Abstract
Background
Trichome patterning in Arabidopsis thaliana is governed by three types of activators, R2R3MYB, bHLH and WD40 proteins, and six R3MYB inhibitors. Among the inhibitors TRIPTYCHON (TRY) seems to fulfill a special function. Its corresponding mutants produce trichome clusters whereas all other inhibitors are involved in trichome density regulation.
Results
To better understand the role of TRY in trichome patterning we analyzed its transcriptional regulation. A promoter analysis identified the relevant regulatory region for trichome patterning. This essential region contains a fragment required for a double negative feedback loop such that it mediates the repression of TRY/CPC auto-repression. By transforming single cells of pTRY:GUS lines with p35S:GL1, p35S:GL3 and p35S:TTG1 in the presence or absence of p35S:TRY or p35S:CPC we demonstrate that TRY and CPC can suppress the TRY expression without the transcriptional down regulation of the activators. We further show by promoter/CDS swapping experiments for the R3MYB inhibitors TRY and CPC that the TRY protein has specific properties relevant in the context of both, cluster formation and trichome density.
Conclusions
Our identification of a TRY promoter fragment mediating a double negative feedback loop reveals new insight in the regulatory network of the trichome patterning machinery. In addition we show that the auto-repression by TRY can occur without a transcriptional down regulation of the activators, suggesting that the differential complex formation model has a biological significance. Finally we show that the unique role of TRY among the inhibitors is a property of the TRY protein.
Background
Trichome patterning in Arabidopsis thaliana has become a well-studied model system to understand cell-cell communication in the context of two-dimensional pattern formation in plants [1-3]. Trichomes are formed in the basal part of young leaves [4]. The trichome position is not correlated with any recognizable leaf structures and clonal analysis excluded a cell lineage mechanism [5,6]. For these reasons, it is widely accepted that patterning is mediated by cellular interactions between initially equivalent cells [2,3,7].
Genetic screens have identified two classes of mutants governing this process. All patterning genes except for TTG1 have close homologs acting in a partially redundant manner [8-16]. The following summary will only consider the most relevant players as judged by the strength of the mutant phenotypes. One mutant class shows fewer or no trichomes. The corresponding genes are therefore considered positive regulators of trichome formation. The three most important positive regulators are the WD40 protein TRANSPARENT TESTA GLABRA1 (TTG1) [17-19], the R2R3 MYB related transcription factor GLABRA1 (GL1) [20], and the basic helix-loop-helix (bHLH)-like transcription factor GLABRA3 (GL3) [4,21,22].
In the second class, trichome clusters or a higher trichome density indicate a repressive role. The two most important inhibitors are the R3 single-repeat MYB factors TRIPTYCHON (TRY) and CAPRICE (CPC) [12,23]. Although, the two corresponding genes show high sequence similarity and an indistinguishable expression pattern in leaves [12], their mutant phenotypes suggest different modes of action. While the cpc mutant has a higher trichome density, the try mutant shows trichome clusters and a reduction in trichome number [4,12].
The expression pattern of most patterning genes is very similar. Initially, all genes are expressed ubiquitously in the cells at the leaf basis where trichome initials are formed (patterning zone). Later, expression increases in trichomes and disappears in epidermal cells [10-14,16,24-26]. The ubiquitous expression corresponds to the pre-pattern situation in which all cells are equivalent. During this phase the positive and negative regulators are considered to be engaged in regulatory feed-back loops that have several important features including the activation of the inhibitors by the activators, the repression of the activators by the inhibitors and the ability of the inhibitors to move between cells [14,15,27,28]. These create differences between the cells and ultimately result in a pattern of trichome and non-trichome cells [2,3].
After the initial pattern is established leaf growth leads to an increased spacing of trichomes without the formation of new trichomes. As in this phase patterning gene expression has ceased in epidermal pavement cells and increased in trichomes, the loss of trichome initiation competence is most likely due to the absence of activator gene expression. Whether activator gene expression in later leaf stages is generally shut off during leaf maturation or due to lateral repression by TRY and/or the other inhibitors is not clear.
The proposed regulatory feedback loop between the activators and the inhibitors ultimately leads to an auto-repression of the inhibitors. This could in principle be achieved in two ways. As the R3 single repeat MYB inhibitors lack a transcriptional activation region they could bind to promoter elements of the activators thereby preventing the transcription of the activators. As the repressors are activated by the activators the reduction of activator activity leads to reduced inhibitor transcription. Alternatively, the inhibitors could posttranslationally render the activation complex inactive [29]. Yeast two hybrid experiments showed that GL1 and TTG1 bind different regions of the GL3 protein suggesting that they form a trimeric transcriptional activation complex [22]. Binding of TRY or CPC to GL3 was shown to displace GL1 thereby inactivating the complex [14,29]. Although both mechanisms lead to a repression of the inhibitors they differ in their regulation scheme. The transcriptional repression of the activators by binding of TRY and/or CPC to the promoters would create a regulatory feed back loop that involves transcriptional down regulation of the activators. The postulated repression by differential complex formation would establish a shortcut of the regulatory feedback loop as the inhibitors can directly repress their own activation.
In this manuscript, we analyze the transcriptional regulation of TRY during trichome patterning. First, we determined the TRY promoter fragment relevant for TRY function and the specific basal and trichome-specific expression pattern. In addition we identified separate regions that are necessary for an enhancement of the specific expression pattern and showed that these regions are necessary for rescue. Second, we showed that TRY or CPC can repress the TRY expression directly without the transcriptional regulation of the activators by transforming single epidermal cells of pTRY:GUS lines with the three activators and TRY or CPC. Finally, we performed promoter swap experiments with CPC and TRY and tested the ability to rescue the try mutant trichome phenotypes. These experiments revealed specific properties of the TRY protein for clustering and trichome density regulation.
Results
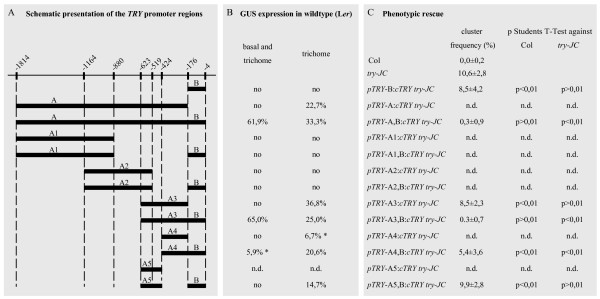
In a previous study, it was shown that a 4.2 kb genomic region containing a 1.8 kb 5' region and a 1.3 kb 3' region is sufficient to rescue the try mutant phenotype [12]. In a first step, we tested whether the 3' region or the introns are relevant for TRY function by transforming try mutant plants with a 1.8 kb 5' region that was fused to the TRY CDS (Figure 1, pTRY-A, B:cTRY try-JC). These plants showed complete rescue of the clustering phenotype indicating that the 1.8 kb 5' region contains all regulatory sequences necessary for the correct TRY expression in the leaf epidermis.
Figure 1.
A series of deletions in the 5' regulatory region of the TRY gene. A) Schematic overview showing the relative positions of TRY promoter fragments with respect to the start codon of the different fragments. Each fragment is fused to the CaMV 35S minimal promoter and either to the GUS coding region or to the TRY CDS followed by the nopaline synthase terminator. Single fragments (A or B) or their fusion were used (A+B). B) Summary of the GUS expression data. We distinguish between the ubiquitous expression called "basal expression" and expression in trichomes. We found two categories, basal and trichome expression and expression only in trichomes. The percentage of analyzed independent T2 lines showing the respective expression category is provided. Data marked with a "*" showed exclusively weak staining as exemplified in Figure 2J. C) Overview of the rescue efficiency. It was determined by the ability to reduce trichome cluster formation in the try-JC mutant. The percentage of clusters relative to the number of trichome initiation sites was calculated on the first four leaves. Statistical difference for each rescue experiment in comparison to Col wild type or to the try mutant is determined through Student's t-test. The difference between the respective two means is significant for P < 0,01.
Expression analysis of TRY promoter fragments
To identify specific regulatory elements, 5' promoter fragments were generated and their regulatory function monitored by fusion to the p35S-minimal promoter and the GUS marker gene (Figure 1A and 2). Because expression of a given construct is variable between different T2 lines we present pictures of the lines with the strongest expression only (Figure 2) and provide the percentage of lines in which the basal expression as well as trichome expression and those in which only the trichome specific expression is found after 24 hours of GUS staining (Figure 1B). Assuming that promoter elements driving a weak expression yield fewer transgenic lines with a strong expression this percentage is taken as an approximation of the expression strength of the promoter fragment under consideration.
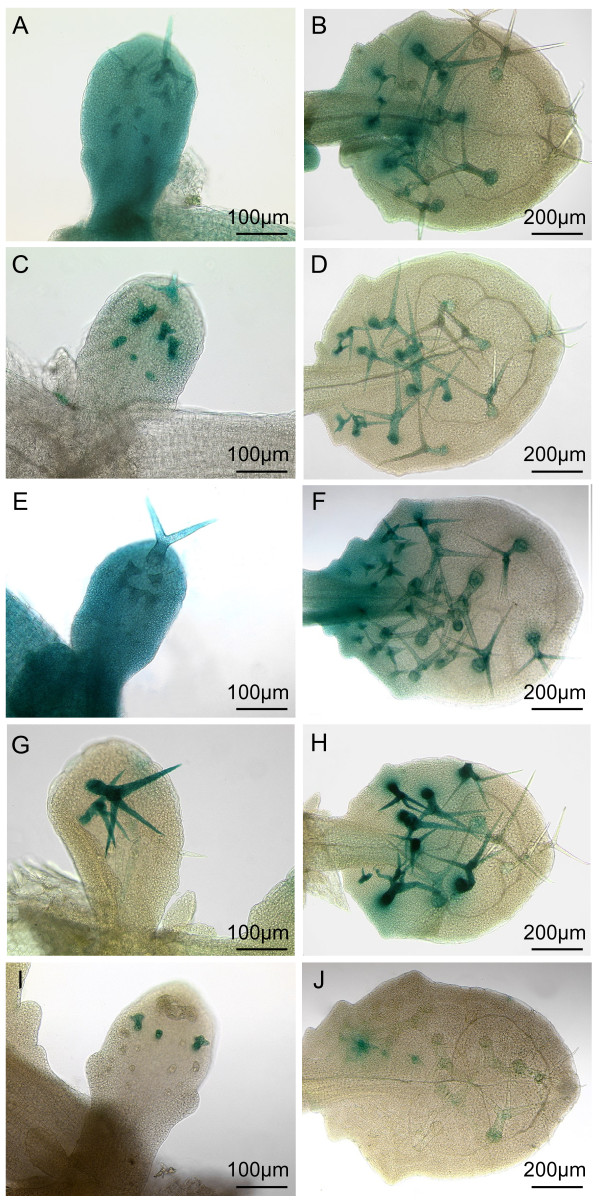
Figure 2.
Expression pattern of the 5' regulatory regions of the TRY promoter as revealed by GUS reporter gene expression. The GUS expression pattern driven by different promoter fragments was monitored on young leaves at stages where new trichomes were still initiated (A, C, E, G and I) and for slightly older leaves in which trichome initiation had already stopped (B, D, F, H and J). Pictures were taken in each case from one of the strongest T2 lines carrying the respective TRY promoter GUS fusion construct: pTRY-A, B:GUS (A, B), pTRY-A:GUS (C, D), pTRY-A3, B:GUS (E, F), pTRY-A4, B:GUS (G, H) and pTRY-A5, B:GUS (I, J). Bars indicate the magnification of the images.
For the expression analysis we initially used a fragment starting immediately upstream of an unique putative TATA Box located 32 base pairs upstream of the possible transcription start as determined by RACE PCR [12] (Figure 1A, pTRY-A). This fragment revealed GUS expression in trichomes, but the ubiquitous expression in young leaf regions (basal expression) observed before [12] was absent (Figure 2C, D). We therefore included the fragment immediately following the A-fragment and stretching to the -4 position relative to the ATG start codon (pTRY-B). pTRY-B represents the 5' UTR identified by Schellmann et al. and includes three possible transcriptional start sides suggested by ESTs (EH866228.1, AV533156.1, AI999616.1) and two putative TATA boxes (TATTA, TATAAA) [12,30-32]. A promoter fragment, pTRY-A, B, combining pTRY-A and pTRY-B revealed also trichome specific expression in 22,7% of the lines but in addition 61,9% of the lines (n = 35) showed the basal expression as well as the expression in trichomes. The pTRY-B fragment alone showed no expression. This indicates that the pTRY-B fragment is essential to enhance or stabilize the expression driven by the pTRY-A fragment.
A further deletion series revealed a minimal promoter region of about 620 bp (pTRY-A3, B). As found for the pTRY-A fragment the pTRY-B region is also necessary in the context of the pTRY-A3 fragment to mediate basal expression (Figure 2E, F). Further 5' deletion of about 200 bp (pTRY-A4, B) revealed trichome specific expression but only weak basal expression. Trichome-specific expression in these lines was only found in advanced stages of trichome development after branch formation (Figure 2G, H). The 200 bp fragment pTRY-A5, B revealed no basal expression and only sometimes a weak irregular expression in trichomes (Figure 2I, J).
These data do not allow to decide whether the pTRY-B fragment enhances both, the basal and trichome specific expression or whether it specifically regulates the basal expression. We therefore compared the expression pattern in pTRY-A3, B and pTRY-A3 at different time points of GUS staining procedure (data not shown). We observed that the GUS staining in young trichomes became detectable in both lines after two hours. While the basal expression in pTRY-A3, B became also detectable after two hours, no basal expression was detectable in pTRY-A3 even after 4 days of GUS staining. These data suggest, that the pTRY-B fragment specifically up-regulates the basal TRY expression in the context of the pTRY-A3 fragment.
Identification of relevant promoter regions by rescue experiments
In order to test their functionality we used various promoter fragments to express the TRY CDS in try-JC mutant plants (Figure 1A, C). In order to avoid the problem that individual transformants may show a wide range of phenotypes we did not select individual lines for analysis in the T2 but directly analyzed the phenotype of T1 plants to hold account on the full phenotypic spectrum. In these experiments both, the pTRY-A, B and the pTRY-A3, B fragments fully rescued the clustering phenotype (Figure 1C). Expression of TRY driven by pTRY-A3, however, had no significant rescue ability (Figure 1C). The smaller fragment (pTRY-A4, B) only partially rescued the try-JC clustering phenotype. Together these data indicate that the pTRY-B fragment is essential for rescue.
Regulation of the TRY promoter by TTG1, GL3, GL1 and TRY or CPC
In a next step we aimed to demonstrate the postulated activation/repression scheme of TTG1, GL3, GL1, TRY and CPC for the minimal TRY promoter fragment. The current models assume that TTG1, GL3 and GL1 can transcriptional activate the inhibitors TRY or CPC and that these in turn repress the activators and thereby also their own expression. The finding that TRY and CPC can compete with GL1 for binding to GL3 [14,29] suggests that the inhibitors can counteract the activity of the activators at the protein level directly. In this case TRY or CPC repress their own expression without a transcriptional repression of the activators.
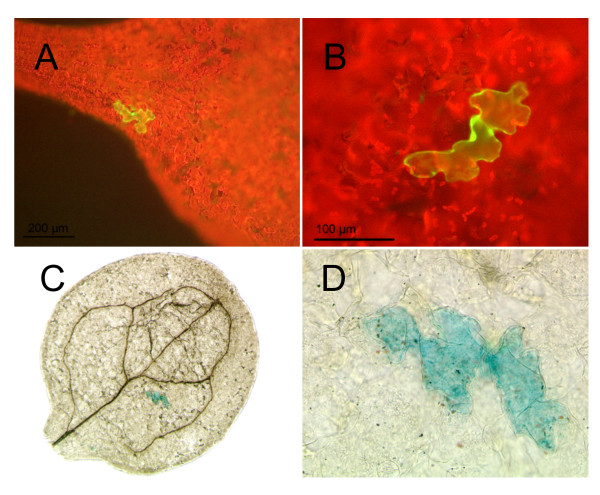
As TRY has been shown to be regulated by the activators in genetic experiments [27] the TRY promoter provides a tool to demonstrate that TRY/CPC can counteract the activators without a transcriptional repression of the activators. We used cotyledons for our analysis as no GUS expression was detected in this organ in pTRY-A3, B:GUS plants (Figure 3C). GL1, GL3 and TTG1 CDS under the control of the p35S promoter were used for transient transformations. In addition to these three constructs we added a p35S:GFP:YFP construct to control the bombardment efficiency. In four independent experiments analyzing each time 100 cells we found on average 68.2 ± 18.0% GUS expressing cells indicating that the simultaneous constitutive expression of TTG1, GL3 and GL1 induces the minimal TRY promoter fragment (Figure 3). Transformation with 35S:GFP:YFP alone revealed no GUS positive cells.
Figure 3.
Transient transformation of pTRY-A3, B:GUS cotyledons. Single epidermal cells of pTRY-A3, B:GUS cotyledons were transiently transformed by particle bombardment. (A) Overview of a cotyledon with a single cell expressing the p35S:GFP:YFP control construct. (B) Higher magnification of a single cell expressing the p35S:GFP:YFP control construct. (C) Overview of a cotyledon with a single cell showing pTRY-A3, B:GUS expression after co-transformation with p35S:GL1, p35S:GL3 and p35S:TTG1. (D) Higher magnification of (C).
In a second step we tested the model whether TRY or CPC can counteract the activity of the activators. Models derived from the finding that TRY and CPC compete with GL1 for binding to GL3 in yeast three hybrid experiments suggest that differential complex formation renders the proposed activator complex inactive [14,29]. We took advantage of the fact that in this experimental setup any indirect repression of TRY by TRY or CPC through the transcriptional repression of the activators is excluded as their expression is under the control of the 35S promoter. In four independent experiments with 100 cells in each experiment we found only 0.2 ± 0.5% or 0.2 ± 0.4% GUS-positive cells when expressing p35S:TRY or p35S:CPC respectively in addition to the three activators. This indicates that TRY and CPC can counteract TRY activation by the three activators without a transcriptional repression of the activators.
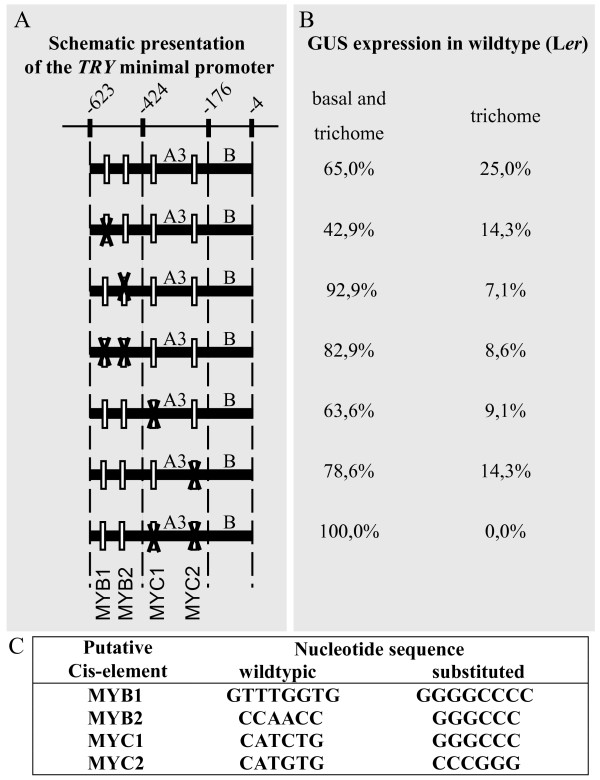
Relevance of MYB and MYC binding sites
Our finding that the activators can activate the TRY promoter in transient expression assays together with the finding that GL3 and GL1 bind to the TRY promoter in ChIP experiments [33,34] prompted us to search for putative MYB and MYC (bHLH factor) binding sites. We found five putative MYB and two putative MYC sites in the pTRY-A3, B fragment that was the minimal promoter fragment for full rescue of the try phenotype (Table 1). Among the five putative MYB sites two seemed most promising as they were identified in the context of regulatory pathways in other plants that are also regulated by TTG1-dependent pathways [35]. In addition the MYB factor binding to these MYB binding sites are in the same clade in the phylogenetic tree as GL1 [36]. We therefore focused on these two putative binding sites. To determine the role of the two selected MYB and the two MYC sites in the regulation of the correct expression pattern we mutated each site individually and both MYB and both MYC sites together (Figure 4C). None of the mutated constructs showed a marked reduction or even absence of pTRY-A3, B:GUS expression (Figure 4B). However, we noted differences such that the MYB1 site has the most positive effect on the basal expression whereas the MYB2 and the MYC sites have a repressive role. As a pTRY-A3, B:TRY construct containing mutations in both MYC sites resulted in a complete rescue of the try mutant clustering phenotype these sites do not appear to be relevant in this context (Additional File 1)
Table 1.
Overview of the identified MYB and MYC binding sites in the 5'-TRY minimal promoter identified by PLACE database
| 5'-TRY- nucleotide sequence |
Position relative to the ATG (start/end) |
Name of the described cis-element | Putative cis-element nucleotide sequence |
Description of the putative cis-element | |
|---|---|---|---|---|---|
| MYB1 | GTTTGGTG | -544/-551 | MYBPLANT | MACCWAMC | Binding of AmMYB305 in Antirrhinum majus to box P from gPAL2 of Phaseolus vulgaris; P box related sequences [48,49] are identified in several promoters of phenylpropanoid biosynthesis related genes (PAL, CHS, CHI, DFR, BZ1) in different plants (Phaseolus vulgaris, Antirrhinum majus, Petunia hybrida, Petroselinum crispum, Arabidopsis thaliana, Zea mays) [50] |
| MYB2 | CCAACC | -531/-536 | MYBPZM | CCWACC | Binding in promoters of A1 and BZ1 genes of phlobaphene pigmentation and flavonoid biosynthesis in Zea mays (factors, e.g. C1, P) [51] |
| MYB3 | TTTGTTA | -607/-613 | MYBGAHV | TAACAAA | Central element of the gibberellin (GA) response complex (GARC) in the high-pI alpha-amylase gene in Hordeum vulgare, binding of GaMYB [52-54]. |
| MYB4 | CCGTT | -153/-157 | MYBCOREATCYCB1 | AACGG | "Myb core" found in the promoter of Arabidopsis thaliana cyclin B1:1 gene [55]. |
| GCCGTTCGT | -150/-158 | v-MYB* | NSYAACGGN | Binding site of the v-MYB oncocgene of the avian myeloblastosis virus [56]. | |
| GGCCGTTCGT | -150/-159 | c-MYB* | NNNAACKGNC | Binding site of the c-MYB, the cellular homolog of v-MYB | |
| MYB5 | TTGAACTTGC | -404/-413 | c-MYB* | NNNAACKGNC | Binding site of the c-MYB, the cellular homolog of v-MYB |
| MYC1 | CATCTG | -399/-404 | MYC CONSENSUSAT |
CANNTG | Binding of AtMYC2 in pAtRD22 (dehydration responsive gene) in Arabidopsis thaliana. |
| MYC2 | CATGTG | -243/-248 | MYC CONSENSUSAT |
CANNTG | Binding of AtMYC2 in pAtRD22 (dehydration responsive gene) in Arabidopsis thaliana [48,49]. |
| MYCATERD1 | CATGTG | Binding of AtNAC to the ERD1 gene (early responsive to dehydration) in dehydrated Arabidopsis thaliana [57,58]. | |||
| MYCATRD22 | CACATG | Binding of AtMYC2 to the RD22 gene (dehydration responsive gene) ) in Arabidopsis thaliana [49]. |
An additional analysis marked by "*" was done by the TRANSFAC database.
Figure 4.
The minimal 5' regulatory region of the TRY gene including the putative analyzed MYB and MYC binding sites and their substitutions. A) Schematic overview showing the relative position of the minimal promoter with respect to the start codon. Each fragment is fused to the CaMV 35S minimal promoter and to the GUS coding region followed by the nopaline synthase terminator. The white boxes symbolize the relative position of the analyzed MYB and MYC binding sites. The black crosses shows which binding site is mutated in the respective fragment. B) Summary of the GUS expression data. We distinguish between the ubiquitous expression called "basal expression" and expression in trichomes. We found two categories, basal and trichome expression and expression only in trichomes. The percentage of analyzed independent T2 lines showing the respective expression category is provided. C) List of the analyzed binding sites with their corresponding wild type nucleotide sequence and the sequence used for base substitution.
The pTRY-B region mediates the repression of the inhibitors repression
In a separate line of experiments we tested, whether reduced pTRY:GUS expression in the absence of the pTRY-B fragment or in the pTRY-A4, B lines is caused by endogenous R3MYB repressor activity. We compared the expression of the pTRY-A3, B, pTRY-A3 and pTRY-A4, B constructs in wild type and the cpc try mutant background (Figure 5). All constructs revealed a strong basal and trichome specific GUS expression. Thus the lack of basal expression in the pTRY-A3 line and the lack of basal and most of the trichome specific expression in the pTRY-A4, B line is rescued in the cpc try mutant. These data suggest that the -424 to -176 fragment (pTRY-A4) promotes the basal expression and that the pTRY-B fragment and the -623 to -424 (pTRY-A5) fragments mediate repression of the inhibitors repression.
Figure 5.
Expression pattern of 5' regulatory regions of the TRY promoter in the cpc try double mutant as revealed by GUS reporter gene expression. The GUS expression pattern driven by different promoter fragments was monitored on young leaves at stages when new trichomes are still initiated. Pictures were taken in each case from one of the strongest T2 lines carrying the respective TRY promoter GUS fusion construct in the cpc-1 try82 double mutant: (A) pTRY-A, B:GUS, (B) pTRY-A3, B:GUS, (C) pTRY-A3:GUS, (D) pTRY-A4, B:GUS.
In order to show that this repression of the inhibitors repression involves the patterning activators we used transient expression assays. The p35S:GL1, p35S:GL3 and p35S:TTG1 constructs were co-bombarded in wild type and cpc try mutants carrying the pTRY-A3, B, pTRY-A3 and pTRY-A4, B constructs (Table 2). In wild type we found GUS-positive cells only in plants carrying the pTRY-A3, B construct (n = 100). By contrast, all constructs revealed GUS-positive cells in the cpc try mutant background indicating that this regulation event involves the trichome patterning activators. Furthermore the percentage of GUS-positive cells per transformed epidermal cells was much higher for pTRY-A3, B transformed cotyledons in cpc try double mutant indicating a stronger activation.
Table 2.
Co-transformation promoter activation assay in Arabidopsis cotyledons
| Ler* | cpc-1 try-82* | |
|---|---|---|
| pTRY-A3, B | 53.7 ± 4.4 | 118.8 ± 18.2 |
| pTRY-A3 | 0.5 ± 0.6 | 80.2 ± 8.2 |
| pTRY-A4, B | 0.0 ± 0.0 | 32.7 ± 3.0 |
* Percentage [%] of GUS activated epidermal cotyledon cells after overnight staining relative to the number of GFP:YFP marked epidermal cotyledon cells from four independent experiments (each experiment included 100 cells).
Specific properties of the TRY protein in the regulation of cluster formation and trichome density
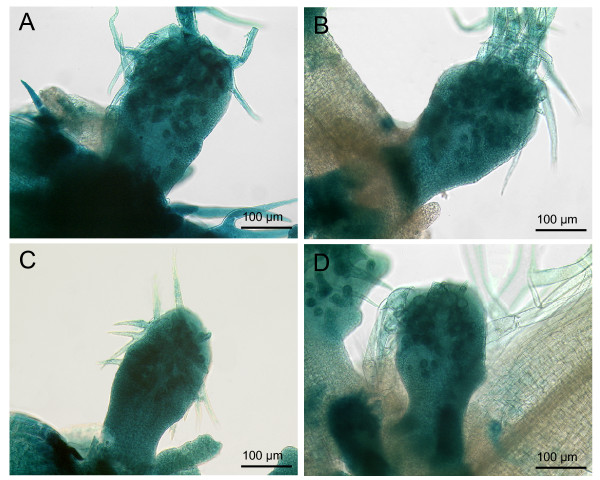
The fact that among the six R3 single repeat MYB inhibitor genes only mutations in TRY lead to a clustering phenotype raised the question, whether the transcriptional regulation of TRY or its protein properties constitute this difference. We therefore compared reciprocal swaps of promoters and CDS of the TRY and CPC genes for their ability to rescue the try mutant. CPC was chosen because it represents the main inhibitor for trichome density regulation and because it has a similar expression pattern including the early ubiquitous and later trichome specific expression. Here we chose 525 bp of the 5' upstream region of the CPC gene, which showed the expected CPC expression in leaves and roots (Additional File 2), and was able to rescue the cpc mutant trichome phenotype when fused to the CPC CDS (Additional File 3). Both combinations containing the CDS of TRY, pTRY:cTRY and pCPC:cTRY, completely rescued the clustering phenotype (Figure 6). By contrast, the combination of the TRY promoter with the CDS of CPC exhibited no significant rescue (Figure 6). This indicates that the specific role of TRY in preventing cluster formation is not based on its transcriptional regulation but on specific protein properties.
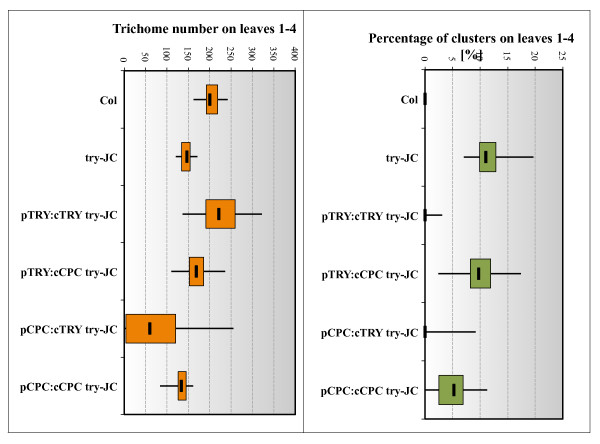
Figure 6.
A box-Whisker-plot of the trichome number and cluster frequency of the TRY/CPC- promoter swap experiments. Monitored are try-JC (n = 30), Col (n = 30), and pTRY:cTRY, pTRY:cCPC, pCPC:cTRY, pCPC:cCPC in try-JC mutant background (each n = 50 T1 plants). As coding sequence the CDS (c) of either TRY or CPC and the promoter pTRY-A3, B and pCPC (-686 to -158) were used. (A) The trichome number is summed for the first four leaves. (B) The percentage of the cluster is calculated relative to the initiation sites for the first four leaves. The boxes themselves contain the middle 50% of the data. The upper line of the box marks the 75th percentile and the lower one the 25th percentile. The line in the box indicates the median value. The ends of the vertical lines indicate the minimum and maximum data values.
We also determined the trichome number in these rescued lines. In this respect try mutants have the opposite effect as all the other inhibitor mutants in showing fewer trichomes than wild type [12]. Expression of TRY under the control of the TRY promoter can significantly rescue the trichome number. By contrast, the pTRY:cCPC construct revealed weak but not significant rescue. When using the CPC promoter we found no rescue with the pCPC:cCPC construct and an overexpression phenotype leading to less or even no trichomes in pCPC:cTRY plants. Thus in summary, we recognized protein-specific properties of TRY in the context of TRY dependent trichome density regulation. The finding that the TRY CDS driven by the CPC promoter could not rescue the try density defect suggests additional relevant differences between the two promoters in this context.
Discussion
In this study we analyzed the transcriptional regulation of TRY to learn more about the unique role of TRY in trichome patterning among the R3MYBs homologs as judged by the clustering phenotype of the try mutants.
Role of the pTRY-B fragment: general enhancer/suppressor or regulator of basal expression
Our promoter analyses revealed an important role of the pTRY-B fragment as it is absolutely necessary for the basal expression in the young leaf and for rescuing the clustering phenotype of the try mutant. It seems to modulate the spatial-temporal expression pattern. Several findings suggest that pTRY-B is specifically required for the basal expression of TRY. First, we never found any basal expression in the absence of the pTRY-B fragment in wild type background. Second, the expression in trichomes is similarly strong (as judged by the GUS staining time course experiments) with and without the pTRY-B fragment and the basal expression co-appears with the trichome specific expression in the presence of the pTRY-B fragment, but is not seen after 4 days without.
The absolute requirement of pTRY-B for rescue of the try clustering phenotype immediately suggests that the basal expression of TRY is relevant for patterning. This finding would be an important piece of support for the current theoretical models [2]. As the pattern is generated in a field of initially equivalent cells, the patterning system needs to start with an initially ubiquitous expression of activators and inhibitors that is necessary for the establishment of a pattern. Thus, according to this scenario the requirement of the pTRY-B fragment and therefore also of the basal expression supports this type of model.
How is the basal TRY expression regulated by the pTRY-B fragment? An answer towards this end comes from our analysis of the pTRY-A3, B:GUS and pTRY-A3:GUS constructs in try cpc mutants. The findings that the pTRY-A3 fragment can be activated by GL1 GL3 and TTG1 in the try cpc double mutants but not in wild type together with the presence of basal expression in pTRY-A3 try cpc plants indicates that the pTRY-B fragment mediates the repression of the TRY repression by TRY or CPC. Thus the apparent requirement of the pTRY-B fragment for basal expression is in fact a double negative regulatory event. The current data suggest that the -424 to -176 fragment is important for turning on the basal expression and that TRY/CPC inhibit this activation with the immediate upstream -623 to -424 and downstream -176 to -4 regions counteracting this inhibition. As the absence of the MYB2 site leads to an increased basal expression it is possible that this site is involved in this regulation loop. Similarly, the higher basal expression upon the deletion of the two MYC sites can be interpreted as a function of these sites in the double negative repression loop.
Self-repression of TRY without transcriptional regulating of the activators
Models explaining trichome formation in Arabidopsis are derived from the activator-inhibitor model formulated by Meinhardt und Gierer [37]. This theoretical model explains pattern formation with two components: an activator activates its own inhibitors and its own expression with the inhibitor being able to move faster than the activator.
When adapting this theoretical model to the biological context there are two possibilities. First, the inhibitor down-regulates its own expression indirectly through the down regulation of the activator. Second, the inhibitor represses its own expression through competitive complex formation [27,29,38]. While we can not exclude the first possibility, our data show that the second scenario is sufficient. We show that the minimal TRY promoter can be ectopically activated by the combined expression of GL1, GL3 and TTG1 in cotyledon cells. As the three activators are expressed under the control of the 35S promoter any transcriptional feed back loops involving these three genes are unlikely to be relevant in this experiment. The repression of the activity of the three activators by TRY or CPC provides evidence that TRY and CPC represses the TRY expression directly rather than through a transcriptional feed back loop involving the activator genes.
Specific properties of TRY protein for patterning
To further understand the molecular nature of the uniqueness of TRY among the six R3-single repeat MYB inhibitors, we used promoter swap experiments with CPC which shares all aspects of the TRY expression pattern as judged by promoter:GUS analysis. This enabled us to study the relevance of the transcriptional regulation and protein function of both genes in the try mutant cluster formation and density phenotypes. We found a different behavior of TRY and CPC proteins in these rescuing experiments such that only TRY protein could rescue the try mutant clustering and density phenotype when expressed under the TRY promoter. A similar situation was found in cpc mutant rescue experiments where the TRY protein expression under the control of the CPC promoter resulted in a stronger overexpression phenotype as compared to the CPC protein [39]. A contribution of TRY specific promoter properties was only found in the context of trichome density regulation. This is contrast to the behavior of ETC3 another homolog of TRY and CPC. The etc3 mutant could be rescued by ETC3 in the same manner by regulation through the ETC3, CPC and TRY promoter, so that the promoters were interchangeable with respect to the trichome density phenotype in the etc3 mutant but not with respect to the try mutant [14]. Thus TRY dependent regulation of trichome density is dependent on both, specific protein properties as well as specific aspects of transcriptional regulation.
The observed differences between TRY and CPC protein functions could in principle be due to various aspects including the protein stability, protein movement and their interaction with other proteins, in particular the bHLH factors. Both proteins have been shown to interact with bHLH factors [10,11,13,14,16,40]. Their interactions, however, seem to differ as CPC binds stronger to GL3 [41] and suppresses the binding of GL1 to GL3 more efficiently than TRY [14]. Different strength in their binding to GL3 is also likely to change the intercellular movement of TRY and CPC [14]. Both proteins have been shown to move between cells and share a 79 bp N-terminal region in which W76 and M78 were shown to be necessary for movement of CPC and are conserved in the TRY protein [27,28,38]. However, TRY protein is lacking the first 9aa that were also be shown to be necessary for CPC movement [28] and could therefore in principle be responsible for a different movement behavior. The most obvious difference between the TRY protein and CPC, ETC1, ETC2, ETC3 and TCL1 is its c-terminal extension of unknown function [41]. While we begin to understand the functional diversification of the R3 single MYB factors in trichome development it is still elusive which properties are responsible for the differences in their requirement for clustering and density control.
Conclusions
In this work we show that the auto-repressive effect of TRY does not require a transcriptional downregulation of the activators suggesting that differential complex formation is biologically relevant. We further show that the unique role of TRY among the inhibitors is a property of the TRY protein. Finally our analysis of the TRY promoter lead to the identification of a 620 bp fragment sufficient to rescue the try mutant phenotype. It contains a fragment that mediates the repression of its own repression suggesting a complex regulation scheme. It is likely, that we are seeing here just the tip of an iceberg, as the transcriptional regulation of TRY has additional complexity at the level of organ specificity involving additional regulatory genes such as the well studied SQUAMOSA PROMOTER BINDING PROTEIN LIKE (SPL) gene [42].
Methods
Plant lines and growth conditions
Plants were grown on soil at 24°C in a 16 h light/8 h dark cycle. Plant transformations were performed by the floral dip method [43]. The transgenic pTRY-A3, B:GUS Ler and cpc-1 try-82 double mutant [12] line was generated by a genetic cross. Complementation experiments were done in try-JC and cpc-1 mutants [23,44] and Col-0 and WS-0 respectively as a control. For the transient transformation of cotyledons pTRY-A3, B:GUS, pTRY-A3:GUS and pTRY-A4, B:GUS lines in Ler or in cpc-1 try-82 double mutant background were used. For these experiments surface sterilized seeds were grown on MS plates containing 1% sucrose and 20 μg/ml Basta for 7 days at 22°C with 16 h light/8 h dark cycle.
Constructs
Construction of the TRY and CPC promoter fragments
Ler pTRY-B was cloned as a HindIII fragment in pGEM-T-easy. All other promoter fragments were cloned in pDONR201 by BP reactions (Invitrogen). Promoter mutations were introduced by PCR based site directed mutagenesis (details are available on request). pENTR1A-w/o-ccdB was created by deleting the EcoRI fragment to take out the Gateway recombination cassette inside the attB sequences of pENTR1A. All fragments were verified by sequencing. (Detailed primer information see Additional File 4).
CaMV 35S minimal promoter GUS and CDS constructs
The basic Gateway destination vector PARB (pANGUS-Gateway RekombinationscasetteA-Basta-resistence) was created in several steps. The CaMV 35S minimal promoter (-46 to +7) fused to the GUS gene from pBT-GUS [45] was cloned as a BamHI and XmaI fragment into pPAM (GenBank AY027531). The Gateway recombination cassette A (Invitrogen) was cloned as a BcuI and SalI fragment in pANGUS (pANGUS-RecA). The kanamycin resistance was replaced by the bar gene with nos-promoter and nos-terminator from pGREEN-Bar as a RsrII and SpeI fragment. The pTRY-B fragment was cloned into the HindIII site of PARB directly in front of the 35S minimal promoter to create PARB-B. PARB-TRY-CDS, PARB-B-TRY-CDS, PARB-CPC-CDS are derivates of PARB and PARB-B in which the GUS gene was replaced by the TRY or CPC CDS (Ler).
Promoter-GUS- and promoter-CDS constructs were generated by LR recombinations (Invitrogen) using the entry clones pCPC-pDONR201, pENTR1A-w/o-ccdB and the different entry clones of the deletion and substitution series of pTRY and the different destination vectors derived from PARB.
Effector constructs for transient co-transformation experiments of Arabidopsis epidermal cotyledon cells
CDS's from Ler were cloned in pENTR1A or by BP recombination in pDONR201. The effector constructs (p35S:GL1, p35S:GL3, p35S:TTG1, p35S:TRY, p35S:CPC) and the control p35S:GFP:YFP were created by LR recombination of the respective entry clones with pAMPAT-GW.
Histochemical analysis and microscopy
GUS activity was assayed as described previously [46]. For light microscopy we used a Leica DMRE microscope. Images were taken with a KY-F70 3-CCD JVC camera and DISKUS software (DISKUS, Technisches Büro). In all experiments 35 independent T2 lines were used for statistical analysis.
Evaluation of the trichome initiation sites and cluster frequency
Trichome initiation sites and the number of trichome clusters were counted on the first four fully expanded leaves on 50 individual T1 plants after Basta selection and on 30 Col-0, WS-0, try-JC and cpc-1 plants. The significance of the difference between complemented plants and either Col or try-JC was tested by Student's T-test (two-tailed distribution and two-sample equal variance, P < 0,01). Trichome density and cluster frequency data in the promoter CDS swap experiment are shown as box-whisker-plots. The quartile function of Excel (Microsoft Office Standard 2007) was used to return the five quartiles for the data sets (minimum value, first quartile (25th percentile), median value, third quartile (75th percentile) and maximum value). The plot itself was created with a free accessible boxplot template (http://www.austromath.at/medienvielfalt/materialien/beschreibendeStatistik/content).
Microprojectile Bombardment
Transient TRY expression analysis was carried out by using the particle bombardment method in Arabidopsis cotyledons [47]. Each set of experiment was done independently at least four times. After bombardment plants were grown for 24 h and the number of transformed cells was determined by the presence of the co-bombarded p35S:GFP:YFP. After overnight GUS staining and tissue clearing the number of GUS stained cells was determined and the percentage of GUS positive cells relative to the transformed cells calculated.
In silico analysis of the TRY promoter
To identify transcription factor binding sites the Plant Cis-acting Regulatory DNA Elements (PLACE, http://www.dna.affrc.go.jp/PLACE/) and the TRANSFAC (TFSEARCH: Searching Transcription Factor Binding Sites (ver 1.3), http://mbs.cbrc.jp/papia/) databases were used.
Abbreviations
TRY: Triptychon; CPC: Caprice; GFP: Green fluorescent protein; GL1: Glabra1; GL3: Glabra3; TTG1: Transparent testa glabra1; YFP: Yellow fluorescent protein; GUS: Glucoronidase; ETC3: Enhancer of triptychon and caprice3; TCL1: Trichomless1; SPL: Squamosa promoter binding protein like (SPL)
Authors' contributions
MP carried out all molecular and genetic studies. MH participated in the design and coordination of the work and wrote the manuscript. All authors have read and approved the final manuscript.
Supplementary Material
Functional relevance of MYC1 and MYC2 sites. A box-Whisker-plot of the trichome number and cluster frequency of the double MYC binding site mutated TRY promoter rescue experiment. For pTRY-A3, B:cTRY and pTRY-A3, B-mutMYC+mutMYC2:cTRY in try-JC mutant background 50 T1 plants are monitored and 30 plants for Col and try-JC. The boxes contain the middle 50% of the data. The upper line of the box marks the 75th percentile and the lower one the 25th percentile. The line in the box indicates the median value. The ends of the vertical lines indicate the minimum and maximum data values.
Expression analysis of the CPC promoter. GUS expression of the 5' regulatory region of the CPC promoter. GUS staining was observed for a young leaf executing trichome patterning (A) or a young leaf already finished trichome patterning (B). In addition a 7 days old primary root grown on MS medium was shown (C). Pictures were taken from one T2 line representative for 35 independent observed lines. Bars as indicated.
Trichome rescue by pCPC:cCPC. A box-Whisker-plot of the trichome number and cluster frequency of the pCPC:cCPC rescue experiment. Monitored are WS-0 (n = 30), cpc-1 (n = 30), and pCPC:cCPC in cpc-1 mutant background (n = 50 T1 plants). The CDS of CPC was expressed under the control of pCPC (-686 to -158). The boxes contain the middle 50% of the data. The upper line of the box marks the 75th percentile and the lower one the 25th percentile. The line in the box indicates the median value. The ends of the vertical lines indicate the minimum and maximum data values.
Primer list. The table shows a list of the relevant primers used for the creation of the constructs.
Contributor Information
Martina Pesch, Email: martina.pesch@uni-koeln.de.
Martin Hülskamp, Email: martin.huelskamp@uni-koeln.de.
Acknowledgements
We thank Birgit Kernebeck for excellent technical assistance. The following vectors were kindly provided: pAMPAT-GW by B. Ülker (MPIZ, Cologne), pANGUS by M. Jakoby (MPIZ, Cologne), pBluescript-GW-RekA by S. Biere (MPIZ Cologne), TRY-CDS-pGEM-T and CPC-CDS-pBluescript by S. Bhylahalli, TRY, GL3 and TTG1 entry clones by U. Herrmann and the pENTR1A-w/o-ccdB by Arp Schnittger. We like to thank Dr. Swen Schellmann for critically reading the manuscript and helpful suggestions. This work was supported by the DFG priority program SFB572.
References
- Hulskamp M. Plant Trichomes: A Model for Cell Differentiation. Nature Reviews Molecular Cell Biology. 2004;5:471–480. doi: 10.1038/nrm1404. [DOI] [PubMed] [Google Scholar]
- Pesch M, Hulskamp M. One, two, three...models for trichome patterning in Arabidopsis? Curr Opin Plant Biol. 2009;12(5):587–592. doi: 10.1016/j.pbi.2009.07.015. [DOI] [PubMed] [Google Scholar]
- Ishida T, Kurata T, Okada K, Wada T. A Genetic Regulatory Network in the Development of Trichomes and Root Hairs. Annu Rev Plant Biol. 2008;59:364–386. doi: 10.1146/annurev.arplant.59.032607.092949. [DOI] [PubMed] [Google Scholar]
- Hulskamp M, Misera S, Jürgens G. Genetic dissection of trichome cell development in Arabidopsis. Cell. 1994;76:555–566. doi: 10.1016/0092-8674(94)90118-X. [DOI] [PubMed] [Google Scholar]
- Larkin JC, Young N, Prigge M, Marks MD. The control of trichome spacing and number in Arabidopsis. Development. 1996;122(3):997–1005. doi: 10.1242/dev.122.3.997. [DOI] [PubMed] [Google Scholar]
- Schnittger A, Folkers U, Schwab B, Jürgens G, Hulskamp M. Generation of a Spacing Pattern: The Role of TRIPTYCHON in Trichome Patterning in Arabidopsis. Plant Cell. 1999;11:1105–1116. doi: 10.1105/tpc.11.6.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkunde R, Pesch M, Hulskamp M. Trichome patterning in Arabidopsis thaliana from genetic to molecular models. Curr Top Dev Biol. 2010;91:299–321. doi: 10.1016/S0070-2153(10)91010-7. [DOI] [PubMed] [Google Scholar]
- Kirik V, Schnittger A, Radchuk V, Adler K, Hulskamp M, Baumlein H. Ectopic expression of the Arabidopsis AtMYB23 gene induces differentiation of trichome cells. Dev Biol. 2001;235(2):366–377. doi: 10.1006/dbio.2001.0287. [DOI] [PubMed] [Google Scholar]
- Kirik V, Lee MM, Wester K, Herrmann U, Zheng Z, Oppenheimer D, Schiefelbein J, Hulskamp M. Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation. Development. 2005;132(7):1477–1485. doi: 10.1242/dev.01708. [DOI] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development. 2003;130(20):4859–4869. doi: 10.1242/dev.00681. [DOI] [PubMed] [Google Scholar]
- Tominaga R, Iwata M, Sano R, Inoue K, Okada K, Wada T. Arabidopsis CAPRICE-LIKE MYB 3 (CPL3) controls endoreduplication and flowering development in addition to trichome and root hair formation. Development. 2008;135(7):1335–1345. doi: 10.1242/dev.017947. [DOI] [PubMed] [Google Scholar]
- Schellmann S, Schnittger A, Kirik V, Wada T, Okada K, Beermann A, Thumfahrt J, Jurgens G, Hulskamp M. TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 2002;21(19):5036–5046. doi: 10.1093/emboj/cdf524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V, Simon M, Wester K, Schiefelbein J, Hulskamp M. ENHANCER of TRY and CPC 2 (ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Mol Biol. 2004;55:389–398. doi: 10.1007/s11103-004-0893-8. [DOI] [PubMed] [Google Scholar]
- Wester K, Digiuni S, Geier F, Timmer J, Fleck C, Hulskamp M. Functional diversity of R3 single-repeat genes in trichome development. Development. 2009;136(9):1487–1496. doi: 10.1242/dev.021733. [DOI] [PubMed] [Google Scholar]
- Wang S, Kwak SH, Zeng Q, Ellis BE, Chen XY, Schiefelbein J, Chen JG. TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis. Development. 2007;134(21):3873–3882. doi: 10.1242/dev.009597. [DOI] [PubMed] [Google Scholar]
- Kirik V, Simon M, Hulskamp M, Schiefelbein J. The ENHANCER OF TRY AND CPC1 (ETC1) gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev Biol. 2004;268:506–513. doi: 10.1016/j.ydbio.2003.12.037. [DOI] [PubMed] [Google Scholar]
- Koornneef M. The complex syndrome of ttg mutants. Arabid Inf Serv. 1981;18:45–51. [Google Scholar]
- Galway ME, Masucci JD, Lloyd AM, Walbot V, Davis RW, Schiefelbein JW. The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev Biol. 1994;166:740–754. doi: 10.1006/dbio.1994.1352. [DOI] [PubMed] [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC. The TTG1 (transparent testa, glabra1) locus which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis encodes a WD40-repeat protein. The Plant Cell. 1999;11(7):1337–1350. doi: 10.1105/tpc.11.7.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD. A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell. 1991;67:483–493. doi: 10.1016/0092-8674(91)90523-2. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Dellaert LWM, VanderVeen JH. EMS- and radiation-induced mutation frequencies at individual loci in Arabidopsis thaliana. Mutat Res. 1982;93:109–123. doi: 10.1016/0027-5107(82)90129-4. [DOI] [PubMed] [Google Scholar]
- Payne CT, Zhang F, Lloyd AM. GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics. 2000;156(3):1349–1362. doi: 10.1093/genetics/156.3.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Tachibana T, Shimura Y, Okada K. Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science. 1997;277(5329):1113–1116. doi: 10.1126/science.277.5329.1113. [DOI] [PubMed] [Google Scholar]
- Larkin JC, Oppenheimer DG, Pollock S, Marks MD. Arabidopsis GLABROUS1 gene requires downstream sequences for function. Plant Cell. 1993;5:1739–1748. doi: 10.1105/tpc.5.12.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Morohashi K, Hatlestad G, Grotewold E, Lloyd A. The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development. 2008;135(11):1991–1999. doi: 10.1242/dev.016873. [DOI] [PubMed] [Google Scholar]
- Koshino-Kimura Y, Wada T, Tachibana T, Tsugeki R, Ishiguro S, Okada K. Regulation of CAPRICE Transcription by MYB Proteins for Root Epidermis Differentiation in Arabidopsis. Plant Cell Physiol. 2005;46(6):817–826. doi: 10.1093/pcp/pci096. [DOI] [PubMed] [Google Scholar]
- Digiuni S, Schellmann S, Geier F, Greese B, Pesch M, Wester K, Dartan B, Mach V, Srinivas BP, Timmer J. et al. A competitive complex formation mechanism underlies trichome patterning on Arabidopsis leaves. Mol Syst Biol. 2008;4:217. doi: 10.1038/msb.2008.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata T, Ishida T, Kawabata-Awai C, Noguchi M, Hattori S, Sano R, Nagasaka R, Tominaga R, Koshino-Kimura Y, Kato T. et al. Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development. 2005;132(24):5387–5398. doi: 10.1242/dev.02139. [DOI] [PubMed] [Google Scholar]
- Esch JJ, Chen M, Sanders M, Hillestad M, Ndkium S, Idelkope B, Neizer J, Marks MD. A contradictory GLABRA3 allele helps define gene interactions controlling trichome development in Arabidopsis. Development. 2003;130:5885–5894. doi: 10.1242/dev.00812. [DOI] [PubMed] [Google Scholar]
- Bernard V, Brunaud V, Lecharny A. TC-motifs at the TATA-box expected position in plant genes: a novel class of motifs involved in the transcription regulation. BMC Genomics 11, 166. BMC Genomics. 2010;11:166. doi: 10.1186/1471-2164-11-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto Y, Kisaka M, Fuse T, Yano M, Ogihara Y. Characterization and functional analysis of three wheat genes with homology to the CONSTANS flowering time gene in transgenic rice. Plant J 36, 82-93. Plant Journal. 2003;36:82–93. doi: 10.1046/j.1365-313X.2003.01859.x. [DOI] [PubMed] [Google Scholar]
- Robert LS, Robson F, Sharpe A, Lydiate D, Coupland G. Conserved structure and function of the Arabidopsis flowering time gene CONSTANS in Brassica napus. Plant Mol Biol. 1998;37:763–772. doi: 10.1023/A:1006064514311. [DOI] [PubMed] [Google Scholar]
- Morohashi K, Grotewold E. A systems approach reveals regulatory circuitry for Arabidopsis trichome initiation by the GL3 and GL1 selectors. PLoS Genet. 2009;5(2):e1000396. doi: 10.1371/journal.pgen.1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K, Zhao M, Yang M, Read B, Lloyd A, Lamb R, Grotewold E. Participation of the Arabidopsis bHLH Factor GL3 in Trichome Initiation Regulatory Events. Plant Physiol. 2007;145(3):736–746. doi: 10.1104/pp.107.104521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay NA, Glover BJ. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005;10(2):63–70. doi: 10.1016/j.tplants.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Quatttrocchio F, Wing J, Woude Kvd, Souer E, Vetten Nd, Mol J, Koes R. Molecular Analysis of the anthocyanin2 gene of Petunia and its role in the evolution of flower color. Plant Cell. 1999;11:1433–1444. doi: 10.1105/tpc.11.8.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt H, Gierer A. Applications of a theory of biological pattern formation based on lateral inhibition. J Cell Sci. 1974;15:321–346. doi: 10.1242/jcs.15.2.321. [DOI] [PubMed] [Google Scholar]
- Wada T, Kurata T, Tominaga R, Koshino-Kimura Y, Tachibana T, Goto K, Marks MD, Shimura Y, Okada K. Role of a positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development. 2002;129(23):5409–5419. doi: 10.1242/dev.00111. [DOI] [PubMed] [Google Scholar]
- Simon M, Lee MM, Lin Y, Gish L, Schiefelbein J. Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev Biol. 2007;311(2):566–578. doi: 10.1016/j.ydbio.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Bernhardt C, Lee MM, Gonzalez A, Zhang F, Lloyd A, Schiefelbein J. The bHLH genes GLABRA3 (GL3) andENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development. 2003;130(26):6431–6439. doi: 10.1242/dev.00880. [DOI] [PubMed] [Google Scholar]
- Wang S, Hubbard L, Chang Y, Guo J, Schiefelbein J, Chen JG. Comprehensive analysis of single-repeat R3 MYB proteins in epidermal cell patterning and their transcriptional regulation in Arabidopsis. BMC Plant Biol. 2008;8:81. doi: 10.1186/1471-2229-8-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N, Cai WJ, Wang S, Shan C-M, Wang LJ, Chen XY. Temporal Control of Trichome Distribution by MicroRNA156-Targeted SPL Genes in Arabidopsis thaliana. Plant Cell. 2010;22:2322–2335. doi: 10.1105/tpc.109.072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S, Bent A. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Larkin JC, Walker JD, Bolognesi-Winfield AC, Gray JC, Walker AR. Allele-specific interactions between ttg and gl1 during trichome development in Arabidopsis thaliana. Genetics. 1999;151:1591–1604. doi: 10.1093/genetics/151.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger-Haussels M, Weisshaar B. Transactivation properties of parsley proline-rich bZIP transcription factors. Plant Journal. 2000;22(1):1–8. doi: 10.1046/j.1365-313x.2000.00687.x. [DOI] [PubMed] [Google Scholar]
- Sessions A, Weigel D, Yanofsky M. The Arabidopsis thaliana MERISTEM LAYER1 promoter specifies epidermal expression in meristems and young primordia. Plant Journal. 1999;20:259–263. doi: 10.1046/j.1365-313x.1999.00594.x. [DOI] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Kernebeck B, Hulskamp M. Mutations in actin-related proteins 2 and 3 affect cell shape development in Arabidopsis. Plant Cell. 2003;15(7):1632–1645. doi: 10.1105/tpc.011676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell. 1997;9:1859–1868. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski RWM, Moyano E, Culianez-Macia FA, Schuch W, Martin C, Bevan M. A flower-specific Myb protein activates transcription of phenylpropanoid biosynthetic genes. EMBO J. 1994;13:128–137. doi: 10.1002/j.1460-2075.1994.tb06242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E, Drummond BJ, Bowen B, Peterson T. The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell. 1994;76:543–553. doi: 10.1016/0092-8674(94)90117-1. [DOI] [PubMed] [Google Scholar]
- Gubler F, Kalla R, Robert JK, Jacobsen JV. Gibberellin-regulated expression of a myb gene in barley aleurone cells: evidence for Myb transactivation of a high-pl alpha-amylase gene promoter. Plant Cell. 1995;7:1879–1891. doi: 10.1105/tpc.7.11.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Raventos D, Keys M, Watts R, Mundy J, Jacobsen JV. Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. Plant Journal. 1999;17:1–9. doi: 10.1046/j.1365-313X.1999.00346.x. [DOI] [PubMed] [Google Scholar]
- Morita A, Umemura T, Kuroyanagi M, Futsuhara Y, Perata P, Yamaguchi J. Functional dissection of a sugar-repressed alpha-amylase gene (Ramy1A) promoter in rice embryos. FEBS letters. 1998;423:81–85. doi: 10.1016/S0014-5793(98)00067-2. [DOI] [PubMed] [Google Scholar]
- Planchais S, Perennes C, Glab N, Mironov V, Inze D, Bergounioux C. Characterization of cis-acting element involved in cell cycle phase-independent activation of Arath;CycB1;1 transcription and identification of putative regulatory proteins. Plant Mol Biol. 2002;50:111–127. doi: 10.1023/a:1016018711532. [DOI] [PubMed] [Google Scholar]
- Biedenkapp H, Borgmeyer U, Sippel A, Klempnauer K-H. Viral myb oncogene encodes a sequence-specific DNA-binding activity. Nature. 1988;335:835–837. doi: 10.1038/335835a0. [DOI] [PubMed] [Google Scholar]
- Simpson SD, Nakashima K, Narusaka Y, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Two different novel cis-acting elements of erd1, a clpA homologous Arabidopsis gene function in induction by dehydration stress and dark-induced senescence. Plant Journal. 2003;33:259–270. doi: 10.1046/j.1365-313X.2003.01624.x. [DOI] [PubMed] [Google Scholar]
- Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16:2481–2498. doi: 10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Functional relevance of MYC1 and MYC2 sites. A box-Whisker-plot of the trichome number and cluster frequency of the double MYC binding site mutated TRY promoter rescue experiment. For pTRY-A3, B:cTRY and pTRY-A3, B-mutMYC+mutMYC2:cTRY in try-JC mutant background 50 T1 plants are monitored and 30 plants for Col and try-JC. The boxes contain the middle 50% of the data. The upper line of the box marks the 75th percentile and the lower one the 25th percentile. The line in the box indicates the median value. The ends of the vertical lines indicate the minimum and maximum data values.
Expression analysis of the CPC promoter. GUS expression of the 5' regulatory region of the CPC promoter. GUS staining was observed for a young leaf executing trichome patterning (A) or a young leaf already finished trichome patterning (B). In addition a 7 days old primary root grown on MS medium was shown (C). Pictures were taken from one T2 line representative for 35 independent observed lines. Bars as indicated.
Trichome rescue by pCPC:cCPC. A box-Whisker-plot of the trichome number and cluster frequency of the pCPC:cCPC rescue experiment. Monitored are WS-0 (n = 30), cpc-1 (n = 30), and pCPC:cCPC in cpc-1 mutant background (n = 50 T1 plants). The CDS of CPC was expressed under the control of pCPC (-686 to -158). The boxes contain the middle 50% of the data. The upper line of the box marks the 75th percentile and the lower one the 25th percentile. The line in the box indicates the median value. The ends of the vertical lines indicate the minimum and maximum data values.
Primer list. The table shows a list of the relevant primers used for the creation of the constructs.