Abstract
FAS/FASL system plays a central role in maintaining peripheral immune tolerance. Human SLE is a prototypic systemic autoimmune disease characterized by expansion of autoreactive lymphocytes. It remains unclear whether a defective FAS/FASL system is involved in the pathogenesis of SLE. In this study, we have discovered a novel nucleotide insertion in FAS mRNA. We demonstrate that this novel FAS mutation occurs at mRNA levels, likely through a site-specific mRNA editing process. The mRNA editing mutation is unique for human FAS because the similar mRNA editing event is absent in other human TNFR family genes with death domains (DR5, DR6, and TNFR1) and in murine FAS. The adenine insertion mutation in the coding region message causes the alteration of human FAS mRNA reading frame. Functionally, cells expressing the edited FAS (edFAS) were refractory to FAS-mediated apoptosis. Surprisingly, cells from SLE patients produced significantly more edFAS products compared to cells from normal healthy controls. Additionally, we demonstrated that persistent engagement of T cell receptor increases human FAS mRNA editing in human T cells. Our data suggest that the site-specific FAS mRNA editing mutation may play a critical role in human immune responses and in the pathogenesis of human chronic inflammatory diseases.
Keywords: FAS, mRNA editing, apoptosis, Systemic Lupus Erythematosus
Introduction
Human FAS antigen (CD95 or APO1; MIM# 134637) is a type I membrane protein and a member of the TNF receptor (TNFR) family. FAS is expressed on many types of immune cells including lymphocytes, neutrophils, and monocytes (Leithauser, et al., 1993). Like other members of the TNFR family, biologically active FAS exists as homotrimeric receptor complexes. The extracellular domains of FAS bind to homotrimeric FAS ligand (FASL; MIM# 134638) complexes. The interaction between FAS and FASL induces the high-order oligomerization of FAS multimers, which initiate a cascade of signaling events leading to the programmed cell death or apoptosis. The FAS/FASL system promotes immune tolerance through the induction of apoptosis and deletion of activated T, B lymphocytes, and macrophages (Ashany, et al., 1995; Elkon and Marshak-Rothstein, 1996; Fukuyama, et al., 1998; Fukuyama, et al., 2002; Ramaswamy and Siegel, 2007; Singer and Abbas, 1994). In mice, mutations in FAS (lpr/lpr) or FASL (gld/gld) lead to spontaneous autoimmune diseases. The critical role of the FAS/FASL system in the maintenance of immune tolerance and prevention of autoimmune disease has been firmly established (Takahashi, et al., 1994; Watanabe-Fukunaga, et al., 1992a). A rare human autoimmune disease, the autoimmune lymphoproliferative syndrome (ALPS or Canale-Smith Syndrome), is associated with the inherited mutations in FAS or FASL (Drappa, et al., 1996; Fisher, et al., 1995; Rieux-Laucat, et al., 1995; Sneller, et al., 1992; Sneller, et al., 1997).
Human SLE is considered as a prototypic systemic autoimmune disease characterized by autoantibody production, immune complex formation, and cell-mediated reactivity against self. SLE can involve multiple organ systems and display very diverse clinical manifestations (Kotzin, 1996). Previously, it has been shown that human autoreactive T cells are resistant to FAS-mediated apoptosis (Zipp, et al., 1997). Therefore, a defective FAS/FASL system may be implicated in the development of human SLE (Kotzin, 1996). Nevertheless, the role of FAS/FASL system in the pathogenesis of SLE remains a conundrum.
In the present study, we have discovered a novel human FAS mRNA mutation in SLE patients. The FAS mRNA mutation results in the production of a defective FAS protein, and our data suggest that this FAS mRNA mutation may play an important role in the pathogenesis of SLE.
Materials and methods
Donors
Anti-coagulated peripheral blood was obtained from healthy normal volunteers and from SLE patients fulfilling the revised ACR criteria for SLE (Tan, et al., 1982). The human studies were reviewed and approved by the Institution Review Board (University of Alabama at Birmingham), and all donors provided written informed consent.
Reagents
All mAbs used were murine origin. CD8-PE, CD3-TC, CD3-FITC, CD4-FITC, CD8-FITC, CD14-FITC, CD19-FITC, CD25-FITC, CD29-FITC, CD69-FITC, Streptavidin-Cy3, and Streptavidin-PE were from Caltag Laboratories (Burlingame, CA). Anti-human FAS mAb (CH-11, mIgM) was purchased from Upstate Biotechnology (Lake Placid, NY). CD95-FITC was from BD PharMingen (San Diego, CA). Anti human FAS (CD95) mAb was purified from supernatant of the murine hybridoma cell line (ATCC#: HB-11726) at UAB (University of Alabama at Birmingham) Epitope Recognition and Immunoreagent Core Facility. Anti human CD3 mAb OKT3 (ATCC#CRL-8001) was prepared by National Cell Culture Center (Minneapolis, MN). Recombinant SuperFAS ligand was obtained from Alexis Biochemicals (San Diego, CA). Human IFNα and IFNγ were purchased from Sigma-Aldrich (St. Louis, MO). Horseradish Peroxidase-conjugated goat anti-mouse IgG(H+L) and donkey anti-rabbit IgG(H+L) were obtained from Jackson ImmunoResearch Lab (West Grove, PA).
Preparation of mixed mononuclear cells (MNC)
Fresh anti-coagulated blood was diluted 1:1 in PBS buffer (Mediatech Inc., Herndon, VA) and centrifuged through a discontinuous two-step Ficoll-Hypaque gradient in 50-ml conical tubes. Mixed mononuclear cells were harvested from the upper and neutrophils from the lower Ficoll-Hypaque interface. Cells were washed three times with PBS (pH 7.4) before being used for mRNA preparations, flow cytometry, and Western Blotting.
Nucleic acid isolation
Human genomic DNA was isolated using the Puregene DNA isolation kit (Gentra Systems, Minneapolis, MN) by following vendor’s instruction. Total RNA was isolated from 107 cells by using TRIzol™ total RNA isolation reagent (Invitrogen, Carlsbad, CA).
RT-PCR and cDNA sequencing
Five μg of total RNA was used for cDNA synthesis with the SuperScript™ preamplification system (Invitrogen). The RT-PCR was performed with 2 μl of cDNA, 200 nM of each primer (sense 5′-GAG GAT TGC TCA ACA AC-3′ and anti-sense 5′-GAT AAA ATG TAC CCA GTA AAA A-3′), 200 μM of dNTPs, 1.5 mM of MgCl2, and 2.5 U of Taq DNA polymerase in a 50-μl reaction volume on an ABI 9700 PCR System using the PCR program (initial holding at 95°C for 3 min, 35 cycles of denaturing at 94°C for 30 s, annealing at 54°C for 45 s, extension at 72°C for 1 min with a final extension at 72°C for 7 min). The PCR products were purified with the QIAquick Gel Extraction Kit (QIAGEN Inc., Chatsworth, CA) and subsequently sequenced on an ABI 377 Sequencer (Applied Biosystems, Inc., Foster City, CA).
Generation of FAS expression constructs
The human FAS expression constructs were generated by cloning whole FAS coding region into the eukaryotic expression vector pcDNA3 (Gibco BRL). The Kpn I/EcoR I-flanked RT-PCR products were amplified from cDNA with upper primer 5′-ACGG GGT ACC GAG GAT TGC TCA ACA AC-3′ (Kpn I site is underlined) and lower primer 5′-CCCG GAA TTC GAT AAA ATG TAC CCA GTA AAA A-3′ (EcoR I site is underlined). The sequences of all FAS expression constructs were confirmed by sequencing from both directions on an ABI 377 Sequencer with ABI BigDye Terminator Cycle Sequencing Kit.
Generation of stable cell lines expressing FAS and edFAS
The 293 cells (ATCC#CRL-1573) were maintained in the DMEM medium supplemented with 10% fetal calf serum (FBS) and L-glutamine (2 mM) in 5% CO2. Transfection reactions were performed in the 100 mm cell culture dishes with the plasmid DNA (24 μg) and 60 μl of Lipofectamine 2000 reagent (Invitrogen). Transfected cells were cultured in DMEM medium supplemented with 10% FBS for two days before the supplement of G418 (1 mg/ml). The polyclonal cells surviving the G418 selection were sorted on a FACS Vantage (BD Biosciences) for equal expression of surface FAS. The human Jurkat T cells (ATCC#TIB-152) were maintained in the RPMI medium supplemented with 10% FBS and L-glutamine (2 mM). Transfections were carried out with the FAS expression construct plasmid DNA (4 μg) and DMRIE-C reagent (12 μl) according to the vendor’s instructions (Invitrogen). Transfected Jurkat T cells were cultured in RPMI medium supplemented with 10% FBS and G418 (1 mg/ml) for the selection of stable transfectants.
Apoptosis assay
Target cells (1×103/well) in 96-well plates were cultured in medium containing the recombinant SuperFAS ligand (20 ng/ml) or anti human FAS mAb CH-11 (2 μg/ml). Cells were continuously cultured at 37 °C. Cell viability was determined using the ATPLite kit (Packard Instruments, Meriden, Connecticut) at various time points. The survival percentages (%) at respective time points were calculated according to vendor’s instruction. Each experiment was performed at least three times in triplicates.
Generation of polyclonal and monoclonal antibodies against edFAS protein
Rabbit antibodies against the unique edFAS polypeptides (SQSLYSCRENSDYHPQGHY) (Fig. 1B) were generated by Research Genetics Inc. using the vendor’s standard protocol (Research Genetics Inc., Huntsville, AL). The rabbit antibodies against the edFAS were purified with the peptide immune affinity columns by the Research Genetics Inc. For the generation of monoclonal antibodies, two BALB/C mice were immunized with the edFAS polypeptides. The monoclonal antibodies specific for the edFAS were produced and purified at the UAB Epitope Recognition and Immunoreagent Core Facility with affinity columns.
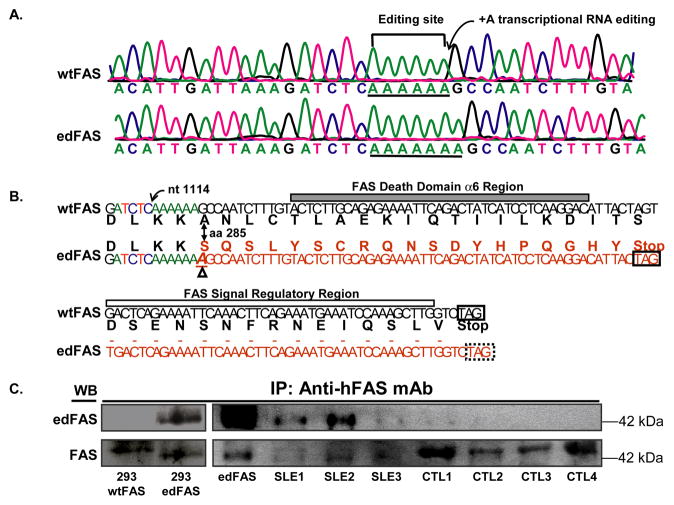
Fig. 1.
Adenine insertion in FAS mRNA leads to the production of novel FAS protein. A). Sequence tracings show an adenine insertion in FAS cDNA. The upper panel shows that the wild-type FAS (wtFAS) cDNA contains a stretch of six adenines (underlined) while the lower panel shows edited FAS (edFAS) cDNA containing seven adenines (underlined) in the same region. B). Schematic representation of nucleotide insertion mutation in FAS mRNA at position 1114 (GenBank accession number X63717). Partial sequences (nucleotide and peptide) of wild-type FAS (wtFAS) and the edited FAS (edFAS) are shown. The peptide sequences are different between wtFAS and edFAS after amino acid codon 285 in FAS cytoplasmic tail. The α6 region of the FAS death domain and the signal regulatory region are absent in the edFAS protein. The frame-shift mutation also creates a novel 19 amino acid peptide in the edFAS. The extra adenine is indicated by Δ sign. C). Detection of mutated FAS (edFAS) protein in SLE patients. Total FAS proteins were immunoprecipitated from cell lysates with anti-human FAS mAb as described in “Materials and Methods”. Rabbit anti- edFAS antibodies failed to react with the wild-type FAS expressed in 293 cells (293 wtFAS). The anti edFAS antibodies reacted with the edFAS expressed in 293 cells (293 edFAS, left upper panel). The edFAS protein was detected by anti-edFAS antibodies in two SLE patients (SLE1 and SLE2). Little or no edFAS protein could be detected in normal control donors (CTL1, CTL2, CTL3, and CTL4). Both edFAS and wtFAS were detected by anti-FAS antibody recognizing FAS extracellular domains (lower panels).
Immunoprecipitation and Western blot analysis
Peripheral blood leukocytes (2×107) were lyzed on ice for 30 min in 1 ml lysis buffer (50 mM Tri-HCl, pH 7.5, 150 mM sodium chloride, 1% Triton X-100, 0.5% sodium deoxycholate, and 1×complete protease inhibitor cocktail (Roche Molecular Biochemicals)). The lysates were centrifuged at 10,000×g for 10 min to pellet the insoluble materials. The resultant supernatants were incubated with 4 μg/ml of anti FAS mAb (huFasM3) for 2 h at 4 °C with gentle agitation. After that, 15 μl of protein G agarose beads were added and the mixture was further incubated with gentle agitation at 4 °C overnight. Immune-complexes bound to protein G beads were collected by centrifugation and washed six times in the lysis buffer. Immunoprecipitated proteins were resolved in 15% SDS-PAGE gels and transferred to nitrocellulose membrane (Sigma-Aldrich) according to manufacturer’s specification. The edFAS protein was detected with rabbit anti edFAS antibodies followed by donkey anti-rabbit Ig/HRP.
Immunofluorescence microscopy
293 cells expressing FAS or edFAS were grown on cover-slips and washed twice with ice cold PBS before being fixed with 3% ultra pure formaldehyde (Tousimis Research Corp, Rockville, MD) in PBS for 45 min at room temperature. Then the cells on cover-slips were permeabilized with 0.2% Triton X-100 in PBS for 3 min and blocked with 20% rabbit normal serum in PBS for 1 hr at room temperature. Cells on the cover slips were then incubated with the primary antibody (affinity purified and biotinylated rabbit anti edFAS antibodies, 2.5 μg/ml) for 1 hr at room temperature. After three washes in PBS, cells were incubated with streptavidin-Cy3. After washing, the slides were mounted with the Mounting Buffer (Molecular Probes) containing DAPI and the slides were analyzed under a under a Leitz Orthoplan microscope with epifluorescence optics.
Intracellular flow cytometry assay
Aliquots of cells of 5×105 cells in 0.1 ml PBS were incubated with saturating concentrations of FITC-conjugated mAb for 30 min at 4 °C followed by two washes. After washing, the cells were fixed and permeabilized with Cytofix/Cytoperm kit (BD Biosciences, San Diego, CA). The biotinylated anti edFAS mAb (clone 1D4, mIgG1) or isotype control mIgG1 (MOPC-21, Sigma-Aldrich) (0.5 μg/ml) were added and the cells were incubated for 30 min. After two washed with Perm/Wash buffer (BD Biosciences), the cells were then incubated with Streptavidin-PE (1.5 μg/ml) at 4 °C for another 30 min. Cells were then analyzed immediately for immunofluorescence intensity using a FACScan (BD Biosciences) after two washes with Perm/Wash buffer.
Determination of edFAS ratios
The percentage of edFAS mRNA relative to the total FAS mRNA was determined by sequencing multiple FAS cDNA clones from each donor. FAS cDNA was directly cloned into pCR2.1-TOPO vector (Invitrogen, Carlsbad, CA). Multiple clones containing FAS cDNA were sequenced using the BigDye sequencing kit. The percentage of the FAS mRNA editing was represented as the ratios of the edFAS clone number versus the total FAS cDNA clone number. In addition, an assay based on Pyrosequencing technology (Pyrosequencing AB, Vallongatan 1, SE-752 28 Uppsala, Sweden) was used. Percentage of edFAS mRNA relative to the total FAS mRNA pool was calculated based on the peak heights by the Pyrosequencing software (Pyrosequencing AB). For the pyrosequencing assay, upper primer 5′-ATG AAG CCA AAA TAG ATG AGA-3′ and the biotin-labeled lower primer 5′Bio-TGA GTC ACT AGT AAT GTC CTT GA-3′ were used for generation of a fragment of 203 bps containing the nucleotide insertion editing site at FAS cDNA position 1114 (GenBank accession number X63717) (Fig. 1). The FAS cDNA fragment was pyrosequenced with the primer 5′-TGA TTA AAG ATC TCA AAA AA-3′. Pyrosequencing was repeated at least three times for each donor.
Induction of FAS mRNA editing in T cells
The human Jurkat T cells were maintained in the RPMI medium supplemented with 10% FBS and L-glutamine (2 mM) in 5% CO2. T cells (5×105/ml) were cultured in RPMI growth medium containing IFNγ (100 U/ml), IFNα (100 U/ml), and anti CD3 mAb (clone OKT3, 5 μg/ml) respectively for 48 hours. The edFAS expression was assayed with intracellular flow cytometry as previously described. In addition, FAS cDNAs from the treated and non-treated T cells were cloned. Similarly, murine T cells (cell lines DOIL10 and DO8W) were treated with hamster anti-murine CD3 mAb 145.2C11 (5 μg/ml) for 48 hour. The murine FAS cDNA was cloned and multiple murine FAS cDNA clones were sequenced.
Statistical analysis
The levels of edFAS products in SLE patients and normal controls were analyzed with Wilcoxon Signed Rank Test. The distributions of edFAS clones in SLE patients and normal controls were analyzed with Chi-Square test. Differences in the apoptosis between cell lines were analyzed with Student’s t test. A P value of less than 0.05 was considered significant.
Results
Identification of FAS mRNA editing mutation
To understand whether FAS or FASL mutations play a role in the pathogenesis of human SLE, we screened the mutations in both FAS and FASL genes. Our earlier study did not reveal any mutation or non-synonymous SNP in the human FASL (Wu, et al., 2002). Subsequently, we examined whether SLE patients carry any FAS mutation(s). We cloned FAS cDNAs from seven SLE patients and seven normal control donors (Table 1). Surprisingly, we observed that a number of FAS cDNA clones contain an adenine insertion mutation at the same location. As shown in Fig. 1A, an adenine (A) is inserted after nucleotide position (nt) 1114 (GenBank accession number X63717). We found that five out of seven SLE patients expressed the adenine insertion (+A) mutation in their FAS cDNAs (Table 1). One SLE patient (SLE26) expressed up to 26% of mutant FAS cDNA clones. Altogether, the mutant FAS clones accounted for 11% of total 244 cDNA clones in seven SLE patients (Table 1). On the other hand, only two mutant FAS cDNA clones out of 231 clones were observed in seven normal controls and the frequency of mutant clones was less than 1% of the total FAS cDNA clones (Table 1) in normal controls.
Table 1.
Distribution of mutant FAS cDNA clones in SLE patients and normal controls
| Donor | cDNA | Genomic DNA | |||
|---|---|---|---|---|---|
| Total clones | Normal clones | Mutant (+A) clones | Frequency of +A clone (%) | +A clone (total clone) | |
| SLE patient* | 244 | 216 | 28 | 11 | 0 (180) |
| SLE07 | 35 | 30 | 5 | 14 | 0 (36) |
| SLE15 | 36 | 36 | 0 | 0 | - |
| SLE22 | 35 | 31 | 4 | 12 | 0 (36) |
| SLE23 | 35 | 34 | 1 | 3 | - |
| SLE24 | 33 | 30 | 3 | 9 | 0 (36) |
| SLE26 | 35 | 26 | 9 | 26 | 0 (36) |
| SLE27 | 35 | 29 | 6 | 17 | 0 (36) |
|
| |||||
| Controls | 231 | 229 | 2 | 1 | 0 (72) |
| CNTL05 | 32 | 32 | 0 | 0 | 0 (36) |
| CNTL08 | 32 | 32 | 0 | 0 | - |
| CNTL12 | 32 | 32 | 0 | 0 | - |
| CNTL20 | 35 | 33 | 2 | 6 | 0 (36) |
| CNTL29 | 33 | 33 | 0 | 0 | - |
| CNTL34 | 33 | 33 | 0 | 0 | - |
| CNTL71 | 34 | 34 | 0 | 0 | - |
The mutant FAS cDNA clones (clones with adenine insertion mutation) was significantly increased in SLE patient group than in the non-SLE controls (2 × 2 contingency table, χ2 = 20.82, p < 0.001, Odds ratio = 14.84).
To determine whether the mutation resided in genomic DNA (somatic gene mutation or inherited polymorphism) or was introduced on the level of mRNA (transcriptional mRNA editing), we cloned the FAS genomic DNA fragment flanking the mutation site and sequenced a total of 216 FAS genomic DNA clones from seven donors (six of them carrying the nucleotide insertion mutation in their FAS cDNA) (Table 1). None of 216 FAS genomic DNA clones carried the same nucleotide insertion mutation observed in their cDNAs, suggesting that the adenine insertion mutation occurs at the transcription (or on mRNA) level through an mRNA editing process.
RNA editing by nucleotide insertion is unique for human FAS
To investigate whether the nucleotide insertion editing event is a generalized phenomenon for TNFR family members, we cloned cDNAs of three other TNFR family members. At least 143 cDNA clones from multiple SLE patients were sequenced for each gene. Nevertheless, we failed to detect any nucleotide insertion editing event in human death receptor 5 (DR5), death receptor 6 (DR6), and TNF receptor 1 (TNFR1) genes. Our data suggest that the nucleotide insertion type of editing mutation is unique for human FAS gene.
Because murine FAS mRNA also contains a homologous region corresponding to the FAS editing site (string of six adenines within its coding region) (Supp. Figure S1), we also examined whether a similar type of FAS mRNA editing occurs in murine T cells. We failed to detect the nucleotide insertion mutation in murine FAS by sequencing 67 murine FAS cDNA clones from murine T cells stimulated with anti-murine CD3 mAb. Furthermore, we failed to detect nucleotide insertion mutation in 71 murine FAS cDNA clones from primary murine T cells (Table 2). Collectively, our data demonstrate the differences between murine FAS and human FAS in their ability to generate new products through mRNA editing mutation.
Table 2.
Distribution of adenine insertion FAS cDNA clones in human and murine T cells
| Cells | Edited clones/Total clones | Editing (%) |
|---|---|---|
| Untreated Jurkat T cells | 2/34 | 6% |
| TCR Stimulated Jurkat T cells | 8/34 | 24% |
| Primary murine T cells | 0/71 | 0% |
| TCR Stimulated murine T cells | 0/67 | 0% |
Detection of edFAS protein
The insertion of adenine causes a FAS reading frame shift, which predicts the production of a truncated FAS protein that truncates the death domain (Fig. 1B) (Huang, et al., 1996). As shown in Fig. 1B, the new reading-frame in FAS would result in the production of a mutant FAS (or edFAS) that has a unique carboxyl terminus (SQSLYSCRENSDYHPQGHY). To confirm the expression of edFAS protein, we generated rabbit polyclonal antibodies against the edFAS polypeptide (unique 19-residue polypeptide) that is absent in the wild-type FAS (wtFAS) protein. The specificity of the rabbit anti edFAS antibodies was verified using the 293 cell lines stably expressing either the edFAS or wtFAS. As shown in Fig. 1C, the affinity-purified rabbit anti edFAS antibodies reacted with a protein band (with the molecular weight around 42 kDa) from the 293 cells stably expressing edFAS (upper left panel). On the other hand, the rabbit antibodies against the edFAS failed to react with the wild-type FAS (wtFAS) proteins (lower left panel, Fig. 1C). The same protein band also reacted with the rabbit antibodies against the FAS extracellular domains, confirming the rabbit anti edFAS antibodies were specific for the edFAS protein. Subsequently, we examined the expression of edFAS protein in mixed mononuclear cells from SLE patients and normal control donors. As shown in upper panel of Fig. 1C, the rabbit anti edFAS antibodies were able to recognize an immunoprecipitated FAS protein band from the mixed mononuclear cells of SLE patients (upper right panel). The same protein band also reacted with anti human FAS antibodies against the extracellular domain of FAS protein (lower right panel, Fig. 1C), verifying the existence of the edFAS protein in human cells. The edFAS protein was obvious in samples from two SLE patients (upper panel, Fig. 1C). On the other hand, almost no edFAS protein was visible in cells from four normal control donors (upper panel, Fig. 1C). Our data suggest that the edFAS protein was mainly produced in cells of SLE patients.
The edFAS is a membrane protein
Because the FAS mRNA mutation only affects the FAS cytoplasmic domain, we speculated that the edFAS may be expressed as a surface receptor, similar to the wild-type FAS (wtFAS). Indeed, as shown in Fig. 2A, both edFAS and wtFAS were detected on the surfaces of 293 cells in a flow cytometry assay, suggesting that the edFAS is a membrane receptor. Immunofluorescence microscopy also confirmed that edFAS is expressed on the cell membrane (Supp. Figure S2).
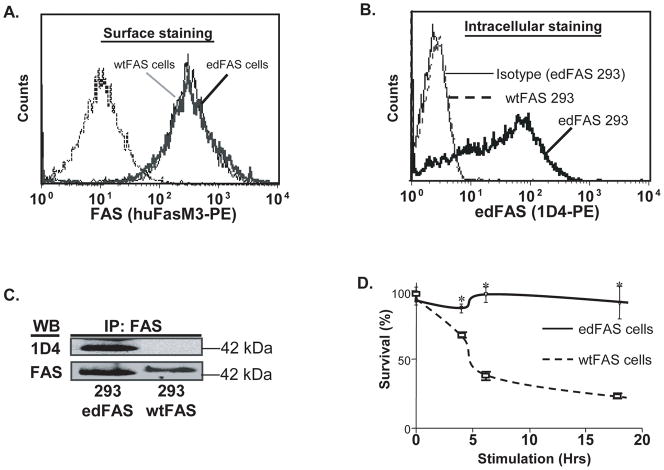
Fig. 2.
The edFAS is deficient in FAS-mediated apoptosis. A). Similar levels of edFAS and wtFAS were expressed on 293 cell surface as detected by the anti-FAS mAb. B). Intracellular flow cytometry assay shows that anti-edFAS mAb (1D4) reacted with edFAS but not with wtFAS expressed in 293 cells. C). The edFAS protein expressed on 293 cells (293 edFAS) could also be detected by anti edFAS mAb (1D4) in a Western blotting assay. No edFAS was detected in 293 cells expressing the wtFAS (293 wtFAS). D). Defective FAS-mediated cell death in 293 cells expressing edFAS. Cell viability was determined with ATPLite Luminescent ATP detection assay. Significant cell death was observed in 293 cells expressing wild-type FAS (wtFAS) after cells were treated with SuperFAS ligand (20 ng/ml) for 4, 6 and 18 hours. No significant cell death occurred in 293 cells expressing edFAS with the same treatment. All experiments have been repeated three times.
Generation and characterization of monoclonal antibodies specific for the edFAS
We obtained four murine hybridoma cell lines (clone# 1A4, 1D4, 1B3, and 4C6) producing monoclonal antibodies against the unique edFAS polypeptide (SQSLYSCRENSDYHPQGHY). Since the differences between the edFAS and wtFAS are within the receptor cytoplasmic tail (Fig. 1B), we developed an intracellular flow cytometry assay for the detection of edFAS. As shown in Fig. 2B, the anti edFAS mAb (clone 1D4) was able to detect the edFAS in 293 cells. On the other hand, the mAb (clone 1D4) failed to react with wtFAS protein in 293 cells. In Western blotting assays, the mAb (1D4) failed to react with the wtFAS protein (upper panel, Fig. 2C). Then again, the mAb (1D4) was able to detect the edFAS protein (upper panel, Fig. 2C). Therefore, mAb 1D4 was specific for edFAS protein. Similar results were obtained with the monoclonal antibodies produced by three other hybridoma lines (1A4, 1B3, and 4C6).
The edFAS is deficient in FAS-mediated apoptosis
Mutations in FAS death domain frequently affect the FAS-mediated apoptosis (Rieux-Laucat, et al., 1995). Since the edFAS protein contains a truncated FAS death domain (Fig. 1B), we speculated that the edFAS is defective in mediating death signal. We used the 293 cell lines stably expressing equivalent amount of the edFAS or wtFAS (Fig. 2A). As shown in Fig. 2D, treatment with SuperFAS Ligand failed to induce apoptosis in the 293 cells expressing the edFAS, suggesting that edFAS with the truncated death domain is unable to mediate death signal. In contrast, treatment with the SuperFAS Ligand induced significant apoptosis in the 293 cells expressing wild type FAS (wtFAS), confirming that the SuperFAS Ligand is biologically active in FAS-mediated cell death. Similarly, treatment with anti-FAS antibody (activating mAb, clone CH-11) failed to induce apoptosis in the 293 cells expressing edFAS while the same treatment induced rapid apoptosis in the 293 cells expressing the wtFAS (data not shown). These data demonstrate that the edFAS cannot trigger FAS-mediated apoptosis in human cells.
Association of FAS mRNA editing with human SLE
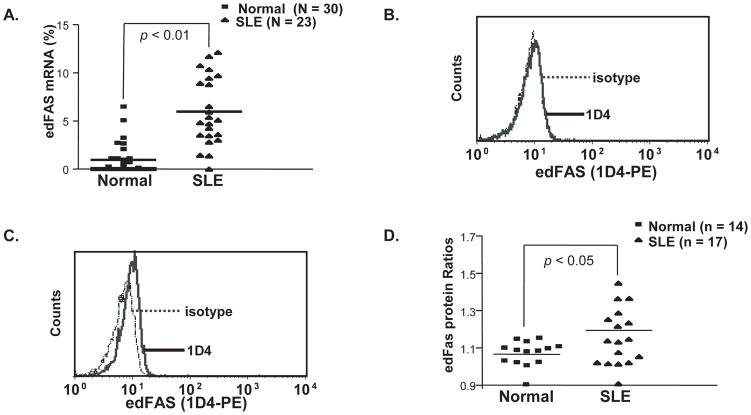
Altered peripheral tolerance is a general feature in autoimmune diseases. To investigate whether FAS mRNA editing mutation is associated with human SLE, we compared the levels of FAS mRNA editing in SLE patients with those in normal control donors. As shown in Table 1, we observed that the total number of mutant FAS cDNA clones (28 out of 216) was significantly higher in SLE patient group than in normal control group (2 out of 229) (χ2 = 20.82, p < 0.001). Additionally, as shown in Fig. 3A, the ratios of mutant FAS cDNAs determined in the Pyrosequencing assay were significantly higher in SLE patients than in the normal controls. Our data indicate the FAS mRNA editing mutation is strongly associated with human SLE.
Fig. 3.
Increased FAS editing mutation products in SLE patients. A). Quantification of edFAS mRNA in SLE patients and normal controls. The percentages of edFAS mRNA relative to the total FAS mRNA were calculated as described in “Materials and Methods”. The percentages of edFAS mRNA were significantly increased in SLE patient groups compared to normal control group (P < 0.01). B). The edFAS protein was undetectable in CD3+ T cells from a normal donor in intracellular flow cytometry using anti edFAS mAb (1D4). C). The edFAS was detected in CD3+ T cells from an SLE patient. D). Different edFAS expression in CD3+ T cells between SLE patients and normal controls. The edFAS protein ratios were calculated as the MFI (mean fluorescence intensity) of anti edFAS mAb (1D4) over the MFI of the mIgG1 isotype control. The edFAS protein production was significantly enhanced in SLE patient groups compared to normal control group (P < 0.05).
Since the difference between the edFAS and wtFAS is in the cytoplasmic tail (Fig. 1B), we used an intracellular flow cytometry assay to determine the levels of edFAS in various cell populations (T cells, B cells, monocytes, and neutrophils). Our data show that the edFAS was mainly expressed in human T cells. As shown in Fig. 3C, we were able to detect edFAS expression in SLE patient CD3+ T cells. On the other hand, almost no edFAS protein could be detected in the CD3+ T cells from a typical normal control donor (Fig. 3B). CD3+ T cells from SLE patients expressed significantly more edFAS protein than those from normal controls (Wilcoxon Signed Rank Test, P < 0.05) (Fig. 3D). We also observed that both CD4+ and CD8+ T cells express edFAS in SLE patients (Supp. Figure S3).
The edFAS inhibits FAS-mediated apoptosis in human T cells
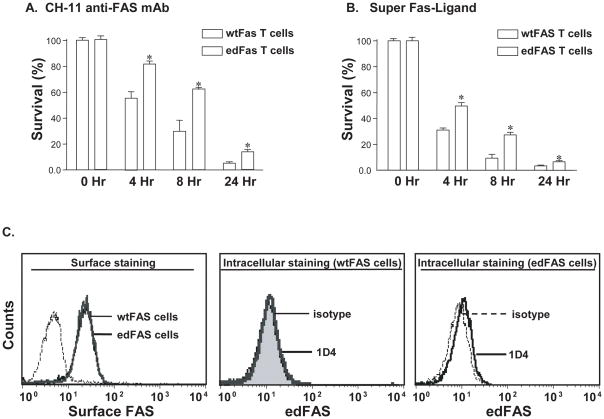
Since the edFAS was mainly expressed in human T cells, we examined the influence of edFAS in FAS-mediated apoptosis in human Jurkat T cells. Jurkat T cells endogenously express high levels of FAS (data not shown). We transfected Jurkat T cells with edFAS or the wild-type FAS (wtFAS) expression constructs. The Jurkat T cells were selected with G-418. Fig. 4C (left panel) shows that FAS expression levels were almost the same between the cells transfected with edFAS and wtFAS. The stable Jurkat T cells transfected with edFAS expressed the detectable amount of the edFAS protein (right panel, Fig. 4C). In contrast, no edFAS protein could be detected in the T cells transfected with wtFAS (middle panel, Fig. 4C). Treatment with anti human FAS antibody (activating mAb, clone CH-11) induced significantly less apoptosis in the Jurkat T cells expressing the edFAS as compared to the T cells transfected with wtFAS, suggesting that edFAS is capable of attenuating the endogenous FAS-mediated apoptosis in T cells (Fig. 4A). Similarly, treatment with SuperFAS ligand induced significantly less apoptosis in the Jurkat T cells expressing edFAS as compared to the T cells expressing wtFAS (Fig. 4B). Collectively, our data demonstrate that the generation of edFAS inhibits the FAS-mediated apoptosis in T cells.
Fig. 4.
edFAS inhibits FAS-mediated apoptosis in T cells. Jurkat T cells, stably expressing FAS constructs, were established by transfection with either the edFAS/pcDNA3 expression construct (edFAS T cells) or the wild-type FAS/pcDNA3 expression construct (wtFAS T cells). Cell viability was determined as described in the section of “Materials and Methods”. All experiments have been repeated at least three times. A). Jurkat T cells were treated with anti FAS mAb (2 μg/ml of CH-11) for 4, 8, and 18 hours. Bars (means ± S.D. of three experiments performed in triplicates) show the percentages of surviving cells after anti FAS antibody treatment. T cells expressing edFAS had significantly higher survival rates than those of the cells expressing only the wtFAS at respective time points (*, P < 0.05). B). Stable T cells expressing edFAS were treated with SuperFAS Ligand (20 ng/ml) for 4, 8, and 18 hours and expression of edFAS significantly inhibited SuperFAS Ligand-induced cell death in Jurkat T cells at respective time points (*, P < 0.05). C). Surface FAS levels detected with extracellular staining were comparable between cells tranfected with edFAS and those with wtFAS (left panel). The edFAS was undetectable in T cells transfected with wtFAS by anti edFAS mAb (1D4) (middle panel). The edFAS expression was detected in T cells transfected with edFAS (right panel).
T cell receptor (TCR) engagement up-regulates the FAS mRNA editing
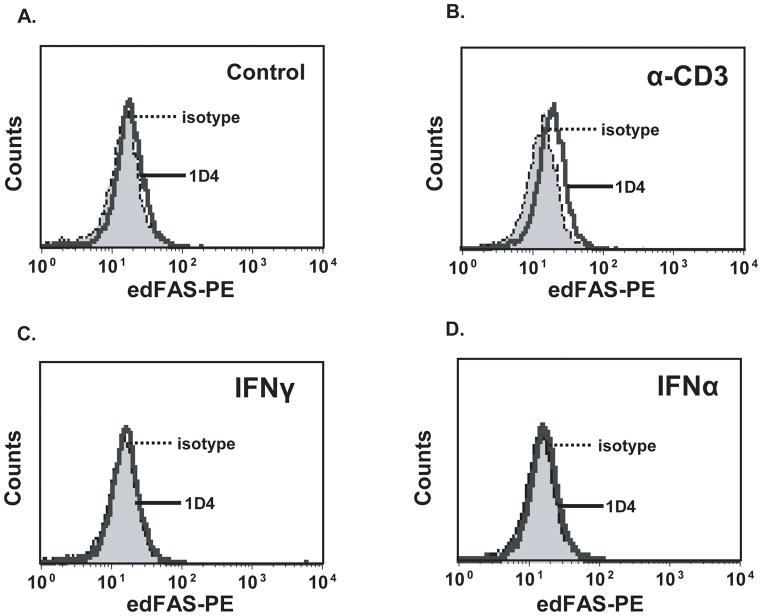
Our earlier data demonstrated that the edFAS protein was mainly detected in human T cells. To investigate the role of cell activation in FAS mRNA editing in T cells, we stimulated human Jurkat T cells with IFNγ, IFNα, and anti CD3 mAb. As shown in Fig. 5, treatment with either IFNγ (Fig. 5C) or IFNα (Fig. 5D) failed to induce the edFAS production. On the other hand, treatment with anti CD3 mAb significantly increased the edFAS production in T cells (Fig. 5B) compared to the non-treated cells (Fig. 5A). To verify the effect of TCR stimulation on FAS mRNA editing, we cloned and sequenced FAS cDNAs from T cells. Table 2 shows that the untreated Jurkat T cells maintain a low level of constitutive FAS mRNA editing (6%). However, after cross-linking of TCR with anti-TCR antibody (OKT3) for 48 hours, Jurkat T cells significantly increased the FAS mRNA editing. The percentage of the mutant FAS cDNA clones accounted for 24% of total FAS cDNA clones (Table 2). Our data suggest that persistent TCR engagement is an important signal leading to the FAS mRNA editing in human T cells. Our data imply that the FAS mRNA editing is a regulated process.
Fig. 5.
Up-regulation of FAS mRNA editing after TCR engagement. Jurkat T cells were treated with anti CD3 mAb (5 μg/ml), or IFNγ (100 U/ml), or IFNα (100 U/ml) for 48 hours. A). Very little edFAS could be detected in non-treated T cell. B). Treatment with anti CD3 mAb for 48 hours significantly increased the edFAS production in T cells. C). Treatment with IFNγ for 48 hours failed to increase the edFAS protein production in T cells. D). Treatment with IFNα for 48 hours did not increase the edFAS protein production in T cells. All experiments were repeated at least three times.
Discussion
Activation-induced cell death (AICD) is a major mechanism to maintain immune homeostasis. AICD occurs in mature T lymphocytes to limit antigen-specific response (Stranges, et al., 2007). After the clearance of antigens and/or pathogens from the host, activated T cells are deleted mainly via the FAS-dependent apoptosis (Alderson, et al., 1995; Brunner, et al., 1995; Dhein, et al., 1995; Zhang, et al., 1999). The apoptosis is initiated by FAS engagement, which triggers the death-inducing signaling complex formation by recruiting FAS-associated death domain (FADD), caspase 8, and caspase 10 proteins. The assembled molecular complex cleaves and activates several downstream proteins in a cascade that leads to apoptotic cell death characterized by structural changes such as DNA fragmentation and chromatin condensation.
The inherited loss-of-function mutations in FAS are directly associated with the human autoimmune lymphoproliferative syndrome (Rieux-Laucat, et al., 1999; Sneller, et al., 1997). The significance of FAS-mediated apoptosis in the maintenance of immune tolerance and prevention of autoimmune disease has been illustrated in animal models as well. The mice carrying mutations in FAS (lpr/lpr mice) or FASL (gld/gld mice) spontaneously develop various SLE-like features including lymphadenopathy, vasculitis, glomerulonephritis, and autoantibody production (Takahashi, et al., 1994; Watanabe-Fukunaga, et al., 1992a). Although rare FAS or FASL mutations have been documented in human SLE (Vaishnaw, et al., 1999; Wu, et al., 1996), no obvious qualitative or quantitative defect due to FAS mutations has been identified in most SLE patients. Therefore, the role of FAS and FAS-mediated apoptosis in the pathogenesis of SLE is not clear.
However, several lines of evidence indicate that FAS may be involved in the defective apoptosis of T cells in SLE. It has been shown that T-cells from SLE patients were resistant to the FAS-mediated apoptosis (Budagyan, et al., 1998). Similarly, CD3-mediated cell death is significantly lower in T cells from SLE patients compared to the cells from normal controls. CD3-mediated AICD could be blocked by anti-FAS mAb, indicating that activated T cells from patients with SLE are relatively resistant to FAS-mediated apoptosis (Kovacs, et al., 1996). Furthermore, T cells from SLE patients have an alteration in the FAS-mediated signal transduction pathway, which leads to the survival of T-lymphocytes in SLE patients (Sakata, et al., 1998). Collectively, these published evidences support the notion that the deficiency in FAS-mediated apoptosis may contribute to the increased numbers of activated autoreactive cells in lupus patients.
In the current study, we uncovered a novel FAS mRNA mutation that occurs mainly in SLE patients. We demonstrate that the distinctive FAS frameshift mutation was a result of FAS mRNA editing. In addition, the FAS mRNA editing mutation leads to the production of a novel defective FAS receptor (edFAS). Most importantly, we observed that the FAS mRNA editing products were significantly increased in SLE patients compared to those in the normal controls. Therefore, FAS mRNA editing mutation may be one of the key mechanisms underlying the abnormal FAS functions in lupus T cells. Because the edFAS with truncated death domain is unable to mediate apoptosis, the expression of edFAS protein may be responsible for the defective apoptosis of autoreactive T lymphocytes observed in SLE patients (Budagyan, et al., 1998; Kovacs, et al., 1996; Sakata, et al., 1998). Consequently, our data support a role for FAS mRNA editing mutation in the pathogenesis of SLE and in the development of autoimmunity.
A single human gene can produce a variety of protein products with related yet distinct functions, which may allow cells of different types or at different developmental stages to perform different tasks. Alternative splicing of pre-mRNAs and alternative polyadenylation site selection are two established forms of intracellular fine-tuning of mRNA as both processes generate diverse gene products from a single gene. Recently, it has become clear that human cells possess yet another mechanism, mRNA editing, to create RNA sequence diversity. Messenger RNA editing is a process defined as the post-transcriptional modifications of mRNA through nucleotide substitutions, insertions, and deletions. Transcript alterations caused by mRNA editing have been described in organisms from unicellular protozoa to humans (Gott and Emeson, 2000). However, aberrant mRNA editing could cause the cells to synthesize dysfunctional proteins or to make an otherwise useful protein at the wrong time. Therefore, the mRNA editing process is tightly regulated. Nevertheless, abnormal mRNA editing is frequently associated with human diseases. For example, the NF1 (Neurofibromatosis type I, a tumor suppressor) mRNA editing results in a truncated NF1 protein lacking tumor-suppressor function (Skuse, et al., 1996). In individuals with a constitutive NF1 mutation, loss of heterozygosity (LOH) that exposes the single mutant allele would lead to tumor formation (Cappione, et al., 1997). The mRNA editing mutations are also associated with some forms of Alzheimer’s disease (van Leeuwen, et al., 1998). Furthermore, the abnormal editing of glutamate receptor GluR-B mRNA is implicated in human malignant gliomas (Maas, et al., 2001).
Our data indicate that FAS mRNA mutation is caused by a site-specific editing process, in which an adenine is inserted at a defined location. In addition, our data suggest that the nucleotide insertion mutation is a FAS gene specific event because no similar mutations were found in other TNFR family members such as DR5, DR6, and TNFR1. Most importantly, FAS mRNA editing results in the production of a defective FAS receptor, which is unable to mediate apoptosis signal. To our knowledge, our current study is the first to report the adenine insertion type of mRNA editing in a human gene. Although the nucleotide sequence of murine FAS cDNA has a high sequence identity (58.5%) to that of human FAS cDNA (Watanabe-Fukunaga, et al., 1992b), we failed to detect the similar type of editing in murine FAS (Table 2). Therefore, it appears as that the adenine nucleotide insertion editing is a site-, gene-, and species-specific event for human FAS (Supp. Figure S1).
We speculate that FAS mRNA editing may be a feedback response to protect cells from ongoing FAS-mediated AICD in the activated T cells since the persistent TCR engagement enhances the FAS mRNA editing. The emergence of edFAS may interfere with the FAS-mediated apoptosis signal through the alteration of AICD threshold in the activated T cells. Therefore, the initiation of FAS mRNA editing may be a critical step in the breakdown of the peripheral immune tolerance. Accordingly, it will be of great interest to investigate whether the FAS mRNA editing plays a universal role in the pathogenesis of various autoimmune or inflammatory diseases.
Although the adenine insertion type of mRNA editing has never been reported with human genes, the adenine insertion mRNA editing has been described in Ebola virus (EBOV) (Volchkov, et al., 2001). The editing site, which consists of seven consecutive adenine residues (AAAAAAA), resembling the EBOV polyadenylation signal (poly(A) signal). Similar to EBOV GP, the human FAS mRNA editing site contains six consecutive adenine residues (AAAAAA) (Fig. 1A). Genes encoding polyadenylated mRNAs depend on their poly(A) signal for the termination of transcription. The poly(A) signal could direct RNA polymerase II to pause and to release partially from the template (Orozco, et al., 2002). Consequently, slippage of the gene transcriptional complex on the RNA template may cause insertion of extra nucleotide. Therefore, we speculate that human FAS uses the poly(A) signal-controlled slippage of the gene transcription complex to generate the edited mRNA products. However, further experiments are required to define the mechanism of human FAS mRNA editing.
Supplementary Material
Acknowledgments
This work was supported by Lupus Research Institute grant (J.W.), National Institutes of Health grants R01-AR33062 (R.P.K.), P60-AR20614 (R.P.K.), P30-AR48311 (R.P.K.), and P01-AR49084 (R.P.K.). The FACS Core Facility of the UAB Arthritis, Musculoskeletal and Autoimmunity Center was supported by Rheumatic Diseases Core Center (P30-AR48311). We thank Dr. Mary Ann Accavitti at UAB Epitome Recognition and Immunoreagent Core (ERIC) Facility for the production and purification of monoclonal antibodies.
Footnotes
Contribution: J.W. and R.P.K. designed experiments, interpreteddata, and wrote the paper. F.X. and K.Q. carried out RT-PCR, PCR, cloning, DNA sequencing, cell viability assay, Western blotting, and flow cytometry. A.W.G. and J.C.E. participated in experiment design and data analysis.
The authors declare that they have no competing financial interest
Supporting Information for this preprint is available from the Human Mutation editorial office upon request (humu@wiley.com)
References
- Alderson MR, Tough TW, Davis-Smith T, Braddy S, Falk B, Schooley KA, Goodwin RG, Smith CA, Ramsdell F, Lynch DH. FAS ligand mediates activation-induced cell death in human T lymphocytes. J Exp Med. 1995;181(1):71–7. doi: 10.1084/jem.181.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashany D, Song X, Lacy E, Nikolic-Zugic J, Friedman SM, Elkon KB. Th1 CD4+ lymphocytes delete activated macrophages through the FAS/APO-1 antigen pathway. Proc Natl Acad Sci U S A. 1995;92(24):11225–9. doi: 10.1073/pnas.92.24.11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner T, Mogil RJ, LaFace D, Yoo NJ, Mahboubi A, Echeverri F, Martin SJ, Force WR, Lynch DH, Ware CF, et al. Cell-autonomous FAS (CD95)/FAS-ligand interaction mediates activation- induced apoptosis in T-cell hybridomas [see comments] Nature. 1995;373(6513):441–4. doi: 10.1038/373441a0. [DOI] [PubMed] [Google Scholar]
- Budagyan VM, Bulanova EG, Sharova NI, Nikonova MF, Stanislav ML, Yarylin AA. The resistance of activated T-cells from SLE patients to apoptosis induced by human thymic stromal cells. Immunol Lett. 1998;60(1):1–5. doi: 10.1016/s0165-2478(97)00128-4. [DOI] [PubMed] [Google Scholar]
- Cappione AJ, French BL, Skuse GR. A potential role for NF1 mRNA editing in the pathogenesis of NF1 tumors. Am J Hum Genet. 1997;60(2):305–12. [PMC free article] [PubMed] [Google Scholar]
- Dhein J, Walczak H, Baumler C, Debatin KM, Krammer PH. Autocrine T-cell suicide mediated by APO-1/(FAS/CD95) Nature. 1995;373(6513):438–41. doi: 10.1038/373438a0. [DOI] [PubMed] [Google Scholar]
- Drappa J, Vaishnaw AK, Sullivan KE, Chu JL, Elkon KB. FAS gene mutations in the Canale-Smith syndrome, an inherited lymphoproliferative disorder associated with autoimmunity [see comments] N Engl J Med. 1996;335(22):1643–9. doi: 10.1056/NEJM199611283352204. [DOI] [PubMed] [Google Scholar]
- Elkon KB, Marshak-Rothstein A. B cells in systemic autoimmune disease: recent insights from FAS- deficient mice and men. Curr Opin Immunol. 1996;8(6):852–9. doi: 10.1016/s0952-7915(96)80015-x. [DOI] [PubMed] [Google Scholar]
- Fisher GH, Rosenberg FJ, Straus SE, Dale JK, Middleton LA, Lin AY, Strober W, Lenardo MJ, Puck JM. Dominant interfering FAS gene mutations impair apoptosis in a human autoimmune lymphoproliferative syndrome. Cell. 1995;81(6):935–46. doi: 10.1016/0092-8674(95)90013-6. [DOI] [PubMed] [Google Scholar]
- Fukuyama H, Adachi M, Suematsu S, Miwa K, Suda T, Yoshida N, Nagata S. Transgenic expression of FAS in T cells blocks lymphoproliferation but not autoimmune disease in MRL-lpr mice. J Immunol. 1998;160(8):3805–11. [PubMed] [Google Scholar]
- Fukuyama H, Adachi M, Suematsu S, Miwa K, Suda T, Yoshida N, Nagata S. Requirement of FAS expression in B cells for tolerance induction. Eur J Immunol. 2002;32(1):223–30. doi: 10.1002/1521-4141(200201)32:1<223::AID-IMMU223>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Gott JM, Emeson RB. Functions and mechanisms of RNA editing. Annu Rev Genet. 2000;34:499–531. doi: 10.1146/annurev.genet.34.1.499. [DOI] [PubMed] [Google Scholar]
- Huang B, Eberstadt M, Olejniczak ET, Meadows RP, Fesik SW. NMR structure and mutagenesis of the FAS (APO-1/CD95) death domain. Nature. 1996;384(6610):638–41. doi: 10.1038/384638a0. [DOI] [PubMed] [Google Scholar]
- Kotzin BL. Systemic lupus erythematosus. Cell. 1996;85(3):303–6. doi: 10.1016/s0092-8674(00)81108-3. [DOI] [PubMed] [Google Scholar]
- Kovacs B, Vassilopoulos D, Vogelgesang SA, Tsokos GC. Defective CD3-mediated cell death in activated T cells from patients with systemic lupus erythematosus: role of decreased intracellular TNF- alpha. Clin Immunol Immunopathol. 1996;81(3):293–302. doi: 10.1006/clin.1996.0192. [DOI] [PubMed] [Google Scholar]
- Leithauser F, Dhein J, Mechtersheimer G, Koretz K, Bruderlein S, Henne C, Schmidt A, Debatin KM, Krammer PH, Moller P. Constitutive and induced expression of APO-1, a new member of the nerve growth factor/tumor necrosis factor receptor superfamily, in normal and neoplastic cells. Lab Invest. 1993;69(4):415–29. [PubMed] [Google Scholar]
- Maas S, Patt S, Schrey M, Rich A. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc Natl Acad Sci U S A. 2001;98(25):14687–92. doi: 10.1073/pnas.251531398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco IJ, Kim SJ, Martinson HG. The poly(A) signal, without the assistance of any downstream element, directs RNA polymerase II to pause in vivo and then to release stochastically from the template. J Biol Chem. 2002;277(45):42899–911. doi: 10.1074/jbc.M207415200. [DOI] [PubMed] [Google Scholar]
- Ramaswamy M, Siegel RM. A FAScinating receptor in self-tolerance. Immunity. 2007;26(5):545–7. doi: 10.1016/j.immuni.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Rieux-Laucat F, Blachere S, Danielan S, De Villartay JP, Oleastro M, Solary E, Bader-Meunier B, Arkwright P, Pondare C, Bernaudin F, et al. Lymphoproliferative syndrome with autoimmunity: A possible genetic basis for dominant expression of the clinical manifestations. Blood. 1999;94(8):2575–82. [PubMed] [Google Scholar]
- Rieux-Laucat F, Le Deist F, Hivroz C, Roberts IA, Debatin KM, Fischer A, de Villartay JP. Mutations in FAS associated with human lymphoproliferative syndrome and autoimmunity. Science. 1995;268(5215):1347–9. doi: 10.1126/science.7539157. [DOI] [PubMed] [Google Scholar]
- Sakata K, Sakata A, Vela-Roch N, Espinosa R, Escalante A, Kong L, Nakabayashi T, Cheng J, Talal N, Dang H. FAS (CD95)-transduced signal preferentially stimulates lupus peripheral T lymphocytes. Eur J Immunol. 1998;28(9):2648–60. doi: 10.1002/(SICI)1521-4141(199809)28:09<2648::AID-IMMU2648>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Singer GG, Abbas AK. The FAS antigen is involved in peripheral but not thymic deletion of T lymphocytes in T cell receptor transgenic mice. Immunity. 1994;1(5):365–71. doi: 10.1016/1074-7613(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Skuse GR, Cappione AJ, Sowden M, Metheny LJ, Smith HC. The neurofibromatosis type I messenger RNA undergoes base-modification RNA editing. Nucleic Acids Res. 1996;24(3):478–85. doi: 10.1093/nar/24.3.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneller MC, Straus SE, Jaffe ES, Jaffe JS, Fleisher TA, Stetler-Stevenson M, Strober W. A novel lymphoproliferative/autoimmune syndrome resembling murine lpr/gld disease. J Clin Invest. 1992;90(2):334–41. doi: 10.1172/JCI115867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneller MC, Wang J, Dale JK, Strober W, Middelton LA, Choi Y, Fleisher TA, Lim MS, Jaffe ES, Puck JM, et al. Clincial, immunologic, and genetic features of an autoimmune lymphoproliferative syndrome associated with abnormal lymphocyte apoptosis. Blood. 1997;89(4):1341–8. [PubMed] [Google Scholar]
- Stranges PB, Watson J, Cooper CJ, Choisy-Rossi CM, Stonebraker AC, Beighton RA, Hartig H, Sundberg JP, Servick S, Kaufmann G, et al. Elimination of antigen-presenting cells and autoreactive T cells by FAS contributes to prevention of autoimmunity. Immunity. 2007;26(5):629–41. doi: 10.1016/j.immuni.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Tanaka M, Brannan CI, Jenkins NA, Copeland NG, Suda T, Nagata S. Generalized lymphoproliferative disease in mice, caused by a point mutation in the FAS ligand. Cell. 1994;76(6):969–76. doi: 10.1016/0092-8674(94)90375-1. [DOI] [PubMed] [Google Scholar]
- Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, Schaller JG, Talal N, Winchester RJ. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25(11):1271–7. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- Vaishnaw AK, Toubi E, Ohsako S, Drappa J, Buys S, Estrada J, Sitarz A, Zemel L, Chu JL, Elkon KB. The spectrum of apoptotic defects and clinical manifestations, including systemic lupus erythematosus, in humans with CD95 (FAS/APO-1) mutations. Arthritis Rheum. 1999;42(9):1833–42. doi: 10.1002/1529-0131(199909)42:9<1833::AID-ANR7>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- van Leeuwen FW, de Kleijn DP, van den Hurk HH, Neubauer A, Sonnemans MA, Sluijs JA, Koycu S, Ramdjielal RD, Salehi A, Martens GJ, et al. Frameshift mutants of beta amyloid precursor protein and ubiquitin-B in Alzheimer’s and Down patients. Science. 1998;279(5348):242–7. doi: 10.1126/science.279.5348.242. [DOI] [PubMed] [Google Scholar]
- Volchkov VE, Volchkova VA, Muhlberger E, Kolesnikova LV, Weik M, Dolnik O, Klenk HD. Recovery of infectious Ebola virus from complementary DNA: RNA editing of the GP gene and viral cytotoxicity. Science. 2001;291(5510):1965–9. doi: 10.1126/science.1057269. [DOI] [PubMed] [Google Scholar]
- Watanabe-Fukunaga R, Brannan CI, Copeland NG, Jenkins NA, Nagata S. Lymphoproliferation disorder in mice explained by defects in FAS antigen that mediates apoptosis. Nature. 1992a;356(6367):314–7. doi: 10.1038/356314a0. [DOI] [PubMed] [Google Scholar]
- Watanabe-Fukunaga R, Brannan CI, Itoh N, Yonehara S, Copeland NG, Jenkins NA, Nagata S. The cDNA structure, expression, and chromosomal assignment of the mouse FAS antigen. J Immunol. 1992b;148(4):1274–9. [PubMed] [Google Scholar]
- Wu J, Metz C, Xu X, Abe R, Gibson AW, Edberg JC, Cooke J, Cooper G, Kimberly RP. A Novel Polymorphic C/EBPbeta element in the FASL Gene Promoter Alters FASL Expression: A Candidate Background Gene for Autoimmunity. J Immunol. 2002 doi: 10.4049/jimmunol.170.1.132. [DOI] [PubMed] [Google Scholar]
- Wu J, Wilson J, He J, Xiang L, Schur PH, Mountz JD. FAS ligand mutation in a patient with systemic lupus erythematosus and lymphoproliferative disease. J Clin Invest. 1996;98(5):1107–13. doi: 10.1172/JCI118892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HG, Su X, Liu D, Liu W, Yang P, Wang Z, Edwards CK, Bluethmann H, Mountz JD, Zhou T. Induction of specific T cell tolerance by FAS ligand-expressing antigen-presenting cells. J Immunol. 1999;162(3):1423–30. [PubMed] [Google Scholar]
- Zipp F, Martin R, Lichtenfels R, Roth W, Dichgans J, Krammer PH, Weller M. Human autoreactive and foreign antigen-specific T cells resist apoptosis induced by soluble recombinant CD95 ligand. J Immunol. 1997;159(5):2108–15. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.