Abstract
Saitohin (STH) is a gene unique to humans and their closest relatives whose function is not yet known. STH contains a single polymorphism (Q7R); the Q allele is human-specific and confers susceptibility to several neurodegenerative diseases. In previous work, we discovered that STH interacts with Peroxiredoxin 6 (Prdx6), a unique member of that family which is bifunctional and whose levels increase in Pick’s disease. In this study, we report that STH also interacts with tau and the non-receptor tyrosine kinase c-Abl (Abl). Furthermore, Abl phosphorylates STH on its single tyrosine residue and STH increases tyrosine phosphorylation by Abl. The effect of Saitohin on Abl-mediated phosphorylation appears to be allele-specific, providing evidence for a new cellular function for STH.
Keywords: Saitohin interactions, primate-specific gene, human-specific allele, MAP tau, tyrosine kinase Abl, allele-specific effects, neurodegeneration
INTRODUCTION
Saitohin (STH), an intronless gene encoding an open reading frame of 128 amino acids, is located in the intron between exons 9 and 10 of the human tau gene (Conrad et al., 2002). It bears no obvious homology to any known protein and its expression pattern is very similar to that of tau. The DNA sequences homologous to the human STH gene reveal an intact, highly conserved open reading frame exclusively in the primates most closely related to humans (chimpanzees, bonobos and gorillas; Conrad et al., 2004; Holzer et al., 2004). Thus, STH is an evolutionary locus that separates humans and their closest relatives from other mammals.
Human STH has a single nucleotide polymorphism that changes glutamine residue 7 to arginine (Q7R; Conrad et al., 2002). The Q allele is the most common in humans but all nonhuman primates are homozygous for the R allele, which makes the Q allele a human-specific marker (Conrad et al., 2004, Holzer et al., 2004). Hence, the R allele is the ancestral haplotypes whereas the Q allele evolved after the hominin lineages separated from the other primates.
Beyond evolution studies, the STH Q allele is over-represented in several neurodegenerative diseases: progressive supranuclear palsy, frontotemporal dementia and Parkinson’s disease (Andreadis, 2011; Wilder et al., 2009). Taken together, these results suggest that identifying binding partners of STH could implicate several proteins and/or pathways as potential contributors in these neurodegenerative diseases as well.
Tau fulfills many roles; among them, axonal microtubule organization and axonal transport. Misregulation of tau splicing and phosphorylation are direct or downstream causes of dementia. In addition to extensive Ser/Thr phosphorylation, tau is also a substrate for src-family non-receptor tyrosine kinases (Lee, 2005; Lebouvier et al., 2009). Specifically, Abl phosphorylates Tyr394 of tau (Derkinderen et al., 2005; Tremblay et al., 2010). Abl shuttles between the nucleus and the cytoplasm and plays a role in numerous cellular processes including cytoskeleton signalling and neuronal function (Moresco and Koleske, 2003; Shaul, 2000; Sirvent et al., 2008). Tau phosphorylated on Tyr394 is found in neurofibrillary tangles and Abl phosphorylation and localization change in Alzheimer’s disease (Jing et al., 2009).
In this study, we demonstrate that STH interacts with tau and Abl, Abl phosphorylates STH on its single tyrosine, and STHQ influences Abl phosphorylation. So STH is a possible entry point for modulating tyrosine phosphorylation and its effect on neurodegeneration.
MATERIALS AND METHODS
Cell culture and transfections
EM4 (human embryonic kidney) cells were maintained in 1:1 DMEM/Ham’s F12; HOG (human oligodendroglioma) and COS (monkey kidney) cells in DMEM; SK-N-SH (human neuroblastoma, henceforth SKN) cells in MEM. All cell media were supplemented with 10% FBS. Cells were transfected when they reached confluence of 40% (if using LT1, Mirus) or 80% (if using lipofectamine 2000, Invitrogen) and harvested 48 hours after transfection.
Expression constructs of STH, tau and Abl
We had previously generated GFP-STHQ by inserting the STHQ cDNA into the BamHI site of EGFP-C1 (BD Biosciences; Conrad et al., 2002) and GFP-STHR by directed mutagenesis of GFP-STHQ (Gao et al 2005; henceforth STHQ and STHR). Using these constructs, we generated several STH mutants: in STHYF, the sole tyrosine residue, Y78, has become a phenylalanine; STH100, STH70 and STH40 contain stop codons at STH residues 102, 74 and 38, respectively; STHD5 contains a deletion of the first 22 amino acids of STH, including Q7.
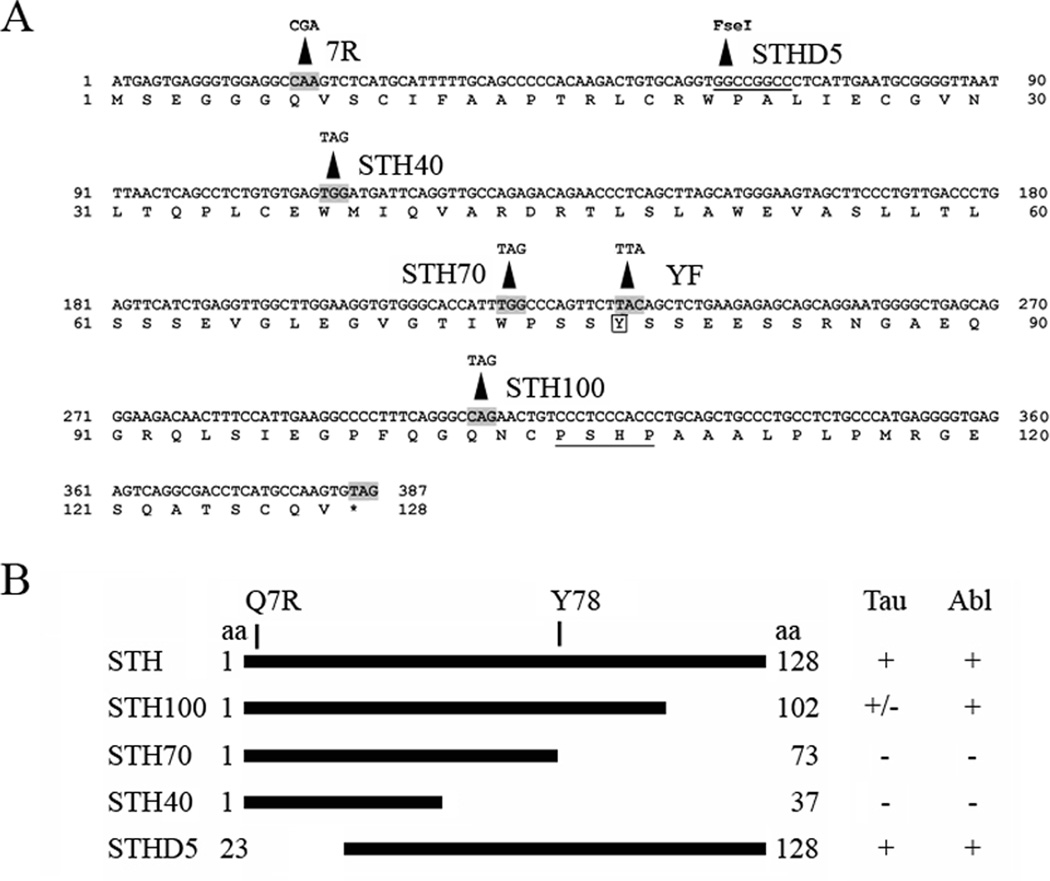
For STHD5 we digested STHQ with EcoRI and FseI, filled the ends with Klenow and did an intramolecular ligation. We created the other mutants by using the QuikChange mutagenesis kit (Stratagene) following the vendor’s instructions, except for extending the DpnI digest overnight. We generated STHYF in both the Q and R background, the deletions in the Q background. The resulting proteins are diagrammed in FIG. 1B and the mutagenic primers are listed in Table 1.
Fig. 1.
(A) The STH nucleotide and amino acid sequence. The Q7R residue and the changes made to create the mutant constructs are indicated. Underlined are the FseI site used for deletion STHD5 and the PXXP motif. The single tyrosine, Y78, is boxed. (B) Schematic representation of the STH deletion constructs. The Q7R, Y78 and beginning and ending amino acids of each construct are indicated. The two columns on the right indicate the strength of their interaction with tau and Abl: +, strongly positive; +/−, weakly positive; −, negative.
Table 1.
Primers
| Name | Nt | Strand Location | Sequence | |
|---|---|---|---|---|
| STH mutagenesis | ||||
| STH100S* | 34 | S | At STH residue 102 | |
| STH70S* | 30 | S | At STH residue 74 | |
| STH40S* | 35 | S | At STH residue 38 | |
| STHYFS* | 34 | S | At STH Y78 | |
| RACE/RT-PCR of STH and RT-PCR of tau | ||||
| STHS | 23 | S | At start of STH ORF | |
| STHN | 23 | A | At end of STH ORF | |
| F-Cel 1 | 21 | S | Upstream of STHS | |
| R-Cel 1 | 21 | A | Downstream of STHN | |
| F-Cel 2 | 21 | S | Upstream of F-Cel 1 | |
| R-Cel 2 | 21 | A | Downstream of R-Cel 1 | |
| HT7S3 | 24 | S | In human tau exon 7 | |
| HT11N | 24 | A | In human tau exon 11 | |
For creating point mutations, we used pairs of these primers and their reverse complements (not shown). Changes from the wild type are boxed; the resulting changes in STH are shown in Fig. 1. In STHS and STHN, the respective start and stop of the STH ORF are also boxed.
Nt=Length, S=sense, A=antisense.
Additionally, we created: GFP-Prdx6 by placing an EcoRI-XhoI fragment with its cDNA into EGFP-C2; and RFP-STHQ and -STHR by inserting the cDNAs into the BamHI site of mRFP-C1 (BD Biosciences). We had already generated FLAG-tau (the fetal 2-3-10- aka 0N3R variant; Luo et al., 2004). For Abl, we placed the wild-type cDNA and its constitutively active PP (P223E/P230E) mutant into the BamHI site of vector pSG5 (Stratagene).
RNA preparation, reverse transcription and PCR
To evaluate if STH can also influence the splicing of endogenous tau exon 10, we transfected STH into SKN cells and prepared RNA by the TRIzol method (Invitrogen). We did reverse transcription (RT) using Superscript II (Invitrogen) at 42 °C for 1 h using random hexamers (Promega), then PCR for 25 cycles using primer pair HT7S3/HT11N (Table 1).
To examine STH levels in brain compartments, we obtained small portions of four AD and four age-matched control cortices and hippocampi from the Brain Bank of McLean Hospital. We homogenized the tissues in TRIzol with a tissue:chloroform:TRIzol ratio of 1:1:10, then prepared RNA according to the manufacturer’s protocol.
Because STH lacks introns, before RT we treated the RNA with RNAase-free DNAase I (Ambion) for 1 h at 37 °C, then heat-inactivated for 15 min at 75 °C. We did RT as we did for tau, then performed quantitative PCR for 21 cycles using primer pair STHS/STHN (Table 1) and the Ambion Quantum kit with a ratio of 2:8 18S primers to 18S competimers.
We calculated the percent inclusion of endogenous exon 10 from a triplicate set of transfections and the ratio of STH to 18S from the four control and AD brain regions by scanning the RT-PCR bands and using the Scanalytics IPLab software.
To map the ends of the STH transcript, we prepared total RNA from HOG cells, then used the Gene Racer kit (Invitrogen) and combinations of primers F-Cel 1 and 2 and R-Cel 1 and 2 (Table 1) according to the vendor's instructions.
Western blotting and co-immunoprecipitations (co-IPs)
We prepared lysates from transfected cells using lysis buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1% Triton X-100) containing Protease Inhibitor and StopPhos phosphatase inhibitor tablets (Roche). Western blots using mouse or rabbit antibodies against GFP (Invitrogen), FLAG (Sigma) and Abl (mouse monoclonal 24-11; Santa Cruz) show that all our constructs express proteins of the correct sizes (Fig. 3 and 4, top and middle panels; Fig. 5, top panel).
Fig. 3.
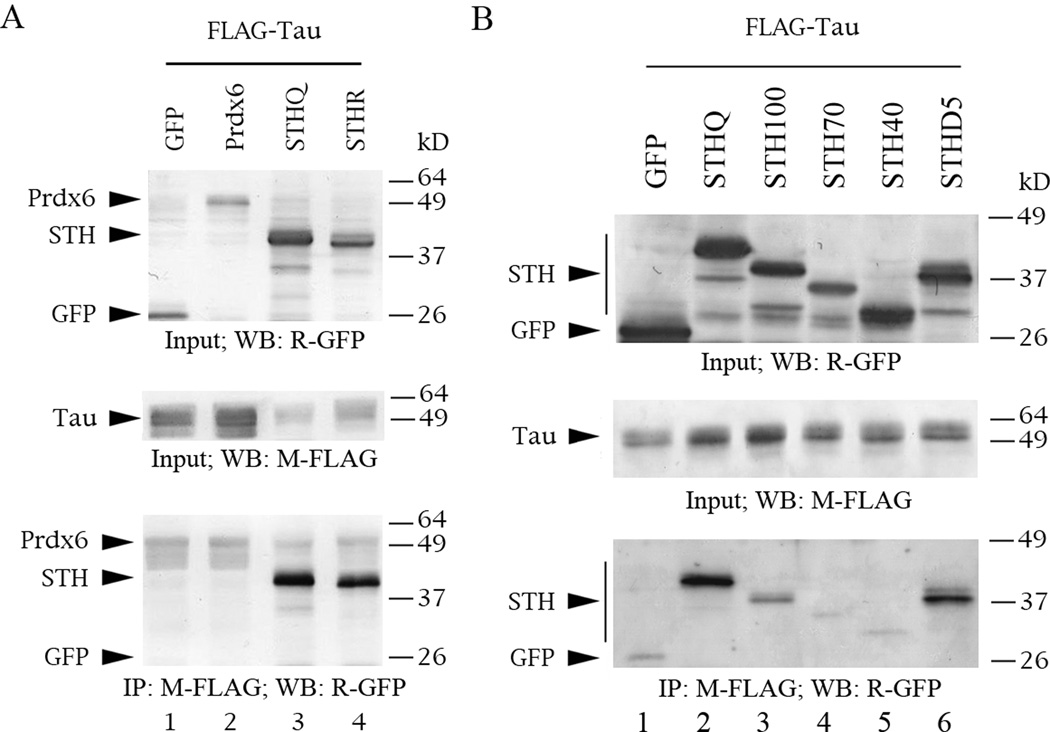
Tau interacts with STH but not with Prdx6, and seems to require the C-terminus of STH for interaction. The Western blots are of protein lysate inputs (top and middle panels) and co-IPs (bottom panels) from co-transfections into COS cells. The lysates were normalized for protein content. Standard markers are indicated on the left of each panel, proteins on the right. The primary antibodies used for IP and/or detection are listed below each panel. M = mouse monoclonal, R = rabbit polyclonal. (A) Co-IPs of FLAG-tau and GFP-STH or -Prdx6. (B) Co-IPs of FLAG-tau and GFP-STHQ deletion constructs.
Fig. 4.
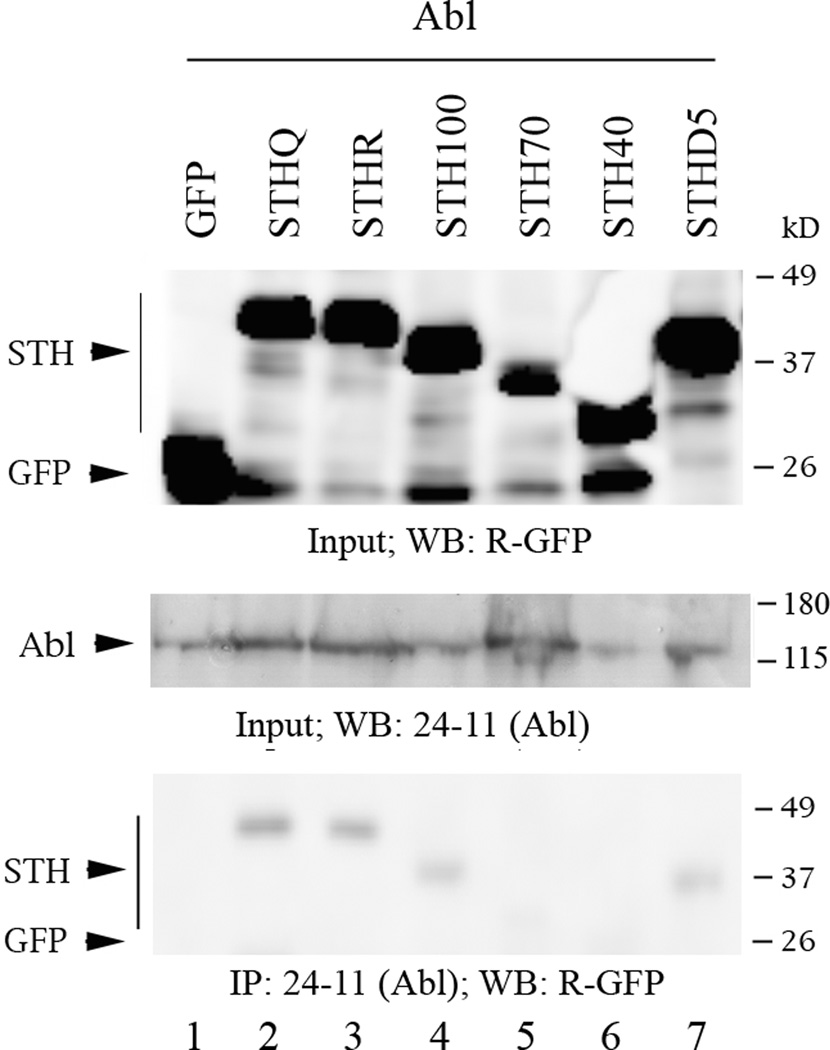
Abl interacts with STH and seems to require the region containing Y78 for interaction. The Western blots are of protein lysate inputs (top and middle panel) and co-IPs (bottom panel) from co-transfections of Abl and GFP-STHQ (full-length and deletions) or GFP-STHR into COS cells. Labeling conventions as the same as in Fig. 3.
Fig. 5.
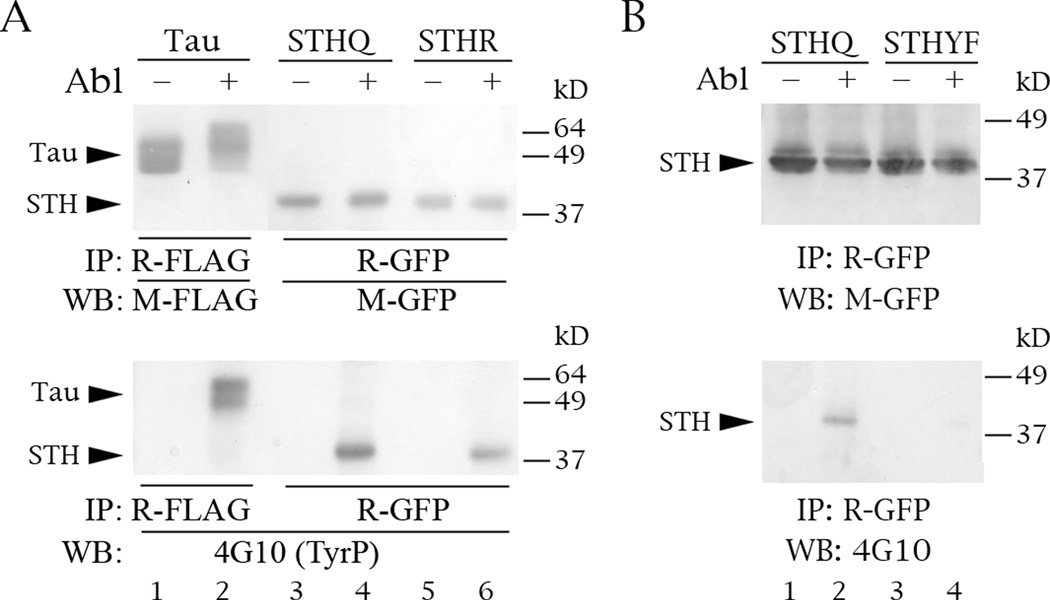
Abl phosphorylates STH on its single tyrosine. The Western blots are of protein lysate inputs (top and middle panels) and IPs (bottom panels) from co-transfections into COS cells. (A) Phosphorylation of FLAG-tau, GFP-STHQ and GFP-STHR by Abl. (B) Abl does not phosphorylate mutant STHYF, in which tyrosine 78 has been changed into a phenylalanine. Labeling conventions as the same as in Fig. 3.
For co-IPs of STH with Abl, we pre-cleared the supernatants by nutating them with protein G agarose (Roche) for 3 hours at 4 °C. We incubated 1 ml of cleared lysate with 1 ug of 24-11 anti-Abl antibody, then with 50 ul of homogeneous protein G agarose with nutation at 4 °C overnight. For co-IPs of STH with FLAG-tau, we incubated 1 ml of lysate with 40 ul of anti-FLAG antibody agarose beads (Sigma) with nutation at 4 °C overnight.
For all co-IPs we washed the complexes 4x with 500 ul of wash buffer (50 mM Tris pH 8.0, 150 mM NaCl) and ran them on 10% SDS-PAGE. To visualize the precipitated proteins, we used rabbit anti-GFP (Invitrogen) and either ECL (Amersham) or Opti-4CN (BioRad).
Phosphorylation assays
To evaluate whether Abl phosphorylates STH, we co-transfected COS cells with Abl plus FLAG-tau or GFP-STH, prepared lysates and precipitated as we did for the co-IPs, except we used 5 ug of rabbit polyclonal anti-FLAG (Sigma) or anti-GFP (Invitrogen) antibody and protein A agarose (Roche). To visualize the phosphorylation status of the precipitated proteins, we used anti-tyrosine antibody 4G10 (mouse monoclonal; Millipore).
To see if STH influences tyrosine phosphorylation, we co-transfected EM4 cells with GFP (used to normalize for transfection efficiency in a Tecan plate reader) plus RFP-STH with or without Abl. We fixed and permeabilized the cells and measured fluorescence in an Odyssey instrument according to the vendor's instructions (LICOR). To track RFP-tagged proteins we used rabbit polyclonal dsRed (Clontech) and anti-rabbit IRDye 800CW (LICOR); to track tyrosine phosphorylation we used 4G10 and anti-mouse Alexa 680 (Invitrogen).
RESULTS
STH is a distinct transcriptional unit
Previous RT-PCR of tissues showed that the expression and localization of STH are largely congruent, but not identical, with those of tau (Conrad et al., 2002). This suggests that STH may be a discrete transcriptional unit. Indeed, the 5' RACE showed a transcriptional start 342 nucleotides upstream of the STH ORF ATG. This is a bona fide start, since the RACE method we used works by capturing the m7G mRNA cap.
The 3' RACE gave a product ending at an AATAAA transcription termination motif 423 nucleotides downstream of the STH ORF stop. There is another AATAAA 1754 nucleotides past the stop. The positions within the AC091628 tau gene contig are: 5' start 112,344, STH ORF 112,686 to 113,072, 3' stops 113,495 and 114,826. Examination of the transcribed 5' UTR of STH by TFSearch (http://www.cbrc.jp/research/db/TFSEARCH.html ) shows that the region proximal to the ORF contains several consensus sites for the GATA family members, whereas the promoter region of tau is rich in GCF and AP-2 consensus sites (Andreadis et al., 1996). Neither promoter has a TATA box but downstream of each is a GT microsatellite.
Tau influences splicing of endogenous tau exon 10
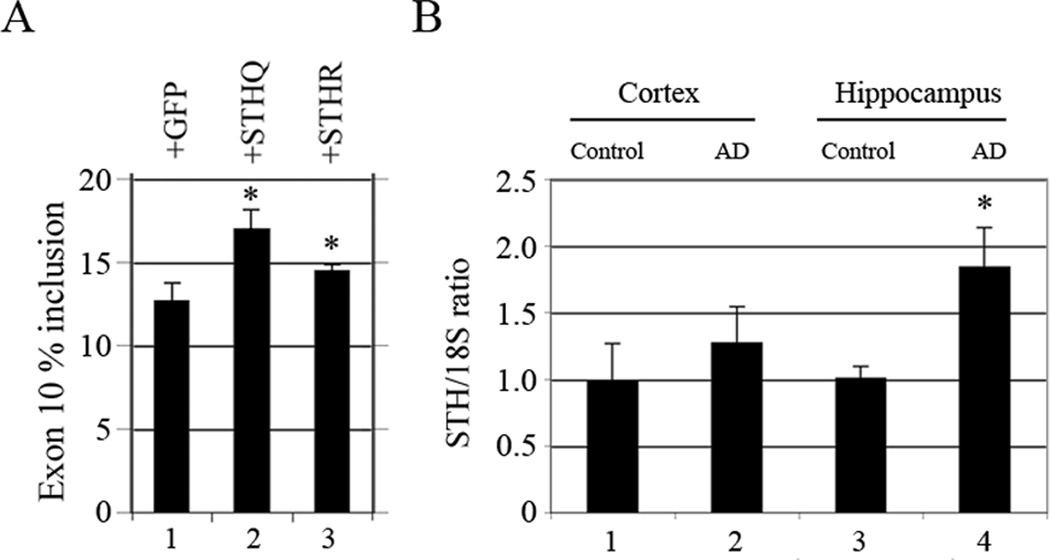
To follow up on our previous finding that STH increases splicing of exon 10 in co-transfected tau constructs (Gao et al., 2005), we examined its effect on endogenous tau. Our results show that STH also increases splicing of endogenous exon 10 in SKN neuroblastoma cells and STHQ does so more than STHR (Fig. 2A, lane 2 versus 3). This finding is congruent with our minigene results, except for one difference: in the minigene context, STHR increased exon 10 splicing more than STHQ (Gao et al., 2005).
Fig. 2.
STH increases splicing of tau exon 10 and increases significantly in AD hippocampus. An asterisk (*) indicates a measurement that reaches statistical significance (p<0.05). (A) Ratio of endogenous tau exon 10 by low-cycle RT-PCR after transfection of GFP-STHQ or -STHR into SKN cells. Primer pair: HT7S3/HT11N. (B) Ratio of STH to 18S by quantitative RT-PCR of total RNA from control and AD hippocampus and cortex. Primer pair: STHS/STHN. 18S:competimer ratio was 2:8.
STH levels increase in AD hippocampus
Because of the genomic location and expression pattern of STH, we deemed it interesting to investigate its levels in brain compartments affected in AD: hippocampus and cortex. The experiments show that STH levels increase in AD cortex but not enough to attain statistical significance (Fig. 2B, lane 2 versus 1). In contrast, STH levels increase significantly in hippocampus (Fig. 2B, lane 4 versus 3). This is particularly intereresting in view of the fact that the hippocampus is affected early in the neurodegeneration process.
STH interacts with tau and Abl, and Abl phosphorylates STH on its single tyrosine residue
Previous work had shown that STH interacts with Abl in vitro and STH residues 91-110 are sufficient for this interaction (Conrad, 2003). To expand these observations to cells, we tested the interaction of our new STH deletion mutants with tau (Fig. 3) and Abl (Fig. 4). The results are summarized in Fig. 1B.
By co-IP, tau does not interact with Prdx6 (Fig. 3A, lane 2) but interacts with both STH alleles at comparable levels (Fig. 3A, bottom panel, lanes 3 and 4). Congruent with this pattern, tau interacts with deletion STHD5 (which lacks residue 7) as strongly as it does with full-length STH (Fig. 3B, bottom panel, lane 6 versus lane 2). Tau binding to mutant STH100 is weak compared to full-length STH (Fig. 3B, lane 3) and there is no binding to mutants STH70 and STH40 (Fig. 3B, lanes 4 and 5). The faint background in lanes 1, 4 and 5 is due to a very weak interaction of GFP with FLAG agarose, which we have observed in other contexts.
In agreement with previous findings, Abl also interacts with STH (Fig. 4, bottom panel, lanes 2 and 3). We occasionally observed weaker binding to STHR than to STHQ, though that pattern was not consistent (data not shown). The interaction of Abl with STH100 and STHD5 is slightly weaker than that with full-length STH (Fig. 4, lanes 4 and 7) and there is no interaction with STH70 or STH40 (Fig. 4, lanes 5 and 6). This is compatible with the earlier findings (Conrad, 2003) but our results indicate that the PXXP motif at STH residues 106-109 is not required for Abl binding.
The obvious next question was whether Abl phosphorylates STH. The single tyrosine of STH is not within a sequence that resembles the consensus of the Abl phosphorylation site. Although there are a number of documented exceptions, the commonly quoted motif is I/V/L-YX2-3-P/F (Songyang et al., 1995), whereas the context of STH Y78 is S-Y-S-S-E-E (Fig. 1A). Nevertheless, Abl phosphorylates both STH alleles, with STHQ phosphorylated slightly more than STHR (Fig. 5A, bottom panel, lanes 4 and 6).
To confirm that Y78 is indeed the Abl target, we changed the tyrosine to a phenylalanine (STHYF). As we expected, Abl no longer phosphorylates STHYF (Fig. 5B, bottom panel, lane 4 versus 2). Interestingly, the location of Y78 correlates with the lack of Abl interaction with deletions STH70 and STH40.
STH increases tyrosine phosphorylation and STHQ does so more than STHR
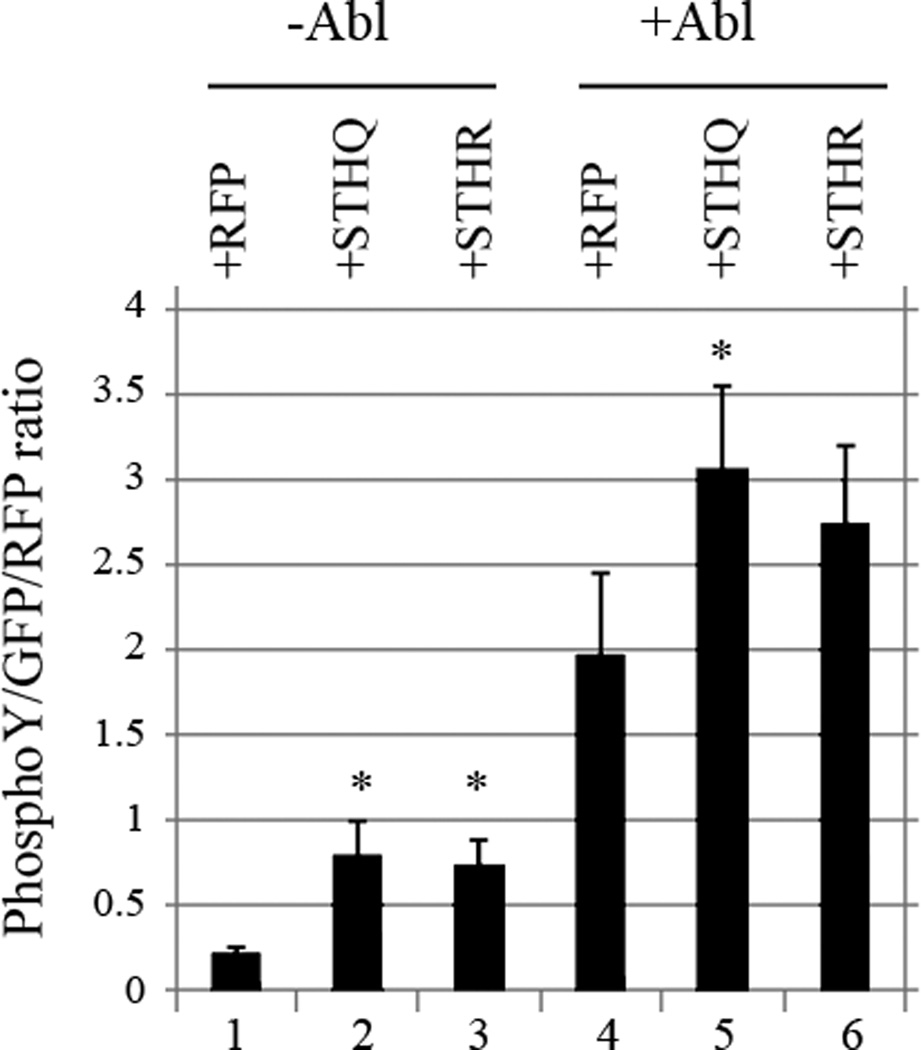
After establishing that STH interacts with Abl, we wanted to find out if it also affects Abl phosphorylation activity. Co-transfections of Abl with GFP affect cell viability, so conventional Westerns are usually not sensitive enough to detect the changes in doubly transfected cells against the background of singly transfected ones. To boost sensitivity, we used the LICOR plate fluorescence method instead. These experiments show that STH increases tyrosine phosphorylation both in the absence (Fig. 6, lanes 2 and 3) and the presence (Fig. 6, lanes 5 and 6) of exogenously added Abl and STHQ does so more than STHR. The difference between the two alleles is particularly pronounced with exogenously added Abl (Fig. 6, lane 5 versus 6).
Fig. 6.
STH increases cellular tyrosine phosphorylation and STHQ does so more than STHR. Levels of tyrosine phosphorylation after co-transfection of Abl and RFP-STHQ or -STHR into EM4 cells. GFP was used to normalize transfection efficiency and LICOR fluorescent methods to detect TyrP and RFP. * = p<0.05.
DISCUSSION
By virtue of its location, restricted evolutionary profile and allele-specific correlations with neurodegenerative diseases, STH is a truly intriguing molecule. Because of its lack of obvious motifs, its function has been elusive. Our previous work showed that STH interacts with Abl in vitro (Conrad, 2003) and with Prdx6 in cells and in vitro in allele-specific fashion (Gao et al., 2005). The present work establishes tau and Abl as additional STH binding partners and gives further hints to the possible role(s) that STH may play in the cell.
Among its many roles, tau promotes neurite outgrowth, organizes axonal microtubules, is involved in kinesin-dependent axonal transport and also appears to be involved in signal transduction in dendritic spines (Ittner and Götz, 2011; Morfini et al., 2009; Wang and Liu, 2007). Tau splicing and phosphorylation modulate tau function and the misregulation of either process results in neurofibrillary tangle formation and neurodegeneration (Andreadis, 2011; Dolan and Johnson, 2010). In particular, misregulation of splicing that leads to altered ratios of tau exon 10 results in tangle-only dementias (Gasparini et al., 2007).
The STH interaction with tau is tantalizing, given that STH is nested in the tau locus, its expression patterns are very similar to those of tau and they partly co-localize (Conrad et al., 2002). The region of interaction appears to be close to the C-terminus of STH. If STH were found to influence the phosphorylation of tau Tyr394 by Abl, this would establish a STH link to neurodegeneration although its exact mechanism would still need to be deciphered.
The increase of tau exon 10 inclusion in the presence of STH is more enigmatic. Since STH is cytosolic (Conrad, 2003; Gao et al., 2005), it must affect splicing of exon 10 by indirect mechanisms. STH might influence the localization or phosphorylation of shuttling splicing factors or their kinases, thereby modulating their activity.
Like tau, tyrosine kinase Abl also performs many roles, including DNA damage response, cell cycle regulation and actin cytoskeleton signal transduction (Moresco and Koleske, 2003; Shaul, 2000; Sirvent et al., 2008). Abl phosphorylation and localization change in Alzheimer’s disease (Jing et al., 2009). Specifically, Abl phosphorylates Tyr394 of tau and this tau species is found in neurofibrillary tangles (Derkinderen et al., 2005; Tremblay et al., 2010).
These connections make the STH/Abl reciprocal effects potentially very relevant: STH appears to be a substrate for Abl, even though its sole tyrosine is not within a canonical Abl phosphorylation sequence. It is possible that Abl affects STH phosphorylation through another tyrosine kinase. Conversely, STH increases Abl-mediated phosphorylation in allele-specific fashion, with the human-specific Q allele showing a stronger effect than the ancestral R.
As mentioned above, STH is cytosolic whereas Abl shuttles between the nucleus and the cytoplasm. One possible mechanism for the effect of STH on Abl is that STH might partition a higher proportion of Abl into the cytoplasm by binding to it. This would result in the increase of cytoplasmic tyrosine phosphorylation. In turn, tyrosine-phosphorylated STH would most likely have a modified activity profile.
A STH-induced shift would not need to be large to cause significant domino effects. In connection with this, it is interesting that our results show a large increase of STH in AD hippocampus. It will be revealing to see if STH levels also increase in tangle-only dementias. Another fascinating commonality is that the splicing regulation of tau exon 10 and the presence of a STH ORF are both species-specific, though the STH species range is far more restricted.
The evidence is circumstantial but highly suggestive that STH, through its allele-specific reciprocal interactions with Prdx6, tau and Abl, may be linked to the cascade of events which lead to neurodegeneration. Chimpanzees, which exclusively have the STH R allele, appear resistant to neurodegeneration whereas the Q allele confers susceptibility to several tangle-only dementias (Conrad, 2002, 2003; Andreadis, 2011). In view of this, it is odd that the ancestral R allele is rare in humans. Perhaps STHQ confers an advantage during development and early life but becomes detrimental in later life. The fact that STH Q allele is unique to humans makes it an invaluable tool to understanding why dementia seems to have singled out our species for preferential treatment.
ACKNOWLEDGMENTS
This study was supported by NIA grants R01 AG018486 and R21 AG028534 to A. A. and The Rosalinde and Arthur Gilbert Foundation/AFAR New Investigator Award in Alzheimer’s Disease, The Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, and Columbia University grants to C. G. C. We wish to thank Dr. Peter Davies in whose laboratory C. G. C. first observed the STH/Abl interaction and Drs. Leslie Griffith and Giulio Superti-Furga for generously sharing the Abl cDNAs.
REFERENCES
- Andreadis A, Wagner B, Broderick JA, Kosik KS. A tau promoter region lacks neuronal specificity. J. Neurochem. 1996;66:2257–2263. doi: 10.1046/j.1471-4159.1996.66062257.x. [DOI] [PubMed] [Google Scholar]
- Andreadis A. Tau splicing and the intricacies of dementia. J. Cell. Physiol. 2011 doi: 10.1002/jcp.22842. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad C, Vianna C, Freeman M, Davies P. A polymorphic gene nested within an intron of the tau gene: implication for Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 2002;99:7751–7756. doi: 10.1073/pnas.112194599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad C. PhD thesis. New York: Albert Einstein College of Medicine; 2003. Saitohin: a polymorphic gene in the tau locus. [Google Scholar]
- Derkinderen P, Scales TM, Hanger DP, Leung KY, Byers HL, Ward MA, Lenz C, Price C, Bird IN, Perera T, Kellie S, Williamson R, Noble W, Van Etten RA, Leroy K, Brion JP, Reynolds CH, Anderton BH. Tyrosine 394 is phosphorylated in Alzheimer's paired helical filament tau and in fetal tau with c-Abl as the candidate tyrosine kinase. J. Neurosci. 2005;25:6584–6593. doi: 10.1523/JNEUROSCI.1487-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan PJ, Johnson GV. The role of tau kinases in Alzheimer's disease. Curr. Opin. Drug Discov. Devel. 2010;13:595–603. [PMC free article] [PubMed] [Google Scholar]
- Gao L, Tse SW, Conrad C, Andreadis A. Saitohin, which is nested in the tau locus and confers allele-specific susceptibility to several neurodegenerative diseases, interacts with Peroxiredoxin 6. J. Biol. Chem. 2005;280:39268–39272. doi: 10.1074/jbc.M506116200. [DOI] [PubMed] [Google Scholar]
- Gasparini L, Terni B, Spillantini MG. Frontotemporal dementia with tau pathology. Neurodegen. Dis. 2007;4:236–253. doi: 10.1159/000101848. [DOI] [PubMed] [Google Scholar]
- Holzer M, Craxton M, Jakes R, Arendt T, Goedert M. Tau gene (MAPT) sequence variation among primates. Gene. 2004;341:313–322. doi: 10.1016/j.gene.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Ittner LM, Götz J. Amyloid-β and tau--a toxic pas de deux in Alzheimer's disease. Nat. Rev. Neurosci. 2011;12:65–72. doi: 10.1038/nrn2967. [DOI] [PubMed] [Google Scholar]
- Jing Z, Caltagarone J, Bowser R. Altered Subcellular Distribution of c-Abl in Alzheimer's Disease. J Alzheimers Dis. 2009;17:409–422. doi: 10.3233/JAD-2009-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebouvier T, Scales TM, Williamson R, Noble W, Duyckaerts C, Hanger DP, Reynolds CH, Anderton BH, Derkinderen P. The microtubule-associated protein tau is also phosphorylated on tyrosine. J. Alzheimers Dis. 2009;18:1–9. doi: 10.3233/JAD-2009-1116. [DOI] [PubMed] [Google Scholar]
- Lee G. Tau and src family tyrosine kinases. Biochim. Biophys. Acta. 2005;1739:323–330. doi: 10.1016/j.bbadis.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Moresco EM, Koleske AJ. Regulation of neuronal morphogenesis and synaptic function by Abl family kinases. Curr Opin Neurobiol. 2003;13:535–544. doi: 10.1016/j.conb.2003.08.002. [DOI] [PubMed] [Google Scholar]
- Morfini GA, Burns M, Binder LI, Kanaan NM, LaPointe N, Bosco DA, Brown RH, Jr, Brown H, Tiwari A, Hayward L, Edgar J, Nave KA, Garberrn J, Atagi Y, Song Y, Pigino G, Brady ST. Axonal transport defects in neurodegenerative diseases. J. Neurosci. 2009;29:12776–12786. doi: 10.1523/JNEUROSCI.3463-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlatterer SD, Tremblay MA, Acker CM, Davies P. Neuronal c-Abl Overexpression Leads to Neuronal Loss and Neuroinflammation in the Mouse Forebrain. J Alzheimers Dis. 2011 doi: 10.3233/JAD-2011-102025. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul Y. c-Abl: activation and nuclear targets. Cell Death Diff. 2000;7:10–16. doi: 10.1038/sj.cdd.4400626. [DOI] [PubMed] [Google Scholar]
- Sirvent A, Benistant C, Roche S. Cytoplasmic signalling by the c-Abl tyrosine kinase in normal and cancer cells. Biol. Cell. 2008;100:617–631. doi: 10.1042/BC20080020. [DOI] [PubMed] [Google Scholar]
- Songyang Z, Carraway KL, Eck MJ, Harrison SC, Feldman RA, Mohammadi M, Schlessinger J, Hubbard SR, Smith DP, Eng C, Lorenzo MJ, Ponder BAJ, Mayer BJ, Cantley LC. Catalytic specificity of protein-tyrosine kinases is critical for selective signalling. Nature. 1995;373:536–539. doi: 10.1038/373536a0. [DOI] [PubMed] [Google Scholar]
- Tremblay MA, Acker CM, Davies P. Tau phosphorylated at tyrosine 394 is found in Alzheimer's disease tangles and can be a product of the Abl-related kinase. Arg. J. Alzheimers Dis. 2010;19:721–733. doi: 10.3233/JAD-2010-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JZ, Liu F. Microtubule-associated protein tau in development, degeneration and protection of neurons. Prog. Neurobiol. 2008;85:148–175. doi: 10.1016/j.pneurobio.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Wider C, Vilariño-Güell C, Jasinska-Myga B, Heckman MG, Soto-Ortolaza AI, Cobb SA, Aasly JO, Gibson JM, Lynch T, Uitti RJ, Wszolek ZK, Farrer MJ, Ross OA. Association of the MAPT locus with Parkinson's disease. Eur. J. Neurol. 2010;17:483–486. doi: 10.1111/j.1468-1331.2009.02847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]