Abstract
There is growing interest in the epigenetic mechanisms that impact human health and disease, including the role of microRNAs (miRNAs). These small (18–25 nucleotide), evolutionarily conserved, non-coding RNA molecules regulate gene expression in a post-transcriptional manner. Several well-orchestered regulatory mechanisms involving miRNAs have been identified, with the potential to target multiple signaling pathways dysregulated in cancer. Since the initial discovery of miRNAs, there has been progress towards therapeutic applications, and several natural and synthetic chemopreventive agents also have been evaluated as modulators of miRNA expression in different cancer types. This review summarizes the most up-to-date information related to miRNA biogenesis, and critically evaluates proposed miRNA regulatory mechanisms in relation to cancer signaling pathways, as well as other epigenetic modifications (DNA methylation patterns, histone marks) and their involvement in drug resistance. We also discuss the mechanisms by which dietary factors regulate miRNA expression, in the context of chemoprevention versus therapy.
Introduction
The discovery of microRNAs (miRNAs) opened a new era in our understanding of gene regulation. These small, evolutionarily conserved, non-coding RNAs were first discovered in Caenorhabditis elegans more than a decade ago during genetic analyses. The first miRNA identified, lin-4, was found to contain complementary sequences in the 3′-untranslated region (UTR) of lin-14 messenger RNA (mRNA). This exciting finding indicated a regulatory mechanism by which lin-4 could modulate mRNA lin-14 translation in C. elegans, thereby mediating temporal pattern formation during development [1, 2]. Regulatory patterns of many other miRNAs have now been recognized in species ranging from viruses to humans [3].
The latest version of miRBase (miRBase Version 16.0) has 1048 miRNA sequences annotated in the human genome, and additional miRNAs are likely to be validated in the future [4–7]. The literature indicates that one-third of these miRNAs are located in 113 gene clusters and, based on the evidence from miRNA profiling data in various tissues and cell lines, these clusters are mostly co-expressed. This observation leads to questions about cistronic expression regulatory patterns in gene clusters. The current understanding is that deregulation of one member of the cluster is accompanied by similar deregulations of other miRNAs from the same cluster. Thus, it would be interesting to ascertain whether one miRNA in a cluster can be regulated independently of others, especially those miRNAs implicated in the pathophysiology of human diseases. MiRNAs are believed to target approximately one-third of human mRNAs, of several protein-coding genes. Due to the differential target binding patterns, a single miRNA may target approximately 200 transcripts simultaneously [8]. Thus, an in-depth analysis of miRNA regulation might provide an effective strategy to control numerous genes simultaneously. The present review focuses on the role of miRNAs in cancer etiology, and provides a synopisis of the associated epigenetic pathways of gene regulation. Therapeutic strategies being implemented to target miRNAs are discussed, including the use of dietary agents and synthetic molecules in several cancers.
Biogenesis and mechanism of action
MiRNAs are naturally-occurring, small, non-coding RNA sequences ~18–25 nucleotides (nt) in length. The biogenesis and processing of the final mature miRNA is a highly regulated process. Long primary miRNA transcripts (pri-miRNAs) containing hundreds to thousands of nt, a 5′cap and a poly (A) tail are produced by RNA Polymerase II [9, 10], with the exception of those within the Alu repeats that are otherwise transcribed by RNA polymerase III [11] from independent genes, the introns of protein-coding genes, or polycistronic regions. The pri-miRNAs contain a single or cluster of miRNAs in a folded hairpin stem structure which is processed in the nucleus by the RNAse III enzyme Drosha, and the double-stranded RNA-binding domain (dsRBD) protein DGCR8 (DiGeorge Critical Region 8)/Pasha [12, 13]. This duo, commonly referred to as the microprocessor complex, cleaves the pri-miRNA into a ~70 nt hairpin precursor known as pre-miRNA [14]. Pre-miRNA is transported to the cytoplasm using a Ran-GTP dependent mechanism by a nuclear export factor, Exportin-5 [15]. The pre-miRNA is processed through a series of cuts by an RNAse III enzyme Dicer, in association with HIV-transactivating response RNA-binding protein (TRBP) and protein kinase dsRNA-dependent activator (PACT) which, in human cells, generates a mature ~18–25 nt miRNA:miRNA* duplex [16, 17]. Under the mediation of Argonaut 2 (Ago2), the RNA duplex is unwound to form two single strands. Ago2, in association with a glycine-tryptophan protein, acts as a key factor in the assembly and function of miR-RISCs (microRNA-RNA-induced silencing complexes). One strand is the mature miRNA or “functional” strand, and the other strand is the immature miRNA* or the “passenger” strand which is degraded [18, 19]. The retained, or mature strand, is the one that has the less stably base-paired 5′ end in the duplex. Thus, thermodynamic stability of the duplex plays a deciding role in the formation of mature miRNA. The miRNA* of the two strands in the duplex is not necessarily a by-product, and in some circumstances has been found to be loaded in the RISC assembly to serve as a functional miRNA [20–22]. The mature miRNA strand is then selectively incorporated into RISC and directs the complex to target mRNA through a poorly defined mechanism [23–25]. Most miRNA genes generate one dominant miRNA species; however, there are many contributing factors regulating the final outcome.
Downregulation of target gene expression by miRNA is mediated by either of two well-studied mechanisms, dictated by the level of complementarity between mRNA and miRNA. First, mature miRNAs with close to perfect complementarity may bind to the 3′UTR of the target mRNA sequence, inducing cleavage and degradation of the transcript by deadenylation and decapping of the mRNA [26]. The second mechanism involves repression of translation, which is most common, due to imperfect sequence complementarity between the miRNA and mRNA [25]. Irrespective of which of these two events predominate, the overall outcome is a reduction in protein encoded by the respective mRNA targets. Emerging evidence suggests that only miRNAs in abundance are able to target a substantial fraction of their available mRNA target sites and significantly impact mRNA stability in several diseases [27–29]. However, the mechanisms controlling mRNA targeting are not completely understood, as low abundance miRNAs also have been reported to synergistically regulate target expression [30].
Another downstream mechanism is the miRNA-directed movement of mRNAs and associated RISC proteins to storage structures such as processing bodies (p-bodies) or cytoplasmic structures [31]. This phenomenon is currently believed to be an outcome of microRNA-based translational repression, rather than a cause of this event [32]. Interestingly, a recent study reported the reversal of miR-induced mRNA repression from these storage bodies and re-entry into polysomes under certain stress conditions [33]. This protective mechanism of repressed mRNAs and their subsequent reversal could be important in several cancer pathways, influencing aggressive tumor recurrence, drug resistance, and metastatic phenotype. Thus, exploring further the underlying mechanisms of miRNA biogenesis will be critical in our understanding of the regulatory patterns of miRNAs, and developing targeted therapeutic strategies.
Different schools of thought exist concerning the specific mechanisms involved in miRNA targeting. One report suggests that specificity in choosing target transcripts is primarily based on the principle of Watson-Crick complementarity between the 3′-UTR of the target mRNA and the nucleotide sequence from position 2 to 8 at the 5′ end of miRNAs, also referred as the “seed region” [34]. Using advanced computational tools, it has been found that multiple miRNAs target the 3′-UTR of a single gene [35]. In a recent interesting finding, researchers reported that some miRNAs had selective binding affinities for 5′-UTR sequence of target genes, a mechanism that appeared to activate gene expression [36, 37]. Another report by Wakiyama et al. demonstrated that efficient miRNA-dependent translation repression requires a m7G-cap along with a poly(A) tail [38]. Since the RISC-miRNA complex plays a major role in directing miRNA to its target sequence, further in-depth analyses are required to understand the specific mechanisms of regulation involved.
An alternative biogenesis mechanism was recently discovered, in which miR-451 was found to enter the RISC by directly loading its precursor pre-miRNA after Drosha processing, skipping the activation steps with Dicer. In this rather unusual processing mechanism, Ago2 was found to replace the activity of Dicer [39]. Interestingly, 27% of tumors across all tissues possess a hemizygous deletion of the Dicer encoding gene. The significance of Dicer in miRNA processing was evidenced via knockdown experiments in vitro and in vivo [40, 41]. For example, reduction of Dicer increased the rate of tumor formation in a K-ras-dependent lung cancer model, and in an Rb-based retinoblastoma model [41, 42]. Recently, Drosha-independent mechanisms were identified, including mirtrons and tailed mirtrons that release pre-miRNAs by splicing and exonuclease trimming. A “mirtron” is defined as a short hairpin intron that uses splicing machinery to bypass Drosha cleavage in initial maturation. Examples include miR-320 and miR-484 [43, 44].
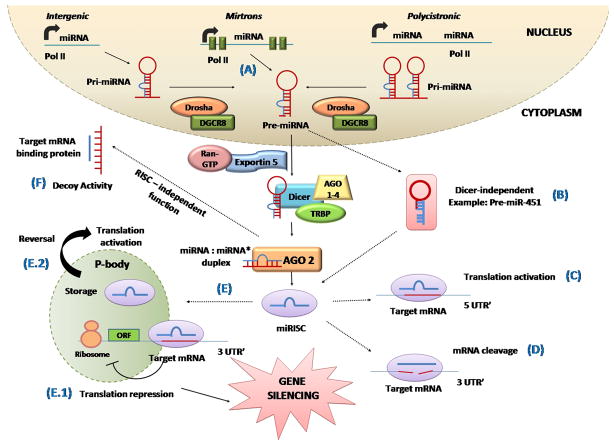
Another interesting mechanism related to miRNA biogenesis and regulation is mediated by SND1 (Staphylococcal nuclease homology domain containing 1), which is one of the components of RISC and is proposed to be involved in gene silencing mechanisms [45]. Johnson et al. reported SND1 was an important component of let-7 directed RISC regulation in RAS signaling [46]. Although the exact role of SND1 is not fully understood, these intriguing findings suggest the possible role of SND1 as a key regulator at both transcriptional and post-transcriptional levels [47]. Also, ribosome recruitment to the mRNA and targeting the mRNA cap structure was found to play a role in inhibition of translational initiation using extracts from mouse Krebs-2 ascites cells [48]. Kiriakidou et al. identified a motif within the Ago2 protein that had significant similarity to a domain of an essential translation initiation factor, eIF4E. It was reported that Ago2 protein competes with eIF4E to bind to the domain and repress initiation of translation [49]. Thus, the complex mechanistic intricacy of miRNAs and their biogenesis pathways (Figure 1) should be taken into consideration when designing therapies.
Figure 1. MicroRNA biogenesis and regulatory pathways.
Pri-miRNAs are transcribed from RNA polymerase II (RNAPII)-specific miRNA genes, from the intronic region of protein coding genes, or from polycistronic transcripts. In the first nuclear step, pri-miRNA is processed into a 70–100 nucleotide precursor hairpin (pre-miRNA) via the Drosha-DGCR8 complex. Pre-miRNA is transported to the cytoplasm through export machinery consisting of Exportin 5 and Ran-GTP. Here, the pre-miRNA is cleaved by another endoribonuclease, Dicer, in partnership with TRBP and Ago proteins, forming a ~20-bp miRNA: miRNA* duplex. After processing, one strand of the duplex is preferentially incorporated with the help of Ago2 into the RISC complex (miRISC), whereas the other “passenger” strand (miRNA*) gets degraded. (A) A few pre-miRNAs are processed directly from short introns (mirtrons), bypassing the Drosha-DGCR8 step. (B) A dicer-independent mechanism, miRNA being cleaved by Ago2 to form a mature miRNA. (C) Some miRNAs bind to the 5′-UTR of the target mRNA and lead to translational activation. (D) Full or near-full complementarity between miRNA and mRNA target facilitates miRISC-directed cleavage of the mRNA target. (E) With low complementarity, miRNA-mediated regulation is carried out by translational repression (E.1). This can occur pre- and/or post-initiation of translation leading to gene silencing. (E.2) Target mRNAs also can be stored in P-bodies, and mechanism reversed by re-entry into polysomes for translation. (F) In a RISC-independent decoy activity, miRNAs can directly bind to proteins, particularly RNA-binding proteins, making them unavailable for binding to their RNA targets.
MicroRNAs and molecular cross-talk in cancer
Cancer is the second leading cause of mortality and is responsible for one in four deaths in the United States [50]. It is a complex, multi-step disease characterized by disruption of the homeostatic balance between cell proliferation and cell death, and uncontrolled clonal expansion leading to tumor formation. Until recently, protein-coding genes were the primary focus of cancer research; however, over the last decade there has been a major paradigm shift with the emerging role of miRNAs and other ‘epigenetic’ mechanisms [51–53]. MiRNAs interact with an estimated 60% of mRNAs through one or more evolutionarily conserved sequences, implicating their role in a wide range of physiological and pathophysiological processes [34, 54].
MiRNAs have been implicated at all stages of cancer, from initiation to tumor promotion and progression, influencing cell proliferation, differentiation, apoptosis, angiogenesis and metastasis [55]. Various miRNAs are up- or down-regulated in human neoplasia, with some overlapping miRNA profiles depending on tissue origin. Previous reports suggested that miRNA expression patterns are tissue-specific and might be a useful tool for classifying human cancers clinically [56, 57]. However, recent reports indicate that miRNA expression levels, rather than specific miRNA identity, characterize normal versus tumor tissue [6].
Genome-wide miRNA profiling has defined miRNA expression and intensity levels in various tumor tissues. Aberrant miRNA expression patterns are often attributed to the presence of miRNAs in regions of chromosomal instability, due to amplification, translocation or deletion events, representing ~52.5% of miRNA genes in cancer-associated regions or fragile sites [58]. Some examples include amplification of the miR-17–92 cluster in lymphomas or its translocation in T cell acute lymphoblastic leukemia [59, 60], amplification of miR-26a in glioblastoma [61], and deletion of an miR-15a/16-1 cluster in a putative tumor suppressor-containing region in B cell lymphoblastic leukemia [62]. There are certain miRNAs that have emerged as prime regulators of key cellular and physiological states in human tumor tissues. For example, miR-21 was found to be consistently upregulated in cancers of the breast, colon, lung, pancreas and stomach, as well as in chronic lymphocytic leukemia (CLL), acute myeloid leukemia (AML), glioblastoma, and myeloma [63]. Members of the let-7 family were found to be downregulated in colon, breast, lung, ovarian, and gastric cancers, suggesting that restoration of let-7 members may be a useful therapeutic approach in these cancers [64–69]. These observations suggested that miRNAs mimic oncogenes or tumor suppressors (Figure 2), due to their respective up- or down-regulated expression patterns in different cancers [70].
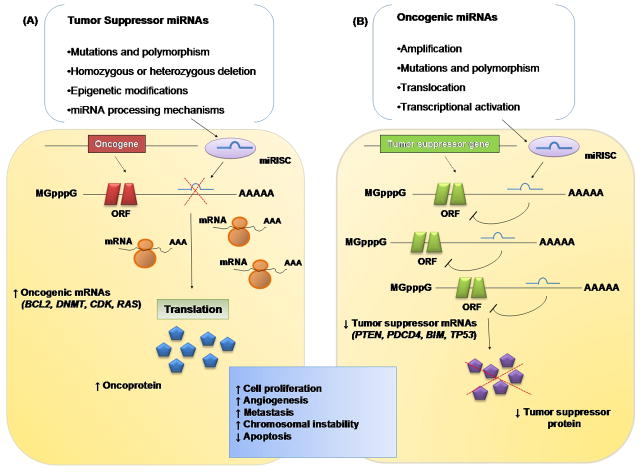
Figure 2. MicroRNAs impacting oncogenic and tumor suppressor pathways.
(A) The reduction or deletion of a “tumor suppressor miRNA” due to mutation, deletion, epigenetic modification, or irregularities in miRNA processing cause inappropriate elevation of miRNA-target oncoproteins, ultimately leading to tumor formation. (B) Amplification or overexpression of an “oncogenic miRNA” inhibits miRNA-targets of vital tumor suppressor genes. The overall outcome is to increase cell proliferation, angiogenesis, and metastasis, or augment chromosomal instability and apoptosis.
Profiling differential expression patterns of miRNAs suggests potential “molecular signatures” that can distinguish histopathologic features in various tissues. For instance, differential miRNA expression profiles clearly defined colon cancer and rectal cancer as two quite distinct pathologies, emphasizing that “associations can be masked when studying them as one disease” [71]. Aberrant expression of 15 miRNAs distinguished tumor from normal tissue in breast cancer patients. Of these, miR-10b, miR-125b, let-7 and miR-145 were consistently downregulated, whereas miR-21 and miR-155 were upregulated in malignant tissue [65]. In another study, metastatic miRNA biomarkers were identified in breast cancer. Reduced metastasis from breast to lung or bone in mice was associated with overexpression of miR-335, miR-126 and miR-206, indicating a possible breast cancer-specific miRNA signature pattern [72]. Using high-throughput screening, several studies have reported deregulation of specific miRNA expression in pancreatic cancer as compared to other tumors. For example, expression levels of miR-375 and miR-376 were significantly higher in mouse pancreas and pancreatic islet cells as compared to brain, heart and liver tissue [73]. One of the most significantly upregulated miRNAs in pancreatic cancer is miR-21, which is considered an ‘oncogenic’ miRNA that may be responsible for chemotherapeutic (gemcitabine) resistance in pancreatic cancer cells [74–76]. The QuantiMir system was used to examine differential expression patterns of 95 miRNAs in 10 pancreatic cancer cell lines and 17 pairs of pancreatic/normal adjacent tissues [77]. This study reported a significant upregulation of miR-196a, miR-190, miR-186, miR-221, miR-222, miR-200b, miR-15b, and miR-95 in most pancreatic cancer tissues and cell lines. Interestingly, upregulation of these eight miRNAs ranged from 70–100% between normal and tumor cells or tissues. MiRNA profiling may be important in determining an effective treatment strategy to deal with this fatal disease, which is otherwise deadly due to its late diagnosis and limited therapeutic options.
Similarly, miRNA signature profiles can be defined in non-small-cell lung cancers (NSCLC) that are useful in distinguishing clinical phenotypes. In a study by Yanaihara et al. five miRNAs (miR-155, miR-17-3p, let-7a-2, miR-145, and miR-21) were found to be differentially expressed in tumor versus normal tissue in lung cancer patients [64]. Importantly, let-7 members were commonly downregulated in lung cancers and appear to serve as a marker of survival in lung cancer patients [78–80]. Taken together, the results imply a pivotal role of miRNAs in the pathogenesis of various human cancers.
Furthermore, miRNA profiles can provide information regarding tumor differentiation and clinical subtypes. For example, certain miRNAs were implicated in late stages of carcinogenesis, and several pro- and anti-angiogenic miRNAs have been identified. Vascular endothelial growth factor (VEGF), an angiogenesis mediator, is known to be regulated by miR-15a-16-1 [81, 82], miR-126 [83, 84], and miR-378 [85] via indirect targeting of various intermediary upstream signaling molecules. Several miRNAs also inhibit cancer cell invasion, adhesion and migration, including miR-122, miR-126, miR-128, miR-146 a/b, miR-31, miR-29c, and the miR-200 family. Conversely, miRNAs that are known to promote metastatic mechanisms include miR-21, miR-10b, miR-155, miR-373 and miR-520c [86]. MiR-29c and let-7g target expression of components of the extra-cellular matrix (ECM) involved in cell adhesion and migration. For example let-7g, a tumor suppressor miRNA, was reported to be poorly expressed in a metastatic hepatocellular carcinoma cell line when compared to normal cells. Forced overexpression of let-7g inhibited cell migration by targeting type I collagen a2 [87]. Overexpression of miR-17 resulted in growth retardation along with reduced cell adhesion, migration and proliferation in the same cancer cell line [88]. Other important regulators of ECM are matrix metalloproteinases (MMPs) that are upregulated by miR-21, miR-221, miR-222 [89, 90] or downregulated by miR-181b, miR-146b [91, 92]. These miRNAs modulate the expression of various genes that regulate invasiveness of cancer cells.
Much is being learned about the downstream targets of miRNAs. However, recent evidence indicates that miRNAs themselves are subject to higher levels of control that regulate both miRNA metabolism and function. One mode of action is the ability to self-regulate. Due to their ability to directly base-pair with various mRNAs, coding for factors involved in biogenesis and regulatory mechanisms, miRNAs can participate in their own transcription mechanisms through feedback loops with specific transcription factors. For example, a regulatory loop between miR-133b and the transcription factor PITX3 controls neuronal differentiation [93]. Another example is provided by let-7, a suppressor of proliferation that can target Dicer mRNA, thereby preventing the upregulation of growth stimulatory miRNAs involved in cancer [94]. Accumulating evidence suggests a role for various oncogenic or tumor suppressor transcription factors such as ras, myc, and p53 in regulating miRNA expression, in a tissue-specific manner. Examples of such regulatory mechanisms include expression of miR-34 and miR-107 families being stimulated by p53 [95, 96], miR-21 regulation modulated by ras [97], and regulation of the miR-17-92 cluster and miR-9 by myc and mycn in lymphoma cells [98] and neuroblastoma cells [99], respectively. SMAD, signal transducer of transforming growth factor-β (TGF-β), and bone morphogenetic factor (BMP), control Drosha-mediated miRNA processing. A SMAD-p68 complex with Drosha is reported to enhance processing and accumulation of miR-21, thereby promoting carcinogenesis [100, 101].
MicroRNA and drug resistance
Cancer therapeutics is an important research area. However, there is room for improvement when viewed in the context of current patient survival rates, and persistent concerns such as intrinsic or acquired drug resistance. Accumulating evidence supports a role for miRNAs in the formation of cancer stem cells, and in the acquisition of an epithelial-mesenchymal transition (EMT) phenotype [102–104]. The mechanisms regulating EMT are known to be closely associated with drug resistance and metastasis. Recent studies have revealed that miRNAs are involved in the development of anti-cancer drug resistance [105, 106]. For example, miR-200, which was earlier reported to be downregulated in various cancers, was also found to be involved in EMT and drug resistance in pancreatic cancer [103]. Re-expression of miRNA-200 downregulated EMT markers such as ZEB1, slug, and vimentin, and enhanced sensitivity to gemcitabine. Similar results were obtained with the suppressor miRNA family, let-7. MiR-200 also was reported to be involved in drug resistance mechanisms in bladder, endometrial, breast, and ovarian cancers [107, 108]. Conversely, oncogenic miR-21 increased chemoresistance by targeting the tumor suppressor protein PDCD4 (programmed cell death 4), thereby causing an upregulation of inhibitors of apoptosis proteins (IAPs) and multidrug-resistant protein-1 (MDR1) in breast cancer cells [109]. MiR-21 has been reported to modulate drug resistance in various other cancers, such as glioblastoma [110], prostate [111], and pancreatic [112] cancers.
According to Garofalo et al.,[113] transfecting NSCLC with anti-miR-221 and -222 resulted in enhanced sensitivity to TRAIL, by modulating p27kip1 and Kit. Similarly, knockdown of miR-221 and -222 in breast cancer cells resulted in enhanced sensitivity to tamoxifen [114]. However, in another report, miR-221 and miR-128b were downregulated in MLL-AF4 ALL and re-expression of these two miRNAs resulted in sensitization of MLL-AF4 ALL cells to glucocorticoids [115]. This suggests the possibility of tissue specificity and synergistic effects modulated by certain miRNAs. In yet another study, transfection of miR-205 in breast cancer cells increased the responsiveness to tyrosine kinase inhibitors of EGFR (gefitinib) and EGFR/HER2 (lapatinib), thereby reducing HER3-mediated drug resistance [116]. Thus, targeted therapy aimed at critical miRNAs involved in drug resistance may help restore efficacy in various cancer treatments.
MicroRNA crosstalk with other epigenetic mechanisms in cancer
Epigenetics is the study of changes in gene expression that are not associated with alterations in DNA sequence. Two important areas of epigenetics focus on histone modifications and DNA methylation [117]. These mechanisms individually or cooperatively regulate cancer signaling pathways, some involving miRNAs [118]. Examples include hypermethylation of CpG islands near the transcriptional start site of miR-34, thereby modulating p53 expression in cancer cells [119]; silencing by methylation of miR-9 loci, which correlates with cancer metastasis [118, 120]; and downregulation of miR-449a, which, in prostate cancer cells, causes overexpression of histone deacetylase 1 (HDAC1) [121]. Conversely, several miRNAs modulate gene expression by altering the methylation machinery or chromatin remodeling factors in cancer cells [122, 123]. Thus, it is intriguing to ponder the complex integrated mechanisms involving DNA methylation, histone modifications, and miRNA profiles.
One of the earlier reports on epigenetic regulation of miRNAs was from studies of bladder cancer [124]. Saito et al. evaluated the effect of simultaneous treatment with 5-Aza-CdR, a potent DNA methylation inhibitor, and 4-phenylbutyric acid (PBA), an HDAC inhibitor. Interestingly, among 17 of 313 miRNAs upregulated, miR-127 was overexpressed almost 50-fold in bladder cancer cells as compared to normal human fibroblasts. Upregulation of miR-127 resulted in downregulation of proto-oncogene BCL6. These and other studies suggest that epigenetic mechanisms can activate tumor suppressor miRNAs (Figure 3).
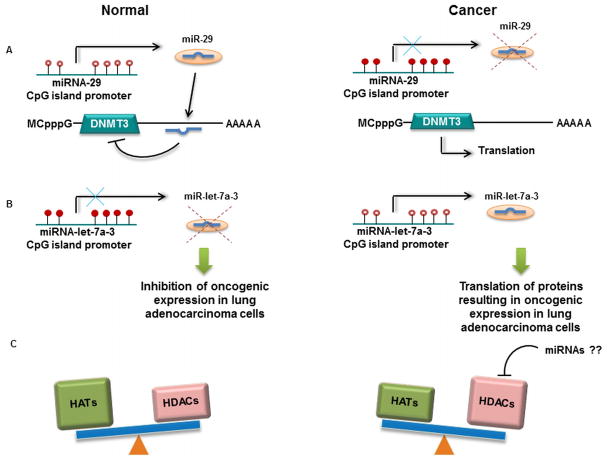
Figure 3. Epigenetic modifications and miRNA regulation.
MiRNAs can impact the epigenetic machinery by regulating key players involved in DNA methylation and chromatin remodeling. (A) Demethylated tumor suppressor miR-29 (open red dots) inhibits translation of cancer-promoting DNMT3s in lung cancer, (B) Hypermethylation of let-7a-3 (closed red dots) in normal cells results in repression of cancer-promoting genes in lung, (C) An integrated loop is becoming apparent between miRNAs, and chromatin remodeling via HATs and HDACs, which influence miRNA expression in the cell.
The effect of DNA methylation on miRNA expression has been investigated by several groups. Lujambio et al. [118] identified miR-148a, miR-34b/c, and miR-9 as commonly silenced in colon cancer cells as compared to normal tissues. Early reports identified miRNA-34a as a target of the tumor suppressor p53 [95, 125, 126], and miR-34a was found to be regulated by DNA methylation, with the silencing mechanism dominating over transactivation by p53. In prostate and pancreatic carcinoma cell lines, silencing of miR-34a by CpG methylation also was observed [127].
Another important silencing mechanism was identified when methylation of miR-148 promoted cancer metastasis in melanoma and breast cancer by upregulating its target gene, TGIF2 [118]. MiR-148-mediated repression of DNMT3b identified high homology binding near the 3′-UTR regions and poly (A) tail, but the exact repressive mechanism remains unclear [128]. Similarly, an inverse correlation was observed between the miR-29 family (especially miR-29a-c) and DNMT3A and DNMT3B in lung cancer cells. Overexpression of miR-29 resulted in re-expression of methylation-silenced tumor suppressor genes such as FHIT and WWOX [122]. Using Dnmt1 and Dnmt3b knockout HCT116 colorectal cancer cells, 18 miRNAs were upregulated > 3-fold in knockout cells, suggesting a CpG island hypermethylation mechanism to silence tumor suppressor miRNAs. One of the important findings was the negative correlation between miR-124a and a bona fide oncogene, cyclin D kinase 6 [129, 130]. In yet another study, let-7a-3 was found to be heavily methylated in normal human tissues as compared to the hypomethylated profile in lung adenocarcinoma samples [131]. Thus, contrary to the paradigm that promoter regions are typically hypermethylated in cancers, hypomethylation might in some circumstances aid in reactivation of let-7a-3 in human lung cancer cells. Interestingly, DNA methylation enzymes such as DNMT1, 3a, and 3b are anticipated to be miRNA targets and vice versa [132, 133]. In a recent study, miR-34b and miR-129-2 were dramatically silenced in gastric carcinogenesis. Epigenetic regulation was implicated by co-treatment with the demethylating agent 5-Aza-dC and the HDAC inhibitor trichostatin A, suggesting a strong association of these miRNAs with poor clinical outcome in gastric cancer patients [134].
Epigenetic regulations are also mediated by histone modifications. The effect of a hydroxamic acid HDAC inhibitor, LAQ824, was evaluated in breast cancer cells. On treatment with LAQ824, a dramatic alteration in miRNA profiles was observed, with 22 miRNAs being upregulated and 5 miRNAs being downregulated [135]. The HDAC inhibitor vorinostat (suberoylanilide hydroxamic acid, SAHA) altered markedly the expression of 31 miRNAs in HCT116 colon cancer cells, as well as downstream targets affecting cell cycle, apoptosis, and differentiation [136]. By comparing HCT116 cells that were p53 wild type versus p53 null, miRNAs were identified that responded to p53 status in cancer cells, including miR-7-1, miR-9, miR-22, miR-30c, miR-32, miR-221 and miR-222 [136]. Other miRNAs associated with specific components of histone modification mechanisms have been identified. For example, miR-449 regulates HDAC1 levels in prostate cancer [121], and HDAC4 is a validated target of miR-1 in hepatocellular carcinomas [137]. These initial studies provide milestones along the path to as yet unexplored aspects of epigenetics and miRNAs in cancer development.
Dietary regulators of microRNAs – potential roles in chemoprevention
In the temporal progression to malignancy, cells accumulate alterations in multiple cellular signaling pathways. Previous attempts to treat cancer often failed due to a “one gene-one target” approach, sometimes referred to as mono-modal therapy. At the same time, the benefits associated with a healthy diet and life style strongly support a multi-modal disease prevention strategy. Various natural dietary chemopreventive agents have been identified, some with well characterized pleiotropic actions in cancer cells. This has led to studies of natural agents that might modulate gene expression by targeting miRNAs, via direct or indirect chemopreventive mechanisms [103, 106, 138–140]. Although there are relatively few such studies at present, this is likely to gain significant attention in the future. Some examples are presented below of dietary or nutritional factors known to impact miRNAs involved in various stages of carcinogenesis, including early chemoprevention versus late-stage therapeutic effects.
MicroRNAs and essential nutritional factors
Vitamin A
Vitamin A is an essential dietary factor involved in vision, reproduction, immune function, cell growth and differentiation. All-trans-retinoic acid (RA), the most biologically active form of Vitamin A, acts as a tumor suppressor in lung, prostate, bladder, liver, breast and pancreatic cancer models [141]. In a recent report [142], 243 miRNAs were examined using microarray analyses in RA-treated human acute promyelocytic leukemia (APL) cells. Interestingly, previously known deregulated miRNAs were differentially expressed upon treatment with RA. In another study using microarrays, several tumor suppressor miRNAs were upregulated upon RA treatment in human APL cells [143]. In the latter study, putative NFκB consensus elements were identified in the upstream genomic region of let-7 cluster following RA treatment.
Vitamin D
Vitamin D and its metabolites, 1,25-dihydroxyvitamin D3 (1, 25D3) and 25-hydroxyvitamin D3 (25(OH)D3), regulate miRNA profiles in different cancers. Treatment of human myeloid leukemia cells with 1,25D3 led to downregulation of miR-181a and miR-181b, resulting in enhanced expression of p27Kip1 and p21Cip1, causing G1 cell cycle arrest [144]. 25(OH)D3 conferred a protective role against cellular stress in breast epithelial cells by modulating p53 and PCNA levels, along with alteration in miR-182 expression [145]. Cancer chemopreventive effects of vitamin D and its metabolites are mediated via binding with its receptor (VDR). MiR-125b was identified as having a potential sequence match in the 3′-UTR region of human VDR mRNA, suggesting a pathway for targeted therapy via VDR downregulation in human cancers [146].
Vitamin E
Deficiency of vitamin E in rats resulted in significant downregulation of tumor suppressor miRNAs in liver. Vitamin E modulated lipid metabolism, inflammation, and other cancer-associated pathways by altering the expression of miR-122 and miR-125b [147].
Vitamin B
Folate is a water-soluble B-vitamin. Hepatocellular carcinoma in rats fed a folate-deficient diet for 54 weeks was associated with increased expression of several miRNAs in tumors, including miR-21, and reduced expression of liver-specific miR-122. Folate replenishment increased suppressor miR-122 levels, and was associated with inhibition of tumorigenesis, suggesting a potential chemoprevention paradigm affecting miRNAs [148]. A similar trend was observed in an in vitro study in which adequate folate in the culture media restored miRNA levels in human lymphoblastoid cells. One of the key miRNAs upregulated in human peripheral blood cells from individuals with low folate intake was miR-222 [149].
Selenium
Selenium deficiency is associated with increased cancer risk. Sodium selenite, an inorganic form of selenium, activates p53 and increases its targets in the miR-34 family [150]. Specific members of miR-34 family, miR-34b and miR-34c, but not miR-34a, were increased significantly in prostate cancer cells treated with sodium selenite. Because of toxicity concerns associated with inorganic and some organic forms of selenium, it will be important for future miRNA studies to examine these compounds on a case-by-case basis, including their metabolites [151–153].
Fatty Acids
n-3-Polyunsaturated fatty acids (n-3 PUFAs) are found in walnuts, fish-oil, soybeans, green leafy vegetables, and seed oils. Protective roles of PUFAs have been documented in various human disease conditions, including cancer [154, 155]. A recent study evaluated the chemopreventive effects of PUFAs on azoxymethane-induced colon cancer in rats. Carcinogen treatment resulted in significant downregulation of five known tumor suppressor miRNAs, which were reversed upon exposure to fish oil. Based on transfection experiments in vitro, tumor suppressor PTEN was found to be targeted by oncogenic miR-21 in human colon cancer cells. Similarly, beta site amyloid precursor protein-cleaving enzyme (BACE-1) was reported as a functional target of tumor suppressor miR-107 and was downregulated in carcinogen-induced tumor tissues versus normal colonic mucosa [156]. This study demonstrated the chemoprotective role of dietary n-3 PUFAs in colon by modulating the miRNA expression pattern in carcinogen-induced rat colon cancer. Short-chain fatty acids which inhibit HDAC activity, such as butyrate, also alter miRNA patterns regulating endodermal differentiation mechanisms, as studied in human embryonic stem cells [157].
MicroRNAs and phytochemicals
Polyphenols
Curcumin
Curcumin, a bioactive ingredient in turmeric, possesses anti-inflammatory, antioxidant, and anti-carcinogenic properties, although such effects are not always realized in vivo [158, 159]. An initial study evaluated miRNA profiles in curcumin-treated pancreatic cancer cells, with evidence for upregulation of 11 miRNAs and downregulation of 18 miRNAs. MiR-22 was upregulated upon curcumin treatment, and the predicted targets were ERα and transcription factor Sp1. MiR-196, an oncogenic miRNA in gastric cancers, was significantly downregulated after curcumin treatment [160]. Due to the low bioavailability of curcumin in vivo, a synthetic analogue (CDF- diflourinated-curcumin) was evaluated in a chemopreventive pancreatic cancer model [112]. Curcumin and its CDF analog, alone or in combination, attenuated expression of miR-200 and miR-21 leading to induction of tumor suppressor PTEN. The CDF analog inhibited sphere forming ability (pancreatospheres) by attenuating cancer stem cell markers and other signaling molecules, via changes in miR-21 and miR-200. These findings suggested a role for certain miRNAs in tumor recurrence in pancreatic cancer, and the effectiveness of the CDF analog as an alternative therapeutic strategy to curcumin parent compound [161]. In a recent study of curcumin and multi-drug resistance, alterations were detected in 342 miRNAs [162]. Significant changes (> 2.5 fold) in various oncogenic and tumor suppressor miRNAs were reported after curcumin treatment. A key target was miR-186*, which promoted apoptosis in cancer cells. Overall, these studies provided support for the idea that diet-induced miRNAs play a role in overcoming drug resistance in cancers.
Resveratrol
Resveratrol is a chemopreventive agent from grapes, mulberries, wine, and peanuts. Effects of resveratrol on colon cancer cells were examined by Tili et al. [163]. Several “signature” miRNAs for colon cancer such as miR-21, miR- 196a, miR- 25, miR-17, and miR-92a-2 were significantly downregulated by resveratrol. Simultaneously, miR-663-mediated regulation of Dicer, PDCD4, PTEN, and TGFβ signaling through the SMAD promoter was observed. This study provided the first insights into resveratrol-mediated miR-663 regulation in colon cancer cells. A resveratrol-induced, miR-663-dependent effect was observed in monocytic cells used to evaluate adaptive and innate immune responses [164]. MiR-663 was reported to target Activator Protein-1 (AP-1) through the Jun signaling pathway. Interestingly, resveratrol also impaired the upregulation of oncogenic miR-155 in a miR-663-dependent manner.
Catechins
Chemopreventive effects of epigallocatechin-3-gallate (EGCG) and other tea catechins have been described in preclinical models for all major sites of cancer development, including colon, prostate, breast, lung, liver, and skin. Mechanistically, EGCG and related catechins target various cancer signaling pathways in a pleiotropic manner; however, clinical efficacy is less clear [165–168]. Recently, miRNAs were included among the molecular targets of EGCG. In human hepatocellular carcinoma cells, one of the 13 miRNAs that was upregulated on EGCG treatment was miR-16, a tumor suppressor miRNA that mediated apoptosis via downregulation of Bcl-2. This mechanistic target was identified based on transfection studies [169]. Further work is needed to elucidate the detailed miRNA “target map” following treatment with EGCG and, equally importantly, by potential chemopreventive metabolites such as the glucuronide and O-methylated forms which constitute the major fractions found in plasma after oral ingestion.
Ellagitannin
Ellagitannins are polymeric polyphenols found in abundance in strawberries, raspberries, almonds, walnuts, and various other fruits and nuts. They were initially characterized for their anti-oxidant and free radical scavenging activity. Anti-inflammatory, anti-tumor promoting, anti-proliferative, and apoptosis-inducing properties also have been identified [170]. A plant grown in Japan and China, Balanophora Japonica MAKINO, contains 1,3-di-O-galloyl-4,6-(s)-HHDP-b-D-glucopyranose. This ellagitannin was examined for anti-proliferative effects in human liver cancer cells, along with profiling of miRNAs [171]. Using a dose- and time-dependent strategy, 17 miRNAs were found to be upregulated and 8 miRNAs were downregulated following treatment of HepG2 cells, including let-7 family members, miR-370, miR-373, and miR-526b. Prediction software and functional analyses identified likely targets with roles in cell proliferation and differentiation; however, the precise mechanisms await further study.
Isoflavones
Soy isoflavones, including genistein, daidzein, and glycitein, have been implicated in anti-carcinogenic mechanisms, via the modulation of estrogen receptor binding in target tissues. Genistein is currently undergoing clinical trials for chemopreventive and therapeutic effects in breast, prostate, bladder, and kidney cancers [172]. Li et al. [103] examined whether isoflavones altered miRNA profiles in pancreatic cancer, and noted a differential effect in gemcitabine-resistant versus gemcitabine-sensitive cancer cells. For example, miRNAs belonging to miR-200 and let-7 families were downregulated in gemcitabine-resistant cells versus gemcitabine-sensitive cells. However, isoflavone treatment increased both miR-200 and let-7 family miRNAs by modulating EMT transcription factors, such as vimentin, slug, and ZEB1. Genistein also upregulated miR-146a in pancreatic cancer cells, inhibiting their invasive potential by downregulating EGFR, NFκB, IRAK-1, and MTA-2 [173].
Another study [174] examined minichromosome maintenance (MCM) genes involved in DNA replication, which are commonly dysregulated in cancer cells. In prostate cancer cells treated with genistein, MCM2 was downregulated by miR-1296. Genistein induced the expression of miR-1296 by up to five-fold, along with cell cycle arrest in S-phase. Chemopreventive effects of genistein on the temporal changes during ovarian cancer progression were assessed using microarray analyses [175]. This was a descriptive study that focused on significant alterations in miRNAs, and awaits further validation of potential targets. Genistein also was investigated in other cancer models, such as human uveal melanoma cells [176]; using both in vitro and in vivo models, miR-27a was found to be downregulated with concomitant upregulation of its target gene, ZBTB10.
Indoles
Cruciferous/Brassica vegetables have received considerable attention due to the chemopreventive properties of the whole food or isolated compounds, such as sulforaphane and indole-3-carbinol (I3C) [177]. Upon ingestion, I3C undergoes acid condensation reactions in the stomach producing a number of oligomers including dimers, trimers, and tetramers. The major compound found in vivo in human plasma is 3,3′-diindolylmethane (DIM) which has been examined for chemoprotective mechanisms in breast, colon, prostate, pancreatic and cervical cancer [178]. In a well-designed study by Izzotti et al. [179], altered miRNA profiles in lung tissue were observed in rats exposed to environmental cigarette smoke. Restoration of miRNAs targeting p53 functions (miR-34b), TGF-β expression (miR-26a), ERBB2 activation (miR-125a), and angiogenesis (miR-10a) was recorded on treatment with five dietary agents, including I3C. Also, as discussed in the studies by Li et al. [103, 173], along with soy isoflavones, DIM influenced EMT via differentially expressed miRNAs in pancreatic cancer cells. Based on these initial reports with I3C and DIM in cancer models, miRNAs appear to be promising molecular targets of dietary indoles, awaiting further mechanistic validation.
Isothiocyanates
Isothiocyanates derived from cruciferous vegetables modulate carcinogen metabolism in different tissues, but likely exert numerous other chemoprotective mechanisms [177, 180]. The effect of phenethyl isothiocyanate (PEITC) on miRNA alterations induced by smoking in rat lung tissue was evaluated by Izzoti et al. [181]. Of the five dietary agents tested, PEITC intervention alone, or in combination with I3C, was the most effective in restoring miRNAs downregulated by exposure to cigarette smoke. Major PEITC-induced miRNA targets were miR-192 (Ras activation); let-7a, let-7c (cell proliferation, angiogenesis, Ras activation); miR-146 (NFκB activation); miR-123, miR-222, (angiogenesis, cell proliferation), and miR-99b (apoptosis). In another study by the same group, miRNA alterations upon exposure to cigarette smoke were investigated in mouse lung and liver tissues [182]. PEITC-induced changes in miRNA expression profiles were more robust in mouse liver (significant > 2-fold downregulation of 9 oncogenic miRNAs and upregulation of 3 tumor suppressor miRNAs) as compared to lung tissue. It would be interesting to evaluate the effect of other dietary isothiocyanates on miRNA expression profiles in cancer models.
Collectively, these studies support a growing interest in the chemopreventive role of dietary agents such as vitamins, fatty acids, trace elements, polyphenols, indoles, and isothiocyanates as modulators of miRNA profiles in cancer etiology and prevention (Figure 4). The latter work will likely dovetail with ongoing research on the therapeutic aspects of miRNAs, as described below.
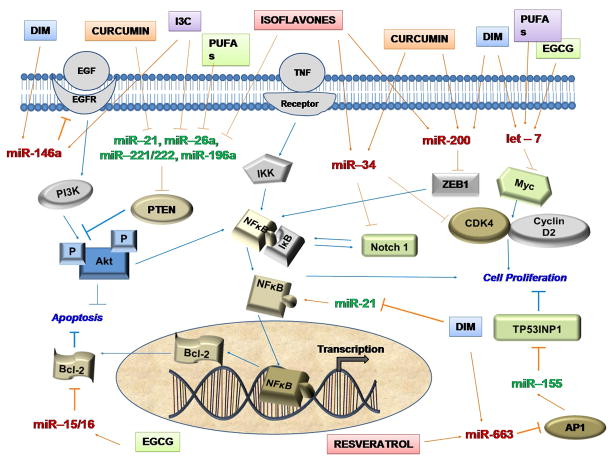
Figure 4. MicroRNA regulation by dietary agents.
Dietary agents such as curcumin, resveratrol, DIM, I3C, EGCG, and ellagitannin modulate miRNAs that regulate cancer signaling pathways.
MicroRNA-based cancer therapeutics
Over the past decade, evidence has accrued on the possibility of using miRNA profiling in the diagnosis of human pathological conditions, including cancer [71]. Through the “fine tuning” of multiple signaling pathways, miRNA-based therapy might restore homeostasis and provide an effective means of regulating the transcriptome or proteome. High-throughput technologies, such as microarrays and genome-wide association studies with deep sequencing, are moving rapidly to enhance miRNA detection. Various therapeutic strategies have thus evolved concurrently with the increased understanding of miRNA regulation and functionality. Most of the strategies are based on the principle of gain- or loss-of-function.
Inhibiting oncogenic miRNAs
The observation that certain miRNAs are commonly upregulated in tumor development provided a basis for investigating “antagomirs” as competitive inhibitors for cancer therapy. These “designer” miRNAs often have good bioavailability and stability, but there are limitations in terms of targeted delivery and toxicity from off-target effects [183, 184]. Another approach is the use of locked nucleic acid (LNA) constructs. These nucleoside analogs possess a ribose ring that is secured by a methylene bridge connecting the 2′-O atom and the 4′-C atom, leading to successful knockdown of specific miRNAs [185, 186]. Although this strategy was found to be effective, again there are concerns regarding potential toxicity and off-target effects. Small molecule inhibitors, such as diazobenzene-1, also have been investigated for their ability to influence oncogenic miRNAs in different cancers [187]. Chemotherapeutic drugs are currently under investigation for their ability to restore miRNAs to a normal phenotype in cancer cells [124, 143].
Another therapeutic strategy is the “miR-mask”’, designed to be completely complementary to a miRNA binding site in the 3′-UTR region of the mRNA being targeted [188]. Despite its apparent specificity, this approach has limited scope due to an inability to target multiple cancer signaling pathways. A slightly more sophisticated approach uses miRNA “sponges”, which contain multiple tandem binding sites corresponding to a miRNA of interest, and exhibit competitive binding with designated targets for a particular miRNA [189]. In a recent study, sponges containing heptameric seed sequences effectively blocked an entire miRNA family, due to their common seed sequence recognition [190]. Despite targeting an entire family of miRNAs, further research is needed to improve delivery and enhance specificity. In a similar vein, liposome-based oligonucleotide-mimics of miRNAs resulted in improved stability and delivery, but had impaired biological activity and enhanced toxicity due to formation of cationic lipids [191–193]. Polymers and nanoparticle-based strategies are generating more promising results in terms of delivery, stability, and reduced toxicity [194, 195].
Upregulating tumor suppressor miRNAs
In most cancers, tumor suppressor miRNAs are repressed or completely absent; thus, reinstating these miRNAs could be therapeutically beneficial [196–198]. Several miRNA mimics have been developed to restore tumor suppressor activity, with successful induction of cell death and inhibition of cell proliferation. So far, these “miR-mimics” have been evaluated in vitro and await experimental validation in vivo. Another approach involves the use of adenovirus-associated vectors, some of which have entered Phase I and II clinical trials [199, 200]. This strategy seeks to upregulate the expression of tumor suppressor miRNAs without integration into the genome and avoiding toxicity. In addition, DNA demethylating agents, HDAC inhibitors, and their combination provides a possible means of restoring expression of suppressor miRNAs in cancers.
Conclusions and future perspectives
There is a pressing need for clinical translation of novel breakthroughs in cancer biology. Enthusiasm abounds for miRNAs as novel gene regulators, with the potential to fine tune physiological processes involved in cellular differentiation and metabolism. Given that the deregulation of miRNA expression is implicated in numerous facets of cancer pathology, we anticipate further interest in miRNAs as novel targets for cancer chemoprevention and therapy.
This review summarized key mechanisms of miRNA biogenesis and the regulatory functions specific to oncogenesis. The emerging role of miRNAs as oncogenic and/or tumor suppressor factors has opened a new avenue for therapeutics, but much work is needed to clarify the mechanisms by which miRNAs regulate their own expression and other signaling pathways. A modest change in the expression of a single miRNA can provoke a cascade that activates several feedback pathways, involving various other miRNAs. Though still in a preliminary stage, several examples exist of the inter-regulatory patterns between promoter regions of miRNAs and various other genes. Profiling studies have provided insights into the complex roles of miRNAs in different clinical situations. For example, miR-195 is upregulated in cardiovascular diseases but downregulated in cancers [201], highlighting the need for careful therapeutic strategies that alleviate one condition without simultaneously exacerbating others.
This review also summarized the broader interest in epigenetics and miRNA regulation, involving cross-talk with DNA methylation patterns and histone modifications. As more studies target miRNAs as a therapeutic strategy, we will gain greater insights into their ability to affect drug resistance mechanisms associated with standard chemotherapeutic drugs. Several therapeutic strategies also have been proposed based on synthetic analogs of miRNAs; however, this field is still in its infancy.
Various dietary agents are now under investigation as modulators of miRNA profiles in cancer, and there is much promise in this area from a chemoprevention standpoint. Effects of natural agents on temporal changes in miRNA profiles during cancer initiation and progression could provide new insights into early biomarkers for cancer chemoprevention. However, issues such as in vivo bioavailability, selective targeting, and the generation of appropriate bioactive metabolites await further examination. Alternative approaches are being investigated, such as synthetic formulations of natural products with enhanced bioavailability, or encapsulation via nanoparticles and liposomes.
As a closing comment, many of the published studies on dietary agents and miRNAs are highly descriptive, and there now exists a clear need to move the research into more detailed, mechanistic areas. This is somewhat analogous to the situation in the 1970s and 1980s in screening antimutagens in vitro – leading journals elected to no longer accept descriptive data without experiments on the associated mechanisms. Such an approach would likely move the field forward in the context of miRNAs involved in cancer chemoprevention. This is important given the growing awareness of the complex regulation of miRNAs and their targets during different stages of cancer development. As we continue to clarify the mechanisms of these interesting gene regulators, there is much optimism that new chemopreventive and therapeutic modalities will be developed along the way.
Table 1. Common cancer-associated miRNAs.
MicroRNAs deregulated in cancers target multiple genes simultaneously. Certain miRNA expression profiles overlap in cancers of different tissues. Please refer to the text and references listed for further details. TS, tumor suppressor-like activity; ONC, oncogenic activity.
| miRNA | Nature | Common targets | Malignancy |
|---|---|---|---|
| miRNA-21 | ONC | PTEN, TPMI, PDCD4, Maspin, NFIB, Timp3, RECK | Colon, Prostate, Lung, Pancreatic, Breast, Liver, Gastric cancers |
| let-7 family | TS | RAS, PRDMI, HMGA2, E2F, c-Myc, cyclin D2 | Colon, Lung, Ovarian, Breast cancers |
| miR-17-92 family | ONC | Tsp I, E2F I, TGFBR2, AIB I, CTGF, BIM, PTEN, CDKN1A | Lung, Colon, Breast, Pancreatic cancers, Lymphomas |
| miR-372/miR-373 | ONC | LATS2 | Testicular Cancer |
| miR-34 | TS | E2F3, Notch1, CDK 4, CDK 5 | Breast, Colon, Pancreatic cancers |
| miR-9 | TS/ONC | Ovarian, Medulloblastoma and Breast cancers | |
| miR-143 | TS | Ras, ERK5 | Gastric, Colon and Prostate cancers |
| miR-155 | ONC | TP53INPI, ATIR, SHIP1, CEBPB | Lung, Breast cancers, CLL, AML |
| miR-26a | TS | CCND2, CCNE2 | Liver cancer |
| miR-200 | TS/ONC | Breast, Bladder, Gastric, Ovarian and Renal cell carcinoma | |
| miR-221/miR-222 | ONC | KIT, p27(Kip1), p57, PTEN | Breast and liver cancer, CLL |
| miR-29 family | TS | MCL-1, CDK6, TCL1, DNMT1, DNMT3 | CLL, Lymphoma, Liver, Lung and Breast cancers |
| miR-15/miR-16 | TS | Bcl-2, Wt-1, MCL1 | Lung and Gastric cancers |
| miR-181 | TS/ONC | TCL 1, E2F5, eIF5A | Early and late stage Colorectal cancer |
Acknowledgments
We thank Mr. Animesh Koya for assistance with the art work. Studies in the authors’ laboratories are supported by National Cancer Institute grants CA090890, CA122959, CA65525, CA122906, and CA80176, and by Award T32 ES007060 from the National Institute of Environmental Health Sciences. The content of this review is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75 (5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 3.Berezikov E, Cuppen E, Plasterk RH. Approaches to microRNA discovery. Nat Genet. 2006;38 (Suppl):S2–7. doi: 10.1038/ng1794. [DOI] [PubMed] [Google Scholar]
- 4.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120(1):21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 5.Shao NY, Hu HY, Yan Z, Xu Y, Hu H, Menzel C, Li N, Chen W, Khaitovich P. Comprehensive survey of human brain microRNA by deep sequencing. BMC Genomics. 2010;11:409. doi: 10.1186/1471-2164-11-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Persson H, Kvist A, Rego N, Staaf J, Vallon-Christersson J, Luts L, Loman N, Jonsson G, Naya H, Hoglund M, et al. Identification of New MicroRNAs in Paired Normal and Tumor Breast Tissue Suggests a Dual Role for the ERBB2/Her2 Gene. Cancer Res. 2011;71(1):78–86. doi: 10.1158/0008-5472.CAN-10-1869. [DOI] [PubMed] [Google Scholar]
- 7.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36(Database issue):D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3(3):e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23(20):4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10 (12):1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13(12):1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 12.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 13.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432(7014):231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 14.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432(7014):235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 15.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10(2):185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans Genes Dev. 2001;15(20):2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293(5531):834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 18.Preall JB, Sontheimer EJ. RNAi: RISC gets loaded. Cell. 2005;123(4):543–545. doi: 10.1016/j.cell.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 19.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11(3):228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 20.Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA. 2010;16(1):43–56. doi: 10.1261/rna.1972910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Czech B, Zhou R, Erlich Y, Brennecke J, Binari R, Villalta C, Gordon A, Perrimon N, Hannon GJ. Hierarchical rules for Argonaute loading in Drosophila. Mol Cell. 2009;36(3):445–456. doi: 10.1016/j.molcel.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okamura K, Liu N, Lai EC. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol Cell. 2009;36(3):431–444. doi: 10.1016/j.molcel.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19(24):2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boese Q, Leake D, Reynolds A, Read S, Scaringe SA, Marshall WS, Khvorova A. Mechanistic insights aid computational short interfering RNA design. Methods Enzymol. 2005;392:73–96. doi: 10.1016/S0076-6879(04)92005-8. [DOI] [PubMed] [Google Scholar]
- 25.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 26.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17(3):118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 27.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hausser J, Landthaler M, Jaskiewicz L, Gaidatzis D, Zavolan M. Relative contribution of sequence and structure features to the mRNA binding of Argonaute/EIF2C-miRNA complexes and the degradation of miRNA targets. Genome Res. 2009;19 (11):2009–2020. doi: 10.1101/gr.091181.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 30.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37(5):495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 31.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7(7):719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007;27 (11):3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125(6):1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 34.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115(7):787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 36.Stark A, Lin MF, Kheradpour P, Pedersen JS, Parts L, Carlson JW, Crosby MA, Rasmussen MD, Roy S, Deoras AN, et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450(7167):219–232. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30(4):460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 38.Wakiyama M, Takimoto K, Ohara O, Yokoyama S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes Dev. 2007;21(15):1857–1862. doi: 10.1101/gad.1566707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465(7298):584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39(5):673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 41.Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J, Kirsch DG, Golub TR, Jacks T. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23 (23):2700–2704. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lambertz I, Nittner D, Mestdagh P, Denecker G, Vandesompele J, Dyer MA, Marine JC. Monoallelic but not biallelic loss of Dicer1 promotes tumorigenesis in vivo. Cell Death Differ. 2010;17(4):633–641. doi: 10.1038/cdd.2009.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22(20):2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28(2):328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caudy AA, Ketting RF, Hammond SM, Denli AM, Bathoorn AM, Tops BB, Silva JM, Myers MM, Hannon GJ, Plasterk RH. A micrococcal nuclease homologue in RNAi effector complexes. Nature. 2003;425(6956):411–414. doi: 10.1038/nature01956. [DOI] [PubMed] [Google Scholar]
- 46.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 47.Li CM, Zheng JG, Du GS. miRNA: a new regulator of gene expression. Yi Chuan. 2004;26(1):133–136. [PubMed] [Google Scholar]
- 48.Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, Biffo S, Merrick WC, Darzynkiewicz E, Pillai RS, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317(5845):1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- 49.Kiriakidou M, Tan GS, Lamprinaki S, De Planell-Saguer M, Nelson PT, Mourelatos Z. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell. 2007;129(6):1141–1151. doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 50.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59(4):225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 51.Sato F, Tsuchiya S, Meltzer SJ, Shimizu K. MicroRNAs and Epigenetics. FEBS J. 2011 doi: 10.1111/j.1742-4658.2011.08089.x. [DOI] [PubMed] [Google Scholar]
- 52.Hatziapostolou M, Iliopoulos D. Epigenetic aberrations during oncogenesis. Cell Mol Life Sci. 2011 doi: 10.1007/s00018-010-0624-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nat Biotechnol. 2010;28(10):1069–1078. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36(Database issue):D149–153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66(15):7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 56.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 57.Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C, Ambros VR, Israel MA. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67(6):2456–2468. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- 58.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tagawa H, Seto M. A microRNA cluster as a target of genomic amplification in malignant lymphoma. Leukemia. 2005;19(11):2013–2016. doi: 10.1038/sj.leu.2403942. [DOI] [PubMed] [Google Scholar]
- 60.Mavrakis KJ, Wolfe AL, Oricchio E, Palomero T, de Keersmaecker K, McJunkin K, Zuber J, James T, Khan AA, Leslie CS, et al. Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nat Cell Biol. 2010;12(4):372–379. doi: 10.1038/ncb2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huse JT, Brennan C, Hambardzumyan D, Wee B, Pena J, Rouhanifard SH, Sohn-Lee C, le Sage C, Agami R, Tuschl T, et al. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009;23(11):1327–1337. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pan X, Wang ZX, Wang R. MicroRNA-21: A novel therapeutic target in human cancer. Cancer Biol Ther. 2011;10(12):1224–1232. doi: 10.4161/cbt.10.12.14252. [DOI] [PubMed] [Google Scholar]
- 64.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 65.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 66.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299(4):425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Motoyama K, Inoue H, Nakamura Y, Uetake H, Sugihara K, Mori M. Clinical significance of high mobility group A2 in human gastric cancer and its relationship to let-7 microRNA family. Clin Cancer Res. 2008;14(8):2334–2340. doi: 10.1158/1078-0432.CCR-07-4667. [DOI] [PubMed] [Google Scholar]
- 68.Yang N, Kaur S, Volinia S, Greshock J, Lassus H, Hasegawa K, Liang S, Leminen A, Deng S, Smith L, et al. MicroRNA microarray identifies Let-7i as a novel biomarker and therapeutic target in human epithelial ovarian cancer. Cancer Res. 2008;68 (24):10307–10314. doi: 10.1158/0008-5472.CAN-08-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boyerinas B, Park SM, Hau A, Murmann AE, Peter ME. The role of let-7 in cell differentiation and cancer. Endocr Relat Cancer. 2010;17(1):F19–36. doi: 10.1677/ERC-09-0184. [DOI] [PubMed] [Google Scholar]
- 70.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 71.Slattery ML, Wolff E, Hoffman MD, Pellatt DF, Milash B, Wolff RK. MicroRNAs and colon and rectal cancer: differential expression by tumor location and subtype. Genes Chromosomes Cancer. 2011;50(3):196–206. doi: 10.1002/gcc.20844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451(7175):147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432(7014):226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 74.Hwang JH, Voortman J, Giovannetti E, Steinberg SM, Leon LG, Kim YT, Funel N, Park JK, Kim MA, Kang GH, et al. Identification of microRNA-21 as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer. PLoS One. 2010;5(5):e10630. doi: 10.1371/journal.pone.0010630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Giovannetti E, Funel N, Peters GJ, Del Chiaro M, Erozenci LA, Vasile E, Leon LG, Pollina LE, Groen A, Falcone A, et al. MicroRNA-21 in pancreatic cancer: correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. 2010;70(11):4528–4538. doi: 10.1158/0008-5472.CAN-09-4467. [DOI] [PubMed] [Google Scholar]
- 76.Moriyama T, Ohuchida K, Mizumoto K, Yu J, Sato N, Nabae T, Takahata S, Toma H, Nagai E, Tanaka M. MicroRNA-21 modulates biological functions of pancreatic cancer cells including their proliferation, invasion, and chemoresistance. Mol Cancer Ther. 2009 doi: 10.1158/1535-7163.MCT-08-0592. [DOI] [PubMed] [Google Scholar]
- 77.Zhang Y, Li M, Wang H, Fisher WE, Lin PH, Yao Q, Chen C. Profiling of 95 microRNAs in pancreatic cancer cell lines and surgical specimens by real-time PCR analysis. World J Surg. 2009;33(4):698–709. doi: 10.1007/s00268-008-9833-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64(11):3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 79.Yu SL, Chen HY, Chang GC, Chen CY, Chen HW, Singh S, Cheng CL, Yu CJ, Lee YC, Chen HS, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13(1):48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 80.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105(10):3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Roccaro AM, Sacco A, Thompson B, Leleu X, Azab AK, Azab F, Runnels J, Jia X, Ngo HT, Melhem MR, et al. MicroRNAs 15a and 16 regulate tumor proliferation in multiple myeloma. Blood. 2009;113(26):6669–6680. doi: 10.1182/blood-2009-01-198408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D’Urso L, Pagliuca A, Biffoni M, Labbaye C, et al. The miR-15a-miR-16–1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14(11):1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 83.Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15(2):272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15(2):261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee DY, Deng Z, Wang CH, Yang BB. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci U S A. 2007;104(51):20350–20355. doi: 10.1073/pnas.0706901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Le XF, Merchant O, Bast RC, Calin GA. The Roles of MicroRNAs in the Cancer Invasion-Metastasis Cascade. Cancer Microenviron. 2010;3(1):137–147. doi: 10.1007/s12307-010-0037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ji J, Zhao L, Budhu A, Forgues M, Jia HL, Qin LX, Ye QH, Yu J, Shi X, Tang ZY, et al. Let-7g targets collagen type I alpha2 and inhibits cell migration in hepatocellular carcinoma. J Hepatol. 2010;52(5):690–697. doi: 10.1016/j.jhep.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shan SW, Lee DY, Deng Z, Shatseva T, Jeyapalan Z, Du WW, Zhang Y, Xuan JW, Yee SP, Siragam V, et al. MicroRNA MiR-17 retards tissue growth and represses fibronectin expression. Nat Cell Biol. 2009;11(8):1031–1038. doi: 10.1038/ncb1917. [DOI] [PubMed] [Google Scholar]
- 89.Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, Krichevsky AM. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28(17):5369–5380. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, Ngankeu A, Taccioli C, Pichiorri F, Alder H, Secchiero P, et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16 (6):498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 91.Wang B, Hsu SH, Majumder S, Kutay H, Huang W, Jacob ST, Ghoshal K. TGFbeta-mediated upregulation of hepatic miR-181b promotes hepatocarcinogenesis by targeting TIMP3. Oncogene. 2010;29(12):1787–1797. doi: 10.1038/onc.2009.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xia H, Qi Y, Ng SS, Chen X, Li D, Chen S, Ge R, Jiang S, Li G, Chen Y, et al. microRNA-146b inhibits glioma cell migration and invasion by targeting MMPs. Brain Res. 2009;1269:158–165. doi: 10.1016/j.brainres.2009.02.037. [DOI] [PubMed] [Google Scholar]
- 93.Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317(5842):1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci U S A. 2008;105(39):14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26 (5):745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.He L, He X, Lowe SW, Hannon GJ. microRNAs join the p53 network--another piece in the tumour-suppression puzzle. Nat Rev Cancer. 2007;7(11):819–822. doi: 10.1038/nrc2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, Olson EN. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18(3):282–293. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435(7043):839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 99.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12(3):247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.David L, Mallet C, Keramidas M, Lamande N, Gasc JM, Dupuis-Girod S, Plauchu H, Feige JJ, Bailly S. Bone morphogenetic protein-9 is a circulating vascular quiescence factor. Circ Res. 2008;102(8):914–922. doi: 10.1161/CIRCRESAHA.107.165530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yu B, Bi L, Zheng B, Ji L, Chevalier D, Agarwal M, Ramachandran V, Li W, Lagrange T, Walker JC, et al. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc Natl Acad Sci U S A. 2008;105(29):10073–10078. doi: 10.1073/pnas.0804218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Peter ME. Regulating cancer stem cells the miR way. Cell Stem Cell. 2010;6(1):4–6. doi: 10.1016/j.stem.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 103.Li Y, VandenBoom TG, 2nd, Kong D, Wang Z, Ali S, Philip PA, Sarkar FH. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;69(16):6704–6712. doi: 10.1158/0008-5472.CAN-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11 (12):1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 105.Zheng T, Wang J, Chen X, Liu L. Role of microRNA in anticancer drug resistance. Int J Cancer. 2010;126(1):2–10. doi: 10.1002/ijc.24782. [DOI] [PubMed] [Google Scholar]
- 106.Sarkar FH, Li Y, Wang Z, Kong D, Ali S. Implication of microRNAs in drug resistance for designing novel cancer therapy. Drug Resist Updat. 2010;13(3):57–66. doi: 10.1016/j.drup.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cochrane DR, Spoelstra NS, Howe EN, Nordeen SK, Richer JK. MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agents. Mol Cancer Ther. 2009;8(5):1055–1066. doi: 10.1158/1535-7163.MCT-08-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Adam L, Zhong M, Choi W, Qi W, Nicoloso M, Arora A, Calin G, Wang H, Siefker-Radtke A, McConkey D, et al. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin Cancer Res. 2009;15(16):5060–5072. doi: 10.1158/1078-0432.CCR-08-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bourguignon LY, Spevak CC, Wong G, Xia W, Gilad E. Hyaluronan-CD44 interaction with protein kinase C(epsilon) promotes oncogenic signaling by the stem cell marker Nanog and the Production of microRNA-21, leading to down-regulation of the tumor suppressor protein PDCD4, anti-apoptosis, and chemotherapy resistance in breast tumor cells. J Biol Chem. 2009;284(39):26533–26546. doi: 10.1074/jbc.M109.027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li Y, Li W, Yang Y, Lu Y, He C, Hu G, Liu H, Chen J, He J, Yu H. MicroRNA-21 targets LRRFIP1 and contributes to VM-26 resistance in glioblastoma multiforme. Brain Res. 2009;1286:13–18. doi: 10.1016/j.brainres.2009.06.053. [DOI] [PubMed] [Google Scholar]
- 111.DeVere White RW, Vinall RL, Tepper CG, Shi XB. MicroRNAs and their potential for translation in prostate cancer. Urol Oncol. 2009;27(3):307–311. doi: 10.1016/j.urolonc.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ali S, Ahmad A, Banerjee S, Padhye S, Dominiak K, Schaffert JM, Wang Z, Philip PA, Sarkar FH. Gemcitabine sensitivity can be induced in pancreatic cancer cells through modulation of miR-200 and miR-21 expression by curcumin or its analogue CDF. Cancer Res. 2010;70(9):3606–3617. doi: 10.1158/0008-5472.CAN-09-4598. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 113.Garofalo M, Quintavalle C, Di Leva G, Zanca C, Romano G, Taccioli C, Liu CG, Croce CM, Condorelli G. MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene. 2008;27(27):3845–3855. doi: 10.1038/onc.2008.6. [DOI] [PubMed] [Google Scholar]
- 114.Zhao JJ, Lin J, Yang H, Kong W, He L, Ma X, Coppola D, Cheng JQ. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem. 2008;283(45):31079–31086. doi: 10.1074/jbc.M806041200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 115.Kotani A, Ha D, Hsieh J, Rao PK, Schotte D, den Boer ML, Armstrong SA, Lodish HF. miR-128b is a potent glucocorticoid sensitizer in MLL-AF4 acute lymphocytic leukemia cells and exerts cooperative effects with miR-221. Blood. 2009;114(19):4169–4178. doi: 10.1182/blood-2008-12-191619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Iorio MV, Casalini P, Piovan C, Di Leva G, Merlo A, Triulzi T, Menard S, Croce CM, Tagliabue E. microRNA-205 regulates HER3 in human breast cancer. Cancer Res. 2009;69(6):2195–2200. doi: 10.1158/0008-5472.CAN-08-2920. [DOI] [PubMed] [Google Scholar]
- 117.Brower V. Epigenetics: Unravelling the cancer code. Nature. 2011;471(7339):S12–13. doi: 10.1038/471S12a. [DOI] [PubMed] [Google Scholar]
- 118.Lujambio A, Calin GA, Villanueva A, Ropero S, Sanchez-Cespedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso MS, Faller WJ, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A. 2008;105(36):13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, Tokino T. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68(11):4123–4132. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- 120.Bandres E, Agirre X, Bitarte N, Ramirez N, Zarate R, Roman-Gomez J, Prosper F, Garcia-Foncillas J. Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer. 2009;125(11):2737–2743. doi: 10.1002/ijc.24638. [DOI] [PubMed] [Google Scholar]
- 121.Noonan EJ, Place RF, Pookot D, Basak S, Whitson JM, Hirata H, Giardina C, Dahiya R. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene. 2009;28(14):1714–1724. doi: 10.1038/onc.2009.19. [DOI] [PubMed] [Google Scholar]
- 122.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104(40):15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Friedman JM, Liang G, Liu CC, Wolff EM, Tsai YC, Ye W, Zhou X, Jones PA. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009;69(6):2623–2629. doi: 10.1158/0008-5472.CAN-08-3114. [DOI] [PubMed] [Google Scholar]
- 124.Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9(6):435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 125.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17(15):1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 126.Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67(18):8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 127.Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Korner H, Knyazev P, Diebold J, Hermeking H. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7(16):2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 128.Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14(5):872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lujambio A, Esteller M. CpG island hypermethylation of tumor suppressor microRNAs in human cancer. Cell Cycle. 2007;6(12):1455–1459. [PubMed] [Google Scholar]
- 130.Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setien F, Casado S, Suarez-Gauthier A, Sanchez-Cespedes M, Git A, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67(4):1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 131.Brueckner B, Stresemann C, Kuner R, Mund C, Musch T, Meister M, Sultmann H, Lyko F. The human let-7a-3 locus contains an epigenetically regulated microRNA gene with oncogenic function. Cancer Res. 2007;67(4):1419–1423. doi: 10.1158/0008-5472.CAN-06-4074. [DOI] [PubMed] [Google Scholar]
- 132.Chuang JC, Jones PA. Epigenetics and microRNAs. Pediatr Res. 2007;61(5 Pt 2):24R–29R. doi: 10.1203/pdr.0b013e3180457684. [DOI] [PubMed] [Google Scholar]
- 133.Chuang CK, Chu DC, Tzou RD, Liou SI, Chia JH, Sun CF. Hypermethylation of the CpG islands in the promoter region flanking GSTP1 gene is a potential plasma DNA biomarker for detecting prostate carcinoma. Cancer Detect Prev. 2007;31(1):59–63. doi: 10.1016/j.cdp.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 134.Tsai KW, Wu CW, Hu LY, Li SC, Liao YL, Lai CH, Kao HW, Fang WL, Huang KH, Chan WC, et al. Epigenetic regulation of miR-34b and miR-129 expression in gastric cancer. Int J Cancer. 2011 doi: 10.1002/ijc.25919. [DOI] [PubMed] [Google Scholar]
- 135.Scott GK, Mattie MD, Berger CE, Benz SC, Benz CC. Rapid alteration of microRNA levels by histone deacetylase inhibition. Cancer Res. 2006;66(3):1277–1281. doi: 10.1158/0008-5472.CAN-05-3632. [DOI] [PubMed] [Google Scholar]
- 136.Shin S, Lee EM, Cha HJ, Bae S, Jung JH, Lee SM, Yoon Y, Lee H, Kim S, Kim H, et al. MicroRNAs that respond to histone deacetylase inhibitor SAHA and p53 in HCT116 human colon carcinoma cells. Int J Oncol. 2009;35(6):1343–1352. doi: 10.3892/ijo_00000452. [DOI] [PubMed] [Google Scholar]
- 137.Datta J, Kutay H, Nasser MW, Nuovo GJ, Wang B, Majumder S, Liu CG, Volinia S, Croce CM, Schmittgen TD, et al. Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008;68(13):5049–5058. doi: 10.1158/0008-5472.CAN-07-6655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 138.Zhang LL, Li L, Wu DP, Fan JH, Li X, Wu KJ, Wang XY, He DL. A novel anti-cancer effect of genistein: reversal of epithelial mesenchymal transition in prostate cancer cells. Acta Pharmacol Sin. 2008;29(9):1060–1068. doi: 10.1111/j.1745-7254.2008.00831.x. [DOI] [PubMed] [Google Scholar]
- 139.Huang J, Plass C, Gerhauser C. Cancer Chemoprevention by Targeting the Epigenome. Curr Drug Targets. 2010 doi: 10.2174/138945011798184155. [DOI] [PubMed] [Google Scholar]
- 140.Davis CD, Ross SA. Evidence for dietary regulation of microRNA expression in cancer cells. Nutr Rev. 2008;66(8):477–482. doi: 10.1111/j.1753-4887.2008.00080.x. [DOI] [PubMed] [Google Scholar]
- 141.Sun SY, Lotan R. Retinoids and their receptors in cancer development and chemoprevention. Crit Rev Oncol Hematol. 2002;41(1):41–55. doi: 10.1016/s1040-8428(01)00144-5. [DOI] [PubMed] [Google Scholar]
- 142.Rossi A, D’Urso OF, Gatto G, Poltronieri P, Ferracin M, Remondelli P, Negrini M, Caporaso MG, Bonatti S, Mallardo M. Non-coding RNAs change their expression profile after Retinoid induced differentiation of the promyelocytic cell line NB4. BMC Res Notes. 2010;3:24. doi: 10.1186/1756-0500-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Garzon R, Pichiorri F, Palumbo T, Visentini M, Aqeilan R, Cimmino A, Wang H, Sun H, Volinia S, Alder H, et al. MicroRNA gene expression during retinoic acid-induced differentiation of human acute promyelocytic leukemia. Oncogene. 2007;26 (28):4148–4157. doi: 10.1038/sj.onc.1210186. [DOI] [PubMed] [Google Scholar]
- 144.Wang X, Gocek E, Liu CG, Studzinski GP. MicroRNAs181 regulate the expression of p27Kip1 in human myeloid leukemia cells induced to differentiate by 1,25-dihydroxyvitamin D3. Cell Cycle. 2009;8(5):736–741. doi: 10.4161/cc.8.5.7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Peng X, Vaishnav A, Murillo G, Alimirah F, Torres KE, Mehta RG. Protection against cellular stress by 25-hydroxyvitamin D3 in breast epithelial cells. J Cell Biochem. 2010;110(6):1324–1333. doi: 10.1002/jcb.22646. [DOI] [PubMed] [Google Scholar]
- 146.Mohri T, Nakajima M, Takagi S, Komagata S, Yokoi T. MicroRNA regulates human vitamin D receptor. Int J Cancer. 2009;125(6):1328–1333. doi: 10.1002/ijc.24459. [DOI] [PubMed] [Google Scholar]
- 147.Gaedicke S, Zhang X, Schmelzer C, Lou Y, Doering F, Frank J, Rimbach G. Vitamin E dependent microRNA regulation in rat liver. FEBS Lett. 2008;582(23–24):3542–3546. doi: 10.1016/j.febslet.2008.09.032. [DOI] [PubMed] [Google Scholar]
- 148.Kutay H, Bai S, Datta J, Motiwala T, Pogribny I, Frankel W, Jacob ST, Ghoshal K. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem. 2006;99(3):671–678. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 149.Marsit CJ, Eddy K, Kelsey KT. MicroRNA responses to cellular stress. Cancer Res. 2006;66(22):10843–10848. doi: 10.1158/0008-5472.CAN-06-1894. [DOI] [PubMed] [Google Scholar]
- 150.Sarveswaran S, Liroff J, Zhou Z, Nikitin AY, Ghosh J. Selenite triggers rapid transcriptional activation of p53, and p53-mediated apoptosis in prostate cancer cells: Implication for the treatment of early-stage prostate cancer. Int J Oncol. 2010;36 (6):1419–1428. doi: 10.3892/ijo_00000627. [DOI] [PubMed] [Google Scholar]
- 151.Nian H, Bisson WH, Dashwood WM, Pinto JT, Dashwood RH. Alpha-keto acid metabolites of organoselenium compounds inhibit histone deacetylase activity in human colon cancer cells. Carcinogenesis. 2009;30(8):1416–1423. doi: 10.1093/carcin/bgp147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lee JI, Nian H, Cooper AJ, Sinha R, Dai J, Bisson WH, Dashwood RH, Pinto JT. Alpha-keto acid metabolites of naturally occurring organoselenium compounds as inhibitors of histone deacetylase in human prostate cancer cells. Cancer Prev Res (Phila) 2009;2(7):683–693. doi: 10.1158/1940-6207.CAPR-09-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dashwood RH. Early detection and prevention of colorectal cancer (review) Oncol Rep. 1999;6(2):277–281. [PubMed] [Google Scholar]
- 154.Williams CD, Whitley BM, Hoyo C, Grant DJ, Iraggi JD, Newman KA, Gerber L, Taylor LA, McKeever MG, Freedland SJ. A high ratio of dietary n-6/n-3 polyunsaturated fatty acids is associated with increased risk of prostate cancer. Nutr Res. 2011;31 (1):1–8. doi: 10.1016/j.nutres.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 155.Gopinath B, Buyken AE, Flood VM, Empson M, Rochtchina E, Mitchell P. Consumption of polyunsaturated fatty acids, fish, and nuts and risk of inflammatory disease mortality. Am J Clin Nutr. 2011 doi: 10.3945/ajcn.110.009977. [DOI] [PubMed] [Google Scholar]
- 156.Davidson LA, Wang N, Shah MS, Lupton JR, Ivanov I, Chapkin RS. n-3 Polyunsaturated fatty acids modulate carcinogen-directed non-coding microRNA signatures in rat colon. Carcinogenesis. 2009;30(12):2077–2084. doi: 10.1093/carcin/bgp245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tzur G, Levy A, Meiri E, Barad O, Spector Y, Bentwich Z, Mizrahi L, Katzenellenbogen M, Ben-Shushan E, Reubinoff BE, et al. MicroRNA expression patterns and function in endodermal differentiation of human embryonic stem cells. PLoS One. 2008;3 (11):e3726. doi: 10.1371/journal.pone.0003726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kumar A, Ahuja A, Ali J, Baboota S. Conundrum and therapeutic potential of curcumin in drug delivery. Crit Rev Ther Drug Carrier Syst. 2010;27(4):279–312. doi: 10.1615/critrevtherdrugcarriersyst.v27.i4.10. [DOI] [PubMed] [Google Scholar]
- 159.Reuter S, Gupta SC, Park B, Goel A, Aggarwal BB. Epigenetic changes induced by curcumin and other natural compounds. Genes Nutr. 2011;6(2):93–108. doi: 10.1007/s12263-011-0222-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sun M, Estrov Z, Ji Y, Coombes KR, Harris DH, Kurzrock R. Curcumin (diferuloylmethane) alters the expression profiles of microRNAs in human pancreatic cancer cells. Mol Cancer Ther. 2008;7(3):464–473. doi: 10.1158/1535-7163.MCT-07-2272. [DOI] [PubMed] [Google Scholar]
- 161.Bao B, Ali S, Kong D, Sarkar SH, Wang Z, Banerjee S, Aboukameel A, Padhye S, Philip PA, Sarkar FH. Anti-Tumor Activity of a Novel Compound-CDF Is Mediated by Regulating miR-21, miR-200, and PTEN in Pancreatic Cancer. PLoS One. 2011;6 (3):e17850. doi: 10.1371/journal.pone.0017850. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 162.Zhang J, Zhang T, Ti X, Shi J, Wu C, Ren X, Yin H. Curcumin promotes apoptosis in A549/DDP multidrug-resistant human lung adenocarcinoma cells through an miRNA signaling pathway. Biochem Biophys Res Commun. 2010;399(1):1–6. doi: 10.1016/j.bbrc.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 163.Tili E, Michaille JJ, Alder H, Volinia S, Delmas D, Latruffe N, Croce CM. Resveratrol modulates the levels of microRNAs targeting genes encoding tumor-suppressors and effectors of TGFbeta signaling pathway in SW480 cells. Biochem Pharmacol. 2010;80 (12):2057–2065. doi: 10.1016/j.bcp.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tili E, Michaille JJ, Adair B, Alder H, Limagne E, Taccioli C, Ferracin M, Delmas D, Latruffe N, Croce CM. Resveratrol decreases the levels of miR-155 by upregulating miR-663, a microRNA targeting JunB and JunD. Carcinogenesis. 2010;31(9):1561–1566. doi: 10.1093/carcin/bgq143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lambert JD, Elias RJ. The antioxidant and pro-oxidant activities of green tea polyphenols: a role in cancer prevention. Arch Biochem Biophys. 2010;501(1):65–72. doi: 10.1016/j.abb.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Thakur VS, Gupta K, Gupta S. The Chemopreventive and Chemotherapeutic Potentials of Tea Polyphenols. Curr Pharm Biotechnol. 2011 doi: 10.2174/138920112798868584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sang S, Lambert JD, Ho CT, Yang CS. The chemistry and biotransformation of tea constituents. Pharmacol Res. 2011 doi: 10.1016/j.phrs.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 168.Larsen CA, Dashwood RH, Bisson WH. Tea catechins as inhibitors of receptor tyrosine kinases: mechanistic insights and human relevance. Pharmacol Res. 2010;62 (6):457–464. doi: 10.1016/j.phrs.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tsang WP, Kwok TT. Epigallocatechin gallate up-regulation of miR-16 and induction of apoptosis in human cancer cells. J Nutr Biochem. 2010;21(2):140–146. doi: 10.1016/j.jnutbio.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 170.Okuda T, Yoshida T, Hatano T. Ellagitannins as active constituents of medicinal plants. Planta Med. 1989;55(2):117–122. doi: 10.1055/s-2006-961902. [DOI] [PubMed] [Google Scholar]
- 171.Wen XY, Wu SY, Li ZQ, Liu ZQ, Zhang JJ, Wang GF, Jiang ZH, Wu SG. Ellagitannin (BJA3121), an anti-proliferative natural polyphenol compound, can regulate the expression of MiRNAs in HepG2 cancer cells. Phytother Res. 2009;23(6):778–784. doi: 10.1002/ptr.2616. [DOI] [PubMed] [Google Scholar]
- 172.Taylor CK, Levy RM, Elliott JC, Burnett BP. The effect of genistein aglycone on cancer and cancer risk: a review of in vitro, preclinical, and clinical studies. Nutr Rev. 2009;67(7):398–415. doi: 10.1111/j.1753-4887.2009.00213.x. [DOI] [PubMed] [Google Scholar]
- 173.Li Y, Vandenboom TG, 2nd, Wang Z, Kong D, Ali S, Philip PA, Sarkar FH. miR-146a suppresses invasion of pancreatic cancer cells. Cancer Res. 2010;70(4):1486–1495. doi: 10.1158/0008-5472.CAN-09-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Majid S, Dar AA, Saini S, Chen Y, Shahryari V, Liu J, Zaman MS, Hirata H, Yamamura S, Ueno K, et al. Regulation of minichromosome maintenance gene family by microRNA-1296 and genistein in prostate cancer. Cancer Res. 2010;70(7):2809–2818. doi: 10.1158/0008-5472.CAN-09-4176. [DOI] [PubMed] [Google Scholar]
- 175.Parker LP, Taylor DD, Kesterson J, Metzinger DS, Gercel-Taylor C. Modulation of microRNA associated with ovarian cancer cells by genistein. Eur J Gynaecol Oncol. 2009;30(6):616–621. [PubMed] [Google Scholar]
- 176.Sun Q, Cong R, Yan H, Gu H, Zeng Y, Liu N, Chen J, Wang B. Genistein inhibits growth of human uveal melanoma cells and affects microRNA-27a and target gene expression. Oncol Rep. 2009;22(3):563–567. doi: 10.3892/or_00000472. [DOI] [PubMed] [Google Scholar]
- 177.Higdon JV, Delage B, Williams DE, Dashwood RH. Cruciferous vegetables and human cancer risk: epidemiologic evidence and mechanistic basis. Pharmacol Res. 2007;55(3):224–236. doi: 10.1016/j.phrs.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Pappa G, Lichtenberg M, Iori R, Barillari J, Bartsch H, Gerhauser C. Comparison of growth inhibition profiles and mechanisms of apoptosis induction in human colon cancer cell lines by isothiocyanates and indoles from Brassicaceae. Mutat Res. 2006;599(1–2):76–87. doi: 10.1016/j.mrfmmm.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 179.Izzotti A, Calin GA, Arrigo P, Steele VE, Croce CM, De Flora S. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 2009;23 (3):806–812. doi: 10.1096/fj.08-121384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Clarke JD, Dashwood RH, Ho E. Multi-targeted prevention of cancer by sulforaphane. Cancer Lett. 2008;269(2):291–304. doi: 10.1016/j.canlet.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Izzotti A, Calin GA, Steele VE, Cartiglia C, Longobardi M, Croce CM, De Flora S. Chemoprevention of cigarette smoke-induced alterations of MicroRNA expression in rat lungs. Cancer Prev Res (Phila) 2010;3(1):62–72. doi: 10.1158/1940-6207.CAPR-09-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Izzotti A, Larghero P, Cartiglia C, Longobardi M, Pfeffer U, Steele VE, De Flora S. Modulation of microRNA expression by budesonide, phenethyl isothiocyanate and cigarette smoke in mouse liver and lung. Carcinogenesis. 2010;31(5):894–901. doi: 10.1093/carcin/bgq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Weiler J, Hunziker J, Hall J. Anti-miRNA oligonucleotides (AMOs): ammunition to target miRNAs implicated in human disease? Gene Ther. 2006;13(6):496–502. doi: 10.1038/sj.gt.3302654. [DOI] [PubMed] [Google Scholar]
- 184.Garzon R, Heaphy CE, Havelange V, Fabbri M, Volinia S, Tsao T, Zanesi N, Kornblau SM, Marcucci G, Calin GA, et al. MicroRNA 29b functions in acute myeloid leukemia. Blood. 2009;114(26):5331–5341. doi: 10.1182/blood-2009-03-211938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452(7189):896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 186.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327(5962):198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Gumireddy K, Young DD, Xiong X, Hogenesch JB, Huang Q, Deiters A. Small-molecule inhibitors of microrna miR-21 function. Angew Chem Int Ed Engl. 2008;47 (39):7482–7484. doi: 10.1002/anie.200801555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318(5848):271–274. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- 189.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4(9):721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Xiao J, Yang B, Lin H, Lu Y, Luo X, Wang Z. Novel approaches for gene-specific interference via manipulating actions of microRNAs: examination on the pacemaker channel genes HCN2 and HCN4. J Cell Physiol. 2007;212(2):285–292. doi: 10.1002/jcp.21062. [DOI] [PubMed] [Google Scholar]
- 191.Dias N, Stein CA. Antisense oligonucleotides: basic concepts and mechanisms. Mol Cancer Ther. 2002;1(5):347–355. [PubMed] [Google Scholar]
- 192.Chiarantini L, Cerasi A, Fraternale A, Millo E, Benatti U, Sparnacci K, Laus M, Ballestri M, Tondelli L. Comparison of novel delivery systems for antisense peptide nucleic acids. J Control Release. 2005;109(1–3):24–36. doi: 10.1016/j.jconrel.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 193.Zuhorn IS, Engberts JB, Hoekstra D. Gene delivery by cationic lipid vectors: overcoming cellular barriers. Eur Biophys J. 2007;36(4–5):349–362. doi: 10.1007/s00249-006-0092-4. [DOI] [PubMed] [Google Scholar]
- 194.Chirila TV, Rakoczy PE, Garrett KL, Lou X, Constable IJ. The use of synthetic polymers for delivery of therapeutic antisense oligodeoxynucleotides. Biomaterials. 2002;23(2):321–342. doi: 10.1016/S0142-9612(01)00125-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AK, Han MS, Mirkin CA. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science. 2006;312(5776):1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 196.Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J, Cheng A, Hall BM, Qualman SJ, Chandler DS, et al. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14(5):369–381. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Xiong Y, Fang JH, Yun JP, Yang J, Zhang Y, Jia WH, Zhuang SM. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology. 2010;51(3):836–845. doi: 10.1002/hep.23380. [DOI] [PubMed] [Google Scholar]
- 198.Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29(5):903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 199.Michelfelder S, Trepel M. Adeno-associated viral vectors and their redirection to cell-type specific receptors. Adv Genet. 2009;67:29–60. doi: 10.1016/S0065-2660(09)67002-4. [DOI] [PubMed] [Google Scholar]
- 200.Aagaard L, Rossi JJ. RNAi therapeutics: principles, prospects and challenges. Adv Drug Deliv Rev. 2007;59(2–3):75–86. doi: 10.1016/j.addr.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Lu M, Zhang Q, Deng M, Miao J, Guo Y, Gao W, Cui Q. An analysis of human microRNA and disease associations. PLoS One. 2008;3(10):e3420. doi: 10.1371/journal.pone.0003420. [DOI] [PMC free article] [PubMed] [Google Scholar]