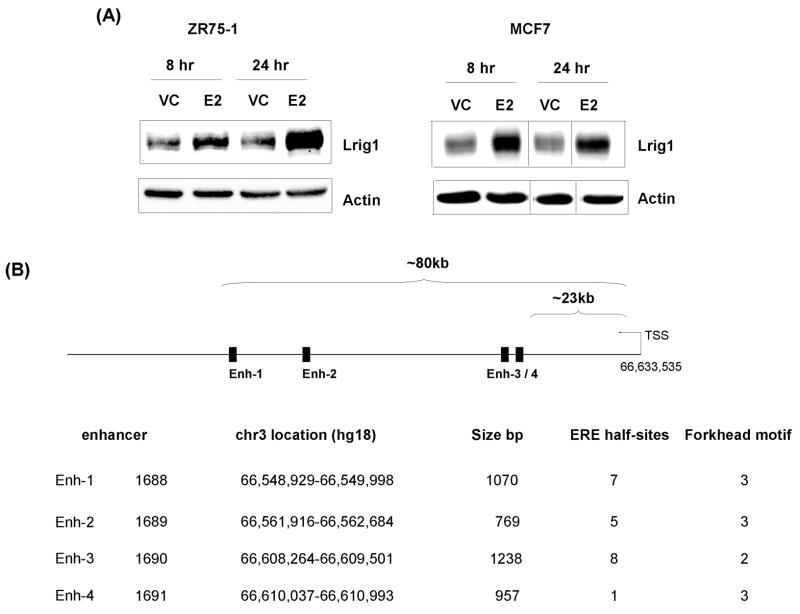
Given the strong correlation between ERα positivity and Lrig1, we next examined whether Lrig1 expression could be stimulated by E2 (17β-estradiol) treatment of ERα-positive breast cancer cells. As shown in Figure 2C, Lrig1 transcript accumulated following E2 treatment in ZR75-1 cells and MCF7 cells (data not shown). The antiestrogens Tamoxifen, which competes with estrogen for ERα binding, and Fulvestrant, which stimulates ERα proteolytic degradation, both antagonized the effects of E2 on Lrig1 (Figure 2D). We also examined the effects of E2 on Lrig1 protein abundance with a representative immunoblot shown in Figure 3A. In both cell lines, E2 stimulated Lrig1 protein accumulation.
Figure 3.
Lrig1 accumulates following E2 stimulation. (A) Hormone starved ZR75-1 (left panel) or MCF7 (right panel) cells were treated with either vehicle control (VC) or 10 nM E2 for either 8 or 24 hours as indicated. Cell lysates were blotted for Lrig1 and actin (loading control). A representative blot is shown. (B) Schematic of the 4 Lrig1 enhancer elements (referred to as enh 1–4 in text) with approximate reference to the furthest downstream transcription start site (TSS) (sequence based on genome version hg18). Table lists size of ERα DNA binding region and ERE and Forkhead motif counts.
Lrig1 is a transcriptional target of ER-α
Since E2 promotes Lrig1 transcript accumulation, we were interested to determine whether Lrig1 is a direct target of ER-α. One essential characteristic of a direct target is ERα binding to regulatory regions within the target gene. To examine whether ERα is found at the Lrig1 locus, we began by mining data from a published genome-wide chromatin immunoprecipitation (ChIP)-on-chip analysis in which 8,525 ERα binding sites were identified in MCF7 cells (33). Probing of this dataset revealed four intronic ER-α binding sites, located from ~ 23 kb to ~ 80 kb from the putative Lrig1 transcription start site (diagrammed in Figure 3B). Each site (present at ~ 1 kb resolution) had at least one ERE (estrogen response element) half site and several forkhead binding motifs. ERE half sites occur frequently in ERα regulated genes. Canonical EREs have been reported in only half of ERα binding sites identified by chromosome-wide mapping; the majority of the remaining sites contained ERE half sites (34). Forkhead binding motifs are enriched at ERα binding sites (34, 35) and the forkhead transcription factor FOXA1 is required for nearly all ERα binding and transcriptional events (36). FOXA1 possesses ATP-independent chromatin remodeling activity and has been called a “pioneer” factor due to its ability to bind silenced chromatin and open it for “business”, enabling the recruitment of other transcription factors such as ERα (37).
The ERα binding sites within the Lrig1 gene are nontraditional in that they are located at significant distance from the promoter and within introns. However, recent work has revealed that ERα binding is found throughout the genome. Chromosome-wide mapping of ERα binding has revealed that many (if not most) binding sites are located at significant distances (> 100 kb) from transcription start sites in ERα-regulated genes (34). Intronic binding of ERα in regulated genes has also been described and is thought to be functionally important for ERα-mediated transcription (38). Interestingly, intronic binding is not unique to ERα as other transcription factors including BARX2 (39) and CREB (40) have been observed to bind to introns within genes which they regulate. Given this, we pursued the four binding sites (designated Enh-1 – Enh-4 in Figure 3B) as potentially functional ERα binding sites with enhancer properties that contribute to E2-mediated Lrig1 expression. We first verified that ERα was recruited to each of the four reported binding sites. Since these sites were mined from MCF7 cell data (33), we extended these findings to ZR75-1 cells, another ERα-positive human breast cancer cell line (Supplementary Figure 4). We then performed ChIP-qPCR analysis using primers flanking each of the ERα-binding ChIP-chip target sites. Xbp1 (X-box binding protein-1) is a well characterized ER-α target and served as a positive control (34, 41). As expected, the first enhancer of Xbp1 (34) demonstrated E2-dependent recruitment of ERα and the coactivator p300 (Figure 4A) (42). E2 also stimulated recruitment of ERα and p300 to each of the four binding sites within Lrig1, with Enh-3 showing the greatest change in occupancy following E2 treatment. The E2-dependent co-recruitment of ERα and p300 lends strong support to the hypothesis that Enh-1-Enh-4 are functionally important regulatory sites.
Figure 4.
Chromatin Immunoprecipitation-qPCR analysis of the Lrig1 enhancer elements. Hormone starved ZR75-1 cells were treated with either vehicle control (VC) or 10 nM E2 for 30 minutes prior to cross-linking. Chromatin immunoprecipitation experiments were carried out using antibodies as indicated and subjected to qPCR with primers against the indicated regions. Enhancer # 1 of Xbp1 served as a positive control. Shown is the mean of three independent replicates with standard deviation. (A) Chromatin immunoprecipitation was performed with antibodies against p300 or ERα. Results are plotted as the fold enrichment of E2 treated cells over vehicle control treated cells. (B) Chromatin immunoprecipitation was performed with antibodies against H3K4me2. Results are plotted as fold enrichment over input DNA for both E2 and control treated cells. (C) Chromatin immunoprecipitation was performed with antibodies against FOXA1. Results are plotted as fold enrichment over input DNA for both E2 and control treated cells.
Histone modifications such as methylation and acetylation are dynamic, each functionally linked to an active or repressed chromatin state (43). Methylation of lysine residue 4 within Histone 3 (H3K4me) characterizes active genes and is distributed across the length of the gene body. Mono- and Di-methylation are associated with functional enhancers while tri-methylation is associated with active promoter regions (44). FOXA1 recruitment to enhancer elements is dependent upon the presence of H3K4me1/me2; reversal of this modification by overexpression of the lysine demethylase LSD1 prevents FOXA1 recruitment (45). To determine whether Enh-1-Enh-4 were modified as would be expected for functional enhancers, we performed ChIP in ZR75-1 cells with an antibody specific for H3K4me2 (Figure 4B). The Xbp1 enhancer showed significant enrichment for H3K4me2, as expected. Enh-1-Enh-4 were all enriched for H3K4me2, with Enh3 demonstrating the most enrichment. H3K4me2 was present before E2 stimulation and did not change significantly following E2 stimulation.
ERα-mediated Lrig1 expression depends on FOXA1
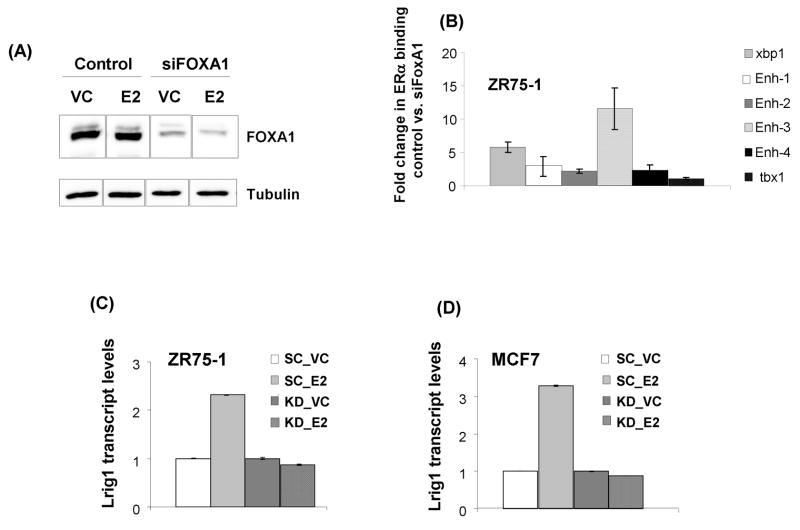
Since FOXA1 is reportedly upstream of nearly all ERα-chromatin interactions (36), we next examined the occupancy of FOXA1 at Enh-1-Enh-4. Given that FOXA1 opens the chromatin and enables ERα recruitment, FOXA1 should be present before E2 stimulation. It has also been reported that FOXA1 dissociates from these regulatory sites following E2 stimulation (36). We observed this pattern for the Xbp1 enhancer and for each of the four Lrig1 elements in ZR75-1 cells (Figure 4C).
We next examined whether ERα recruitment to Lrig1 elements is FOXA1-dependent by knocking down FOXA1 using siRNA. A representative immunoblot of FOXA1 knockdown is shown in Figure 5A. FOXA1 knockdown was confirmed by western blot in all experiments using FOXA1 siRNA. ERα occupancy following E2 stimulation was evaluated by ChIP. Xbp1 was used as a positive control since E2-mediated ERα recruitment to its enhancer is FOXA1-dependent (34). Tbx1, a member of the T-box family of transcription factors (46), is an ERα target that is independent of FOXA1 binding (45) and served as a negative control. ERα occupancy at the Xbp1 enhancer was diminished approximately 5-fold by FOXA1 knockdown, confirming its FOXA1 dependency, while ERα occupancy at the Tbx1 enhancer was unaffected by FOXA1 knockdown, confirming its independence of FOXA1. ERα occupancy at each of the four Lrig1 elements was reduced by FOXA1 knockdown although Enh-4 did not show a statistically significant decrease. Enh-3 showed the greatest reduction in ERα occupancy (~ 10-fold) with FOXA1 knockdown (Figure 5B).
Figure 5.
E2-mediated regulation of Lrig1 is FOXA1-dependent. (A) Representative western blot of FOXA1 knockdown efficiency. Hormone starved ZR75-1 cells were treated with either scramble control or FOXA1 siRNA. The following day the medium was changed to include either vehicle control (VC) or 10 nM E2 for an additional 24 hours. Cell lysates were collected at 48 hours post-transfection. (B) Hormone starved ZR75-1 cells were treated with either scramble control or FOXA1 siRNA 48 hours prior to treatment with 10 nM E2 or VC for 30 minutes followed by cross-linking. Chromatin immunoprecipitation was performed with antibodies against ERα and subjected to qPCR with primers against the indicated regions. Xbp1 enhancer 1 served as a positive control while the Tbx1 enhancer (FOXA1-independent) served as a negative control. Results are plotted as fold change in ERα occupancy, comparing scramble control and FOXA1 siRNA treatment. Shown is the mean of three independent replicates with standard deviation. Hormone starved ZR75-1 (C) and MCF7 (D) cells were treated with either scramble control or siRNA to FOXA1. Cells were then treated with either vehicle control (VC) or E2 for 8 hours before harvesting. Lrig1 transcript abundance was measured using Taqman real-time qPCR. Experiments were performed in triplicate and repeated at least three times with a representative experiment shown.
To examine the functional impact of FOXA1 on Lrig1 expression, we examined Lrig1 transcript accumulation following E2 stimulation +/− FOXA1 siRNA. As shown in Figure 5C and D, the E2-dependent increase in Lrig1 transcript abundance was negated by FOXA1 knockdown in both ZR75-1 and MCF7 cells. Together with the evidence presented thus far, these data demonstrate that Lrig1 is a direct transcriptional target of ERα and its expression is dependent upon FOXA1-mediated ERα recruitment to one or more enhancer elements. Primary transcriptional targets of ERα do not require new protein synthesis for regulation by E2 (since all necessary factors are pre-existing in cells) and the induction of these targets is maintained in the presence of protein synthesis inhibitors such as cycloheximide. Review of a recent microarray analysis of E2-regulated genes in MCF7 cells (47) reveals that Lrig1 induction by E2 is unaffected by cycloheximide, defining Lrig1 as a primary transcriptional target.
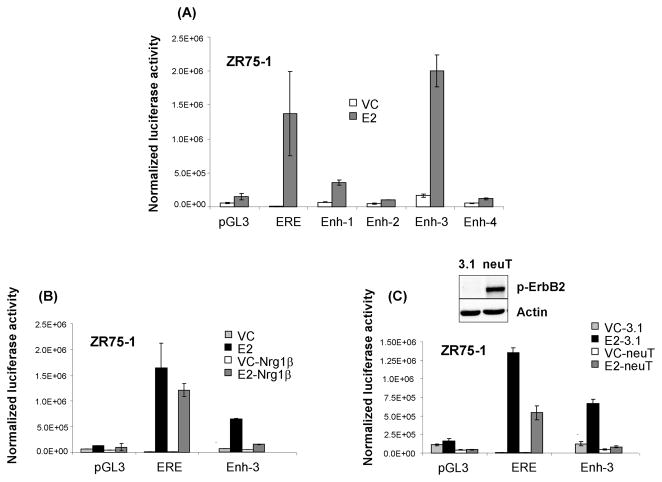
Luciferase reporter assays are useful tools in determining whether a putative DNA regulatory element has the capacity to drive transcription. This approach was recently used by Carroll et al (34) to demonstrate that distal ERα binding domains have enhancer characteristics. Using this approach, we cloned the four Lrig1 elements individually into the pGL3 luciferase vector and transfected these constructs into hormone-starved ZR75-1 cells. The pCMX vector expressing β-galactosidase was co-transfected as a control. Cells were treated with vehicle control or E2 and assayed for firefly luciferase activity. Luciferase activity was normalized to β-galactosidase activity to control for transfection efficiency (48). The 3x-ERE-luc vector (containing canonical estrogen response elements (49)) served as a positive control. As shown in Figure 6A, the ERE positive control produced robust E2-dependent luciferase activity, as expected (note: vehicle control is graphed for the ERE but is very small). The Enh-2 and Enh-4 elements did not demonstrate activity beyond the background activity of the pGL3 parental vector. However, both Enh-1 and more notably, the Enh-3 element, drove E2-dependent luciferase activity that was significantly higher than the pGL3 parental vector. These results indicate that as individual elements, Enh-1 and Enh-3 have enhancer activity. Although Enh-2 and Enh-4 do not show activity in this assay, they may contribute to E2 regulation of Lrig1 expression in the endogenous chromatin context.
Figure 6.
ErbB2 activation suppresses Lrig1 transcriptional output. All panels: ZR75-1 cells were transiently transfected with the pGL3-SV40 luciferase reporter vector (pGL3) or pGL3-SV40 containing enhancer elements 1–4. The β-galactosidase encoding pCMX vector was co-transfected and served as an internal control for transfection efficiency. 3X-ERE-Luc served as a positive control for E2 responsiveness. (A) Cells were hormone starved and treated with either vehicle control (VC) or 10 nM E2 for 18 hours. Cells were then assayed for firefly luciferase and β-galactosidase activity. Results shown are firefly luciferase activity normalized to β-galactosidase activity. Shown are representative experiments performed in triplicate with standard deviation. For (B), cells were treated as in (A) and in addition, cells were treated with VC or E2 plus 10 nM Nrg1β. For (C), cells were additionally transfected with either pcDNA3.1 vector control or NeuT expressing vector before treatment and assay as in (A).
ErbB2 suppresses Lrig1 by disrupting regulation by ERα
In prior studies from our lab, we found that Lrig1 is down-regulated in ErbB2 positive tumors when compared to ErbB2 negative tumors (3). In ERα-positive MCF7 and T47D breast cancer cells, we observed that ErbB2 activation suppressed Lrig1 transcript and protein. Conversely, ErbB2 knockdown in MCF7 cells led to Lrig1 accumulation (3). Both experiments were done in hormone replete media, indicating that ErbB2 activation has a dominant effect on Lrig1 expression. Since ERα and ErbB2 have been found to negatively regulate one another (33, 50), we hypothesized that ErbB2 may suppress Lrig1 by antagonizing ERα-mediated Lrig1 induction. To explore this, we examined the impact of ErbB2 activation on E2-dependent Lrig1 transcriptional activity using the luciferase reporter assay in ZR75-1 cells. ErbB2 signaling was stimulated either by Neuregulin-1β or by ectopic expression of a constitutively active point mutant of ErbB2, NeuT (51, inset of Figure 6C). Neuregulin-1β activates ErbB2 by promoting its heterodimerization with other family members, predominantly ErbB3. In both cases, ErbB2 activation significantly decreased E2-dependent reporter activity from the Enh-3 element (Figure 6B & C). Since ErbB2 has been found to decrease ER-α protein expression (50), these results are likely a consequence of ErbB2-mediated ERα downregulation. Indeed, blotting of lysates confirmed that ErbB2 activation decreased ERα expression (data not shown). Therefore, one mechanism by which ErbB2 suppresses Lrig1 is by downregulating ERα, antagonizing ERα-mediated Lrig1 transcription. These data are in agreement with our prior findings that a) ErbB2-positive tumors have lower Lrig1 transcript than ErbB2- negative tumors and b) ErbB2 activation down-regulates Lrig1 transcript (Supplementary Figure 5).
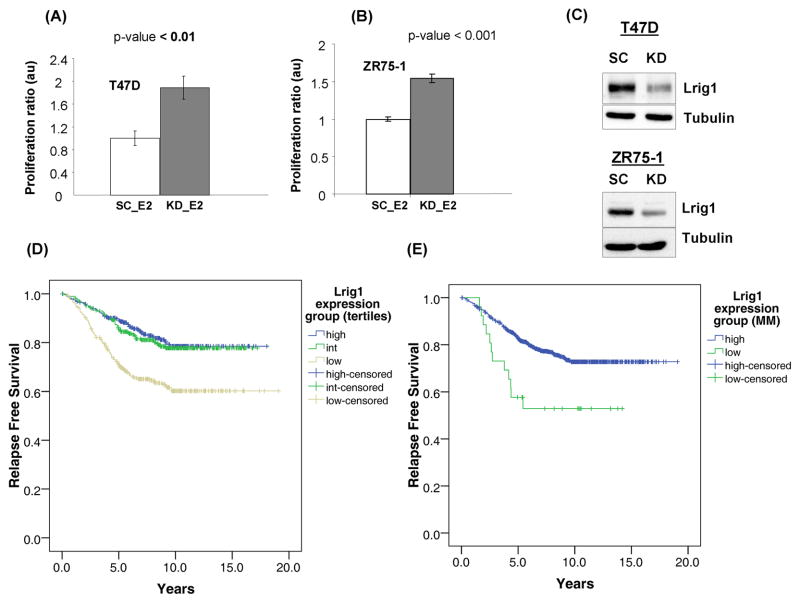
Lrig1 limits E2-dependent breast cancer cell growth and correlates with longer relapse-free survival in ER-positive breast cancer
Prior studies have provided evidence that Lrig1 functions as a growth suppressor although Lrig1 function may vary depending on tissue context and other variables (14, 15). The role of Lrig1 in ERα-positive breast cancer is currently unknown. We hypothesized that induction of Lrig1 provides an anti-growth signal, limiting the overall proliferative response to E2. While the ultimate outcome of ERα signaling is cellular proliferation, this outcome is a balance of growth (feed forward) and anti-growth (negative feedback) signals. To determine the role of Lrig1 induction in E2-driven tumor cell growth, T47D and ZR75-1 breast cancer cells (which both demonstrate E2-dependent Lrig1 induction) were treated with E2 +/− Lrig1 siRNA and subjected to an MTT assay (Figure 7A & B). Western blotting confirmed the efficient silencing of Lrig1 by siRNA in both cell lines (Figure 7C). The growth of E2-stimulated cells treated with scramble control (Lrig1 induction by E2 intact) was normalized to 1.0. The growth of E2-stimulated cells treated with Lrig1 siRNA (Lrig1 induction by E2 disrupted) was then compared to scramble control cells. As shown in Figure 7A & B, Lrig1 silencing augmented the E2-dependent growth of both T47D and ZR75-1 cells, demonstrating that the induction of Lrig1 acts to limit estrogen-dependent tumor cell growth. These data strongly suggest that in ER-positive breast cancer, Lrig1 functions as an ERα regulated growth suppressor.
Figure 7.
Lrig1 suppresses growth of ER-positive breast cancer cells and correlates with prolonged relapse-free survival. Hormone starved T47D (A) and ZR75-1 (B) cells were treated with either scramble control or Lrig1 siRNA and then treated with E2 for 48 hours. Cell viability was measured using the MTT assay. The growth of E2 treated/scramble control cells was normalized to 1.0. (C) Representative western blot depicting efficiency of Lrig1 knockdown. (D) and (E) Association between Lrig1 expression and relapse-free survival (RFS) was determined. Patients were divided into groups based on Lrig1 expression levels. Two grouping systems were used: (D) tertiles; (E) cutoffs determined by mixed model (MM) clustering. Kaplan-Meier survival analysis was then performed for RFS with these expression groups as a factor. Significant survival differences between the groups were determined by log rank (Mantel-Cox) test (linear trend for factor levels). Events beyond 10 years were censored.
Resistance to anti-estrogens such as Tamoxifen arises when cells acquire the ability to grow in an estrogen-independent manner. To determine whether Lrig1 loss promotes estrogen-independent growth, Lrig1 was knocked down in ZR75-1 cells and cell growth under hormone starved conditions was evaluated. Interestingly, Lrig1 depletion enhanced estrogen-independent growth, suggesting that Lrig1 loss may contribute to anti-estrogen resistance (Supplementary Figure 6A). However, Lrig1 depletion did not increase the growth of MCF7 cells under identical conditions (data not shown), suggesting that cellular context may be an important modulator. Loss of ERα is one means by which cells become less dependent on estrogen for growth. To determine whether Lrig1 loss decreased ERα expression, lysates from control and knock-down cells were blotted for ERα. Lrig1 loss had no effect on ERα expression (Supplementary Figure 6B), strongly suggesting that loss of Lrig1 enhances estrogen-independent growth by other means. Since Lrig1 has been found to limit receptor tyrosine kinase signaling (1–6), Lrig1 loss may drive estrogen-independent growth through increased signaling.
To expand upon and investigate the clinical relevance of our findings, we examined Lrig1 expression in ERα-positive breast tumor specimens from publicly available gene expression studies (listed in Supplementary Table 1, 21–29). Nine studies were combined and normalized as described in the methods section. 858 patients (ER-pos/LN-neg/ErbB2-neg) passed all filtering steps and were included in our analysis. Lrig1 expression was significantly associated with 5year (p=1.919E-8) and 10year (p=8.109E-6) relapse status by MU test. Patients with relapse had lower LRIG1 expression on average. Patients were also divided into groups based on Lrig1 expression levels. Two grouping systems were used: (1) tertiles or (2) cutoffs as determined by mixed model (MM) clustering. Kaplan-Meier survival analysis was then performed for relapse-free-survival (RFS) with these expression groups as a factor. Patients grouped into the low expression group had significantly worse RFS than intermediate or high expression groups by log rank test for both tertiles grouping (p=1.782E-7, Figure 7D) and MM grouping (p=0.004, Figure 7E). Since relapse occurs when tumor growth resumes after a period of apparent dormancy, these data provide strong support for the role of Lrig1 as a growth suppressor.
Our data demonstrate that ERα plays a key role in Lrig1 expression in breast cancer cells. Despite this, there is heterogeneity in Lrig1 expression, with a subset of ER-positive patients displaying low Lrig1 expression and earlier relapse. This suggests that there are other factors which compromise the integrity of the ERα-Lrig1 loop, such as ErbB2 activation (Figure 6). Although the patient cohort examined in this study is ErbB2-negative (ErbB2 locus not amplified), this does not exclude ErbB2 signaling which may arise for other reasons such as autocrine growth factor production or ErbB2 cross-talk with other receptors. In agreement with this, we previously found that activation of modest (non-amplified) levels of ErbB2 in ER-positive T47D and MCF7 cells suppresses Lrig1 (3).
Discussion
Since its initial cloning in 1996 (52) and its later discovery as a negative regulator of receptor tyrosine kinases (1 – 6), Lrig1 has emerged as an important player in cellular signaling and as a putative tumor suppressor (7). Despite this, the mechanisms which govern Lrig1 expression and loss in tumors have remained uncharacterized. Lrig1 is structurally homologous to Kekkon-1, a negative regulator of the Drosophila EGF receptor/DER (53), and was initially proposed to be a Kekkon-1 ortholog (7). Kekkon-1 is transcriptionally induced by DER activation, acting in a negative feedback loop to regulate DER-dependent oogenesis (53). As a putative Kekkon-1 ortholog, it was anticipated that Lrig1 would be induced by ErbB receptor signaling and one study (2) reported that Lrig1 accumulated following EGF stimulation of Hela cells. Nevertheless, in breast cancer cells, we have found that ErbB2 activation suppresses rather than induces Lrig1 (3, Supplementary Figure 5). Our findings are supported by data mined from a microarray study in MCF7 cells in which expression of constitutively active ErbB2 led to decreased Lrig1 transcript (54, GSE3542). Differences between Lrig1 and Kekkon-1 may be reconciled in part by the fact that Drosophila Lambik, rather than Kekkon-1, is the closest Lrig1 relative (55). Very little is known regarding Lambik function although it was recently found capable of substituting for SMA-10, the putative Lrig ortholog in C-elegans, strongly suggesting that Lambik functions in a “Lrig-like” manner (56).
Lrig1 expression, when considered across all cancer types investigated to date, is heterogeneous, with over-expression reported in prostate and colorectal cancer (14, 15) and under-expression reported in renal, cervical and breast cancers (3, 12, 13). Even within a particular tumor type such as prostate cancer, Lrig1 has been correlated with good or bad prognosis in different patient cohorts (15). This variation in expression has led to the proposal that Lrig1 may function as a “double edged sword”, switching between tumor suppressor/promoter in a manner dependent upon cellular context (57). Since Lrig1 plays an important role in the regulation of membrane receptor stability, its tumor suppressor or promoter function may track with the profile of receptors it regulates, which in turn may vary with cellular/tissue context. In breast cancer, Lrig1 function is unclear although accumulating evidence (3, 4 and this study) supports the concept that it functions as a growth suppressor. However, copy number of the Lrig1 gene was observed to be moderately increased in one study of ErbB2-positive tumors, which suggests that Lrig1 could have a growth promoting role (58). On the other hand, in our prior study which also included ErbB2-positive tumors (3), Lrig1 transcript and protein were significantly decreased suggesting that Lrig1 transcript and protein are selected against even if the gene dosage is increased as Ljuslider and colleagues found (58).
To gain insight into the role of Lrig1 in breast cancer, we compared the expression pattern of Lrig1 in various subtypes of breast cancer. Our analysis reveals that ERα-positive breast cancer is enriched for Lrig1 transcript and protein relative to ERα-negative breast cancer. In this study, we uncover a molecular mechanism which drives elevated Lrig1 expression in ERα-positive breast cancer, demonstrating that Lrig1 is a direct transcriptional target of ERα/FOXA1. Our cell culture experiments suggest that ERα contributes significantly to Lrig1 expression in the ER-positive setting since hormone starved cells express modest Lrig1 levels and E2 stimulation gives a robust increase in Lrig1 expression. Interestingly, Lrig1 protein is dramatically increased following E2 stimulation while changes in mRNA are more modest, suggesting that Lrig1 may also be post-transcriptionally modulated by E2/ERα. The induction of Lrig1 by ERα has clinical significance since ERα-positive breast cancer patients with intermediate/high Lrig1 (which likely reflects ERα-driven expression) have significantly longer relapse-free survival. Factors which disrupt this pathway, such as activation of ErbB2 or other RTKs which repress ERα function (50), would lead to lower Lrig1 levels, accelerated tumor growth and worsened prognosis. This is reflected in the increased growth of tumor cell lines when Lrig1 induction by ERα is disrupted with siRNA. Examination of a recent microarray study indicates that expression of constitutively active Raf-1 in MCF7 cells dramatically down-regulates both ERα and Lrig1 transcript (54, GSE3542). Since Raf-1 is a common downstream target of RTKs, this suggests that RTK activation will suppress rather than induce Lrig1, as we have found. Furthermore, evaluation of data from Bhat- Nakshatri et al (59) indicates that expression of constitutively active Akt in MCF7 cells displaces ERα from the Enh-1 and Enh-3 Lrig1 enhancer elements, the two elements that demonstrated E2-dependent activity in the reporter assay (Figure 6). Therefore, activation of both the Raf/Mek/MapK and PI3K/Akt pathways contributes to suppression of Lrig1 by RTK signaling, through effects on ERα. Tumor/growth suppressor proteins are down-regulated in tumors by a variety of mechanisms including epigenetic silencing (60) and loss of heterozygosity (61). While this study explores the transcriptional suppression of Lrig1 by oncogenic signaling, additional mechanisms may contribute to Lrig1 down-regulation.
Collectively, our data lend support to the hypothesis that in ERα-positive breast cancer, Lrig1 functions as an important growth suppressor and restrains E2-dependent tumor cell growth, prolonging relapse-free survival. In addition, our data suggest that in some cellular contexts, Lrig1 loss may contribute to estrogen-independent growth and acquisition of resistance to anti-estrogens. A “threshold” level of Lrig1 expression is necessary to realize the benefits with respect to prolonged relapse free survival as patients with both intermediate and high levels of expression segregated from those with low expression. Since ErbB2 activation down-regulates Lrig1, our results suggest that ErbB2 inhibitors such as Lapatinib may be one strategy to restore Lrig1 to tumors with low expression. Our data also suggests that Lrig1 may be useful as a prognostic marker in ERα-positive breast cancer although further studies are necessary to explore this. In summary, Lrig1 is expressed at low levels in ErbB2-positive (3) and ERα-negative breast cancer (this study) but enriched in ERα-positive cancer due to direct transcriptional regulation by ERα. Induction of Lrig1 limits E2-dependent tumor cell growth and correlates with prolonged relapse-free survival in the ERα-positive setting.
Supplementary Material
Acknowledgments
We thank Dr. Hongwu Chen from UC Davis for providing the 3X-ERE-luc construct.
This work was supported by NIH grants CA118384 (C.S.), GM068994 (K.C.) and by the Director, Office of Science, Office of Biological & Environmental Research, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231, by the National Institutes of Health, National Cancer Institute grants P50 CA 58207and by the Stand Up To Cancer-American Association for Cancer Research Dream Team Translational Cancer Research Grant SU2C-AACR-DT0409 (JG). The content of the information does not necessarily reflect the position or the policy of the Government, and no official endorsement should be inferred. O.G. is supported by the Canadian Institutes of Health Research. H.R. is a recipient of a Department of Defense Breast Cancer Research Program pre-doctoral fellowship.
Footnotes
Conflicts of Interest: Joe W. Gray serves on Scientific Advisory Boards for Agendia, Cepheid, KromaTiD and New Leaf ventures.
References
- 1.Laederich MB, Funes-Duran M, Yen L, Ingalla E, Wu X, Carraway KL, 3rd, et al. The leucine- rich repeat protein LRIG1 is a negative regulator of ErbB family receptor tyrosine kinases. J Biol Chem. 2004 Nov 5;279(45):47050–6. doi: 10.1074/jbc.M409703200. [DOI] [PubMed] [Google Scholar]
- 2.Gur G, Rubin C, Katz M, Amit I, Citri A, Nilsson J, et al. LRIG1 restricts growth factor signaling by enhancing receptor ubiquitylation and degradation. EMBO J. 2004 Aug 18;23(16):3270–81. doi: 10.1038/sj.emboj.7600342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller JK, Shattuck DL, Ingalla EQ, Yen L, Borowsky AD, Young LJ, et al. Suppression of the negative regulator LRIG1 contributes to ErbB2 overexpression in breast cancer. Cancer Res. 2008 Oct 15;68(20):8286–94. doi: 10.1158/0008-5472.CAN-07-6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shattuck DL, Miller JK, Laederich M, Funes M, Petersen H, Carraway KL, 3rd, et al. LRIG1 is a novel negative regulator of the Met receptor and opposes Met and Her2 synergy. Mol Cell Biol. 2007 Mar;27(5):1934–46. doi: 10.1128/MCB.00757-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ledda F, Bieraugel O, Fard SS, Vilar M, Paratcha G. Lrig1 is an endogenous inhibitor of Ret receptor tyrosine kinase activation, downstream signaling, and biological responses to GDNF. J Neurosci. 2008 Jan 2;28(1):39–49. doi: 10.1523/JNEUROSCI.2196-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stutz MA, Shattuck DL, Laederich MB, Carraway KL, 3rd, Sweeney C. LRIG1 negatively regulates the oncogenic EGF receptor mutant EGFRvIII. Oncogene. 2008 Sep 25;27(43):5741–52. doi: 10.1038/onc.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hedman H, Nilsson J, Guo D, Henriksson R. Is LRIG1 a tumour suppressor gene at chromosome 3p14.3. Acta Oncol. 2002;41(4):352–4. doi: 10.1080/028418602760169398. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki Y, Miura H, Tanemura A, Kobayashi K, Kondoh G, Sano S, et al. Targeted disruption of LIG-1 gene results in psoriasiform epidermal hyperplasia. FEBS Lett. 2002 Jun 19;521(1–3):67–71. doi: 10.1016/s0014-5793(02)02824-7. [DOI] [PubMed] [Google Scholar]
- 9.Jensen KB, Collins CA, Nascimento E, Tan DW, Frye M, Itami S, et al. Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis. Cell Stem Cell. 2009 May 8;4(5):427–39. doi: 10.1016/j.stem.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jensen KB, Watt FM. Single-cell expression profiling of human epidermal stem and transit-amplifying cells: Lrig1 is a regulator of stem cell quiescence. Proc Natl Acad Sci U S A. 2006 Aug 8;103(32):11958–63. doi: 10.1073/pnas.0601886103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jensen KB, Jones J, Watt FM. A stem cell gene expression profile of human squamous cell carcinomas. Cancer Lett. 2008 Dec 8;272(1):23–31. doi: 10.1016/j.canlet.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomasson M, Hedman H, Guo D, Ljungberg B, Henriksson R. LRIG1 and epidermal growth factor receptor in renal cell carcinoma: a quantitative RT--PCR and immunohistochemical analysis. Br J Cancer. 2003 Oct 6;89(7):1285–9. doi: 10.1038/sj.bjc.6601208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindstrom AK, Ekman K, Stendahl U, Tot T, Henriksson R, Hedman H, et al. LRIG1 and squamous epithelial uterine cervical cancer: correlation to prognosis, other tumor markers, sex steroid hormones, and smoking. Int J Gynecol Cancer. 2008 Mar-Apr;18(2):312–7. doi: 10.1111/j.1525-1438.2007.01021.x. [DOI] [PubMed] [Google Scholar]
- 14.Ljuslinder I, Golovleva I, Palmqvist R, Oberg A, Stenling R, Jonsson Y, et al. LRIG1 expression in colorectal cancer. Acta Oncol. 2007;46(8):1118–22. doi: 10.1080/02841860701426823. [DOI] [PubMed] [Google Scholar]
- 15.Thomasson M, Wang B, Hammarsten P, Dahlman A, Persson JL, Josefsson A, et al. LRIG1 and the liar paradox in prostate cancer: A study of the expression and clinical significance of LRIG1 in prostate cancer. Int J Cancer. Dec 2; doi: 10.1002/ijc.25820. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 16.Slamon DJ, Godolphin W, Jones LA, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–12. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 17.Funes M, Miller JK, Lai C, Carraway KL, 3rd, Sweeney C. The mucin Muc4 potentiates neuregulin signaling by increasing the cell-surface populations of ErbB2 and ErbB3. J Biol Chem. 2006;281:19310–9. doi: 10.1074/jbc.M603225200. [DOI] [PubMed] [Google Scholar]
- 18.O’Geen H, Nicolet CM, Blahnik K, Green R, Farnham PJ. Comparison of sample preparation methods for ChIP-chip assays. Biotechniques. 2006;41:577–80. doi: 10.2144/000112268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffith OL, Montgomery SB, Bernier B, et al. ORegAnno: an open-access community-driven resource for regulatory annotation. Nucleic Acids Res. 2008;36 (Database issue):D107–13. doi: 10.1093/nar/gkm967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chin K, DeVries S, Fridlyand J, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–41. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 21.Ivshina AV, George J, Senko O, et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66:10292–301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Klijn JG, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–9. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 23.Loi S, Haibe-Kains B, Desmedt C, et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25:1239–46. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 24.Desmedt C, Piette F, Loi S, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007;13:3207–14. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 25.Symmans WF, Hatzis C, Sotiriou C, et al. Genomic index of sensitivity to endocrine therapy for breast cancer. J Clin Oncol. 2010;28:4111–9. doi: 10.1200/JCO.2010.28.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Sieuwerts AM, McGreevy M, et al. The 76-gene signature defines high-risk patients that benefit from adjuvant tamoxifen therapy. Breast Cancer Res Treat. 2009;116:303–9. doi: 10.1007/s10549-008-0183-2. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt M, Bohm D, von Torne C, et al. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 2008;68:5405–13. doi: 10.1158/0008-5472.CAN-07-5206. [DOI] [PubMed] [Google Scholar]
- 28.Sotiriou C, Wirapati P, Loi S, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–72. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 29.Miller LD, Smeds J, George J, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A. 2005;102:13550–5. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gong Y, Yan K, Lin F, et al. Determination of oestrogen-receptor status and ERBB2 status of breast carcinoma: a gene-expression profiling study. Lancet Oncol. 2007;8:203–11. doi: 10.1016/S1470-2045(07)70042-6. [DOI] [PubMed] [Google Scholar]
- 31.Dai M, Wang P, Boyd AD, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hurtado A, Holmes KA, Geistlinger TR, et al. Regulation of ERBB2 by oestrogen receptor- PAX2 determines response to tamoxifen. Nature. 2008;456:663–6. doi: 10.1038/nature07483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carroll JS, Liu XS, Brodsky AS, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 35.Laganiere J, Deblois G, Lefebvre C, Bataille AR, Robert F, Giguere V. From the Cover: Location analysis of estrogen receptor alpha target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc Natl Acad Sci U S A. 2005;102:11651–6. doi: 10.1073/pnas.0505575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet. 2011;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–89. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 38.Lin Z, Reierstad S, Huang CC, Bulun SE. Novel estrogen receptor-alpha binding sites and estradiol target genes identified by chromatin immunoprecipitation cloning in breast cancer. Cancer Res. 2007;67:5017–24. doi: 10.1158/0008-5472.CAN-06-3696. [DOI] [PubMed] [Google Scholar]
- 39.Stevens TA, Iacovoni JS, Edelman DB, Meech R. Identification of novel binding elements and gene targets for the homeodomain protein BARX2. J Biol Chem. 2004;279:14520–30. doi: 10.1074/jbc.M310259200. [DOI] [PubMed] [Google Scholar]
- 40.Impey S, McCorkle SR, Cha-Molstad H, et al. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–54. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 41.Lacroix M, Leclercq G. About GATA3, HNF3A, and XBP1, three genes co-expressed with the oestrogen receptor-alpha gene (ESR1) in breast cancer. Mol Cell Endocrinol. 2004;219:1–7. doi: 10.1016/j.mce.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 42.Vo N, Goodman RH. CREB-binding protein and p300 in transcriptional regulation. J Biol Chem. 2001;276:13505–8. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- 43.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 44.Lupien M, Brown M. Cistromics of hormone-dependent cancer. Endocr Relat Cancer. 2009;16:381–9. doi: 10.1677/ERC-09-0038. [DOI] [PubMed] [Google Scholar]
- 45.Lupien M, Eeckhoute J, Meyer CA, et al. FoxA1 translates epigenetic signatures into enhancer- driven lineage-specific transcription. Cell. 2008;132:958–70. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Packham EA, Brook JD. T-box genes in human disorders. Hum Mol Genet. 2003;12(Spec No 1):R37–44. doi: 10.1093/hmg/ddg077. [DOI] [PubMed] [Google Scholar]
- 47.Bourdeau V, Deschenes J, Laperriere D, Aid M, White JH, Mader S. Mechanisms of primary and secondary estrogen target gene regulation in breast cancer cells. Nucleic Acids Res. 2008;36:76–93. doi: 10.1093/nar/gkm945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Louie MC, Revenko AS, Zou JX, Yao J, Chen HW. Direct control of cell cycle gene expression by proto-oncogene product ACTR, and its autoregulation underlies its transforming activity. Mol Cell Biol. 2006;26:3810–23. doi: 10.1128/MCB.26.10.3810-3823.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zou JX, Revenko AS, Li LB, Gemo AT, Chen HW. ANCCA, an estrogen-regulated AAA+ ATPase coactivator for ERalpha, is required for coregulator occupancy and chromatin modification. Proc Natl Acad Sci U S A. 2007;104:18067–72. doi: 10.1073/pnas.0705814104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parra-Palau JL, Pedersen K, Peg V, et al. A major role of p95/611-CTF, a carboxy-terminal fragment of HER2, in the down-modulation of the estrogen receptor in HER2-positive breast cancers. Cancer Res. 2010;70:8537–46. doi: 10.1158/0008-5472.CAN-10-1701. [DOI] [PubMed] [Google Scholar]
- 51.Bargmann CI, Hung MC, Weinberg RA. Multiple independent activations of the neu oncogene by a point mutation altering the transmembrane domain of p185. Cell. 1986;45:649–57. doi: 10.1016/0092-8674(86)90779-8. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki Y, Sato N, Tohyama M, Wanaka A, Takagi T. cDNA cloning of a novel membrane glycoprotein that is expressed specifically in glial cells in the mouse brain. LIG-1, a protein with leucine-rich repeats and immunoglobulin-like domains. J Biol Chem. 1996;271:22522–7. doi: 10.1074/jbc.271.37.22522. [DOI] [PubMed] [Google Scholar]
- 53.Ghiglione C, Carraway KL, 3rd, Amundadottir LT, Boswell RE, Perrimon N, Duffy JB. The transmembrane molecule kekkon 1 acts in a feedback loop to negatively regulate the activity of the Drosophila EGF receptor during oogenesis. Cell. 1999;96:847–56. doi: 10.1016/s0092-8674(00)80594-2. [DOI] [PubMed] [Google Scholar]
- 54.Creighton CJ, Hilger AM, Murthy S, Rae JM, Chinnaiyan AM, El-Ashry D. Activation of mitogen-activated protein kinase in estrogen receptor alpha-positive breast cancer cells in vitro induces an in vivo molecular phenotype of estrogen receptor alpha-negative human breast tumors. Cancer Res. 2006;66:3903–11. doi: 10.1158/0008-5472.CAN-05-4363. [DOI] [PubMed] [Google Scholar]
- 55.MacLaren CM, Evans TA, Alvarado D, Duffy JB. Comparative analysis of the Kekkon molecules, related members of the LIG superfamily. Dev Genes Evol. 2004;214:360–6. doi: 10.1007/s00427-004-0414-4. [DOI] [PubMed] [Google Scholar]
- 56.Gumienny TL, Macneil L, Zimmerman CM, et al. Caenorhabditis elegans SMA-10/LRIG is a conserved transmembrane protein that enhances bone morphogenetic protein signaling. PLoS Genet. 2010;6:e1000963. doi: 10.1371/journal.pgen.1000963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hedman H, Henriksson R. LRIG inhibitors of growth factor signalling - double-edged swords in human cancer? Eur J Cancer. 2007;43:676–82. doi: 10.1016/j.ejca.2006.10.021. [DOI] [PubMed] [Google Scholar]
- 58.Ljuslinder I, Golovleva I, Henriksson R, Grankvist K, Malmer B, Hedman H. Co-incidental increase in gene copy number of ERBB2 and LRIG1 in breast cancer. Breast Cancer Res. 2009;11:403. doi: 10.1186/bcr2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bhat-Nakshatri P, Wang G, Appaiah H, et al. AKT alters genome-wide estrogen receptor alpha binding and impacts estrogen signaling in breast cancer. Mol Cell Biol. 2008;28:7487–503. doi: 10.1128/MCB.00799-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Estecio MR, Issa JP. Dissecting DNA hypermethylation in cancer. FEBS Lett. 2010 doi: 10.1016/j.febslet.2010.12.001. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rothenberg SM, Settleman J. Discovering tumor suppressor genes through genome-wide copy number analysis. Curr Genomics. 2010;11:297–310. doi: 10.2174/138920210791616734. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.