Abstract
Sjögren-Larsson syndrome (SLS) is an autosomal recessive disorder characterized by ichthyosis, mental retardation, spasticity and mutations in the ALDH3A2 gene for fatty aldehyde dehydrogenase, an enzyme that catalyzes the oxidation of fatty aldehyde to fatty acid. More than 70 mutations have been identified in SLS patients, including small deletions or insertions, missense mutations, splicing defects and complex nucleotide changes. We now describe 2 SLS patients whose disease is caused by large contiguous gene deletions of the ALDH3A2 locus on 17p11.2. The deletions were defined using long distance inverse PCR and microarray-based comparative genomic hybridization. A 24-year-old SLS female was homozygous for a 352-kb deletion involving ALDH3A2 and 4 contiguous genes including ALDH3A1, which codes for the major soluble protein in cornea. Although lacking corneal disease, she showed severe symptoms of SLS with uncommon deterioration in oral motor function and loss of ambulation. The other 19-month-old female patient was a compound heterozygote for a 1.44-Mb contiguous gene deletion and a missense mutation (c.407C>T, P136L) in ALDH3A2. These studies suggest that large gene deletions may account for up to 5% of the mutant alleles in SLS. Geneticists should consider the possibility of compound heterozygosity for large deletions in patients with SLS and other inborn errors of metabolism, which has implications for carrier testing and prenatal diagnosis.
Keywords: ALDH3A1, fatty aldehyde, ichthyosis, mutation, SLC47A1, ULK2
INTRODUCTION
Sjögren-Larsson syndrome (SLS; OMIM 270200) is an autosomal recessive inborn error of metabolism caused by mutations in the ALDH3A2 gene for fatty aldehyde dehydrogenase (FALDH) [1,2]. Clinical features of SLS include ichthyosis, spastic diplegia or tetraplegia, mental retardation, seizures and a distinctive retinal crystalline maculopathy characterized by perifoveal glistening white dots. The ichthyosis is usually congenital in onset and is often pruritic in nature. Neurological symptoms of mental retardation and spasticity typically develop by the 2nd year of life and present with delay in achieving motor and cognitive milestones. The symptoms of SLS vary from mild to profound, and are generally non-progressive.
FALDH catalyzes the oxidation of fatty aldehyde to fatty acid, and is a necessary component of the fatty alcohol:NAD+ oxidoreductase enzyme complex that catalyzes the sequential oxidation of fatty alcohol to fatty acid [3,4,5]. SLS patients consequently have elevated fatty alcohols in plasma, urine and cultured cells [6-9]. The symptoms of SLS are thought to arise directly or indirectly from accumulation of fatty aldehyde, fatty alcohol or related lipid products in the skin and brain [10].
The ALDH3A2 gene is located on chromosome 17p11.2. More than 70 mutations have been discovered in SLS patients, including small deletions or insertions, missense mutations, splicing defects and complex mutations composed of deletion/insertions and nucleotide substitutions [11]. Intragenic deletions of one or more exons have also been rarely described [12-14]. Most mutations in SLS are private, and many patients have been found to be homozygous due to consanguinity or founder effects.
We now present 2 unique SLS patients in whom the disease was caused by unusually large deletions involving ALDH3A2 and surrounding genes on chromosome 17p11.2.
MATERIALS AND METHODS
The Institutional Review Board at the University of Nebraska Medical Center approved this research, and all subjects consented to the study.
Patient Descriptions
Patient 1
This 24-year-old female was born at 32 weeks gestation to consanguineous first-cousin Pakistani parents. She was noted to have erythematous dry scaly skin at birth, but had no collodion membrane. She developed several seizures at 6 weeks of age, which were associated with hypocalcemia. Delays in achieving motor milestones and speech were noted in infancy. After 2 febrile illnesses at 6-7 months of age, she lost the ability to roll over and her limbs became very stiff. A brain CT at 9 months of age showed moderate cerebral atrophy. However, she began to sit unsupported and crawl at about 18 months. Physical examination at 3 years of age showed developmental delay, spastic diplegia and generalized ichthyosis. Speech consisted of occasionally saying “mama” and “baba”. Laboratory studies were normal, including karyotype, EEG, electroretinogram, thyroid function tests, plasma phytanic acid, urine amino acids and metabolic screen. Her ichthyosis responded well to etretinate therapy, which was subsequently switched to isotretinoin and then discontinued several years later because of concerns about retinoid toxicity. By 10 years of age, she was no longer speaking and began having difficulty swallowing. Physical exam showed a pruritic, generalized ichthyosis along with spastic diplegia, leg contractures and ankle clonus (Fig 1A). She had photophobia and avoided bright lights. She was able to ambulate only with a walker using a crouched gait. Brain MRI revealed bilateral, symmetrical abnormal T2-weighted signal involving the fronto-parietal and superior periventricular regions. Several months later she developed increased drooling with tongue protrusion and dysphagia. Oral motor incoordination and loss of swallowing slowly progressed and necessitated placement of a gastrostomy tube at 12 years of age. Bilateral hamstring lengthening was also performed at this age. At 14 years, she was noted to have spastic tetraplegia and be non-ambulatory. At 17 years of age, ophthalmologic examination under sedation showed chronic blepharitis, but no cataracts, retinal or corneal abnormalities. The diagnosis of SLS was confirmed by demonstrating FALDH deficiency (8% of mean normal activity) in cultured fibroblasts.
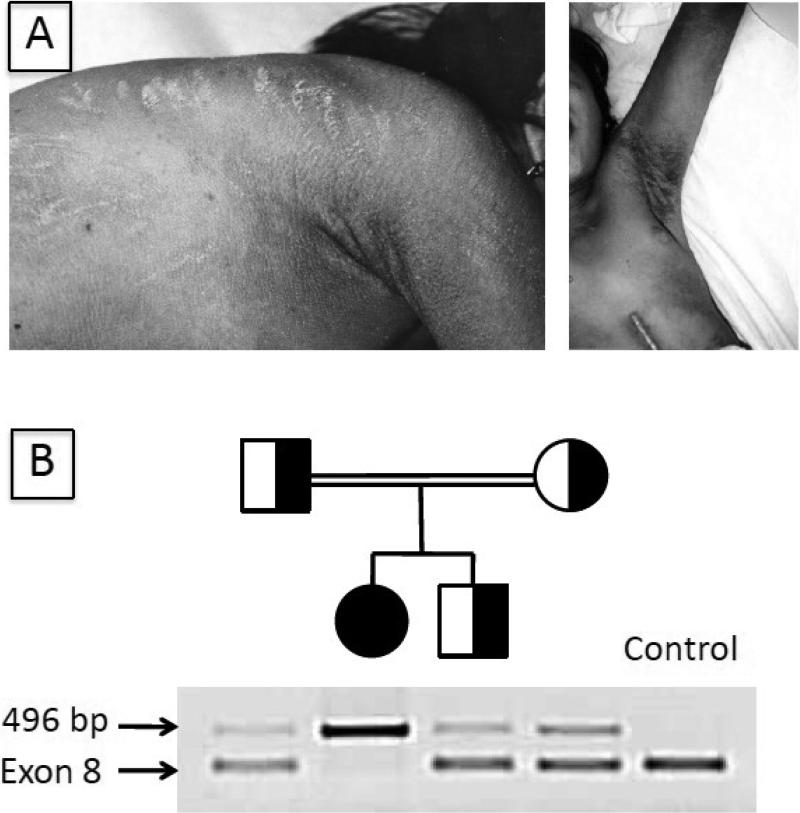
Fig 1.
Phenotypic appearance and mutation detection in Patient 1. A. Left photo: At 14 years of age, note the generalized pruritic ichthyosis with excoriations on the trunk. Right photo: Dark scales in the axillary region and an abdominal scar from gastrostomy tube placement are evident. B. Multiplex PCR amplification of the deletion mutation in genomic DNA generated a 496 PCR product from Patient 1, her mother and father, and heterozygous brother. A control PCR product (exon 8) was produced using DNA from the parents and heterozygous brother, but not from Patient 1.
Patient 2
This 19-month-old female infant was born at 36 weeks gestation to a 27-year-old mother and non-consanguineous father. Mother was of Irish and Cherokee Indian descent; father was English and American Indian. At birth, the infant was noted to have a collodion membrane with shiny, “tight” skin that transformed into dry, scaling skin after 2-3 weeks. Over time, she began scratching continually, often to the point of bleeding. At 10 months of age, a skin biopsy revealed hyperkeratosis, papillomatosis and acanthosis.
At 12 months of age, physical examination revealed height, weight, and head circumference within normal age limits. Significant findings included ichthyotic skin, ears adherent to the scalp bilaterally, and shortened fifth fingers bilaterally. She was noted to have spastic diplegia. Truncal hypotonia prevented her from sitting up for more than short periods of time. A Denver II Developmental Assessment found her to have social, language, fine motor, and gross motor abilities consistent with an age of 9, 7, 6, and 4 months old, respectively. Ophthalmologic examination revealed no ocular or visual abnormalities, including no evidence of macular glistening white dots. Her skin was dry with hyperkeratosis or scaling on the extremities, trunk, neck, palms, and soles (Fig 2D). The hyperkeratosis was most severe on the patient's legs, forearms, neck and trunk, where it was reminiscent of lamellar ichthyosis. Her hair and fingernails were normal. At 19 months of age, the patient suffered a tonic-clonic seizure, which subsequently recurred on several occasions.
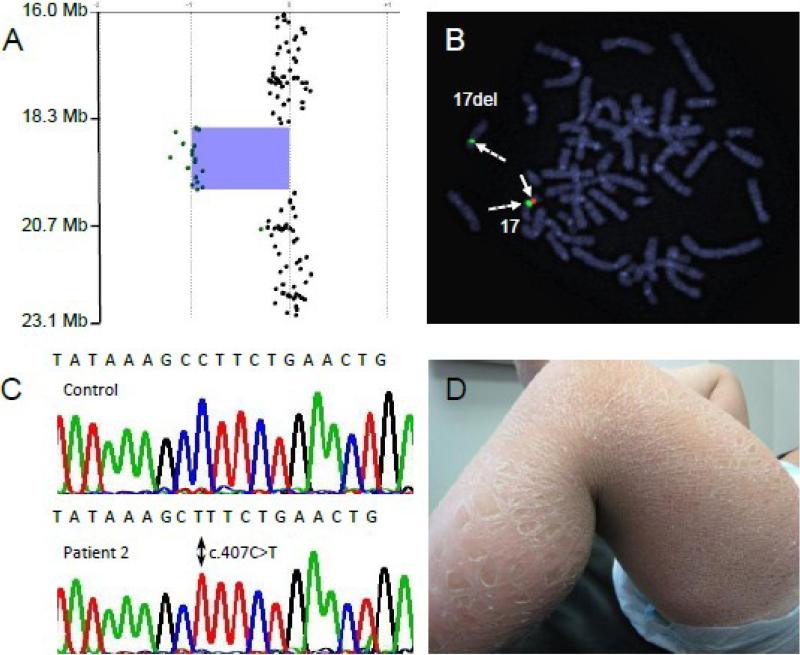
Fig 2.
Genetic analysis and phenotypic appearance of Patient 2. A: Microarray analysis of chromosome 17p11.2 showing heterozygous deletion. B: FISH using a chromosome 17-specific centromeric probe (green signal, dashed arrows) identifies both 17 chromosomes. The RP11-147i5 probe (red) corresponds to the deleted region and hybridizes to only the non-deleted chromosome 17 (solid arrow). C. DNA sequencing chromatogram demonstrates a hemizygous c.407C>T (p.P136L) mutation in exon 3 of the ALDH3A2 gene. D. Note the variable appearance of the ichthyosis on the leg of patient 2 with large lamellar-like scales on the lower leg and smaller scales on the thigh.
Fibroblast Culture
Cultured skin fibroblasts were grown from skin biopsies in Dulbecco's minimal essential medium containing 10% fetal bovine serum, penicillin, and streptomycin at 37°C in an atmosphere of 5% CO2. The cells were collected by trypsinization and washed twice with phosphate-buffered saline. Cell pellets were stored at -70°C for DNA isolation.
FALDH Assay
Fibroblasts were homogenized and assayed for FALDH activity using octadecanal as substrate as described [5], except that all reactions were run at 37° C in a 96-well fluorescent plate reader (Molecular Dynamics) in a total volume of 0.38 ml. Cell protein was determined according to Lowry et al [15]. Enzyme activity was determined as pmol/min/mg cell protein, and expressed as percentage of normal mean enzyme activity.
DNA Isolation
Genomic DNA was purified from cultured fibroblasts and blood using the Wizard Genomic DNA Purification kit (Promega). Buccal DNA was collected from controls and family members for PCR as described [16].
Deletion Characterization by Long Distance Inverse-PCR (LDI-PCR)
LDI-PCR was performed essentially as described by Willis et al [17]. A PstI restriction enzyme cut site was identified in the DNA sequence flanking one end of the deletion. One microgram of genomic DNA was therefore digested with 0.5 Unit of PstI (Patient 1) in a total volume of 25 μl according to the manufacturer's instructions (New England Biolabs). The DNA digest was purified using a QIAquick PCR Purification Kit (Qiagen). Circular DNA was created by ligating 660-700 ng digested DNA with 30 Units of T4 ligase (Promega) in a total volume of 660-700 μl overnight at 25° C. The ligated DNA was purified by a double ethanol precipitation and resuspended in TE buffer.
Using oligonucleotide primers within the known DNA sequence flanking the restriction enzyme cut site, we inversely amplified the circular DNA using the Expand Long Template PCR System (Roche). All PCR reactions were performed in a final volume of 50 μl. The reactions contained 1 Unit Taq polymerase, 0.05 mM dNTPs, 12-20 ng purified ligated DNA, 17.5 mM MgCl2, and 15-20 ng of each primer (primer sequence is available by request). The amplification occurred in three steps: 1) an initial denaturation at 94° C for 5 min; 10 cycles of denaturation at 94° C for 30 sec, annealing at 64° C for 1 min, and elongation at 72° C for 4 min; 2) 10 cycles of denaturation at 94° C for 30 sec, annealing at 64° C for 1 min, and elongation at 72° C for 5 min; 3) 20 cycles of denaturation at 94° C for 30 sec, annealing at 64° C for 1 min, and elongation at 72° C for 6 min; a final extension at 72° C for 1 min. The resulting PCR products were separated on a 1% agarose gel, purified with a MinElute Spin Column and sequenced.
Genomic DNA Amplification across the Deletion in Patient 1
After identifying deletion breakpoints by LDI-PCR, primers were designed to amplify across the breakpoint in undigested genomic DNA (Table 1). PCR amplification of the patient-specific DNA fragment was multiplexed with amplification of exon 8 of ALDH3A2 as a control. The PCR contained 1 U Taq polymerase, 0.05 mM dNTPs, 50 ng of DNA, 17.5 mM MgCl2, 10 ng of the patient-specific primers, and 20 ng of the exon 8 primers (Table 1). The cycling conditions consisted of 1) an initial denaturation at 94° C for 5 min; 2) 30 cycles of denaturation at 94° C for 30 sec, annealing at 68° C for 1 min, and elongation at 72° C for 1 min; 3) a final extension at 72° C for 5 min.
Table 1.
Oligonucleotide primers used in this study.
| DNA Product | Primer | Oligonucleotide Primer Sequence |
|---|---|---|
| Patient 1 | Forward | 5’-TGTGTAACCTTGCAGATTCCTAGGTTC-3’ |
| Genomic | ||
| Deletion | ||
| Reverse | 5’-ATCTCAGTGGAAATCTGGACAGTGACAC -3’ | |
| ALDH3A2 | Forward | 5’-TTGACACATAACTGAGCACACAGCCCTC-3’ |
| Exon 8 | ||
| Reverse | 5’-AGCAGCCCATACAATCCACTCATGA-3’ | |
| ALDH3A2 | Forward | 5’-GAGCTGCAGAAATAATTGGGAGTACCTAGC-3’ |
| Exon 3 for Detection of c.407C>T | ||
| Reverse | 5’-CTTGGCTGTATTTTCACTCAGTTC-3’ |
DNA Sequencing
LDI-PCR products and ALDH3A2 exons amplified from genomic DNA [13] were sequenced using the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, Inc, Foster City, CA) and an ABI 377A sequencer.
Screening for the c.407C>T (P136L) Mutation in Patient 2
The c.407C>T (P136L) mutation in exon 3 creates a new HindIII restriction enzyme cut site. To screen for this mutation, a 102 bp fragment from exon 3 containing the mutation was amplified from genomic DNA and digested with HindIII. The PCR contained 0.75 U Taq polymerase, 2.5 mM dNTPs, 17.5 mM MgCl2, 1 μl buccal DNA and 10 ng of each primer (Table 1) in reaction volume of 50 μl. The cycling conditions were 1) initial denaturation at 95° C for 7 min, 2) 40 cycles of 95° C for 30 sec, 62° C for 1 min and 72° C for 1 min, 3) final extension at 72° C for 5 min. The PCR product (0.25 μg) was digested in 50 μl reaction volume containing Buffer 2 and 0.5 Unit HindIII overnight at 37° C according to the manufacturer's instructions (New England Biolabs). The digestion products were separated on a 3% agarose gel. The c.407C>T mutation produces a HindIII cut site with generation of a 80bp fragment, whereas the wild-type DNA is undigested.
Array Comparative Genomic Hybridization (CGH) and Fluorescence In Situ Hybridization (FISH)
CGH was performed using a whole genome 44K custom oligonucleotide array (Agilent Technologies, Inc) as described [18]. The deletion was confirmed via FISH, using a bacterial artificial chromosome corresponding to the deleted region (RP11-147i5) and alpha satellite DNA from chromosome 17. Probes were labeled and hybridized to metaphase chromosomes as described previously [18].
RESULTS
Patient 1
Patient 1 was first suspected to have a large gene deletion when ALDH3A2 exons could not be amplified from genomic DNA. Southern blot analysis subsequently confirmed a complete deletion of the gene. The deletion boundaries were initially narrowed down by generating PCR products from flanking DNA sequences and sequentially walking closer to the deletion until no PCR products were produced; however, attempts to amplify across the deletion using multiple combinations of primers were unsuccessful. We therefore used LDI-PCR to determine the precise deletion breakpoints. This method depends on knowing DNA sequence flanking one end of the deletion breakpoint. Restriction enzymes that cut within the known sequence (and presumably somewhere on the other sides of the deletion breakpoints) were used to generate DNA fragments that were subsequently ligated into circular DNA. Primers situated within the known DNA sequence were then used to inversely amplify across the deleted DNA region in the circular DNA and locate the DNA sequence from the other side of the deletion to determine the precise breakpoints.
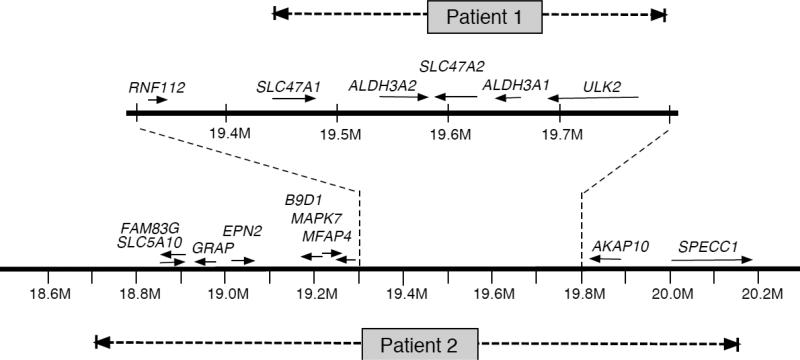
Using this approach, a 4.1 kb PCR product was produced by LDI-PCR of the patient's circularized DNA, and sequencing of the PCR product identified the deletion breakpoints on chromosome 17 at nucleotide 19,446,110 and nucleotide 19,798,450 (in GRCh37/hg19 of the human genomic assembly). This 352 kb deletion includes ALDH3A2, ALDH3A1, ULK2, SLC47A1 and SLC47A2 (Fig 3). The homozygous deletion was confirmed by array CGH (data not shown).
Fig 3.
ALDH3A2 genetic locus at chromosome 17p11.2 showing the large deletions in the patients. The horizontal solid lines indicate chromosome DNA with an expanded region delimited by the vertical dashed lines. Arrows represent orientation of genes from 5’ to 3’ direction. Horizontal dashed lines correspond to deleted regions in Patient 1 and Patient 2. Nucleotide positions are numbered in megabases (M).
Patient 1 was a child of consanguineous parents. Using primers that flanked the deletion breakpoints, a 496 bp PCR product was amplified from the patient's genomic DNA and her parents, thus confirming the parental carrier status for this deletion, but not from controls (Fig 1B). A control PCR product of exon 8 was produced using DNA from the parents and her unaffected brother, but not from the patient, consistent with her homozygous genotype.
Patient 2
Because of the patient's constellation of clinical findings, CGH analysis was initially undertaken using a 44K oligonucleotide genomic microarray. Patient 2 was found to carry a heterozygous 1.44 Mb interstitial deletion of 17p11.2 [chr17:18,716,455 - 20,160,197] in NCBI36/hg18 of the human genomic assembly that spans 15 genes, including ALDH3A1 and ALDH3A2 (Fig 2A, Fig 3). The deletion lies within the recurrent Smith-Magenis syndrome (SMS) deletion region, but does not include the complete SMS critical region. FISH analysis confirmed the heterozygous deletion (Fig 2B).
Sequence analysis of the patient's only remaining ALDH3A2 gene copy identified a novel hemizygous missense mutation (c.407C>T, P136L) in exon 3 (Fig 2C). This mutation generates a new HindIII restriction enzyme cut site. By screening a PCR amplicon of exon 3 and digesting with HindIII, we did not detect the mutation in 50 unrelated Caucasian control subjects, suggesting that it is not a common polymorphism. The P136 amino acid residue is invariantly conserved among FALDH proteins in vertebrate species ranging from zebra fish to humans and undoubtedly has a critical function in the protein. In this regard, the P136 residue in human FALDH corresponds to P138 in a related rat class 3 ALDH protein, for which the crystalline structure has been reported [19]. P138 of the rat protein initiates a kink between the β2 β-strand and the αB α-helical domain within the Rossmann fold, which is important for NAD binding.
Investigation of the parents by microarray and DNA sequencing revealed that the patient's father carried the deletion mutation and her mother was heterozygous for the P136L mutation.
DISCUSSION
The SLS patients reported here have unusually large deletions, including the entire ALDH3A2 gene on chromosome 17p11.2. Previously reported intragenic deletions in SLS have ranged from 1 or 2 nucleotides to as large as 6 kb [11], but no complete gene deletions have been reported. Our patients carried contiguous gene deletion alleles that included ALDH3A2 and multiple flanking genes. Indeed, this region of chromosome 17 is associated with large de novo contiguous gene mutations that are a well-known marker of SMS [20], which is caused by haploinsufficiency for the RAI1 gene [21].
Patient 1 represents a unique “experiment in nature”. She is homozygous for a contiguous gene mutation that deletes ALDH3A2 and 4 neighboring genes, including ALDH3A1, ULK2, SLC47A1 and SLC47A2. ALDH3A1 encodes an aldehyde dehydrogenase that is expressed at high levels in stomach and comprises up to 40% of the total soluble protein in cornea [22]. The enzyme oxidizes medium-chain aliphatic aldehydes [23], and is thought to eliminate toxic aldehydes generated during lipid peroxidation [24]. However, its unusual abundance in cornea suggest that the protein may also have a non-catalytic role in absorbing UV light and protecting the underlying lens from damage [25]. A naturally occurring strain of mice (SWR/J) that is deficient in ALDH3A1 enzyme activity [26] and Aldh3a1-/- gene knockout mice [25] are abnormally susceptible to cataract formation when exposed to UV light. The ULK2 gene codes for a UNC-51-like serine/threonine protein kinase that is widely expressed in tissues [27] and is involved in cell signaling pathways for apoptosis [28], autophagy [29,30], axonal outgrowth and endocytosis [31], and fibroblast growth factor receptor substrates [32]. The SLC47A1 and SLC47A2 genes encode multidrug and toxin extrusion proteins (MATE1 and MATEK-2, respectively) that are localized in the kidney and are important for actively secreting certain organic cationic molecules and drugs, including metformin and cimetidine, from the blood into the urine [33,34,35]. Non-synonymous polymorphisms in the genes may be responsible for decreasing, or even eliminating, renal clearance of these ionic drugs and thereby affect drug pharmacokinetics and toxicity [35,36]. Mice naturally lack the SLC47A2 gene, but genetic knockout of the murine SLC47A1 gene results in a profound reduction in metformin excretion and increased blood concentrations of this drug [37].
Although missing ALDH3A2 and 4 additional genes, the clinical phenotype of Patient 1 was within the spectrum seen in other SLS patients, albeit at the more severe end. Some of her symptoms, however, may originate from the deleted neighboring genes. Her oral motor dysfunction and excessive drooling is not usually seen in SLS, and most patients do not require a gastrostomy tube for feeding. Despite lacking the ALDH3A1 protein, she did not exhibit corneal or lens opacities. Her long-standing photophobia and squinting, which are common features of SLS patients, may have limited her ocular exposure to UV light and prevented eye damage. The absence of perimacular glistening white dots, which are often seen in SLS, indicates that her photophobia does not arise from light hitting and refracting off these abnormal structures. Without the MATE1 and MATEK-2 transporters, Patient 1 should be at higher risk for adverse effects of MATE-dependent drugs. The lack of ULK2 would seem to impair several cell signaling pathways and may contribute to her more severe SLS phenotype with progressive loss of speech, oral motor function and worsening spasticity. Most SLS patients have a static disease with little or no symptom progression over time. Additional subtle phenotypic effects of her deleted genes could be missed on her clinical background of severe SLS. Moreover, the extent to which the clinical phenotype of Patient 2 is influenced by haploinsufficiency for the many deleted genes is also not known.
The genetic analysis of Patient 2 underscores the importance for considering large gene deletions in patients with ostensibly homozygous ALDH3A2 mutations and the need for parental testing to confirm carrier status. If this patient had only undergone standard mutation analysis by sequencing genomic exons amplified by PCR, her genotype could have been initially misidentified as homozygous P136L. In the absence of parental testing, a heterozygous deletion or duplication would have gone undetected, and subsequent genetic testing of family relatives or prenatal diagnosis could have led to erroneous results. Mutation analysis of her parents, however, would have raised the possibility of a co-existing deletion mutation in the patient and prompted further DNA analyses by array CGH or high-density single nucleotide polymorphism (SNP) chips.
The frequency of large gene deletions in SLS is not precisely known. We are aware, however, of two other SLS patients who also carry unique large gene deletions that involve the complete ALDH3A2 gene or most of it (unpublished observations). Together with the two mutations reported here, these large deletions account for approximately 5% of the mutant alleles reported so far.
Large deletion mutations have been detected in several other inborn errors of metabolism, including Canavan disease [38-40], cystinosis [41], steroid sulfatase deficiency [42-44], X-linked adrenoleukodystrophy [45], phenylketonuria [46], pyruvate dehydrogenase deficiency [47] and ornithine transcarbamylase deficiency [44,49]. Although CGH and SNP arrays are rarely done for the evaluation of suspected metabolic disorders, they are becoming a standard diagnostic test for patients with non-specific developmental delay. Our findings in SLS suggest that these tests also have utility for detecting contiguous gene deletions in an increasing number of inborn errors of metabolism.
ACKNOWLEDGMENTS
This work was supported by grant AR044552 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health, and by the Sjögren-Larsson Syndrome Research Fund at the University of Nebraska.
Abbreviations
- CGH
comparative genomic hybridization
- FALDH
fatty aldehyde dehydrogenase
- FISH
fluorescence in situ hybridization
- LDI-PCR
long distance inverse-PCR
- OMIM
Online Mendalian Inheritance in Man
- SLS
Sjögren-Larsson syndrome
- SMS
Smith-Magenis syndrome
- SNP
single nucleotide polymorphism
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sjögren T, Larsson T. Oligophrenia in combination with congenital ichthyosis and spastic disorders. Acta Psychiatr. Neurol. Scand. 1957;32(suppl 113):1–113. [PubMed] [Google Scholar]
- 2.De Laurenzi V, Rogers GR, Hamrock DJ, Marekov LN, Steinert PM, Compton JG, et al. Sjögren-Larsson syndrome is caused by mutations in the fatty aldehyde dehydrogenase gene. Nat. Genet. 1996;12:52–7. doi: 10.1038/ng0196-52. [DOI] [PubMed] [Google Scholar]
- 3.Rizzo WB, Craft DA. Sjögren-Larsson syndrome. Deficient activity of the fatty aldehyde dehydrogenase component of fatty alcohol:NAD+ oxidoreductase in cultured fibroblasts. J. Clin. Invest. 1991;88:1643–8. doi: 10.1172/JCI115478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rizzo WB, Dammann AL, Craft DA. Sjögren-Larsson syndrome. Impaired fatty alcohol oxidation in cultured fibroblasts due to deficient fatty alcohol:nicotinamide adenine dinucleotide oxidoreductase activity. J. Clin. Invest. 1988;81:738–44. doi: 10.1172/JCI113379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kelson TL, Secor McVoy JR, Rizzo WB. Human liver fatty aldehyde dehydrogenase: microsomal localization, purification, and biochemical characterization. Biochim. Biophys. Acta. 1997;1335:99–110. doi: 10.1016/s0304-4165(96)00126-2. [DOI] [PubMed] [Google Scholar]
- 6.Rizzo WB, Dammann AL, Craft DA, Black SH, Tilton AH, Africk D, et al. Sjögren-Larsson syndrome: inherited defect in the fatty alcohol cycle. J. Pediatr. 1989;115:228–34. doi: 10.1016/s0022-3476(89)80070-8. [DOI] [PubMed] [Google Scholar]
- 7.Rizzo WB, Craft DA. Sjögren-Larsson syndrome: accumulation of free fatty alcohols in cultured fibroblasts and plasma. J. Lipid Res. 2000;41:1077–81. [PubMed] [Google Scholar]
- 8.Willemsen MA, de Jong JG, van Domburg PH, Rotteveel JJ, Wanders RJ, Mayatepek E. Defective inactivation of leukotriene B4 in patients with Sjögren-Larsson syndrome. J. Pediatr. 2000;136:258–60. doi: 10.1016/s0022-3476(00)70113-2. [DOI] [PubMed] [Google Scholar]
- 9.Rizzo WB, Craft DA, Somer T, Carney G, Trafrova J, Simon M. Abnormal fatty alcohol metabolism in cultured keratinocytes from patients with Sjögren-Larsson syndrome. J. Lipid Res. 2008;49:410–9. doi: 10.1194/jlr.M700469-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rizzo WB. Sjögren-Larsson syndrome: molecular genetics and biochemical pathogenesis of fatty aldehyde dehydrogenase deficiency. Mol. Genet. Metab. 2007;90:1–9. doi: 10.1016/j.ymgme.2006.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rizzo WB, Carney G. Sjögren-Larsson syndrome: diversity of mutations and polymorphisms in the fatty aldehyde dehydrogenase gene (ALDH3A2) Hum. Mutat. 2005;26:1–10. doi: 10.1002/humu.20181. [DOI] [PubMed] [Google Scholar]
- 12.Sillén A, Anton-Lamprecht I, Braun-Quentin C, Kraus CS, Sayli BS, Ayuso C, et al. Spectrum of mutations and sequence variants in the FALDH gene in patients with Sjögren-Larsson syndrome. Hum. Mutat. 1998;12:377–84. doi: 10.1002/(SICI)1098-1004(1998)12:6<377::AID-HUMU3>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 13.Rizzo WB, Carney G, Lin Z. The molecular basis of Sjögren-Larsson syndrome: mutation analysis of the fatty aldehyde dehydrogenase gene. Am. J. Hum. Genet. 1999;65:1547–60. doi: 10.1086/302681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kraus C, Braun-Quentin C, Ballhausen WG, Pfeiffer RA. RNA-based mutation screening in German families with Sjögren-Larsson syndrome. Eur. J. Hum. Genet. 2000;8:299–306. doi: 10.1038/sj.ejhg.5200453. [DOI] [PubMed] [Google Scholar]
- 15.Lowry OH, Rosebough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–75. [PubMed] [Google Scholar]
- 16.Richards B, Skoletsky J, Shuber AP, Balfour R, Stern RC, Dorkin HL, et al. Multiplex PCR amplification from the CFTR gene using DNA prepared from buccal brushes/swabs. Hum. Mol. Genet. 1993;2:159–63. doi: 10.1093/hmg/2.2.159. [DOI] [PubMed] [Google Scholar]
- 17.Willis TG, Jadayel DM, Coignet LJ, Abdul-Rauf M, Treleaven JG, Catovsky D, Dyer MJ. Rapid molecular cloning of rearrangements of the IGHJ locus using long-distance inverse polymerase chain reaction. Blood. 1997;90:2456–64. [PubMed] [Google Scholar]
- 18.Baldwin EL, Lee JY, Blake DM, Bunke BP, Alexander CR, Kogan AL, et al. Enhanced detection of clinically relevant genomic imbalances using a targeted plus whole genome oligonucleotide microarray. Genet. Med. 2008;10:415–29. doi: 10.1097/GIM.0b013e318177015c. [DOI] [PubMed] [Google Scholar]
- 19.Liu ZJ, Sun YJ, Rose J, Chung YJ, Hsiao CD, Chang WR, et al. The first structure of an aldehyde dehydrogenase reveals novel interactions between NAD and the Rossmann fold. Nat. Struct. Biol. 1997;4:317–26. doi: 10.1038/nsb0497-317. [DOI] [PubMed] [Google Scholar]
- 20.Vlangos CN, Yim DK, Elsea SH. Refinement of the Smith-Magenis syndrome critical region to approximately 950kb and assessment of 17p11.2 deletions. Are all deletions created equally? Mol. Genet. Metab. 2003;79:134–41. doi: 10.1016/s1096-7192(03)00048-9. [DOI] [PubMed] [Google Scholar]
- 21.Slager RE, Newton TL, Vlangos CN, Finucane B, Elsea SH. Mutations in RAI1 associated with Smith-Magenis syndrome. Nat. Genet. 2003;33:466–8. doi: 10.1038/ng1126. [DOI] [PubMed] [Google Scholar]
- 22.Pappa A, Sophos NA, Vasiliou V. Corneal and stomach expression of aldehyde dehydrogenases: from fish to mammals. Chem. Biol. Interact. 2001;130-132:181–91. doi: 10.1016/s0009-2797(00)00233-7. [DOI] [PubMed] [Google Scholar]
- 23.Pappa A, Estey T, Manzer R, Brown D, Vasiliou V. Human aldehyde dehydrogenase 3A1 (ALDH3A1): biochemical characterization and immunohistochemical localization in the cornea. Biochem. J. 2003;376:615–23. doi: 10.1042/BJ20030810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Townsend AJ, Leone-Kabler S, Haynes RL, Wu Y, Szweda L, Bunting KD. Selective protection by stably transfected human ALDH3A1 (but not human ALDH1A1) against toxicity of aliphatic aldehydes in V79 cells. Chem. Biol. Interact. 2001;130-132:261–73. doi: 10.1016/s0009-2797(00)00270-2. [DOI] [PubMed] [Google Scholar]
- 25.Lassen N, Bateman JB, Estey T, Kuszak JR, Nees DW, Piatigorsky J, et al. Multiple and additive functions of ALDH3A1 and ALDH1A1: cataract phenotype and ocular oxidative damage in Aldh3a1(-/-)/Aldh1a1(-/-) knock-out mice. J. Biol. Chem. 2007;282:25668–76. doi: 10.1074/jbc.M702076200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shiao T, Tran P, Siegel D, Lee J, Vasiliou V. Four amino acid changes are associated with the Aldh3a1 locus polymorphism in mice which may be responsible for corneal sensitivity to ultraviolet light. Pharmacogenetics. 1999;9:145–53. [PubMed] [Google Scholar]
- 27.Yan J, Kuroyanagi H, Tomemori T, Okazaki N, Asato K, Matsuda Y, et al. Mouse ULK2, a novel member of the UNC-51-like protein kinases: unique features of functional domains. Oncogene. 1999;18:5850–9. doi: 10.1038/sj.onc.1202988. [DOI] [PubMed] [Google Scholar]
- 28.Yang MH, Yoo KH, Yook YJ, Park EY, Jeon JO, Choi SH, et al. The gene expression profiling in murine cortical cells undergoing programmed cell death (PCD) induced by serum deprivation. J. Biochem. Mol. Biol. 2007;40:277–85. doi: 10.5483/bmbrep.2007.40.2.277. [DOI] [PubMed] [Google Scholar]
- 29.Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell. Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan EY, Longatti A, McKnight NC, Tooze SA. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol. Cell. Biol. 2009;29:157–71. doi: 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou X, Babu JR, da Silva S, Shu Q, Graef IA, Oliver T, et al. Unc-51-like kinase 1/2-mediated endocytic processes regulate filopodia extension and branching of sensory axons. Proc. Natl. Acad. Sci. U. S. A. 2007;104:5842–7. doi: 10.1073/pnas.0701402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Avery AW, Figueroa C, Vojtek AB. UNC-51-like kinase regulation of fibroblast growth factor receptor substrate 2/3. Cell Signal. 2007;19:177–84. doi: 10.1016/j.cellsig.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 33.Masuda S, Terada T, Yonezawa A, Tanihara Y, Kishimoto K, Katsura T, et al. Identification and functional characterization of a new human kidney-specific H+/organic cation antiporter, kidney-specific multidrug and toxin extrusion 2. J. Am. Soc. Nephrol. 2006;17:2127–35. doi: 10.1681/ASN.2006030205. [DOI] [PubMed] [Google Scholar]
- 34.Tanihara Y, Masuda S, Sato T, Katsura T, Ogawa O, Inui K. Substrate specificity of MATE1 and MATE2-K, human multidrug and toxin extrusions/H(+)-organic cation antiporters. Biochem. Pharmacol. 2007;74:359–71. doi: 10.1016/j.bcp.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 35.Meyer zu Schwabedissen HE, Verstuyft C, Kroemer HK, Becquemont L, Kim RB. Human multidrug and toxin extrusion 1 (MATE1/SLC47A1) transporter: functional characterization, interaction with OCT2 (SLC22A2), and single nucleotide polymorphisms. Am. J. Physiol. Renal Physiol. 2010;298:F997–F1005. doi: 10.1152/ajprenal.00431.2009. [DOI] [PubMed] [Google Scholar]
- 36.Kajiwara M, Terada T, Ogasawara K, Iwano J, Katsura T, Fukatsu A, et al. Identification of multidrug and toxin extrusion (MATE1 and MATE2-K) variants with complete loss of transport activity. J. Hum. Genet. 2009;54:40–6. doi: 10.1038/jhg.2008.1. [DOI] [PubMed] [Google Scholar]
- 37.Tsuda M, Terada T, Mizuno T, Katsura T, Shimakura J, Inui K. Targeted disruption of the multidrug and toxin extrusion 1 (MATE1) gene in mice reduces renal secretion of metformin. Mol. Pharmacol. 2009;75:1280–6. doi: 10.1124/mol.109.056242. [DOI] [PubMed] [Google Scholar]
- 38.Zeng BJ, Wang ZH, Torres PA, Pastores GM, Leone P, Raghavan SS, Kolodny EH. Rapid detection of three large novel deletions of the aspartoacylase gene in non-Jewish patients with Canavan disease. Mol. Genet. Metab. 2006;89:156–63. doi: 10.1016/j.ymgme.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 39.Kaya N, Imtiaz F, Colak D, Al-Sayed M, Al-Odaib A, Al-Zahrani F, et al. Genome-wide gene expression profiling and mutation analysis of Saudi patients with Canavan disease. Genet. Med. 2008;10:675–84. doi: 10.1097/gim.0b013e31818337a8. [DOI] [PubMed] [Google Scholar]
- 40.Caliebe A, Vater I, Plendl H, Gesk S, Siebert R. A 439 kb-sized homozygous deletion in 17p13.3 leading to biallelic loss of the ASPA as cause of Canavan disease detected by SNP-array analysis. Mol. Genet. Metab. 2010;99:184–5. doi: 10.1016/j.ymgme.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 41.Touchman JW, Anikster Y, Dietrich NL, Maduro VV, McDowell G, Shotelersuk V, et al. The genomic region encompassing the nephropathic cystinosis gene (CTNS): complete sequencing of a 200-kb segment and discovery of a novel gene within the common cystinosis-causing deletion. Genome Res. 2000;10:165–73. doi: 10.1101/gr.10.2.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paige DG, Emilion GG, Bouloux PM, Harper JI. A clinical and genetic study of X-linked recessive ichthyosis and contiguous gene defects. Br J Dermatol. 1994;131:622–9. doi: 10.1111/j.1365-2133.1994.tb04972.x. [DOI] [PubMed] [Google Scholar]
- 43.Weissörtel R, Strom TM, Dörr HG, Rauch A, Meitinger T. Analysis of an interstitial deletion in a patient with Kallmann syndrome, X-linked ichthyosis and mental retardation. Clin. Genet. 1998;54:45–51. doi: 10.1111/j.1399-0004.1998.tb03692.x. [DOI] [PubMed] [Google Scholar]
- 44.van Steensel MA, Vreeburg M, Engelen J, Ghesquiere S, Stegmann AP, Herbergs J, et al. Contiguous gene syndrome due to a maternally inherited 8.41 Mb distal deletion of chromosome band Xp22.3 in a boy with short stature, ichthyosis, epilepsy, mental retardation, cerebral cortical heterotopias and Dandy-Walker malformation. Am. J. Med. Genet A. 2008;146A:2944–9. doi: 10.1002/ajmg.a.32473. [DOI] [PubMed] [Google Scholar]
- 45.Corzo D, Gibson W, Johnson K, Mitchell G, LePage G, Cox GF, et al. Contiguous deletion of the X-linked adrenoleukodystrophy gene (ABCD1) and DXS1357E: a novel neonatal phenotype similar to peroxisomal biogenesis disorders. Am. J. Hum. Genet. 2002;70:1520–31. doi: 10.1086/340849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mallolas J, Vilaseca MA, Pavia C, Lambruschini N, Cambra FJ, Campistol J, et al. Large de novo deletion in chromosome 12 affecting the PAH, IGF1, ASCL1, and TRA1 genes. J Mol. Med. 2001;78:721–4. doi: 10.1007/s001090000160. [DOI] [PubMed] [Google Scholar]
- 47.Singer BH, Iyer RK, Kerr DS, Ahmad A. Deletion at chromosomal band Xp22.12-Xp22.13 involving PDHA1 in a patient with congenital lactic acidosis. Mol. Genet. Metab. 2010;101:87–9. doi: 10.1016/j.ymgme.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 48.Arranz JA, Madrigal I, Riudor E, Armengol L, Milà M. Complete deletion of ornithine transcarbamylase gene confirmed by CGH array of X chromosome. J. Inherit. Metab. Dis. 2007;30:813. doi: 10.1007/s10545-007-0578-y. [DOI] [PubMed] [Google Scholar]
- 49.Balasubramaniam S, Rudduck C, Bennetts B, Peters G, Wilcken B, Ellaway C. Contiguous gene deletion syndrome in a female with ornithine transcarbamylase deficiency. Mol. Genet. Metab. 2010;99:34–41. doi: 10.1016/j.ymgme.2009.08.007. [DOI] [PubMed] [Google Scholar]