Abstract
Background
Campylobacter jejuni is a leading foodborne pathogen worldwide. Despite the fastidious nature of C. jejuni growth, increasing numbers of human campylobacteriosis suggest that C. jejuni may possess unique mechanisms to survive under various stress conditions. C. jejuni possesses only three sigma factors (FliA, RpoD, and RpoN) and lacks stress-defense sigma factors. Since FliA and RpoD are dedicated to flagella synthesis and housekeeping, respectively, in this study, we investigated the role of RpoN in C. jejuni's defense against various stresses.
Results
Survivability of an rpoN mutant was compared with the wild-type C. jejuni under various stress conditions. While the growth of the rpoN mutant was as comparably as that of the wild type in shaking cultures, the rpoN mutant exhibited significant survival defects when cultured statically. The rpoN mutant was more sensitive to osmotic stress (0.8% NaCl) with abnormally-elongated cell morphology. Compared to the wile type, the rpoN mutant was more susceptible to acid stress (pH 5) and more resistant to hydrogen peroxide. However, the rpoN mutation had little effect on the resistance of C. jejuni to alkaline pH, heat, cold and antimicrobials.
Conclusions
The results demonstrate that RpoN plays an important role in C. jejuni's defense against various stresses which this bacterial pathogen may encounter during transmission to and infection of humans.
Background
Campylobacter is a leading cause of human gastroenteritis and is annually responsible for estimated 400-500 million cases of human infection worldwide [1]. Among Campylobacter species, C. jejuni is the major human-pathogenic species, accounting for more than 90% of human campylobacteriosis [2,3]. Human C. jejuni infections are primarily caused by the consumption of contaminated poultry, because the spillage of intestinal content containing a large number of C. jejuni during slaughter can contaminate cooling water, knives and poultry meat in the processing plant [4]. During transmission of C. jejuni from animals, primarily poultry, to humans, this important zoonotic foodborne pathogen encounters various stresses, such as non-growth temperatures, starvation, hypo- and hyper-osmotic stress, and desiccation [5,6]. Despite the well-known fact that Campylobacter is a fastidious bacterium, human campylobacteriosis cases have significantly increased presumably due to the ability of this pathogen to survive under harsh environmental conditions [7-10] in addition to its low infectious dose (400~800 bacteria) [11]. For example, high genetic diversity of Campylobacter spp. and the ability to transform into a viable-but-non-culturable state may enhance its adaptability to unfavorable growth conditions [7,8]. Additionally, biofilm formation and stringent response also contribute to the survival of Campylobacter under stress conditions [9,10]. However, the molecular mechanisms for stress resistance are still largely unknown in Campylobacter.
In many bacterial species, alternative sigma factors play an important role in regulation of stress-defense genes under hostile environmental conditions [12]. Because a sigma factor can coordinate gene transcription in response to environmental stimuli, many bacteria possess multiple alternative sigma factors, some of which are often dedicated to stress responses. For example, RpoS is a sigma factor important for adaptive responses in many Gram-negative pathogens, and RpoS mutations in Escherichia coli, Salmonella, Pseudomonas and Vibrio significantly impair bacterial ability to resist various stresses, such as starvation, low pH, oxidative stress, hyperosmolarity, heat and cold [13-17]. In E. coli, RpoS is involved in resistance to high osmolarity in stationary-phase cells and survival in cold-shock by regulating one set of RpoS-dependent genes, including otsA and otsB, which are necessary for synthesis of internal trehalose as an osmoprotectant and important for survival at low temperature [18,19]. In addition, RpoS controls the acid resistance in E. coli by modulating gadC, a gene involved in the glutamate-dependent low pH-resistance, hdeAB, encoding pH-regulated periplasmic chaperons, and cfa, a gene for cycloporpane fatty acid synthesis [20]. As another stress-response sigma factor, RpoE regulates extracytoplasmic functions related to sensing and responding to bacterial periplasmic and extracellular environmental changes, which contributes to heat- and oxidative stress resistance in many Gram-negative bacteria, including E. coli, Pseudomonas and Salmonella [21,22]. The RpoE mutation in Salmonella reduces bacterial survival and growth in macrophages by the loss of RpoE-dependent gene expression such as htrA, a gene required for oxidative stress resistance [23,24]. To defend from extracytoplasmic stress and generate unfolded envelope proteins caused by stress shock, additionally, RpoE regulates the expression of genes encoding periplasmic folding catalysts, proteases, biosynthesis enzymes for lipid A and components of the cell envelope [25]. The alternative transcription factor sigma B is known to play a central role in gene expression regulation in response to nutrient starvation and environmental stresses, including exposure to acid, ethanol, and heat in Gram-positive bacteria, Listeria and Bacillus [12,17]. The sigma factor B regulon in Gram-positive bacteria also include genes involved in the stress response, such as catalases, intracellular proteases and efflux pumps [26].
Although alterative sigma factors involved in stress defense are available in many bacteria, the C. jejuni genome sequence revealed that C. jejuni does not possess stress-related sigma factors and has only three sigma factors (RpoD, FliA, and RpoN) [27]. RpoD and FliA are known to be dedicated to the transcription of housekeeping and flagella biosynthesis genes, respectively. RpoN is involved in the transcription of genes of flagella biosynthesis [28]; thus, the rpoN mutation affects the formation of flagellar secretory apparatus [29], and the secretion of virulence proteins (e.g., Cia proteins) via the flagella export apparatus [30]. In addition, RpoN plays an important role in bacterial motility, colonization and invasion abilities directly or indirectly in C. jejuni [31]. Since RpoN is involved in the regulation of genes required for virulence, stress resistance and nitrogen fixation in many bacteria, we hypothesized that RpoN may function as an alternative sigma factor associated with stress resistance in C. jejuni. In this work, we investigated the effect of rpoN mutation on the resistance of C. jejuni under various environmental stresses.
Results
Survival defects of the rpoN mutant
After construction of an rpoN mutant and a complementation strain, bacterial motility was determined to verify the success of the rpoN mutation, because an rpoN mutation is known to make Campylobacter aflagellate and non-motile [32,33]. Consistently, the rpoN mutant showed significant defects in motility with complete restoration by complementation (Additional file 1, Figure S1). To examine if an rpoN mutation affects the growth of C. jejuni, bacterial growth was measured at different temperatures with or without shaking. The growth of the rpoN mutant was comparable to that of the wild type in broth cultures with shaking (Figure 1A); however, the rpoN mutant showed significant growth defects, when it was cultured without shaking, and this growth defect in static cultures was completely restored in the complementation strain as determined by measuring the optical density (Figure 1B). To verify if the difference of OD value between the wild type and the rpoN mutant can be related to bacterial viability, viable cells were also counted under the same condition. Without shaking, the viable cell counts of the wild type and the complementation strain were slightly reduced until 24 hr, and then the wild type and the complementation strain started to grow. In contrast, the viable cell counts of the rpoN mutant continued to reduce during the whole period of culture (Figure 1B), suggesting that the rpoN mutation resulted in survival defects. The survival defect of the rpoN mutant in the static culture was observed at both 37°C and 42°C (data not shown). These results show that RpoN affects the survival of C. jejuni under aeration-limited static culture conditions.
Figure 1.
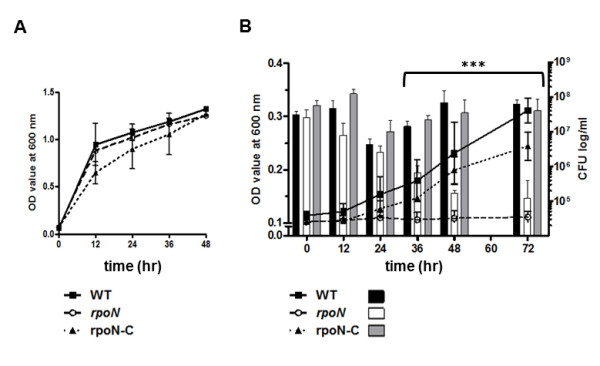
Growth of the rpoN mutant under different aeration conditions. The C. jejuni strains were microaerobically cultured in MH broth at 42°C with shaking at 180 rpm (A) and without shaking (B). At the described time intervals, the optical density at 600 nm was measured, and viable cells were counted in static culture condition (without shaking) by plating serially-diluted C. jejuni cultures on MH agar plates. The results are the mean ± standard deviation of three independent experiments. ***: P < 0.001; the significance of results was statistically analyzed by two-way ANOVA analysis of variance with Bonferroni's post-tests at a 95% confidence interval using Prism software (version 5.01; GraphPad Software Inc., USA).
Susceptibility of the rpoN mutant to osmotic stress
Due to the hypersensitivity of Campylobacter to various osmolytes [34,35], NaCl was used as an osmolyte to investigate the susceptibility of the rpoN mutant to osmotic stress in this study. When grown on Mueller-Hinton (MH) agar plates containing a high concentration (0.8%) of NaCl, the rpoN mutant exhibited significant growth defects (Figure 2A). The colony size of the rpoN mutant on MH agar plates was extremely small even after incubation for two days compared to the wild type (data not shown), suggesting the rpoN mutant suffers more osmotic stress than the wild type under the same stress condition. We used transmission electron microscopy (TEM) to investigate bacterial morphology under the osmotic stress. Interestingly, 79.3 ± 9.0% of rpoN mutant cells were abnormally elongated after exposure to osmotic stress. The rpoN mutant was approximately several times longer (approximately > 5 μm) than the wild type in the presence of 0.8% NaCl, and the morphological change in the rpoN mutant was restored by complementation (Figure 2B).
Figure 2.
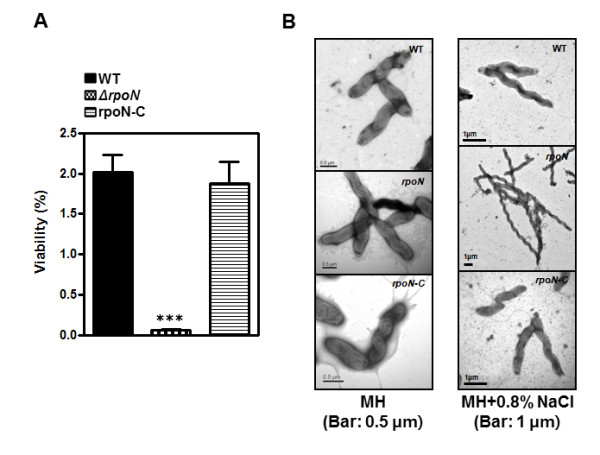
Changes in viability and morphology under osmotic stress. (A) Viable cell counts of the rpoN mutant on MH agar pates containing 0.8% NaCl after incubation for 24 hr. Results are expressed as the mean ± standard deviation of three independent experiments. ***: P < 0.001; the significance of results was statistically analyzed by one-way ANOVA analysis of variance with Dunnett test at a 99.9% confidence intervals using Prism software (version 5.01; GraphPad Software Inc.). (B) Electron microphotographs of WT and mutants grown under high osmotic stress conditions (MH agar pates containing 0.8% NaCl). Bacteria were examined by EF-TEM with negative staining with 0.2% uranyl acetate. Each scale bar of the normal and 0.8% NaCl conditions correspond to 0.5 μm and 1 μm, respectively.
Susceptibility of the rpoN mutant to pH stress
While the optimal pH range for the growth of C. jejuni is 6.5-7.5, C. jejuni can still survive at pH 5.5 - 8.5 [5]. Resistance of the rpoN mutant to acid stress was assessed by growing on MH agar plates at pH 5.5. The acid stress tests showed that the viability of the rpoN mutant was substantially reduced at pH 5.5 compared to the wild type (Figure 3). In contrast, alkali stress (pH 8.5) did not make any differences in viability between the wild type and the rpoN mutant (Additional file 2, Figure S2A). These results suggest that rpoN contributes to C. jejuni's resistance to acidic stress, but not to alkali stress.
Figure 3.
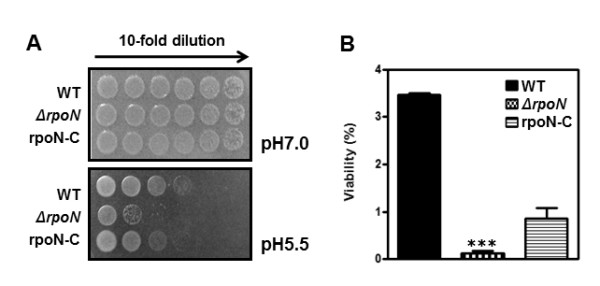
Effect of the rpoN mutation on acid stress resistance. (A) Growth of the rpoN mutant under different pH conditions was examined by dotting 10 μl of serially-diluted bacterial cultures. The results are representative of three independent experiments with similar results. (B) Viable cell counts on MH agar with different pH after 24 hr incubation. The % viability is expressed as mean ± standard deviation of three independent experiments. ***: P < 0.001; the significance of results was statistically analyzed by one-way ANOVA using Prism software (version 5.01; GraphPad Software Inc.).
Resistance of the rpoN mutant to oxidative stress
The oxidative stress resistance of the rpoN mutant was examined by growing on MH agar plates containing 1 mM hydrogen peroxide. Although the rpoN mutant is more sensitive to osmotic and acid stresses than the wild type, the rpoN mutant was more resistant to hydrogen peroxide than the wild type (Figure 4), and the susceptibility was restored to the wild-type level by complementation (Figure 4).
Figure 4.
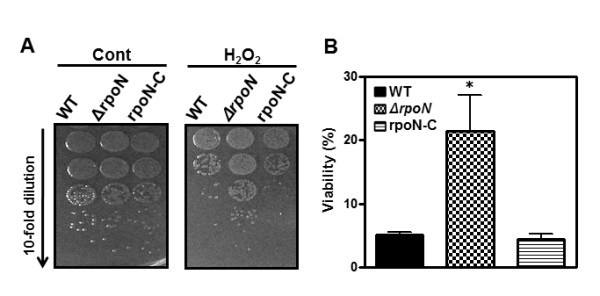
Resistance of the rpoN mutant to hydrogen peroxide. After treatment with hydrogen peroxide (H2O2) for 1 hr, changes in viability were determined by dotting 10 μl of bacterial culture (A) or by plating culture aliquots on MH agar plates to count viable cells (B). The data (A) are representative of three independent experiments with similar results. The % viability (B) is expressed as mean ± standard deviation of three independent experiments. The significance of results was P < 0.05 indicated by an asterisk (Prism software version 5.01; GraphPad Software Inc.).
Effects of an rpoN mutation on resistance to heat, cold and antimicrobials
Cold and heat stress was generated by exposure to -20°C and 55°C, respectively, and made little difference in viability between the rpoN mutant and the wild type (Additional file 2, Figure S2B). In addition, an rpoN mutation did not affect C. jejuni's resistance to antimicrobials, such as erythromycin, cefotaxime, gentamicin, polymyxin B, rifampicin and ampicillin (Additional file 3, Table S1).
Discussion
Many bacterial pathogens have multiple sigma factors to regulate gene expression efficiently in response to environmental changes [12]. In this work, the role of RpoN was investigated under various stress conditions. Notably, significant survival defects were observed when the rpoN mutant was grown statically (Figure 1), whereas the growth of the rpoN mutant was comparable to that of the wild type in shaking cultures. To assess if the survival defect of the rpoN mutant in static cultures would be mediated by the motility defect by the rpoN mutation, we compared the growth of a flaA mutant with the wild type under the same culture condition; however, the flaA mutant grew as comparably as the wild type (data not shown). This suggests that the survival defect of the rpoN mutant under the static culture condition was not caused by its loss of motility. Instead, the survival defects of the rpoN mutant may be related to the ability to respire under oxygen-limited conditions, because the levels of oxygen dissolved in broth media are lower in static culture than shaking culture. C. jejuni rarely encounters an active aeration system in its natural habitat (e.g., poultry intestines), which may be more similar to static culture than shaking culture. The rpoN mutation significantly impairs C. jejuni's ability to colonize the intestines of chicken because of poor attachment of the aflagellated rpoN mutant to the epithelial cells in the intestines [32,36]. In addition to the loss of motility by the rpoN mutation, the survival defects in the static culture condition may also be responsible for the colonization defect of the rpoN mutant. Molecular mechanisms of the survival defect in the rpoN mutant are currently being investigated in our group.
Because RpoN is known to be important for osmotolerance in some bacteria, such as Listeria monocytogenes [37], resistance to osmotic stress was compared between the rpoN mutant and the wild type. NaCl is a common food additive used to inhibit microbial growth, and significantly impairs the culturability of Campylobacter at concentrations greater than 2.0% [38]. In this work, the growth of C. jejuni was substantially inhibited even by 0.8% NaCl (Figure 2A). TEM analysis showed that the wild-type C. jejuni was slightly elongated at high (0.8%) NaCl concentration, whereas the rpoN mutant was significantly elongated compared to the wild type at the same NaCl concentration (Figure 2B). The morphological change was completely restored by complementation (Figure 2B), suggesting the active involvement of RpoN in this morphological change of C. jejuni under osmotic stress. Morphological abnormalities of the rpoN mutant indicate that the rpoN mutant is more stressed than the wild type under the same osmotic stress condition (Figure 2). Morphological changes by osmotic stress have also been reported in other bacteria. Aeromonas hydrophila, a Gram-negative rod-shaped bacterium, becomes elongated or turns to spherical forms by exposure to high NaCl concentrations [39]. In Lactobacillus casei, high NaCl concentrations affect the size of bacterial cell and cell-wall modification, and the alteration of the cell wall increases antimicrobial susceptibility [40]. Although the genetic response of C. jejuni to high and low osmotic conditions has not been well studied yet, it has been reported that the rod spiral C. jejuni turns to coccoid forms when grown in nutrient media with low osmolality [34]. The previous report plus our findings demonstrate that both hyper- and hypo-osmotic stress abnormally alters the morphology of C. jejuni. This may probably result from changes in intracellular ion concentrations by (de-)hydration under osmotic stress and may influence bacterial gene expression; however, understanding its molecular mechanisms still awaits further investigation.
The rpoN mutant was highly susceptible to acidic stress (pH 5.5) compared to wild type (Figure 3), whereas the growth of both the rpoN mutant and the wild type was similarly reduced under alkaline conditions (pH 8.5; Additional file 2, Figure S2A). Recently, an extensive screening of a transposon mutant library revealed that the adaptation of C. jejuni to acidic pH requires a number of genes mediating various cellular processes, including those involved in motility, metabolism, stress response, DNA repair and surface polysaccharide biosynthesis [41]. Interestingly, mutations of motility-associated genes, such as flgR and fliD, impaired the growth of C. jejuni at low pH [41]. Based on this previous report, the increased susceptibility to acid stress in the rpoN mutant may be associated with the motility defect of the rpoN mutant.
Reactive oxygen species are inevitably produced by aerobiosis and cause damages to biomolecules, such as proteins, DNA and membranes [42]. As a microaerophile, C. jejuni requires oxygen for growth, though atmospheric level of oxygen is toxic to the cell. Various factors are known to mediate oxidative stress resistance in C. jejuni, including SodB (superoxide dismutase), KatA (catalase), AhpC (alkyl hydroperoxide reductase), Dps (DNA-binding protein from starved cells), the multidrug efflux pump CmeG, and PerR [43,44]. In this work, the rpoN mutant was more resistant to H2O2 than the wild type, and complementation restored the H2O2 susceptibility to the wild-type level (Figure 4). This is similar to the case of PerR; the perR mutation increased C. jejuni's resistance to H2O2 by derepressing katA [45]. It is unknown if RpoN is functionally related to PerR. However, the 16 RpoN-regulated genes which were predicted by in silico analysis in C. jejuni do not contain the oxidative stress resistance genes and perR [46]; thus, it appears that the change in H2O2 susceptibility by an rpoN mutation can be indirect in C. jejuni. It has been reported that the rpoN mutation makes the C. jejuni morphology less spiral [32,33], suggesting RpoN affects the formation of the typical rod-spiral morphology of C. jejuni. In our study, we also observed that the rpoN mutant is slightly less-spiral compared to the wild type, and osmotic (NaCl) stress resulted in obvious morphological changes in the rpoN mutant (Figure 2B). The morphological changes caused by the rpoN mutation can be accompanied by the alteration of bacterial membrane and cell wall, and would possibly result in permeability changes. H2O2 is non-ionic and freely passes through membranes. Thus, the rpoN mutation may interfere with the permeability of H2O2 and confer resistance to H2O2; however, this possibility will be examined in future studies.
Conclusions
As a zoonotic foodborne pathogen, C. jejuni encounters various environmental stresses during transmission and infection, such as changes in osmolarity, temperature and the high acidic pH in the stomach; only the bacteria that survive in these deleterious stresses can reach human hosts. Thus, the ability of C. jejuni in stress resistance can be considered an important factor associated with food safety. This work clearly demonstrated that RpoN plays an important role in the resistance of C. jejuni to various stresses. Compared to the wild type, the rpoN mutant was more susceptible to osmotic stress (0.8% NaCl) and acidic pH. Interestingly, the rpoN mutation rendered C. jejuni more resistant to H2O2 than the wild type. Notably, the rpoN mutant exhibited significant survival defects in the static culture conditions. Although understanding of molecular mechanisms for stress tolerance may exceed the scope of our present work, in this study, we provided new insights into the role of RpoN, one of the three sigma factors of C. jejuni, in the survivability of this bacterial pathogen under various stress conditions.
Methods
Bacterial strains, plasmids, and culture conditions
C. jejuni 81-176 was used in this study. The strains, plasmids, and primers used in this study are listed in Table 1. C. jejuni 81-176 and its derivatives were routinely grown at 42°C on MH agar plates or MH broth with shaking at 180 rpm under microaerobic condition (6% O2, 7% CO2, 4% H2, and 83% N2) adjusted by the MART (Anoxomat™, Mart Microbiology B.V, Netherlands). To investigate the effect of rpoN disruption on C. jejuni growth, C. jejuni was cultured in 50 ml MH broth either in conical tubes without shaking or in Erlenmeyer flasks with shaking. Occasionally, culture media were supplemented with kanamycin (50 μg ml-1) or chloramphenicol (10 μg ml-1) where required.
Table 1.
Bacterial strains, plasmids and primers used in this study
| Strains, plasmid and primers | Description | Source |
|---|---|---|
| E. coli | ||
| DH5α | F', Φ80 dlacZΔM15, endA1, recA1, hsdR17 (rk-, mk+), supE44, thi1, Δ(lacZYA-argF)U169, deoR, λ- | Invitrogen |
| C. jejuni | ||
| 81-176 FMB1116 FMB2017 |
wild type, clinical isolate rpoN::cat rpoN complementation using pFMBComC-rpoN |
[51] In this study In this study |
| Plasmids | ||
| pUC19 pUC-rpoN pUC-rpoN::cat pUC19-16Sto23S pFMBComC pFMBComC-rpoN |
Cloning and suicide vector, Ampr pUC19 carrying rpoN and flanking region pUC19 carrying rpoN::cat pUC19 carrying fragments of genes encoding 16s and 28s rRNAs Kanr cassette cloned into pUC19-16Sto23S pFMB carrying rpoN |
NEB In this study In this study In this study In this study In this study |
| Primers | ||
| rpoN_F rpoN_R catF(SmaI) catR(SmaI) rpoNC_F(XbaI) rpoNC_R(XbaI) |
ATGATAAGGGTAAGAATTATTTTGAT AAAAAAATCACGCAAGGGGATAAGTCCTC GTGTTCCTTTCCCGGGTAATTGCG GATAAAAACCCGGGGGAACTAAAGGG TTGCAACCAATctAGACGTGAAAAAG AAGTGTAATGGGTTTtcTAGATTGATAG |
|
Construction of the rpoN mutant and rpoN-complemented strain
C. jejuni mutants were constructed with C. jejuni 81-176 as the parental strain by performing electroporation of suicide plasmids [47]. The antibiotic resistant genes used to construct mutants were prepared as followed; a chloramphenicol resistance cassette (cat) was amplified from pRY112 using primers of catF(SmaI) and catR(SmaI), and Vent Polymerase (New England Biolabs). To construct C. jejuni FMB1116, a DNA fragment containing rpoN and flanking region was amplified using primers rpoN_F and rpoN_R, and then ligated into SmaI-digested pUC19. The resultant plasmid was digested with SmiI, and then cat cassette was inserted into that digested site. The orientation of the cat cassette was confirmed by sequencing, and the plasmid in which the orientation of cat cassette was same to rpoN was designated as pUC-rpoN::cat. This plasmid was used as a suicide plasmid to construct C. jejuni FMB1116. For the rpoN complementation, an extra copy of rpoN was integrated into the chromosome by the methodology reported elsewhere [48]. Briefly, a DNA fragment containing rpoN and its putative promoter region was amplified with rpoNC_F(XbaI) and rpoNC_R(XbaI) primers. The PCR product was digested with XbaI and cloned into pFMB, which carries rRNA gene cluster and a kanamycin resistance cassette. The constructed plasmid was delivered to the bacterial cell, FMB1116, by electroporation.
Transmission electron microscopy
Bacterial cell suspension of each C. jejuni cultured on MH agar plate with or without NaCl was absorbed onto a 400 mesh carbon-coated grid, negatively stained with 0.2% aqueous uranyl acetate (pH4.0), and observed in an EF-TEM (LIBRA 120, Carl Zeiss, Hamburg, Germany) at an accelerating voltage of 80 kV.
Viability tests under various stress conditions
C. jejuni strains were inoculated into MH broth to an OD at 600 nm (OD600) of 0.1. After culturing to the early mid log phase (about 5 hr), OD600 was adjusted to 0.2. The aliquots of bacterial cells were exposed to several different stress conditions. The resistance to osmotic and pH shock was measured by culturing serially-diluted bacterial cells for 24 hr on MH agar plates containing 0.8% NaCl or at pH levels of 5.5 and 7.5. To test the susceptibility to oxidative stress, C. jejuni strains were exposed to the final concentration of 1 mM of H2O2 under microaerophilic condition for 1 hr. For heat and cold stresses, bacterial cells were incubated at 55°C and -20°C for 15 min or 1 hr, respectively. After exposure to each stress conditions, serially diluted culture aliquots were plated on MH agar and incubated at 42°C for 2 days to count viable cells. The viability was presented in percentage compared with the CFU of the sample without being exposed to stress.
Antimicrobial susceptibility tests
Minimal inhibitory concentrations (MICs) and minimal bactericidal concentrations (MBCs) of erythromycin, cefotaxime, gentamicin, polymyxin B, rifampicin and ampicillin were determined by a microtitre broth dilution method as described previously [49,50].
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SH, BJ, JY, and SR conceived and designed the study. SH carried out the experimental work and wrote the manuscript. JY designed the mutant construction. SH, BJ, and SR analyzed and interpreted the data. SR and BJ revised the manuscript critically for important intellectual content. All authors read and approved the final manuscript.
Supplementary Material
Figure S1. Loss of motility in the rpoN mutant. The diameter of each motility zone was measured after 36 hr incubation of C. jejuni strains on 0.4% motility agar plates at 42°C.
Figure S2. Effect of the rpoN mutation on C. jejuni's resistance to alkali, heat and cold stresses. (A) Resistance to alkali stress. The growth under different pHs was examined by dotting 10 μl of serially-diluted bacterial cultures. pH 7 was used as a control. (B) Heat and cold resistance. Bacteria were exposed to 55°C and -20°C. After exposure, the viability changes were measured by dotting 10 μl of bacterial cultures on MH agar plates.
Table S1. Antimicrobial susceptibility of the rpoN mutant.
Contributor Information
Sunyoung Hwang, Email: skyborn7@snu.ac.kr.
Byeonghwa Jeon, Email: bjeon@upei.ca.
Jiae Yun, Email: yja97@snu.ac.kr.
Sangryeol Ryu, Email: sangryu@snu.ac.kr.
Acknowledgements
We thank Dr. Qijing Zhang (Iowa State University, USA) for providing C. jejuni 81-176. This work was supported by the grant (A09058009010000100) to SR from the Korean Health Technology R&D Project, the Ministry for Health, Welfare and Family Affairs, Republic of Korea. Sunyoung Hwang is a recipient of the graduate fellowship provided by the Ministry of Education through the Brain Korea 21 Project.
References
- Ruiz-Palacios GM. The health burden of Campylobacter infection and the impact of antimicrobial resistance: playing chicken. Clin Infect Dis. 2007;44(5):701–703. doi: 10.1086/509936. [DOI] [PubMed] [Google Scholar]
- Gillespie IA, O'Brien SJ, Penman C, Tompkins D, Cowden J, Humphrey TJ. Demographic determinants for Campylobacter infection in England and Wales: implications for future epidemiological studies. Epidemiol Infect. 2008;136(12):1717–1725. doi: 10.1017/S0950268808000319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman CR, Neimann J, Wegener HC, Tauxe RV. In: Campylobacter. 2. Nachamkin I, Blaser MJ, editor. Washington, D.C.: American Society for Microbiology; 2000. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations; pp. 121–138. [Google Scholar]
- FAO/WHO. Microbiological risk assessment series No12. Geneva.: Food and Agriculture organization of the United Nations/World health organization; 2009. Risk assessment of Campylobacter spp. in broiler chickens: Technical Report; p. 132. [Google Scholar]
- Murphy C, Carroll C, Jordan KN. Environmental survival mechanisms of the foodborne pathogen Campylobacter jejuni. J Appl Microbiol. 2006;100(4):623–632. doi: 10.1111/j.1365-2672.2006.02903.x. [DOI] [PubMed] [Google Scholar]
- Park SF. In: Campylobacter Moecular and Cellular Biology. Ketley JM, Konkel ME, editor. Norfolk, U.K.: Horison Bioscience; 2005. Campylobacter jejuni stress responses during survival in the food chain and colonization. [Google Scholar]
- Alter T, Scherer K. Stress response of Campylobacter spp. and its role in food processing. J Vet Med B Infect Dis Vet Public Health. 2006;53(8):351–357. doi: 10.1111/j.1439-0450.2006.00983.x. [DOI] [PubMed] [Google Scholar]
- Tangwatcharin P, Chanthachum S, Khopaibool P, Griffiths MW. Morphological and physiological responses of Campylobacter jejuni to stress. J Food Prot. 2006;69(11):2747–2753. doi: 10.4315/0362-028x-69.11.2747. [DOI] [PubMed] [Google Scholar]
- Reuter M, Mallett A, Pearson BM, van Vliet AH. Biofilm formation by Campylobacter jejuni is increased under aerobic conditions. Appl Environ Microbiol. 2010;76(7):2122–2128. doi: 10.1128/AEM.01878-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor EC, Wells DH, MacKichan JK, Falkow S. The Campylobacter jejuni stringent response controls specific stress survival and virulence-associated phenotypes. Mol Microbiol. 2005;56(1):8–27. doi: 10.1111/j.1365-2958.2005.04525.x. [DOI] [PubMed] [Google Scholar]
- Young KT, Davis LM, Dirita VJ. Campylobacter jejuni: molecular biology and pathogenesis. Nat Rev Microbiol. 2007;5(9):665–679. doi: 10.1038/nrmicro1718. [DOI] [PubMed] [Google Scholar]
- Schwab U, Hu Y, Wiedmann M, Boor KJ. Alternative sigma factor sigmaB is not essential for Listeria monocytogenes surface attachment. J Food Prot. 2005;68(2):311–317. doi: 10.4315/0362-028x-68.2.311. [DOI] [PubMed] [Google Scholar]
- Dong T, Schellhorn HE. Role of RpoS in virulence of pathogens. Infect Immun. 2010;78(3):887–897. doi: 10.1128/IAI.00882-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Chen J, Liu R, Zhang XH, Jiang YA. Mutation of rpoS gene decreased resistance to environmental stresses, synthesis of extracellular products and virulence of Vibrio anguillarum. FEMS Microbiol Ecol. 2009;70(2):130–136. doi: 10.1111/j.1574-6941.2009.00713.x. [DOI] [PubMed] [Google Scholar]
- Stockwell VO, Hockett K, Loper JE. Role of RpoS in stress tolerance and environmental fitness of the phyllosphere bacterium Pseudomonas fluorescens strain 122. Phytopathology. 2009;99(6):689–695. doi: 10.1094/PHYTO-99-6-0689. [DOI] [PubMed] [Google Scholar]
- Vasudevan P, Venkitanarayanan K. Role of the rpoS gene in the survival of Vibrio parahaemolyticus in artificial seawater and fish homogenate. J Food Prot. 2006;69(6):1438–1442. doi: 10.4315/0362-028x-69.6.1438. [DOI] [PubMed] [Google Scholar]
- Kazmierczak MJ, Wiedmann M, Boor KJ. Alternative sigma factors and their roles in bacterial virulence. Microbiol Mol Biol Rev. 2005;69(4):527–543. doi: 10.1128/MMBR.69.4.527-543.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoebel DM, Hokamp K, Last MS, Dorman CJ. Compensatory evolution of gene regulation in response to stress by Escherichia coli lacking RpoS. PLoS Genet. 2009;5(10):e1000671. doi: 10.1371/journal.pgen.1000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandror O, DeLeon A, Goldberg AL. Trehalose synthesis is induced upon exposure of Escherichia coli to cold and is essential for viability at low temperatures. Proc Natl Acad Sci USA. 2002;99(15):9727–9732. doi: 10.1073/pnas.142314099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman SR, Small PL. Identification of sigma S-dependent genes associated with the stationary-phase acid-resistance phenotype of Shigella flexneri. Mol Microbiol. 1996;21(5):925–940. doi: 10.1046/j.1365-2958.1996.00058.x. [DOI] [PubMed] [Google Scholar]
- Thongboonkerd V, Vanaporn M, Songtawee N, Kanlaya R, Sinchaikul S, Chen ST, Easton A, Chu K, Bancroft GJ, Korbsrisate S. Altered proteome in Burkholderia pseudomallei rpoE operon knockout mutant: insights into mechanisms of rpoE operon in stress tolerance, survival, and virulence. J Proteome Res. 2007;6(4):1334–1341. doi: 10.1021/pr060457t. [DOI] [PubMed] [Google Scholar]
- Testerman TL, Vazquez-Torres A, Xu Y, Jones-Carson J, Libby SJ, Fang FC. The alternative sigma factor sigmaE controls antioxidant defences required for Salmonella virulence and stationary-phase survival. Mol Microbiol. 2002;43(3):771–782. doi: 10.1046/j.1365-2958.2002.02787.x. [DOI] [PubMed] [Google Scholar]
- Kazmierczak MJ, Wiedmann M, Boor KJ. Alternative sigma factors and their roles in bacterial virulence. Microbiol Mol Biol Rev. 2005;69(4):527–543. doi: 10.1128/MMBR.69.4.527-543.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller C, Bang IS, Velayudhan J, Karlinsey J, Papenfort K, Vogel J, Fang FC. Acid stress activation of the sigma(E) stress response in Salmonella enterica serovar Typhimurium. Mol Microbiol. 2009;71(5):1228–1238. doi: 10.1111/j.1365-2958.2009.06597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba BM, Gross CA. Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol Microbiol. 2004;52(3):613–619. doi: 10.1111/j.1365-2958.2003.03982.x. [DOI] [PubMed] [Google Scholar]
- van Schaik W, Abee T. The role of sigmaB in the stress response of Gram-positive bacteria -- targets for food preservation and safety. Curr Opin Biotechnol. 2005;16(2):218–224. doi: 10.1016/j.copbio.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S. et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403(6770):665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- Hendrixson DR, Akerley BJ, DiRita VJ. Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol Microbiol. 2001;40(1):214–224. doi: 10.1046/j.1365-2958.2001.02376.x. [DOI] [PubMed] [Google Scholar]
- Hendrixson DR, DiRita VJ. Transcription of sigma54-dependent but not sigma28-dependent flagellar genes in Campylobacter jejuni is associated with formation of the flagellar secretory apparatus. Mol Microbiol. 2003;50(2):687–702. doi: 10.1046/j.1365-2958.2003.03731.x. [DOI] [PubMed] [Google Scholar]
- Konkel ME, Klena JD, Rivera-Amill V, Monteville MR, Biswas D, Raphael B, Mickelson J. Secretion of virulence proteins from Campylobacter jejuni is dependent on a functional flagellar export apparatus. J Bacteriol. 2004;186(11):3296–3303. doi: 10.1128/JB.186.11.3296-3303.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando U, Biswas D, Allan B, Willson P, Potter AA. Influence of Campylobacter jejuni fliA, rpoN and flgK genes on colonization of the chicken gut. Int J Food Microbiol. 2007;118(2):194–200. doi: 10.1016/j.ijfoodmicro.2007.07.038. [DOI] [PubMed] [Google Scholar]
- Fernando U, Biswas D, Allan B, Willson P, Potter AA. Influence of Campylobacter jejuni fliA, rpoN and flgK genes on colonization of the chicken gut. Int J Food Microbiol. 2007. [DOI] [PubMed]
- Jagannathan A, Constantinidou C, Penn CW. Roles of rpoN, fliA, and flgR in expression of flagella in Campylobacter jejuni. J Bacteriol. 2001;183(9):2937–2942. doi: 10.1128/JB.183.9.2937-2942.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reezal A, McNeil B, Anderson JG. Effect of low-osmolality nutrient media on growth and culturability of Campylobacter species. Appl Environ Microbiol. 1998;64(12):4643–4649. doi: 10.1128/aem.64.12.4643-4649.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle MP, Roman DJ. Response of Campylobacter jejuni to sodium chloride. Appl Environ Microbiol. 1982;43(3):561–565. doi: 10.1128/aem.43.3.561-565.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrixson DR, DiRita VJ. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol Microbiol. 2004;52(2):471–484. doi: 10.1111/j.1365-2958.2004.03988.x. [DOI] [PubMed] [Google Scholar]
- Okada Y, Okada N, Makino S, Asakura H, Yamamoto S, Igimi S. The sigma factor RpoN (sigma54) is involved in osmotolerance in Listeria monocytogenes. FEMS Microbiol Lett. 2006;263(1):54–60. doi: 10.1111/j.1574-6968.2006.00405.x. [DOI] [PubMed] [Google Scholar]
- Jackson DN, Davis B, Tirado SM, Duggal M, van Frankenhuyzen JK, Deaville D, Wijesinghe MA, Tessaro M, Trevors JT. Survival mechanisms and culturability of Campylobacter jejuni under stress conditions. Antonie Van Leeuwenhoek. 2009;96(4):377–394. doi: 10.1007/s10482-009-9378-8. [DOI] [PubMed] [Google Scholar]
- Pianetti A, Battistelli M, Citterio B, Parlani C, Falcieri E, Bruscolini F. Morphological changes of Aeromonas hydrophila in response to osmotic stress. Micron. 2009;40(4):426–433. doi: 10.1016/j.micron.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Piuri M, Sanchez-Rivas C, Ruzal SM. Cell wall modifications during osmotic stress in Lactobacillus casei. J Appl Microbiol. 2005;98(1):84–95. doi: 10.1111/j.1365-2672.2004.02428.x. [DOI] [PubMed] [Google Scholar]
- Reid AN, Pandey R, Palyada K, Whitworth L, Doukhanine E, Stintzi A. Identification of Campylobacter jejuni genes contributing to acid adaptation by transcriptional profiling and genome-wide mutagenesis. Appl Environ Microbiol. 2008;74(5):1598–1612. doi: 10.1128/AEM.01508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atack JM, Kelly DJ. Oxidative stress in Campylobacter jejuni: responses, resistance and regulation. Future Microbiol. 2009;4(6):677–690. doi: 10.2217/fmb.09.44. [DOI] [PubMed] [Google Scholar]
- Kelly DJ. The physiology and metabolism of Campylobacter jejuni and Helicobacter pylori. Symp Ser Soc Appl Microbiol. 2001. pp. 16S–24S. [DOI] [PubMed]
- Jeon B, Wang Y, Hao H, Barton YW, Zhang Q. Contribution of CmeG to antibiotic and oxidative stress resistance in Campylobacter jejuni. J Antimicrob Chemother. 2011;66(1):79–85. doi: 10.1093/jac/dkq418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet AH, Baillon ML, Penn CW, Ketley JM. Campylobacter jejuni contains two fur homologs: characterization of iron-responsive regulation of peroxide stress defense genes by the PerR repressor. J Bacteriol. 1999;181(20):6371–6376. doi: 10.1128/jb.181.20.6371-6376.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo CD, Taboada E, Nash JH, Lanthier P, Kelly J, Lau PC, Verhulp R, Mykytczuk O, Sy J, Findlay WA. et al. Genome-wide expression analyses of Campylobacter jejuni NCTC11168 reveals coordinate regulation of motility and virulence by flhA. J Biol Chem. 2004;279(19):20327–20338. doi: 10.1074/jbc.M401134200. [DOI] [PubMed] [Google Scholar]
- van Vliet AHM, Wood AC, Henderson J, Wooldridge K, Ketley JM. In: Methods in Microbiology (vol 27) Bacterial Pathogenesis. Williams P, Ketley JM, Salmond G, editor. San Diego: Academic press; 1998. Genetic Manipulation of enteric Campylobacter species; pp. 407–419. [Google Scholar]
- Karlyshev AV, Wren BW. Development and application of an insertional system for gene delivery and expression in Campylobacter jejuni. Appl Environ Microbiol. 2005;71(7):4004–4013. doi: 10.1128/AEM.71.7.4004-4013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews JM. Determination of minimum inhibitory concentrations. J Antimicrob Chemother. 2001;48(Suppl 1):5–16. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- Jeon B, Muraoka W, Scupham A, Zhang Q. Roles of lipooligosaccharide and capsular polysaccharide in antimicrobial resistance and natural transformation of Campylobacter jejuni. J Antimicrob Chemother. 2009;63(3):462–468. doi: 10.1093/jac/dkn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black RE, Levine MM, Clements ML, Hughes TP, Blaser MJ. Experimental Campylobacter jejuni infection in humans. J Infect Dis. 1988;157(3):472–479. doi: 10.1093/infdis/157.3.472. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Loss of motility in the rpoN mutant. The diameter of each motility zone was measured after 36 hr incubation of C. jejuni strains on 0.4% motility agar plates at 42°C.
Figure S2. Effect of the rpoN mutation on C. jejuni's resistance to alkali, heat and cold stresses. (A) Resistance to alkali stress. The growth under different pHs was examined by dotting 10 μl of serially-diluted bacterial cultures. pH 7 was used as a control. (B) Heat and cold resistance. Bacteria were exposed to 55°C and -20°C. After exposure, the viability changes were measured by dotting 10 μl of bacterial cultures on MH agar plates.
Table S1. Antimicrobial susceptibility of the rpoN mutant.


